Abstract
When homologous microsatellites are compared between species, significant differences in mean length are often noted. A dominant cause of these length differences is ascertainment bias due to selection for maximum repeat number and repeat purity when the markers are being developed. However, even after ascertainment bias has been allowed for through reciprocal comparisons, significant length differences remain, suggesting that the average microsatellite mutation rate differs between species. Two classes of mechanism have been proposed: rapid evolution of enzymes involved in the generation and repair of slippage products (enzyme evolution model) and heterozygote instability, whereby interchromosomal events at heterozygous sites offer extra opportunities for mutations to occur (heterozygote instability model). To examine which of these hypotheses is most likely, we compared ascertainment bias and species length differences between humans and chimpanzees in autosomal and Y chromosomal microsatellites. We find that levels of ascertainment bias are indistinguishable, but that interspecies length differences are significantly greater for autosomal loci compared with haploid Y chromosomal loci. Such a pattern is consistent with predictions from the heterozygote instability model and is not expected under models of microsatellite evolution that do not include interchromosomal events such as the enzyme evolution model.
MICROSATELLITES are sequences of repeated short (1–5 bp) motifs used commonly as genetic markers due to their high levels of slippage-generated repeat copy number variation (Levinson and Gutman 1987; Schlötterer and Tautz 1992). However, despite intense study, many aspects of microsatellite evolution remain unresolved. For example, although most mutations involve the gain or loss of a single repeat unit, some studies report a tendency for gains in length to outnumber losses (Amos et al. 1996; Kayser et al. 2000; Dupuy et al. 2004), while others report either unbiased mutations (Brinkmann et al. 1998; Xu et al. 2000) or a tendency to decline in length (Sajantila et al. 1999) especially among very long microsatellites (Xu et al. 2000; Huang et al. 2002). Mutation rates and other properties appear to depend critically on the structure of the microsatellite, with repeat number and the degree of interruption both being important (Brinkmann et al. 1998; Kayser et al. 2000, 2004; Huang et al. 2002). Such structural variation may help to explain the contradictory nature of some of these studies (for overview see Ellegren 2004).
An alternative view on the mutation process can be gained by exploiting the fact that primer sets designed for one species often amplify homologous loci in related species, a process referred to as cross-species amplification. Early studies using this approach found an unexpected pattern in which microsatellites in one species were often consistently longer than their homologs in a related species. Thus, human microsatellites are usually longer than their chimpanzee homologs (Rubinsztein et al. 1995; Amos and Rubinsztein 1996; Amos et al. 1996), microsatellites in sheep are longer than in cows (Crawford et al. 1998), and barn swallow loci are longer than in other related birds (Primmer and Ellegren 1998).
Unfortunately, length comparisons between species are complicated by an ascertainment bias (Ellegren et al. 1995, 1997; Zhu et al. 2000; Webster et al. 2002; Amos et al. 2003). During marker development, highly polymorphic loci are normally preferred and, since the degree of polymorphism and microsatellite length are positively correlated (e.g., Kayser et al. 2004), this usually means that loci are chosen to be as long as possible in the target species. As a consequence, there is a tendency for markers to be longer and thus more polymorphic in the species from which they were originally cloned, relative to homologous products in related species they are applied to. In principle, ascertainment bias can be canceled out by carrying out a reciprocal analysis whereby a balanced set of loci are selected from genomic microsatellite libraries cloned from each of the species concerned using the same selection criteria. In the small number of studies where this has been done, both ascertainment bias and consistent length differences between species appear to be present. For example, microsatellites are longer in humans compared with chimpanzees (Cooper et al. 1998; Webster et al. 2002), sheep relative to cattle (Crawford et al. 1998), and Drosophila melanogaster compared with D. simulans (Amos et al. 2003).
If, over and above any length differences due to ascertainment bias, homologous microsatellites tend to be longer in one of two closely related species, this implies both a biased mutation process and variation in the genomewide microsatellite mutation rate. To date, two models have been proposed to account for a change in the genomewide mutation rate: first, the enzyme evolution model according to which a mutation in any of a number of enzymes involved in DNA replication or mismatch repair could change the rate at which slippage events either arise or are removed (Rubinsztein et al. 1999) and second, the heterozygote instability model according to which heterozygous loci might have a higher mutation rate due to mismatch repair events that occur during synapsis (Amos et al. 1996; Amos and Harwood 1998). Under the heterozygote instability model, demographic events such as population expansion or hybridization that increase heterozygosity would also increase the genomewide rate of microsatellite mutations.
One way to distinguish between these two models is to compare the behavior of diploid and haploid microsatellites, specifically autosomal markers and those carried by the nonrecombining portion of the Y chromosome. Under the enzyme evolution model these two classes should behave similarly. In contrast, under the heterozygote instability model, changes in genomewide mutation rate are driven by changes in heterozygosity and, since markers on the nonrecombining portion of the Y are haploid, such a mechanism cannot apply. Until recently, the number of known human Y chromosomal microsatellite markers was only ∼50 and very few of them had been successfully detected in chimpanzees (Gusmao et al. 2002), too few to conduct meaningful statistical analyses. Recently, 166 additional human Y chromosomal microsatellite markers were identified by taking advantage of the human genome sequence (Kayser et al. 2004), and 139 of them have been tested in nonhuman primate species, including chimpanzees (Erler et al. 2004). Furthermore, with the recent availability of human and chimpanzee genome sequences (http://www.genome.ucsc.edu) it is now possible to compare both species at the sequence level.
In a recent study, Vowles and Amos (2006) examined both ascertainment bias and species-pair length differences by identifying large numbers of pairs of homologous autosomal microsatellite sequences in the human and chimpanzee genome sequences. Here, the fact that each locus is represented only by a single allele appears more than compensated for by the large number of loci that can be studied, and they were able to explore how both ascertainment bias and interspecies length differences varied with microsatellite length. This work confirmed previous reports that both ascertainment bias and length differences are present, showing that the dominant component is formed from an ascertainment bias that rises steeply with increasing length in the species where the locus was discovered (the “focal” species). In contrast, interspecies length differences appear to vary little with length and average ∼2.5 bp longer in humans.
In this study, we extend the methods of Vowles and Amos (2006) to compare autosomal and Y chromosomal microsatellites and specifically ask whether the human–chimpanzee length difference seen for autosomal loci also applies to haploid markers. This analysis is augmented by data from a large panel of human-derived Y microsatellites genotyped and sequenced in both humans and chimpanzees.
MATERIALS AND METHODS
Database comparisons:
With the availability of the human genome sequence and draft chimpanzee genome sequence, it is now possible to estimate both ascertainment bias and species-specific length differences for homologous microsatellites in human and chimpanzee using database searches (Vowles and Amos 2006). The process emulates the way in which cross-species amplifications are conducted in that a series of loci are identified in one genome, referred to as the focal species, and then compared with the lengths of their homologs in a second “nonfocal” species. The focal and nonfocal species are then reversed and the process repeated. Given the abundance of sequence data available, it is possible not only to look at, for example, all loci carrying >12 uninterrupted repeats, but also to extend the analysis to treat each possible length class independently.
Using the University of California Santa Cruz (UCSC) Genome Browser web interface, we searched for homologous pairs of loci present in both humans (Homo sapiens, July 2003 assembly) and chimpanzees (Pan troglodytes, November 2003 assembly). Overall homology between the two genomes is determined by masking repeat sequences and linking the best alignments for short stretches of genomic sequence to form a chain (Kent et al. 2003). A file containing established chains is available to download and was used to identify homologous pairs of loci using a visual basic program that we wrote. Our program searches through the human microsatellite loci identified by the tandem repeat finder (TRF) (Benson 1999) and returns a subset of loci where homologs are present in both species, along with their database coordinates. Selection criteria for TRF were set at a minimum repeat tract length of eight repeats for each of the different repeat motif classes di-, tri-, tetra-, and pentanucleotides and defining a minimum repeat tract length in base pairs of 16, 24, 32, and 40, respectively.
To make sure that only nonrecombining Y chromosomal loci were used in the analysis, loci in the two pseudoautosomal regions of the Y chromosome were excluded, the location of the boundaries being determined by identifying the locations of genes that were known to recombine with the X chromosome (Graves et al. 1998). Loci with homologs elsewhere might also be able to recombine so these were also identified using BLAT, a program on Genome Browser that identifies approximate sequence matches to its own index of the genome and returns quality scores of detailed alignments for good matches. BLAT returned a small number of matches with high similarity scores, 133 from loci selected in humans and 32 from loci selected in chimpanzees. These were also excluded from the analysis.
Using the pairs of coordinates generated by our program, sequences were downloaded comprising the microsatellite plus 200 bases of flanking sequences either side for the focal species and 300 bases either side for the nonfocal species. The larger window for nonfocal loci allows for possible insertion or deletion events. Sequences >500 bases were removed from the analysis as they typically contained large tracts of N's. In addition, sequences mapping to nonhomologous chromosomes were also removed. To ensure maximum comparability and as a further test of homology we then performed an “electronic” amplification by designing arbitrary primer sequences and discarding loci where the nonfocal locus had three or more mismatches in either primer. Repeat length was defined as the total length of the repeat tract (in number of repeats) allowing interruptions of equal length to or shorter than the repeat unit size.
Calculation of ascertainment bias and length difference:
Given data from reciprocal measurements of microsatellite length (in number of repeats) between two species where both species are in turn treated as focal, both ascertainment bias, Ab, and any absolute length difference between species, D, can be estimated as follows,
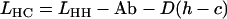 |
(1) |
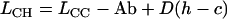 |
(2) |
(Amos et al. 2003), where LXY is the length of loci derived from species X and measured in species Y, and D(h − c) is the average difference in length of loci between humans and chimpanzees expressed as excess length in humans. Rearranging, we obtain
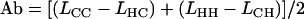 |
(3) |
 |
(4) |
Standard errors cannot be calculated separately for Ab and D(h − c) but are instead estimated as a joint value (Figure 1). Consequently, all standard errors presented for Ab and D(h − c) are highly conservative, maximum values that assume that the second quantity is estimated with zero error. In a large study of autosomal loci, we have shown that ascertainment bias increases dramatically with repeat copy number in the focal species (Vowles and Amos 2006). Consequently, although ascertainment bias and length differences can be estimated for any reciprocally tested loci, when sample sizes permit, it is preferable to estimate both quantities separately for each repeat number or length class.
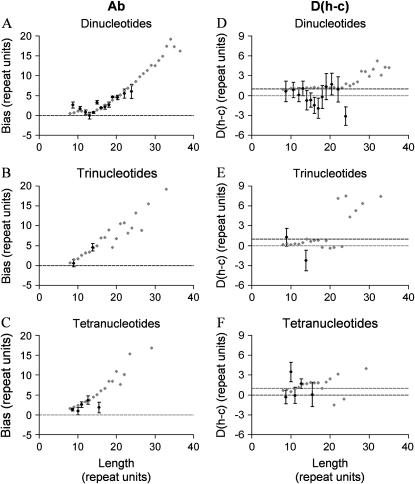
Figure 1.—
Comparison of ascertainment bias, Ab, and residual length difference (in number of repeat units) between humans and chimpanzees expressed as excess length in humans, D(h − c), for Y chromosomal microsatellites. For comparison, we include data obtained from a parallel study of autosomal microsatellites where the same approach was applied. (A–C) Dependence of Ab on focal species microsatellite length for a range of repeat types: solid symbols, Y chromosome microsatellites; shaded symbols, autosomal microsatellites; horizontal dashed line indicates Ab = 0. (D–F) Dependence of D(h − c) on focal species microsatellite length for a range of repeat types. Again, solid symbols, Y chromosome microsatellites; shaded symbols, autosomal microsatellites; horizontal dashed lines are included for D(h − c) = 0 and D(h − c) = 1. Where <10 loci are found for a given repeat length, data are combined into length classes with a minimum membership of 10. Error bars are one standard error of the mean. There were too few pentanucleotide loci identified on the Y chromosome to calculate meaningful values of Ab and D(h − c).
To test for a difference in D(h − c) between Y chromosome and autosomal microsatellites a general linear model (GLM) was fitted. D(h − c) was modeled with two predictor variables: D(h − c) = length + chromosome, where length was a continuous variable and chromosome a factor with two levels (i.e., Y and autosomal). GLM analysis was performed using the statistical software package R (R core development team, version 2.0.0).
DNA sequence analysis:
To provide a comparison with the database analyses, we also explored a large panel of Y chromosome microsatellites by direct PCR amplification. In a previous study, 166 new Y chromosomal microsatellites were identified from screening the nonrecombining part of the human Y chromosome sequence using the tandem repeat finder track on the UCSC Genome Table Browser (H. sapiens, April 2002 assembly). Loci were accepted that had repeat units of size 3–6 bp with eight or more repeats, showed male specificity, and revealed genetic variability in a panel of eight human males from diverse origins on the basis of Y chromosome single-nucleotide polymorphisms (Y-SNPs) (Kayser et al. 2004). In addition, 136 of these loci were analyzed in various nonhuman primate species, including the central chimpanzee (P. troglodytes troglodytes) (Erler et al. 2004). Chimpanzee samples were made available by Svante Pääbo of the Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany, from cells obtained from individuals held at the Centre International de Recherches Medicales de Franceville (CIRMF), Gabon, and donated by E. Jean Wickings, Unité de Génétique des Ecosystèmes Tropicaux, CIRMF.
To guard against changes in length caused by insertions/deletions of flanking sequence DNA and the possibility of amplifying nonhomologous loci, we sequenced one allele from each of 56 Y short tandem repeat (STR) loci that yielded male-specific products in the central chimpanzee in the previous study (Erler et al. 2004). DNA was extracted and PCR amplification was carried out as described elsewhere (Erler et al. 2004). Male specificity was reconfirmed by parallel amplification of a female central chimpanzee sample. In a few cases the female sample yielded a PCR product, but this always differed in size compared with the male-specific product. For DNA sequence analysis, PCR products were purified enzymatically with the ExoSAP clean-up method. Wherever multiple bands were produced, the male-specific band was gel purified with the QIAquick gel extraction kit (QIAGEN, Hilden, Germany). Purified PCR products were sequenced using the ABI Big Dye Terminator cycle sequencing ready reaction kit (PE Applied Biosystems, Foster City, CA) according to the recommendations of the manufacturer and analyzed with an ABI 3700 sequencer (PE Applied Biosystems), whereby all positions were determined from both DNA strands. The sequence trace files were analyzed and aligned with the program SEQMAN II version 5.00 (DNASTAR, Madison, WI). All positions were confirmed by manual inspection of the trace files.
To increase the number of pairs of homologous human and chimpanzee microsatellites where the sequence is known in both species, loci amplified in central chimpanzees by Erler et al. (2004) were also identified in the chimpanzee genome sequence on the UCSC Genome Browser (November 2003 assembly). The browser displays regions of sequence homology between the human and chimpanzee genome sequences so coordinates of homologous chimpanzee loci could be identified using coordinates of human loci (see below). In this way, 10 further chimpanzee microsatellite sequences were identified, giving a total of 66 sequences. For all 66 chimpanzee loci, data on genetic diversity based on seven unrelated male central chimpanzees as well as information on human DNA sequence and human genetic variation based on eight unrelated male individuals were available from our previous studies (Erler et al. 2004; Kayser et al. 2004). All microsatellite alleles were expressed according to their repeat copy number as deduced from DNA sequences and PCR product fragment lengths assuming that the flanking sequences remain constant in length across individuals of the same species.
RESULTS
Table 1 gives the total number of Y microsatellites found on the nonrecombining portion of the Y in humans and chimpanzees where the homologous loci could also be identified in the other species and where copies were not detected elsewhere in the genome. Data are classified according to repeat type, namely di-, tri-, tetra-, and pentanucleotides. Larger repeat sizes were encountered too infrequently to be useful here.
TABLE 1.
Numbers of Y chromosomal microsatellites identified
| No. of loci selected from humans using genome browser
|
No. of loci selected from chimpanzees using genome browser
|
No. of loci amplified by PCR in humans and chimpanzees
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Motif size | With chimpanzee homologs | Loci in PAR or with duplicates elsewhere in the genome | Total useda | With human homologs | Loci in PAR or with duplicates elsewhere in the genome | Total useda | Where sequences were also identified | Polymorphic in chimpanzeesab | Polymorphic in humansac |
| 2 | 197 | 64 | 133 | 102 | 13 | 89 | 0 | 0 | 0 |
| 3 | 35 | 21 | 14 | 24 | 9 | 15 | 19 | 11 | 13 |
| 4 | 90 | 38 | 52 | 63 | 18 | 45 | 40 | 30 | 35 |
| 5 | 18 | 10 | 8 | 6 | 5 | 1 | 7 | 3 | 6 |
| Total | 340 | 133 | 207 | 195 | 45 | 150 | 66 | 44 | 54 |
We include only loci found in both species in the nonrecombining portion of the Y chromosome (outside the pseudoautosomal regions, PAR) and without homologs elsewhere in the genome.
Based on seven unrelated chimpanzee males (Erler et al. 2004).
Based on eight unrelated human males (Kayser et al. 2004).
The largest sample of homologous human and chimpanzee Y microsatellites are those found by database searching (see Table 1). Figure 1 displays graphical summaries of both ascertainment bias and D(h − c) for di-, tri-, and tetranucleotide motifs. Wherever possible, each length class was treated separately but where this was not possible due to low sample sizes, length classes were combined to groups of similar length loci with minimum group membership = 10. For the most part, this meant that dinucleotide motifs were placed in single length classes while tri- and tetranucleotides had to be combined into more heterogeneous groups. In each part, equivalent results for a parallel analysis of autosomal loci are presented for comparison.
Ascertainment bias appears more or less indistinguishable between autosomal and Y microsatellites, particularly among the dinucleotides where sample sizes are larger (Figure 1A). In contrast, the difference in length between humans and chimpanzees after allowing for ascertainment bias, D(h − c), appears more variable and difficult to interpret. Focusing on the dinucleotide microsatellites where most data are available, it appears that, despite the extra variability, D(h − c) is lower for Y microsatellites than for equivalent autosomal loci. By inspection, 11 of 13 length classes have lower D(h − c), which is significant using a simple binomial test (P = 0.022, two-tailed). For a more formal analysis we fitted a general linear model with D(h − c) as the response variable and length and chromosome (autosome or Y) as explanatory factors. The term chromosome is significant at P = 0.023, indicating that the autosomes and Y differ (Table 2). However, none of the transformations we tried yielded normally distributed residuals so this result needs to be treated with caution. Finally, while autosomal D(h − c) is consistently significantly greater than zero (t = 4.3, 29 d.f., P < 0.001), Y chromosomal D(h − c) does not differ significantly from zero with either a binomial test (7 above zero: 6 below, not significant) or a t-test (t = 0.28, 12 d.f., P > 0.05).
TABLE 2.
Results for the test of the model D(h − c) = chromosome + length, where chromosome is a factor with two levels to indicate whether the microsatellites are from the nonrecombining Y chromosome or from autosomal chromosomes, while length is a continuous variable measured in repeat units
| d.f. | Sum of squares | Mean squares | F-ratio | P(>F) | Significance | |
|---|---|---|---|---|---|---|
| Chromosome | 1 | 243 | 243 | 5.2045 | 0.02253 | * |
| Length | 1 | 3,287 | 3,287 | 70.4725 | 0.00000 | *** |
| Error | 83,355 | 3,887,898 | 47 |
Significance is shown at *P < 0.05 and at ***P < 0.001.
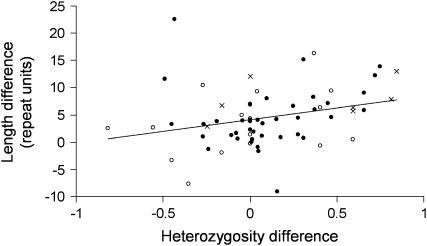
Consistent and nonartifactual length differences between homologous microsatellites in related species suggest both a biased mutation process and a difference in the genomewide mutation rate between the species concerned. Under an unbiased mutation process, differences in rate will affect only the variance of the length difference and will not alter its expectation of zero. Similarly, a biased mutation process will create a consistent length difference only if more mutations occur in one lineage relative to another. Consequently, a correlation between difference in heterozygosity and difference in length between pairs of homologous loci will be indicative of a biased mutation process and of rate variation between lineages. To test this expectation, we further analyzed 66 loci through PCR amplification (see Table 1). Plotting difference in mean length against difference in “heterozygosity” (calculated as 1 − Σ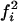 , where fi is the frequency of the ith allele at the locus) we found a significant positive correlation (r = 0.29, t = 2.35, d.f. = 62, P = 0.022; Figure 2), providing support for an upwardly biased mutation process and hence in agreement with most studies of autosomal loci (Amos et al. 1996; Primmer et al. 1998; Cooper et al. 1999; Ellegren 2000).
, where fi is the frequency of the ith allele at the locus) we found a significant positive correlation (r = 0.29, t = 2.35, d.f. = 62, P = 0.022; Figure 2), providing support for an upwardly biased mutation process and hence in agreement with most studies of autosomal loci (Amos et al. 1996; Primmer et al. 1998; Cooper et al. 1999; Ellegren 2000).
Figure 2.—
Correlation of difference in length (in number of repeat units) and difference in heterozygosity of homologous human and chimpanzee Y chromosomal microsatellites. The 64 homologous microsatellites analyzed included 17 tri- (open circles), 40 tetra- (solid circles), and 7 pentanucleotide repeats (crosses). From the originally amplified 66 loci two outliers (not shown; at coordinates 0.027, 40.56 and 0.037, 49.13), which correspond to two extremely long repeat tracts in humans (∼60), were excluded from calculation of both the best-fit line and the correlation coefficient (see text) since they contained multiple interruptions, suggesting that the underlying mutations did not occur via slippage. However, the correlation is significant irrespective of whether the two data points are excluded or included. The best-fit regression line is shown as a solid line.
DISCUSSION
In this article we have tested the hypothesis that interchromosomal events may be involved in microsatellite evolution by comparing ascertainment bias and interspecies length differences between loci sampled from the autosomes and Y chromosomes in humans and chimpanzees. We find that length difference and variability difference are positively correlated, indicating a biased mutation process. In addition, using genome sequence data, we find that while ascertainment bias is more or less identical between microsatellites on the autosomes and those on the Y chromosome, the residual length difference left after controlling for ascertainment bias is significantly greater for autosomal loci compared with Y chromosomal loci. Furthermore, while the residual length difference is significantly greater than zero for autosomal loci, it is indistinguishable from zero for loci on the Y chromosome.
Several studies report unbiased mutations at microsatellite loci (Brinkmann et al. 1998; Xu et al. 2000, although in this study they also found an increased rate of contractions with increased repeat number, with the rate of expansions remaining constant). However, in an unbiased process where ascertainment bias has been removed statistically by reciprocal testing, length differences between species should not occur. That such length differences are found therefore argues that the mutation process is biased, probably in favor of expansions. In our current study we have been able to garner enough data to test Y chromosome microsatellites separately. We find a positive correlation between difference in variability and difference in length among homologous pairs of human and chimpanzee microsatellites. Such patterns are difficult to explain without a biased mutation process and consequently lend further support to studies that report positive mutation biases (Kayser et al. 2000; Dupuy et al. 2004).
Using large numbers of homologous pairs of loci identified in the human and chimpanzee genomes sequences, we were able to determine how ascertainment bias varies with repeat number. We find a pattern where bias increases dramatically with repeat number, seen most clearly for dinucleotide repeat loci because of their greater abundance. Interestingly, the bias profile is indistinguishable from that produced in an earlier study where we analyzed data for autosomal microsatellites (Vowles and Amos 2006), suggesting that the causes of ascertainment bias are similar, regardless of the chromosome they reside on.
Having controlled for ascertainment bias we were then able to determine whether any residual inherent length difference remained. We found that Y chromosomal microsatellites show no evidence of a length difference, suggesting that the mutation rate of Y chromosome microsatellites does not differ between the lineages leading to humans and chimpanzees. This result contrasts with our previous study of autosomal loci where a clear length difference in favor of humans was observed. Unfortunately, despite much increased sample sizes of loci, our total sample size remains modest, particularly in comparison with the vast numbers of autosomal loci that are available, and consequently statistical power is low. However, it is notable that the number of length classes where the interspecies length difference is smaller for the Y chromosomal microsatellites than for autosomal loci is significantly larger than expected by chance. Thus, while the mutation rate at autosomal loci is greater in humans than in chimpanzees, for Y chromosomal microsatellites the mutation rates are similar and we cannot reject the hypothesis that they are identical.
An important problem with autosome–Y chromosome comparisons is the limitation on the absolute number of loci that can be studied. Until recently, only a handful of Y chromosomal microsatellites were available, but even with the numbers we use here extracted from the human genome sequence, coverage is imperfect. Most notably, repeat motifs longer than dinucleotides do not have sufficient representation to explore differences among length classes. Such comparisons are important, since ascertainment bias in particular varies greatly with repeat copy number. A second lack of coverage is that of long dinucleotide loci. For autosomal loci, the length difference between humans and chimpanzees is small but consistent over short to moderate length loci but increases among longer (>25 repeat) loci (Figure 1D). This size range coincides with the range where data for Y chromosomal loci are rare or lacking. Thus, although it is clear that the length difference between humans and chimpanzees is smaller for Y as opposed to autosomal microsatellites in the length range where most markers lie, we cannot be sure what happens among longer loci.
We originally set out to test the contrasting predictions of two models capable of explaining short-term changes in the genomewide mutation rate of microsatellites, the enzyme evolution model and the heterozygote instability model. The pattern we find, where autosomal microsatellites reveal a length difference between humans and chimpanzees not seen at haploid, Y chromosome loci is at odds with the enzyme evolution model because any changes in the enzymes involved with DNA replication would likely influence autosomal and Y chromosome mutation rates equally. However, a difference between autosomal and haploid microsatellites as observed in our study is expected under the heterozygote instability model where heterozygous sites are more mutable. Here, demographic processes, such as the population expansion experienced by humans, that tend to increase heterozygosity, will also stimulate a genomewide increase in mutation rate. How big this effect may be remains to be determined and will require a combination of modeling and data from other comparisons between species to elucidate. The final possibility that should be considered is whether our findings might be influenced by natural selection. Balancing selection seems impossible due to the haploid nature of the Y chromosome but, although it is generally assumed that differential selection does not influence Y chromosome diversity, some instances have been reported where Y chromosome genotypes appear associated with human phenotypes (see summary in Jobling and Tyler Smith 2003). However, such instances are rare, controversial, and involve Y-SNPs in single populations, not Y chromosome microsatellites as used in our study. Moreover, Y microsatellites evolve very fast relative to the strength of selection indicated by Y-SNPs with a mutation rate ∼100,000 times higher than that of Y-SNPs (Kayser et al. 2000; Thomson et al. 2000). Overall, therefore, it seems unlikely that natural selection significantly influences our findings.
Acknowledgments
We thank E. Jean Wickings, Svante Pääbo, and Katrin Köhler for providing chimpanzee samples as well as Antje Weihmann and Barbara Höffner for expert technical assistance in DNA sequencing. M.K. is grateful to Mark Stoneking for providing laboratory facilities at Max Planck Institute for Evolutionary Anthropology at Leipzig as well as for his general support. This work was supported by a Natural Environment Research Council studentship and a Cambridge Philosophical Society research studentship to E.J.V. as well as a studentship by the German National Merit Foundation (Studienstiftung des Deutschen Volkes) to D.K. Additionally, this study received support from the Max Planck Society and from the Leverhulme Trust.
References
- Amos, W., and J. Harwood, 1998. Factors affecting levels of genetic diversity in natural populations. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 353: 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos, W., and D. C. Rubinsztein, 1996. Microsatellites are subject to directional evolution. Nat. Genet. 12: 13–14. [DOI] [PubMed] [Google Scholar]
- Amos, W., S. J. Sawcer, R. W. Feakes and D. C. Rubinsztein, 1996. Microsatellites show mutational bias and heterozygote instability. Nat. Genet. 13: 390–391. [DOI] [PubMed] [Google Scholar]
- Amos, W., C. M. Hutter, M. D. Schug and C. F. Aquadro, 2003. Directional evolution of size coupled with ascertainment bias for variation in Drosophila microsatellites. Mol. Biol. Evol. 20: 660–662. [DOI] [PubMed] [Google Scholar]
- Benson, G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann, B., M. Klintschar, F. Neuhuber, J. Huhne and B. Rolf, 1998. Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am. J. Hum. Genet. 62: 1408–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, G., D. C. Rubinsztein and W. Amos, 1998. Ascertainment bias cannot entirely account for human microsatellites being longer than their chimpanzee homologues. Hum. Mol. Genet. 7: 1425–1429. [DOI] [PubMed] [Google Scholar]
- Cooper, G., N. J. Burroughs, D. A. Rand, D. C. Rubinsztein and W. Amos, 1999. Markov chain Monte Carlo analysis of human Y-chromosome microsatellites provides evidence of biased mutation. Proc. Natl. Acad. Sci. USA 96: 11916–11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, A. M., S. M. Kappes, K. A. Paterson, M. J. deGotari, K. G. Dodds et al., 1998. Microsatellite evolution: testing the ascertainment bias hypothesis. J. Mol. Evol. 46: 256–260. [DOI] [PubMed] [Google Scholar]
- Dupuy, B. M., M. Stenersen, T. Egeland and B. Olaisen, 2004. Y-chromosomal microsatellite mutation rates: differences in mutation rate between and within loci. Hum. Mutat. 23: 117–124. [DOI] [PubMed] [Google Scholar]
- Ellegren, H., 2000. Heterogeneous mutation processes in human microsatellite DNA sequences. Nat. Genet. 24: 400–402. [DOI] [PubMed] [Google Scholar]
- Ellegren, H., 2004. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5: 435–445. [DOI] [PubMed] [Google Scholar]
- Ellegren, H., C. R. Primmer and B. C. Sheldon, 1995. Microsatellite evolution—directionality or bias. Nat. Genet. 11: 360–362. [DOI] [PubMed] [Google Scholar]
- Ellegren, H., S. Moore, N. Robinson, K. Byrne, W. Ward et al., 1997. Microsatellite evolution—a reciprocal study of repeat lengths at homologous loci in cattle and sheep. Mol. Biol. Evol. 14: 854–860. [DOI] [PubMed] [Google Scholar]
- Erler, A., M. Stoneking and M. Kayser, 2004. Development of Y-chromosomal microsatellite markers for nonhuman primates. Mol. Ecol. 13: 2921–2930. [DOI] [PubMed] [Google Scholar]
- Graves, J. A. M., M. J. Wakefield and R. Toder, 1998. The origin and evolution of the pseudoautosomal regions of human sex chromosomes. Hum. Mol. Genet. 7: 1991–1996. [DOI] [PubMed] [Google Scholar]
- Gusmao, L., A. Gonzalez-Neira, C. Alves, M. Lareu, S. Costa et al., 2002. Chimpanzee homologous of human Y specific STRs—a comparative study and a proposal for nomenclature. Forensic Sci. Int. 126: 129–136. [DOI] [PubMed] [Google Scholar]
- Huang, Q. Y., F. H. Xu, H. Shen, H. Y. Deng, Y. J. Liu et al., 2002. Mutation patterns at dinucleotide microsatellite loci in humans. Am. J. Hum. Genet. 70: 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling, M. A., and C. Tyler-Smith, 2003. The human Y chromosome: an evolutionary marker comes of age. Nat. Rev. Genet. 4: 598–612. [DOI] [PubMed] [Google Scholar]
- Kayser, M., L. Roewer, M. Hedman, L. Henke, J. Henke et al., 2000. Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am. J. Hum. Genet. 66: 1580–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser, M., R. Kittler, A. Erler, M. Hedman, A. C. Lee et al., 2004. A comprehensive survey of human Y-chromosomal microsatellites. Am. J. Hum. Genet. 74: 1183–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W. J., R. Baertsch, A. Hinrichs, W. Miller and D. Haussler, 2003. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. USA 100: 11484–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson, G., and G. A. Gutman, 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4: 203–221. [DOI] [PubMed] [Google Scholar]
- Primmer, C. R., and H. Ellegren, 1998. Patterns of molecular evolution in avian microsatellites. Mol. Biol. Evol. 15: 997–1008. [DOI] [PubMed] [Google Scholar]
- Primmer, C. R., N. Saino, A. P. Moller and H. Ellegren, 1998. Unraveling the processes of microsatellite evolution through analysis of germ line mutations in barn swallows Hirundo rustica. Mol. Biol. Evol. 15: 1047–1054. [Google Scholar]
- Rubinsztein, D. C., W. Amos, J. Leggo, S. Goodburn, S. Jain et al., 1995. Microsatellite evolution—evidence for directionality and variation in rate between species. Nat. Genet. 10: 337–343. [DOI] [PubMed] [Google Scholar]
- Rubinsztein, D. C., B. Amos and G. Cooper, 1999. Microsatellite and trinucleotide-repeat evolution: evidence for mutational bias and different rates of evolution in different lineages. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 354: 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajantila, A., M. Lukka and A. C. Syvanen, 1999. Experimentally observed germline mutations at human micro- and minisatellite loci. Eur. J. Hum. Genet. 7: 263–266. [DOI] [PubMed] [Google Scholar]
- Schlötterer, C., and D. Tautz, 1992. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 20: 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, R., J. K. Pritchard, P. D. Shen, P. J. Oefner and M. W. Feldman, 2000. Recent common ancestry of human Y-chromosomes: evidence from DNA sequence data. Proc. Natl. Acad. Sci. USA 97: 7360–7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowles, E. J., and W. Amos, 2006. Quantifying ascertainment bias and species-specific length differences in human and chimpanzee microsatellites using genome sequences. Mol. Biol. Evol. 23(3): 598–607. [DOI] [PubMed] [Google Scholar]
- Webster, M. T., N. G. C. Smith and H. Ellegren, 2002. Microsatellite evolution inferred from human-chimpanzee genomic sequence alignments. Proc. Natl. Acad. Sci. USA 99: 8748–8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., M. Peng, Z. Fang and X. P. Xu, 2000. The direction of microsatellite mutations is dependent upon allele length. Nat. Genet. 24: 396–399. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., D. C. Queller and J. E. Strassmann, 2000. A phylogenetic perspective on sequence evolution in microsatellite loci. J. Mol. Evol. 50: 324–338. [DOI] [PubMed] [Google Scholar]