Abstract
The extent of linkage disequilibrium (LD) is an important factor in designing association mapping experiments. Unlike other plant species that have been analyzed so far for the extent of LD, cultivated potato (Solanum tuberosum L.), an outcrossing species, is a highly heterozygous autotetraploid. The favored genotypes of modern cultivars are maintained by vegetative propagation through tubers. As a first step in the LD analysis, we surveyed both coding and noncoding regions of 66 DNA fragments from 47 accessions for single nucleotide polymorphism (SNP). In the process, we combined information from the potato SNP database with experimental SNP detection. The total length of all analyzed fragments was >25 kb, and the number of screened sequence bases reached almost 1.4 million. Average nucleotide polymorphism (θ = 11.5 × 10−3) and diversity (π = 14.6 × 10−3) was high compared to the other plant species. The overall Tajima's D value (0.5) was not significant, but indicates a deficit of low-frequency alleles relative to expectation. To eliminate the possibility that an elevated D value occurs due to population subdivision, we assessed the population structure with probabilistic statistics. The analysis did not reveal any significant subdivision, indicating a relatively homogenous population structure. However, the analysis of individual fragments revealed the presence of subgroups in the fragment closely linked to the R1 resistance gene. Data pooled from all fragments show relatively fast decay of LD in the short range (r2 = 0.208 at 1 kb) but slow decay afterward (r2 = 0.137 at ∼70 kb). The estimate from our data indicates that LD in potato declines below 0.10 at a distance of ∼10 cM. We speculate that two conflicting factors play a vital role in shaping LD in potato: the outcrossing mating type and the very limited number of meiotic generations.
THE development of new cultivars is a lengthy process that can be expedited if the genes for desirable traits are mapped and tagged with molecular markers. Recently, the association mapping method became an important tool in plant genetics. The method exploits observed biodiversity in existing material without the need to develop new mapping populations. The association mapping method was successfully applied to map genes in several plant species, including potato (Gebhardt et al. 2004; Simko 2004; Simko et al. 2004a,b). The power and resolution of association mapping depends on the extent of linkage disequilibrium (LD) in mapping populations. LD is characterized as the nonrandom association of alleles at different loci and can be affected by most of the processes observed in population genetics, including mating pattern, frequency of recombination, and population history (Flint-Garcia et al. 2003; Rafalski and Morgante 2004). From plant species, LD has been studied most extensively in Arabidopsis (Nordborg et al. 2002) and maize (Remington et al. 2001; Tenaillon et al. 2001; Ching et al. 2002); however, little is known about LD in potato.
So far, almost all of the LD studies on plants have been performed on highly homozygous material developed by repeated selfing. Unlike other investigated plant species, cultivated potato (Solanum tuberosum L.) is a highly heterozygous autotetraploid (2n = 4x = 48) with complex polysomic inheritance. Although the species is self-compatible, Simmonds and Smartt (1999) classify potato as an outcrosser because it suffers from severe inbreeding depression that prevents development of homozygous lines. The heterogenous genotype of modern cultivars is fixed by vegetative propagation through tubers. Due to the narrow genetic base (Love 1999) and vegetative mode of propagation, most of the cultivars are highly related to each other (Simko 2004) and are separated by only a few meiotic generations (Gebhardt et al. 2004). Conversely, wild Solanum is a highly diverse group of species with reference to their ploidy and mating type.
Single nucleotide polymorphism (SNP) is a single-point mutation in which one nucleotide is substituted with another at a particular locus. To discover SNPs in a specific DNA region, several representative genotypes must be sampled from the target population and their sequences compared. SNPs are markers of choice in association studies owing to their abundance, amenability to high-throughput screening, and usually biallelic status. Recently, Rickert et al. (2003) screened a part of the potato genome for the presence of SNPs that could be used for tagging pathogen resistance loci. They applied pyrosequencing and the single nucleotide primer extension method to detect polymorphic loci in a panel of 17 tetraploid and 11 diploid genotypes. All sequences from this study, including SNPs position, were deposited into the publicly available PoMaMo database (Meyer et al. 2005).
Here we report the results from initial assessment of LD in potato. To estimate the extent of LD, we surveyed loci that include both coding and noncoding regions of the potato genome. In the process, we combined available information from the PoMaMo database with experimental SNP detection. Our goal was to provide initial information about LD pattern in potato that would help in prospective association studies.
MATERIALS AND METHODS
Plant material:
A set of 47 potato accessions was analyzed for the presence of nucleotide variation. This set consisted of 1 monoploid, 17 diploid, and 29 tetraploid accessions (Table 1). Most of the accessions originated from S. tuberosum, but the presence of other Solanum species (S. berthaultii, S. chacoense, S. kurtzianum, S. phureja, S. tarijense, S. vernei, S. yungasense) is evident in the known pedigrees. Monoploid and diploid accessions included in the analysis represent material used in the resistance-breeding programs; tetraploid accessions correspond to diversity of cultivated potato (S. tuberosum). The analyzed set also includes major genetic contributors of the germplasm for prominent cultivars (Love 1999).
TABLE 1.
Accessions used in the SNP and LD analysis
| Accession | Ploidy | Pedigree | Reference |
|---|---|---|---|
| 1-3/84 | 1× | Solanum phureja | Varrieur (2002) |
| B11-A | 2× | S. berthaultii | Bonierbale et al. (1994) |
| BCT-61 | 2× | S. berthaultii, S. tuberosum | Bonierbale et al. (1994) |
| DG81 | 2× | S. chacoense, S. tuberosum, S. yungasense | Rickert et al. (2003) |
| DG83 | 2× | S. chacoense, S. tuberosum, S. yungasense | Rickert et al. (2003) |
| G87 | 2× | S. kurtzianum, S. tarijense, S. tuberosum, S. vernei | Rickert et al. (2003) |
| HH1-9 | 2× | S. tuberosumb | Bonierbale et al. (1994) |
| M200-30D | 2× | S. berthaultii, S. tuberosum | Bonierbale et al. (1994) |
| P3 | 2× | S. tuberosumb | Rickert et al. (2003) |
| P6/210a | 2× | S. tuberosumb | Rickert et al. (2003) |
| P18 | 2× | S. tuberosumb | Rickert et al. (2003) |
| P38 | 2× | S. tuberosumb | Rickert et al. (2003) |
| P40 | 2× | S. tuberosumb | Rickert et al. (2003) |
| P41 | 2× | S. tuberosumb | Rickert et al. (2003) |
| P50 | 2× | S. tuberosumb | Rickert et al. (2003) |
| P54 | 2× | S. tuberosumb | Rickert et al. (2003) |
| Sph | 2× | S. phureja | Rickert et al. (2003) |
| USW22-30 | 2× | S. tuberosumb | Bonierbale et al. (1994) |
| Atlantic | 4× | S. tuberosum | Named variety |
| B0718-3 | 4× | S. tuberosum | USDA breeding line |
| Bintje | 4× | S. tuberosum | Named variety |
| Cherokee | 4× | S. tuberosum | Named variety |
| Desiree | 4× | S. tuberosum | Named variety |
| Katahdin | 4× | S. tuberosum | Named variety |
| Kennebec | 4× | S. tuberosum | Named variety |
| NKA1 | 4× | S. tuberosumc | Rickert et al. (2003) |
| NKA2 | 4× | S. tuberosumc | Rickert et al. (2003) |
| NKA3 | 4× | S. tuberosumc | Rickert et al. (2003) |
| NKA4 | 4× | S. tuberosumc | Rickert et al. (2003) |
| NKA5 | 4× | S. tuberosumc | Rickert et al. (2003) |
| NKA6 | 4× | S. tuberosumc | Rickert et al. (2003) |
| NKA7 | 4× | S. tuberosumc | Rickert et al. (2003) |
| NKA8 | 4× | S. tuberosumc | Rickert et al. (2003) |
| Pontiac | 4× | S. tuberosum | Named variety |
| Russet Burbank | 4× | S. tuberosum | Named variety |
| SR1 | 4× | S. tuberosumc | Rickert et al. (2003) |
| SR10 | 4× | S. tuberosumc | Rickert et al. (2003) |
| SR11 | 4× | S. tuberosumc | Rickert et al. (2003) |
| SR12 | 4× | S. tuberosumc | Rickert et al. (2003) |
| SR2 | 4× | S. tuberosumc | Rickert et al. (2003) |
| SR3 | 4× | S. tuberosumc | Rickert et al. (2003) |
| SR4 | 4× | S. tuberosumc | Rickert et al. (2003) |
| SR5 | 4× | S. tuberosumc | Rickert et al. (2003) |
| SR6 | 4× | S. tuberosumc | Rickert et al. (2003) |
| Superior | 4× | S. tuberosum | Named variety |
| USDA 41956 | 4× | S. tuberosum | USDA breeding line |
| USDA ×96-56 | 4× | S. tuberosum | USDA breeding line |
Accession used for the BAC library construction.
Dihaploid material (2n = 2x = 24) derived from tetraploid S. tuberosum. The dihaploids as well as most of the diploid potato species are self-incompatible.
Exact pedigree information is not available, but the accession likely originates from an S. tuberosum cross.
Detection of nucleotide variation:
To assess the polymorphisms in potato, 66 fragments distributed across all potato chromosomes were surveyed. SNPs were detected experimentally by sequencing or in silico by analyzing potato sequences deposited in the PoMaMo database (Meyer et al. 2005). Sequences from potato accession cited by Rickert et al. (2003) in Table 1 originate from the online database; sequences from all other accessions were generated in our laboratory.
Total genomic DNA was extracted from fresh in vitro plants using the DNeasy plant mini-prep kit (QIAGEN, Valencia, CA). Primers and conditions to amplify DNA fragments were the same as described in the PoMaMo database. The StVe1 locus was amplified according to specifications in Simko et al. (2004b). PCR products were sequenced directly or cloned with the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) first, if necessary. Direct sequencing of PCR products was carried in the absence of insertions and deletions (indels). When indels were present, PCR amplicons were cloned and 12 colonies/tetraploid accession or 4 colonies/diploid accession were sequenced (Simko 2004). Amplicons from the monoploid (2n = 1x = 12) accession were used to identify DNA fragments containing paralogs, and such fragments were excluded from further data analysis. Nucleotide variations detected experimentally and in silico were then combined and analyzed with PolyBayes SNP detection software (Marth et al. 1999). To discern true allelic variations from sequencing errors, PolyBayes considers alignment depth, the base quality, and base composition to calculate probability that the sequences represent true variants. This approach also helps eliminate PCR errors, unless they are occurring systematically in the exact same DNA region. Sequence variants were considered to be true SNPs when the PolyBayes probability score exceeded 0.99. Since all singletons had scores <0.99, they were excluded from linkage disequilibrium analysis. Similarly, insertions and deletions were observed, but not used in data analyses.
Data analysis:
The level of genetic variation at the nucleotide level was estimated as nucleotide polymorphism (θ, Watterson 1975) and nucleotide diversity (π, Tajima 1983). Watterson's θ is based on the number of segregating sites, while Tajima's π is based on the pairwise differences between sequences in the sample. To test the neutrality of mutations, we employed Tajima's D test (Tajima 1989) that is based on differences between π and θ. Haplotypes in each fragment were identified from the cloned and sequenced PCR products or inferred with Haploview software (Barrett et al. 2005) if amplicons were sequenced directly.
Surveyed fragments originated from RFLP markers, BAC library insertions, and known genes for which sequences were available in the PoMaMo database in March 2005. The average insert size in the BAC library is ∼70 kb and surveyed fragments corresponded to sequenced insert ends (Rickert et al. 2003). Fragments were included in the data analysis if sequence information for all alleles was available from at least 10 different accessions. Description of individual fragments and their positions on the potato molecular linkage map is available in the PoMaMo database; StVe1 is described in Simko et al. (2004b).
To identify fragments coding functional sequence, all fragments were compared with the existing Solanaceae expressed sequence tag (EST) and plant protein databases (NCBI: http://www.ncbi.nlm.nih.gov; SGN: http://www.sgn.cornell.edu; and TIGR: http://www.tigr.org). The region was considered a putative coding region if the scores from the EST (blastn) and protein (blastx) query were at least 200 and 100, respectively.
Chromatograms were viewed and aligned with BioEdit (Hall 1999) and Clustal X (Thompson et al. 1997). Analyses of genetic variation were carried out using DnaSP sequence polymorphism software (Rozas and Rozas 1999). Linkage disequilibrium (r2 and D′) between two loci in the genome was calculated with Haploview (Barrett et al. 2005). In five cases, three different alleles per locus were detected in a few accessions. Since Haploview cannot handle this type of data, the loci were “diploidized” and the least frequent allele was discarded. Decay of LD with distance was estimated from a logarithmic trend line fit to the data (Hamblin et al. 2004; Hyten 2005). Population structure was evaluated using probabilistic statistics implemented in the program Structure (Pritchard et al. 2000). Distances between surveyed loci were calculated from their respective positions on the molecular linkage map in the PoMaMo database (http://gabi.rzpd.de).
RESULTS
Sequence polymorphism:
To detect DNA sequence polymorphisms we surveyed 66 fragments with length (including indels) in a range between ∼100 and ∼1100 bp. Three-quarters of the fragments were between 250 and 650 bp long, 15% were <250 bp, and 10% were >650 bp. Due to either failure of primers to amplify product or missing data in the PoMaMo database, not all accessions were always informative; therefore, the sample size for individual fragments differs. The total length of all analyzed amplicons was >25 kb, and the number of screened sequence bases reached almost 1.4 million (Table 2). In total, we detected 1145 sequence variants, of which ∼95% were nucleotide substitutions and the remaining 5% were indels. The most frequently observed types of nucleotide substitutions were biallelic transitions (C ↔ T, A ↔ G) followed by biallelic transversions (A ↔ T, G ↔ T, A ↔ C, G ↔ C). The transition/transversion (TI/TV) ratio was close to 1.5, almost three times higher than would be expected if all nucleotide exchanges happen with the same frequency. On average, one (biallelic) SNP was observed every 24 bp or every 23 bp if rare tri- and tetra-allelic substitutions are considered.
TABLE 2.
Estimates of nucleotide variation
| Parameter | Value | Frequency |
|---|---|---|
| No. of loci screened | 66 | |
| Total length of amplicons in base pairs | 25,138 | |
| No. of bases of sequence screened | 1,397,461 | |
| No. of all sequence variants | 1,145 | 1/22 bp |
| No. of indels | 57 | 1/441 bp |
| No. of nucleotide substitutions | 1,088 | 1/23 bp |
| No. of biallelic nucleotide substitutions | 1,055 | 1/24 bp |
| Transitions/transversions (TI/TV) ratio | 1.48 | |
| No. of triallelic nucleotide substitutions | 30 | 1/838 bp |
| No. of tetra-allelic nucleotide substitutions | 3 | 1/8379 bp |
| Nucleotide polymorphism (θ × 10−3), mean | 11.5 | |
| Nucleotide polymorphism (θ × 10−3), coding region | 10.1 | |
| Nucleotide polymorphism (θ × 10−3), nonsynonymous level of diversity | 6.3 | |
| Nucleotide polymorphism (θ × 10−3), synonymous level of diversity | 14.9 | |
| Nucleotide polymorphism (θ × 10−3), noncoding region | 11.9 | |
| Nucleotide polymorphism ratio, coding/noncoding | 0.85 | |
| Nucleotide polymorphism ratio, nonsynonymous/synonymous | 0.42 | |
| Nucleotide diversity (π × 10−3), mean | 14.6 | |
| Nucleotide diversity (π × 10−3), coding region | 11.2 | |
| Nucleotide diversity (π × 10−3), nonsynonymous level of diversity | 9.3 | |
| Nucleotide diversity (π × 10−3), synonymous level of diversity | 18.0 | |
| Nucleotide diversity (π × 10−3), noncoding region | 15.8 | |
| Nucleotide diversity ratio, coding/noncoding | 0.71 | |
| Nucleotide diversity ratio, nonsynonymous/synonymous | 0.52 | |
| Tajima's D, mean | 0.5 | |
| Tajima's D, coding region | 0.2 | |
| Tajima's D, noncoding region | 0.9 |
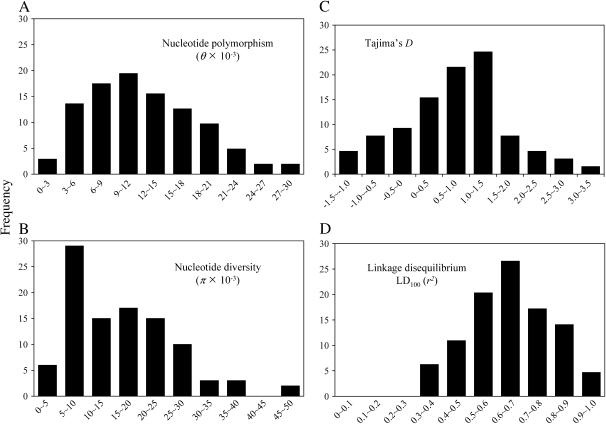
In general, nucleotide polymorphism (θ = 11.5 × 10−3) and diversity (π = 14.6 × 10−3) were high in the analyzed part of the potato genome. The values for polymorphism ranged from 1.9 × 10−3 to 29.3 × 10−3 and for diversity from 1.6 × 10−3 to 45.2 × 10−3 (Figure 1, A and B). Both nucleotide polymorphism and diversity was higher in noncoding than in coding regions. Within coding regions, synonymous levels of diversity were more than twice as common as nonsynonymous levels of diversity (Table 2). Tajima's test of neutrality of mutations revealed a significant departure from neutral expectations in 9% of the analyzed fragments (Figure 1C). All of these fragments showed positive D values indicating a deficit of low-frequency alleles relative to expectation. The mean value of D for all fragments was 0.5, but the value was generally higher in noncoding than in coding regions (0.9 and 0.2, respectively, Table 2).
Figure 1.—
Distribution of (A) nucleotide polymorphism (θ × 10−3), (B) nucleotide diversity (π × 10−3), (C) Tajima's D, and (D) linkage disequilibrium (r2) within 100 bp (LD100) in the surveyed fragments.
Linkage disequilibrium:
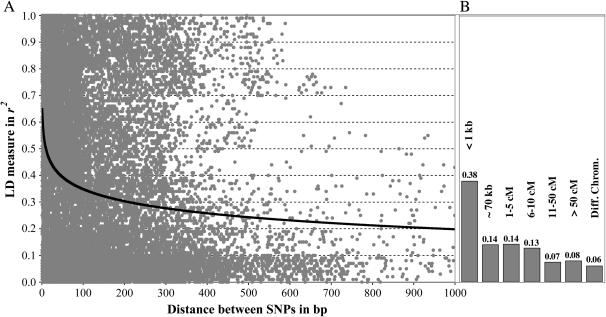
LD between two loci in a genome can be estimated by a number of statistics, of which the most common are r2 and D′. Both statistics have a range from 0 (equilibrium) to 1 (disequilibrium). Although neither r2 nor D′ performs extremely well with small sample sizes, we used the r2 statistic, as it is indicative of how the marker might correlate with the allele of interest (Flint-Garcia et al. 2003). Since most of the fragments are <1 kb long, the analysis reveals disequilibrium patterns at a short distance, ≤1 kb. The r2 value pooled over the entire data set shows a gradual decline in LD as a function of distance and reaches a value of ∼0.21 at 1 kb (Figure 2A). To observe decay of LD over distances >1 kb, we calculated r2 between polymorphic loci detected in different fragments, but originating from the same BAC clone. Since the average insert size in the BAC library is ∼70 kb (Rickert et al. 2003) and surveyed fragments corresponded to insert ends, an approximate distance between two loci within the same BAC clone can be calculated. In addition, the chromosomal location of all analyzed fragments is known (PoMaMo database) and therefore distance (in centimorgans) between two polymorphic loci from different BAC clones can be inferred. Average r2 between two loci separated by ∼70 kb was 0.14, which is substantially smaller than average values detected for the short-range (≤1 kb) LD (0.38). Additional analyses showed progressive decay of LD, and loci separated by >50 cM had an r2 value of 0.08 only. The lowest LD was observed between unlinked loci from different chromosomes (r2 = 0.06, Figure 2B).
Figure 2.—
Decay of linkage disequilibrium (r2) as a function of distance between two polymorphic sites. Pooled data from all surveyed fragments were used to estimate the (A) short-range (≤1 kb) and (B) long-range linkage disequilibrium. Note that distances for the long-range LD are in kilobase pairs (kb) and centimorgans (cM). The last column indicates LD between polymorphic loci from different chromosomes.
Population structure:
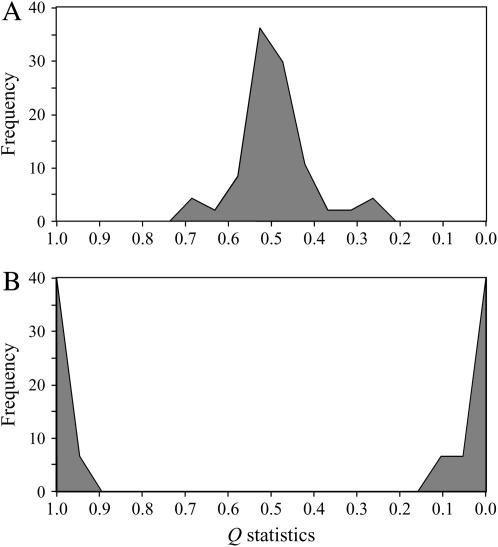
We did not observe population stratification in the surveyed set of accessions when all DNA fragments were included in the analysis together (Figure 3A). Similar results were obtained when each chromosome was analyzed separately. The only case of evident population structure was detected in the BA121o1-T7 BAC clone when stratification analysis was carried on individual fragments (Figure 3B).
Figure 3.—
Inferred population structure for the set of potato accessions. (A) Results based on SNPs from all surveyed fragments. (B) Results based on SNPs from the BA121o1-T7 fragment only. Q statistics indicate the proportion of an accession's genome that belongs to the first of two possible subgroups (K = 2).
DISCUSSION
Nucleotide variation:
There is a substantial level of variation in fragments that were included in this study. Nucleotide substitution in potato—1 SNP/23 bp in this study and 1 SNP/21 bp detected by Rickert et al. (2003)—translates into ∼1 SNP/87 bp (∼1/θ) between pairs of randomly selected sequences. This level of polymorphism is higher than was observed in many other cultivated plant species. For example, there is 1 SNP/60 bp in aspen (Ingvarsson 2005), 104 bp in maize (Tenaillon et al. 2001), 130 bp in sugar beet (Schneider et al. 2001), 232 bp in rice (Nasu et al. 2002), 435 bp in sorghum (Hamblin et al. 2004), and 1030 bp in soybean (Zhu et al. 2003). When potato nucleotide polymorphism (θ) and diversity (π) are compared with other crops where both coding and noncoding regions were analyzed, total polymorphism in potato (θ = 11.5 × 10−3) is similar to that in maize (9.6 × 10−3, Tenaillon et al. 2001), but ∼12-fold larger than that in soybean (0.97 × 10−3, Zhu et al. 2003). Similarly, the total nucleotide diversity (π = 14.6 × 10−3) in potato is larger than that in the sugar beet (7.6 × 10−3, Schneider et al. 2001), maize elite lines (6.3 × 10−3, Ching et al. 2002), and soybean (1.25 × 10−3, Zhu et al. 2003). Although definitively not all, at least a part of such high polymorphism in potato may be explained by mating system. It has been observed before that outcrossing species have higher levels of sequence variation than selfing species (Pollak 1987). For example, Baudry et al. (2001) found that self-incompatible Lycopersicon species are up to 40 times more variable than self-compatible species. Similarly, nucleotide variation was substantially reduced in self-pollinating Leavenworthia species (Liu et al. 1999). Bamberg and del Rio (2004) compared four wild Solanum species for level of genetic diversity on the basis of evaluation of RAPD markers. Outcrossing diploid species had substantially greater genetic diversity than both tetraploid and diploid selfing species. Even higher diversity was observed in outcrossing tetraploid species, suggesting that not only mating type but also ploidy plays a role in population diversity.
The ratio of transitions to transversions in potato (1.48) was on par with sugar beet (1.63, Schneider et al. 2001) but larger than in soybean (0.93, Zhu et al. 2003). Assuming complete randomness of mutations, the expected TI/TV ratio would be 1:2 or 0.5. A clear bias toward transitions indicates that each type of transitional change (purine ↔ purine, pyrimidine ↔ pyrimidine) is produced almost three times more often than each type of transversional change (purine ↔ pyrimidine).
The ratio of nucleotide diversity in coding and noncoding sequences (0.71) was higher than that observed in Arabidopsis (0.38 calculated by Zhu et al. 2003 from other experiments), soybean (0.45, Zhu et al. 2003), and maize (0.65, Tenaillon et al. 2001). It is possible that the higher ratio observed in potato is indicative of regulatory or splicing functions of noncoding perigenic sequence (Cargill et al. 1999). Another plausible explanation is that sorting of surveyed sequences into the coding and noncoding regions in silico was not always accurate, leading to an increased ratio. To test accuracy of the sorting, the in silico approach was applied on known functional genes surveyed in this study. All of the tested fragments were correctly classified, indicating that the method identifies coding regions well. Conversely, we cannot dismiss the possibility of false-positive results, although the combination of two threshold values (200 for blastn and 100 for blastx) should reduce misclassification of the noncoding regions.
In the coding region of analyzed fragments, we observed a relatively high frequency of synonymous mutations when compared to nonsynonymous mutations. The ratio between nonsynonymous and synonymous polymorphism (0.42) suggests a natural selection that eliminates mutations resulting in deleterious amino acid replacement. This ratio is close to 0.38 observed in soybean (Zhu et al. 2003), 0.34 in both Arabidopsis (calculated from Olsen et al. 2002 by Zhu et al. 2003) and the maize Dwarf8 gene (Thornsberry et al. 2001), but considerably higher than 0.29 in aspen (Ingvarsson 2005), 0.26 in sorghum (Hamblin et al. 2004), or 0.23 in maize chromosome 1 (Tenaillon et al. 2001). The ratio of nonsynonymous to synonymous polymorphism observed in potato suggests a relatively low level of purifying selection in comparison with other plant species. This may be due to the autotetraploid nature of cultivated potato, in which deleterious alleles are masked by the extra genomes. We found a strong correlation (r = 0.91, P < 0.001) between the synonymous and nonsynonymous levels of diversity in individual fragments. This correlation may be caused by dissimilar mutation rates in the surveyed fragments.
To test the neutrality of mutations and to provide information about possible population structure Tajima's D was calculated across all surveyed fragments. The overall D value was relatively high (0.5), although not significant. Positive D indicates a deficit of low-frequency alleles relative to expectations. This could be due to a population bottleneck, population subdivision, or balancing selection (Ching et al. 2002). The value in potato is between those detected in sorghum (0.30, Hamblin et al. 2004) and soybean (1.08, Zhu et al. 2003), both of which show a significant bottleneck in their population history. To eliminate the possibility that elevated D value occurs due to population subdivision, we assessed population structure with the probabilistic statistic suggested by Pritchard et al. (2000). When SNPs from all chromosomes were included into the analysis, no significant subdivision was observed (Figure 3A) indicating a relatively homogenous population. However, the analysis of individual fragments revealed the presence of two separate subgroups in BA121o1-T7 (Figure 3B). Perhaps, because of population subdivision, this fragment has the largest D value (3.2) of all surveyed fragments (Figure 1C). When Tajima's D was calculated separately for the two subgroups the value decreased to −0.2 and −0.4, respectively, providing additional evidence of population structure in this fragment. Examination of the two subgroups indicated that one of them includes accessions with the R1 gene for race-specific resistance to Phytophthora infestans, while the other one contains accessions without R1. It was observed previously that the BA121o1 clone is located in the R1 gene area (Ballvora et al. 2002). Interestingly, polymorphism of the BA121o1-T7 fragment is 20-fold smaller among accessions with the resistance gene than among those that do not carry the gene. It appears that the BA121o1-T7 fragment in the first group is under strong selective pressure. The selective pressure can target the fragment either directly or indirectly through genetic association with the R1 gene. However, information about pedigree and resistance response is too limited to make a reliable conclusion regarding this hypothesis.
Linkage disequilibrium:
Mating system influences population size and effective rate of recombination in plants. Even if selfing species may have an increased recombination rate per meiosis, selfing increases homozygosity, thereby limiting the number of heterozygotes that can be shuffled by recombination. For this reason, selfing dramatically reduces the effective recombination rate (Nordborg 2000) and LD in predominantly selfing species generally extends over a longer physical distance. Authors studying selfing species observed LD extending for >150 kb in Arabidopsis (Nordborg et al. 2002), ∼100 kb in rice (Garris et al. 2003), and >50 kb in soybean (Zhu et al. 2003). Conversely, LD in outcrossing maize (Remington et al. 2001) and aspen (Ingvarsson 2005) declines to a negligible level (r2 < 0.1), usually within 1 kb. Extent of LD in potato appears to be between these two groups; r2 at 1 kb was 0.208 and declined to 0.137 at physical distance of ∼70 kb. It seems that after an initial relatively fast decline, decay of LD slows and is not as dramatic as in some other outcrossing species. This could be because of the vegetative mode of propagation that leads to a very limited number of meiotic generations separating S. tuberosum accessions. When Gebhardt et al. (2004) compared genotypes from the German potato GenBank they found that almost 40% of the accessions were separated from each other by only one meiotic generation.
However, LD can also be affected by origin of the analyzed population. Hyten (2005) compared four different soybean populations on level of LD decline. While in the domesticated Asian Glycine max population LD did not decline along the 500-kb sequenced region, the wild G. soja population had large LD decline with LD block size averaging 12 kb. Comparable observations were made in maize (Tenaillon et al. 2001) and aspen (Ingvarsson 2005). It would be interesting to make a similar comparison in potato. There is enormous variability in ploidy, mating type, and effective population size in wild species, primitive cultivated species, and modern cultivated varieties. Unfortunately, the present set does not allow such comparison, since most of the accessions originate from S. tuberosum and also because the origin of many of the accessions is not completely known. We hypothesize that differences in LD extent among various potato populations are considerable. For example, S. tuberosum is a vegetative propagated species that went through domestication that created a bottleneck in the effective population size (e.g., Simko 2004 and the citations herein), was subject of artificial selection at a number of production- and resistance-related genes, and shows a high level of coancestry among modern cultivars (Love 1999) that are separated by only a few meiotic generations (Gebhardt et al. 2004).
All of these factors slow the decay of LD and indicate that LD extent in cultivated S. tuberosum should generally be longer than in outcrossing wild potato species. Conversely, some of the wild potatoes are predominantly selfing species (e.g., diploid S. verrucosum and tetraploid S. fendleri) with a low level of diversity (Bamberg and del Rio 2004), and thus the extent of LD in these species might be relatively long. Another factor affecting LD extent in cultivated potato is its autotetraploid nature that may allow accumulation of recessive mutations at a higher rate. High mutation rate generally decreases LD, but LD around newly created mutated alleles remains high until dissipated by recombination (Rafalski and Morgante 2004).
Conclusions:
Our assessment of genomewide LD used 66 DNA fragments from both coding and noncoding regions that were distributed across the potato genome. Analysis of these fragments indicates relatively high nucleotide variation in potato as compared to other plant species. Initial data relating to the decay of LD suggest that LD in potato is less extensive than that in selfing Arabidopsis or soybean, but longer than that in outcrossing maize or aspen. Assuming only the biallelic nucleotide substitutions with equal frequency of distribution and ∼100 alleles per locus, a statistical significance of the observed allelic association between two polymorphic loci can be detected (by Fisher's exact test) if r2 > 0.13. A value this high (on the average) was still seen at the distance of 1–5 cM (r2 = 0.142, Figure 2B), indicating that the association test can be possibly used at a relatively long distance. Yet, this estimate of LD extent is based on pooled data only and differences among genomic regions could be substantial, as illustrated by the range of r2 values (0.32–0.95) at the distance of ≤100 bp (LD100, Figure 1D). Therefore it is essential to analyze additional genomic regions and populations, representing the high variability observed in potato. Populations of selfing potato species might have a long LD and be good candidates for genomewide association mapping, while populations of outcrossing species will likely show a short LD and be suited more for high-resolution mapping. A set of populations with a range of LD will be a vital tool for gene mapping in potato with the association mapping approach.
Acknowledgments
The authors thank W. De Jong and four anonymous reviewers for valuable suggestions, and R. Veilleux for monoploid potato genotypes. This project was supported in part by the Agricultural Research Sciences potato research program.
References
- Ballvora, A., M. R. Ercolano, J. Weiss, K. Meksem, C. A. Bormann et al., 2002. The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 30: 361–371. [DOI] [PubMed] [Google Scholar]
- Bamberg, J. B., and A. H. del Rio, 2004. Genetic heterogeneity estimated by RAPD polymorphism of four tuber-bearing potato species differing by breeding system. Am. J. Potato Res. 81: 377–383. [Google Scholar]
- Barrett, J. C., B. Fry, J. Maller and M. J. Daly, 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- Baudry, E., C. Kerdelhue, H. Innan and W. Stephan, 2001. Species and recombination effects on DNA variability in the tomato genus. Genetics 158: 1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonierbale, M. W., R. L. Plaisted, O. Pineda and S. D. Tanksley, 1994. QTL analysis of trichome-mediated insect resistance in potato. Theor. Appl. Genet. 87: 973–987. [DOI] [PubMed] [Google Scholar]
- Cargill, M., D. Altshuler, J. Ireland, P. Sklar, K. Ardlie et al., 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22: 231–238. [DOI] [PubMed] [Google Scholar]
- Ching, A., K. S. Caldwell, M. Jung, M. Dolan, O. S. Smith et al., 2002. SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genet. 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint-Garcia, S. A., J. M. Thornsberry and E. S. Buckler, 2003. Structure of linkage disequilibrium in plants. Ann. Rev. Plant Biol. 54: 357–374. [DOI] [PubMed] [Google Scholar]
- Garris, A. J., S. R. McCouch and S. Kresovich, 2003. Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.). Genetics 165: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt, C., A. Ballvora, B. Walkemeier, P. Oberhagemann and K. Schüler, 2004. Assessing genetic potential in germ plasm collections of crop plants by marker-trait association: a case study for potatoes with quantitative variation of resistance to late blight and maturity type. Mol. Breed. 13: 93–102. [Google Scholar]
- Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Hamblin, M. T., S. E. Mitchell, G. M. White, W. Gallego, R. Kukatla et al., 2004. Comparative population genetics of the panicoid grasses: sequence polymorphism, linkage disequilibrium and selection in a diverse sample of Sorghum bicolor. Genetics 167: 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten, D. L., 2005. Genetic diversity and linkage disequilibrium in wild soybean, landraces, ancestral, and elite soybean populations. Ph.D. Thesis, University of Maryland, College Park, MD.
- Ingvarsson, P. K., 2005. Nucleotide polymorphism and linkage disequilbrium within and among natural populations of European aspen (Populus tremula L., Salicaceae). Genetics 169: 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F., D. Charlesworth and M. Kreitman, 1999. The effect of mating system differences on nucleotide diversity at the phosphoglucose isomerase locus in the plant genus Leavenworthia. Genetics 151: 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, S. L., 1999. Founding clones, major contributing ancestors, and exotic progenitors of prominent North American potato cultivars. Am. J. Potato Res. 76: 263–272. [Google Scholar]
- Marth, G. T., I. Korf, M. D. Yandell, R. T. Yeh, Z. J. Gu et al., 1999. A general approach to single-nucleotide polymorphism discovery. Nat. Genet. 23: 452–456. [DOI] [PubMed] [Google Scholar]
- Meyer, S., A. Nagel and C. Gebhardt, 2005. PoMaMo: a comprehensive database for potato genome data. Nucleic Acids Res. 33: D666–D670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu, S., J. Suzuki, R. Ohta, K. Hasegawa, R. Yui et al., 2002. Search for and analysis of single nucleotide polymorphisms (SNPs) in rice (Oryza sativa, Oryza rufipogon) and establishment of SNP markers. DNA Res. 9: 163–171. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., 2000. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics 154: 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., J. O. Borevitz, J. Bergelson, C. C. Berry, J. Chory et al., 2002. The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 30: 190–193. [DOI] [PubMed] [Google Scholar]
- Olsen, K. M., A. Womack, A. R. Garrett, J. I. Suddith and M. D. Purugganan, 2002. Contrasting evolutionary forces in the Arabidopsis thaliana floral developmental pathway. Genetics 160: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak, E., 1987. On the theory of partially inbreeding finite populations. 1. Partial selfing. Genetics 117: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens and P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski, A., and M. Morgante, 2004. Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends Genet. 20: 103–111. [DOI] [PubMed] [Google Scholar]
- Remington, D. L., J. M. Thornsberry, Y. Matsuoka, L. M. Wilson, S. R. Whitt et al., 2001. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 98: 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert, A. M., J. H. Kim, S. Meyer, A. Nagel, A. Ballvora et al., 2003. First-generation SNP/InDel markers tagging loci for pathogen resistance in the potato genome. Plant Biotechnol. J. 1: 399–410. [DOI] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Schneider, K., B. Weisshaar, D. C. Borchardt and F. Salamini, 2001. SNP frequency and allelic haplotype structure of Beta vulgaris expressed genes. Mol. Breed. 8: 63–74. [Google Scholar]
- Simko, I., 2004. One potato, two potato: haplotype association mapping in autotetraploids. Trends Plant Sci. 9: 441–448. [DOI] [PubMed] [Google Scholar]
- Simko, I., S. Costanzo, K. G. Haynes, B. J. Christ and R. W. Jones, 2004. a Linkage disequilibrium mapping of a Verticillium dahliae resistance quantitative trait locus in tetraploid potato (Solanum tuberosum) through a candidate gene approach. Theor. Appl. Genet. 108: 217–224. [DOI] [PubMed] [Google Scholar]
- Simko, I., K. G. Haynes, E. E. Ewing, S. Costanzo, B. J. Christ et al., 2004. b Mapping genes for resistance to Verticillium albo-atrum in tetraploid and diploid potato populations using haplotype association tests and genetic linkage analysis. Mol. Genet. Genomics 271: 522–531. [DOI] [PubMed] [Google Scholar]
- Simmonds, N. W., and J. Smartt, 1999. Principles of Crop Improvement. Blackwell Science, Oxford.
- Tajima, F., 1983. Evolutionary relationship of DNA-sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M. I., M. C. Sawkins, A. D. Long, R. L. Gaut, J. F. Doebley et al., 2001. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc. Natl. Acad. Sci. USA 98: 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry, J. M., M. M. Goodman, J. Doebley, S. Kresovich, D. Nielsen et al., 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28: 286–289. [DOI] [PubMed] [Google Scholar]
- Varrieur, J. M., 2002. AFLP marker analysis of monoploid potato. Ph.D. Thesis, Virginia Polytechnic Institute, Blackburg, VA.
- Watterson, G. A., 1975. Number of segregating sites in genetic models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. L., Q. J. Song, D. L. Hyten, C. P. Van Tassell, L. K. Matukumalli et al., 2003. Single-nucleotide polymorphisms in soybean. Genetics 163: 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]