Abstract
Twelve populations of Escherichia coli evolved in and adapted to a glucose-limited environment from a common ancestor. We used two-dimensional protein electrophoresis to compare two evolved clones, isolated from independently derived populations after 20,000 generations. Exceptional parallelism was detected. We compared the observed changes in protein expression profiles with previously characterized global transcription profiles of the same clones; this is the first time such a comparison has been made in an evolutionary context where these changes are often quite subtle. The two methodologies exhibited some remarkable similarities that highlighted two different levels of parallel regulatory changes that were beneficial during the evolution experiment. First, at the higher level, both methods revealed extensive parallel changes in the same global regulatory network, reflecting the involvement of beneficial mutations in genes that control the ppGpp regulon. Second, both methods detected expression changes of identical gene sets that reflected parallel changes at a lower level of gene regulation. The protein profiles led to the discovery of beneficial mutations affecting the malT gene, with strong genetic parallelism across independently evolved populations. Functional and evolutionary analyses of these mutations revealed parallel phenotypic decreases in the maltose regulon expression and a high level of polymorphism at this locus in the evolved populations.
A key challenge in evolutionary biology is to decipher the relationship between genotype and phenotype, in particular for those traits associated with increased fitness (Lewontin 1974). Understanding evolutionary adaptation requires identification of the target genes of natural selection, which is a difficult task especially for complex traits in environments where any number of physiological pathways might be important. Moreover, most organisms are not easy to analyze and manipulate genetically; even most model organisms, for which there exists a wealth of genetic information, have large and complex genomes that make such analyses very difficult. However, significant contributions to characterizing the genotype-to-phenotype relationships that underlie evolutionary adaptations have been achieved in recent years by focusing on cases of parallel and convergent changes.
Parallel evolution is defined as the independent evolution of the same trait in different lineages (Futuyma 1986). Independent populations of various types of organisms—from bacteriophages and bacteria to insects and vertebrates—have in many studies been shown to evolve parallel phenotypic traits under similar environmental conditions (Vasi et al. 1994; Losos et al. 1998; Rainey and Travisano 1998; Ferea et al. 1999; Wichman et al. 1999; Huey et al. 2000; Nosil et al. 2002; Nachman et al. 2003; Zhang 2003; Schluter 2004; Colosimo et al. 2005). The statistically predictable relationship between environmental features and the evolution of organismal traits is unlikely to arise by genetic drift and instead strongly implicates natural selection. Thus, parallel and convergent changes in phenotypic traits are widely regarded as hallmarks of adaptive evolution.
Molecular studies focusing on specific phenotypic traits have also sometimes been successful in identifying the genetic basis of the parallel evolution of these phenotypic characters. For example, parallel evolution of resistance to heavy metals, insecticides, and drugs led to the discovery of the molecular changes underlying these adaptations in organisms as diverse as plants, insects, and viruses (Schat et al. 1996; Crandall et al. 1999; Daborn et al. 2002). Most such studies of parallel molecular evolution have focused on the genes underlying a few traits, such as resistance, that reflect adaptation to a specific selective agent or other environmental factor. However, fitness itself is a complex phenotypic trait that emerges from all the interactions among the molecular components of an entire organism. Therefore, it is of considerable interest, as well as a great challenge, to assess the extent of parallel evolution for as many traits and genes as possible in a given system.
The development of tools to measure global gene expression profiles offers the new opportunity to measure simultaneously hundreds or thousands of expression-level phenotypic traits. These measurements are possible either at the level of messenger RNAs by using DNA macroarrays (DeRisi et al. 1997) or at the protein level with two-dimensional proteomic electrophoresis (Lee and Lee 2003). A model system in which both sets of data could be obtained and compared would, on the one hand, advance our understanding of the overall extent and importance of parallelism during the evolutionary process and, on the other hand, provide new insight into the relative contribution of different levels of gene regulation to parallel changes. Both methodologies could also be used, alone or in combination, to help reveal the type and identity of the underlying mutations. The strength of such an analysis depends on the assumption that any parallel changes observed are independent and not based on common ancestry. This assumption can be fulfilled with certainty in experiments where replicate microbial populations independently evolve from the same founding ancestral genotype (Elena and Lenski 2003). The availability of genomic resources for the study organism also facilitates further research in identifying the molecular mechanisms underlying adaptation to particular environments on the basis of the pattern of parallel phenotypic changes.
A clone of Escherichia coli B was used as the common ancestor to found the longest-running evolution experiment (Lenski 2004). Twelve independent populations were initiated from this ancestor and propagated by daily serial transfer in the same defined glucose-limited environment for >20,000 generations (Lenski et al. 1991; Lenski and Travisano 1994; Cooper and Lenski 2000). Over time, the populations diverged genetically from their ancestor and genetic diversity accumulated within the evolving populations (Papadopoulos et al. 1999). All the replicate populations achieved more or less parallel gains in competitive fitness, indicative of substantial adaptation, on the basis of competition experiments with marked variants of the ancestor in the same glucose-limited environment (Lenski and Travisano 1994; Cooper and Lenski 2000). Several other phenotypic aspects also evolved in parallel, including cell size (Lenski et al. 1998), growth parameters (Vasi et al. 1994), catabolic functions (Cooper and Lenski 2000), and DNA topology (Crozat et al. 2005). Moreover, analyses of gene expression profiles using DNA macroarrays for 2 of the populations revealed parallel changes in the transcription of 59 genes after 20,000 generations of experimental evolution (Cooper et al. 2003). The genetic bases of some of these parallel phenotypic changes were investigated and mutations discovered in genes involved in ribose utilization (Cooper et al. 2001), DNA topology (Crozat et al. 2005), and the stringent response (Cooper et al. 2003). In each of these cases, relevant mutations were found in many or all of the independently evolved populations, implying parallel molecular evolution. By constructing isogenic strains that differed by single mutations and performing competition experiments in the same glucose-limited environment of the evolution experiment, 4 of these genes were demonstrated to be targets of natural selection (Cooper et al. 2001, 2003; Crozat et al. 2005): spoT, involved in the metabolism of the signaling molecule ppGpp (Cashel et al. 1996); topA, encoding topoisomerase I (Wang 1971); fis, encoding a histone-like protein (Dorman and Deighan 2003); and the rbs ribose-utilization operon (Abou-Sabé et al. 1982). However, summing the fitness gains caused by the mutations in these 4 genes explains only about one-third of the average fitness increase after 20,000 generations of evolution, indicating that more beneficial mutations await discovery, consistent with an estimate of ∼10–20 beneficial mutations substituted in each population (Lenski 2004). By contrast to these cases of parallel genetic changes, sequencing 36 gene regions chosen at random found no mutations at all except in 4 populations that evolved mutator phenotypes (Sniegowski et al. 1997; Cooper et al. 2003; Lenski et al. 2003).
In this study, we analyzed the extent of parallel change at the protein level in two evolved clones, each from an independently evolved population, by performing two-dimensional protein gel electrophoresis. Their proteomic profiles were compared with one another and with the profile obtained for their common ancestor. The two evolved clones that we analyzed were the same ones that had been investigated previously using DNA macroarrays (Cooper et al. 2003), which allowed us to compare the inferences on the basis of transcriptional and proteomic profiles. Although we observed impressive correspondence between these two approaches in terms of an important suite of parallel changes involving a global regulatory pathway, our study also identified an additional target of natural selection involving a lower level of regulation that affected 8 of the 12 populations.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions:
A single clone of E. coli B (Lederberg 1966; Schneider et al. 2002) was used as the common ancestor to found 12 populations that were subsequently propagated in a constant glucose-limited minimal medium for >20,000 generations (Lenski et al. 1991; Lenski and Travisano 1994; Cooper and Lenski 2000; Lenski 2004). Six populations, designated Ara − 1 to Ara − 6, were initiated from an Ara− variant of the ancestor (REL606), whereas the six other populations, designated Ara + 1 to Ara + 6, were initiated from an Ara+ variant of the same ancestor (REL607). The arabinose utilization capacity was used as an internal marker in competition experiments and was demonstrated to be neutral in the selection environment (Lenski et al. 1991; Lenski and Travisano 1994). At 500-generation intervals throughout the evolution experiment, mixed samples from each of the 12 populations were conserved as glycerol suspensions at −80°. Individual evolved clones were isolated as needed by plating these mixed samples on tetrazolium–arabinose plates (Lenski et al. 1991). Two evolved clones, designated REL8593A and REL9282A (Cooper and Lenski 2000; Cooper et al. 2003), were isolated at 20,000 generations from two focal populations, Ara − 1 and Ara + 1, respectively, and were used to characterize their proteomic profiles in comparison with the ancestral profile. Many other evolved clones were isolated from the various populations throughout the experimental generations to sequence the malT gene and analyze the dynamics of mutational substitutions in that gene.
Strain BW3216, isolated after 280 generations of glucose-limited growth in a chemostat (Notley-McRobb and Ferenci 1999), was used to isolate a malT mutant allele that shows constitutive upregulation of the maltose regulon. Strain BW2951 is an MC4100 derivative that carries a lamB–lacZ transcriptional fusion marked with kanamycin resistance (Notley and Ferenci 1995). Strain pop7164 carries a malT deletion allele (Schreiber et al. 2000) and was used as a control in immunoblotting experiments.
For cloning experiments, we used E. coli JM109 (Yanisch-Perron et al. 1985) with standard procedures (Sambrook et al. 1989) or TOP10 (Invitrogen, San Diego) according to the supplier's recommendations. Plasmids pCRII-Topo (Invitrogen) and pKO3 (Link et al. 1997) were used for cloning experiments and allelic replacements, respectively.
Strains were grown in Davis minimal (DM) medium supplemented with 25 μg/ml glucose (DM25) as in the evolution experiment (Lenski et al. 1991) or with 250 μg/ml glucose (DM250) or were grown in rich LB medium (Sambrook et al. 1989). The same minimal medium except with maltose substituted for glucose was used to assess maltose utilization capacity. Unless otherwise stated, DM25 or DM250 means that glucose was the carbon source. Chloramphenicol (30 μg/ml) or kanamycin (50 μg/ml) were added as needed.
Genetic techniques and β-galactosidase assay:
The lamB–lacZ transcriptional fusion was moved into the ancestral or derived backgrounds by P1 transduction, using P1vir (Miller 1992), strain BW2951 as donor, and selecting for kanamycin resistance. The β-galactosidase activities were assayed by use of o-nitrophenyl β-d-galactopyranoside as a substrate and were calculated as micromolars of o-nitrophenol per minute per milligram of cellular protein (Miller 1992). All activity values reported are the average of three independent experiments.
Protein preparation and two-dimensional polyacrylamide gel electrophoresis:
Cultures were incubated at 37° for 24 hr in DM250 medium. Cell pellets were harvested by centrifugation at 5000 × g, washed once with 50 mm Tris, pH 8, resuspended in 200 μl of 50 mm Tris, 0.2 m DTT, 0.3% SDS (w/v), and 1 mm EDTA, and boiled for 5 min. Twenty microliters of 50 mm Tris, pH 7.5, 50 mm MgCl2, containing 1 mg/ml DNase I and 0.25 mg/ml RNase A (Roche), were added to the cell extracts and the resulting suspensions were incubated for 10 min at 4°. Finally, proteins were solubilized by adding 800 μl of buffer containing 10 m urea, 4% NP40 (v/v), 0.1 m DTT, and 2.2% ampholines with a pH range from 3 to 10 (v/v) (Amersham Pharmacia). Protein samples were stored at −20°.
Two-dimensional polyacrylamide gel electrophoresis (PAGE) was performed using a Multiphor II system (Amersham Pharmacia) for isoelectrofocalization and the Protean II xi cell (Bio-Rad, Hercules, CA) for SDS–PAGE. Dry strips (laboratory made, 18 cm, pH range from 4 to 8) were hydrated in the protein samples (500 μg) prepared in 2 m thiourea, 5 m urea, 1.6% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (w/v), 100 μm DTT, and 0.8% ampholines (v/v). Isoelectrofocalizations were run until pH equilibrium was reached (Lelong and Rabilloud 2003). These strips were equilibrated for 10 min in Tris, pH 6.8, containing 2.5% SDS (w/v), 30% glycerol (v/v), 6 m urea, and 50 mm DTT. This step was repeated in the same buffer, with DTT replaced by 100 mm iodoacetamide. For the second-dimension electrophoresis, SDS–PAGE was performed as described (Laemmli 1970), using 12% acrylamide gels, with the following modifications: cathode and anode migration buffers consisted of 50 mm Tris, 0.1% SDS (w/v) with 200 mm taurine, and 380 mm glycine, respectively. Proteins were stained with either Coomassie brilliant blue R-250 or silver.
Analysis of two-dimensional protein gels:
Quantitative analysis of the protein gels was performed using Melanie II software (Genebio, Geneva, Switzerland). The global protein profile of the three strains (two evolved, one ancestral) was performed in triplicate from three independent cultures of each strain. Each three-part set of comparisons was performed in parallel on the same day and with the same media and solutions to produce gels with similar background and signal levels for quantitative analysis. This approach allowed us to achieve excellent reproducibility by making gel parameters maximally consistent for each set of comparisons. Our quantitative analysis proceeded in three steps as follows. First, for every protein spot on each gel, we calculated its relative abundance as the ratio between its volume and the total volume of all spots on the same gel. Volume is defined as a function of optical density integrated over the spot area. Second, changes in protein expression of each evolved clone were standardized using the ancestor as a reference. Thus, a value of 2.0 means that the relative abundance of a particular protein was twice as high in the evolved clone as in its ancestor, whereas a value of 0.5 indicates that the relative abundance in the evolved clone was only half that in the ancestor. Third, the mean and standard deviations of the standardized values were computed for each spot from the three replicate gels, and the data were analyzed statistically using the R-package 0.5.13 software. In particular, two-tailed t-tests were used to determine whether the expression of a particular protein in an evolved clone was greater or less than 1 (i.e., the null expectation in the absence of any evolutionary change) using P < 0.05 as the criterion for statistical significance. Although our statistical analyses used data that were standardized relative to the total abundance integrated over all protein spots, for purposes of illustration we identify two proteins, AhpC and GapA, as “internal standards” because their relative abundance was nearly constant over all three genetic backgrounds and all three sets of gels.
Mass spectrometry peptide sequencing:
Protein spots were manually excised from Coomassie-blue-stained 2-D gels, oxidized with 7% H2O2, and subjected to in-gel tryptic digestion. For matrix-assisted laser desorption ionization–time of flight–mass spectrometry (MALDI–TOF–MS), peptides were mixed with matrix solution [α-cyano-4-hydroxycinnamic acid at half saturation in 60% acetonitrile/0.1% trifluoroacetic acid (v/v)] and analyzed with a MALDI–TOF mass spectrometer (Autoflex, Bruker Daltonik, Bremen, Germany) in reflector/delayed extraction mode over a mass range of 0–4200 Da. Consecutive automatic searches against the Swissprot Trembl database were performed for each sample using version 1.9 of Mascot software. For nano-liquid chromatography–tandem mass spectrometry (nano-LC–TMS/MS), peptides were extracted with formic acid and acetonitrile and injected into a CapLC (Waters, Milford, MA) nanoLC system that was directly coupled to a QTOF Ultima mass spectrometer (Waters). MS and MS/MS data were acquired and processed automatically using MassLynx 4.0 software (Waters). Consecutive searches against the Swissprot Trembl database were performed for each sample using Mascot 1.9.
Electrophoresis and immunoblot analyses of proteins:
Cells were grown in triplicate in DM250 medium for the time indicated. Cell pellets were resuspended in lysis buffer (70 mm Tris, pH 7.4, 1 mm EDTA, 10% glycerol, 1 mm DTT) before sonication. The lysates were centrifuged at 10,000 × g for 30 min at 4°, and total protein concentrations were determined using the Bradford protein assay kit (Bio-Rad) and bovine serum albumin as a standard. Equal amounts of protein samples were analyzed by SDS–polyacrylamide gel electrophoresis. Prestained protein standards (Amersham, Buckinghamshire, UK) were used for molecular weight estimation. Immunoblot analyses of proteins electrotransferred (Bio-Rad) onto polyvinylidene difluoride membranes (Amersham Pharmacia) were performed with polyclonal antibodies raised against LamB (courtesy of A. Meinke, Intercell AG), and monoclonal antibodies raised against RpoA as an internal standard (courtesy of M. Cashel). The blots were developed using nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate (Sigma, St. Louis) systems.
Strain construction:
Each evolved malT allele was recombined into the chromosome of the ancestral clone using suicide plasmid pKO3, which has a temperature-sensitive replication origin (Link et al. 1997), as described previously (Crozat et al. 2005). Briefly, the evolved malT alleles were cloned into pKO3, and the resulting plasmids were electrotransformed into the ancestor clone. Integration of the evolved allele into the ancestral malT gene was selected by plating transformed cells on chloramphenicol–LB agar and incubating at high temperature. Chloramphenicol-resistant cells were subsequently plated on sucrose-containing LB agar to select for plasmid loss, as the plasmid carries sacB, which renders cells sensitive to killing by sucrose. Sucrose-resistant and chloramphenicol-sensitive plasmid-free clones were then screened for the presence of the evolved malT alleles either by a PCR–restriction fragment length polymorphism (PCR–RFLP) approach using PauI (Euromedex) to distinguish between ancestral and evolved alleles, in the case of the Ara − 1 population, or by characterizing the length of the PCR product containing the malT ancestral or evolved allele, in the case of the Ara + 1 population. All constructed strains were also “deconstructed” such that the evolved alleles were replaced by their ancestral counterparts. Reversion of the relevant phenotypes (fitness, Western blots) was always confirmed in these deconstructed strains, indicating the absence of secondary mutations during the strain construction process. The same strategy was used to move the upregulated constitutive malT allele from BW3216 into the ancestor.
Fitness assays:
Isogenic ancestral strains with the introduced evolved malT alleles competed against the ancestor carrying the original malT allele and the opposite neutral Ara marker (Lenski et al. 1991). Each pairwise competition was replicated sixfold, with competitions performed in the same medium as the evolution experiment for 6 days with 1:100 daily transfers to allow detection of small fitness effects. Fitness assays thus encompass all the same phases of population growth that occurred in the evolution experiment, including lag phase, exponential growth, and stationary phase (Vasi et al. 1994). Immediately prior to each assay, the competitors were separately acclimated to the same regime and then transferred together from stationary-phase cultures into fresh medium (with 1:200 dilution each and thus 1:100 combined). Samples were taken immediately after mixing at day 0 and again after 6 days of competition to measure the abundance of the competitors; at both times, all the cells are in stationary phase (24 hr after the previous transfer). From the initial and final cell counts and the known dilution factors, we calculated the realized (net) population growth of each competitor. Fitness was then calculated as the ratio of the realized growth rates of the two strains during their direct competition, and t-tests were performed to evaluate whether the measured fitness values differed significantly from the null hypothetical value of 1.
Measurement of the mutation rate from Mal+ to Mal−:
A Luria–Delbrück fluctuation test (Luria and Delbrück 1943) was performed to estimate the mutation rate from Mal+ to Mal− using 60 replicate cultures. Independent cultures were each founded from a small number (∼100) of cells of the Mal+ ancestor in flasks containing 10 ml of DM25. After 24 hr, each culture was diluted, several hundred cells were spread on tetrazolium–maltose indicator plates, and Mal+ (pink) and Mal− (red) colonies were counted. These data yielded the total number of cells and the frequency of Mal− mutants in each culture, from which the rate of mutation from Mal+ to Mal− was estimated (Lea and Coulson 1949; Ma et al. 1992).
Evolutionary dynamics of malT substitutions:
The approximate time of origin and the subsequent dynamics of the malT mutations that were substituted in each focal population were investigated using a PCR–RFLP strategy with clones isolated from samples frozen at various generations during the long-term evolution experiment. For population Ara − 1, numerous clones from generations 2500, 3000, 4000, and 5000 were subjected to PCR using primers ODS273 (5′-GTCGCTGCTGGAAGAGTCGC-3′) and ODS304 (5′-ACCACGGGCCAGCGAGCATT-3′). The presence or absence of the evolved malT mutation was determined by PauI digestion of the PCR products. For population Ara + 1, numerous clones from generations 4000, 5000, 7000, and 10,000 were subjected to PCR using primers ODS291 (5′-CAGCCCCTGACGCTCAATCT-3′) and ODS302 (5′-GCAGAATACCCACTCAGCCC-3′). The size of the PCR product was used to score the malT allele as either ancestral or evolved.
Evolutionary dynamics of the growth capacity on maltose:
Numerous clones were isolated from population Ara + 1 at generations 2000, 3000, 4000, 5000, and 7000 (for the last three time points, the same clones as those analyzed to ascertain the dynamics of the malT substitutions were used) and from population Ara − 6 at generations 2000, 3000, 4000, 5000, 6000, 7000, and 9000. All clones were grown at 37° in DM medium supplemented with 250 μg/ml maltose to score their growth ability using the following criteria. Growth within 8 hr of incubation was interpreted as a Mal+ phenotype, whereas no growth even after 24 hr indicated the inability to use maltose as the sole carbon source (Mal−). Also, growth within 24 hr but without any detectable growth after 8 hr was interpreted as the occurrence of suppressor mutations in those clones restoring growth (MalS; see results). In all cases, growth was characterized by a sustained increase in viable cell counts and optical density.
RESULTS
Global protein profiles of ancestral and evolved clones:
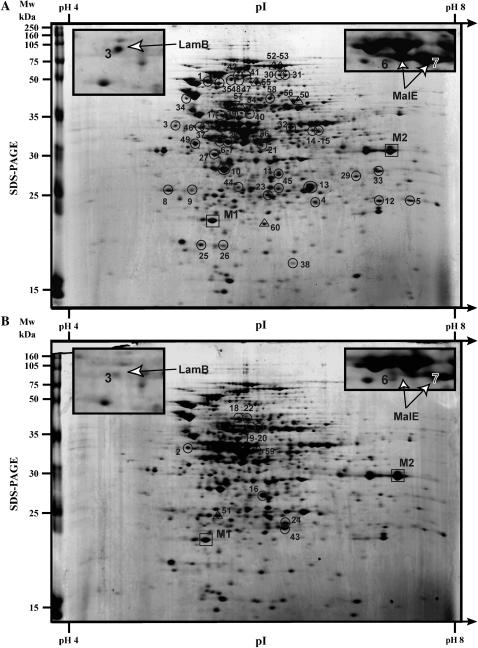
Protein extracts were prepared from three sets of independent cultures of three clones: the ancestral strain (REL606) and two independently evolved clones isolated after 20,000 generations, one from population Ara − 1 and the other from population Ara + 1. For each set of independent cultures, the three strains were grown in parallel in DM250 medium for 24 hr at 37°, during which time they reached stationary phase. Equal amounts of protein from each culture were subjected to two-dimensional electrophoresis. Such protein gels typically allow the visualization of ∼300–400 protein spots, representing 6–10% of the entire coding capacity of the E. coli genome, and we observed similar numbers in our gels. Figure 1 shows a pair of representative gels comparing the ancestor and one evolved clone. Quantitative analysis of the three replicate sets of gels for all three clones was performed using the Melanie II software (Genebio), as described in materials and methods. We first standardized the abundance of each protein spot relative to the total abundance of all spots on the same gel. We then expressed any evolved change in the expression of a given protein by calculating the ratio of its standardized abundance in an evolved clone to that in the ancestor, using the values obtained in the same gel set. The three replicate sets of gels thus gave three estimates of the extent of change in expression level for a given protein in each evolved clone, and these estimates were used to test whether a change was statistically significant. Table 1 provides the identity of all the protein spots whose relative abundance changed significantly in one or both evolved clones. It also includes two reference proteins whose abundances were quite constant among the ancestral and evolved clones and over all three replicate sets of gels. A total of 60 spots showed significant changes in one or both of the evolved clones. However, 12 proteins were each identified in two different spots, where the duplicate forms represent post-translational modifications or degradation products; in all 12 cases, the two different forms showed very similar evolutionary changes. Thus, 48 proteins showed significant changes in expression in one or both populations. Thirty-eight of these proteins changed in both independently evolved clones and, strikingly, all 38 had evolved in the same direction relative to the ancestral expression level, indicating strong evolutionary parallelism. Parallel changes across lineages are often indicative of adaptive evolution, and thus these 38 proteins are of particular interest. Seven parallel changes reflect increased expression of proteins in the evolved clones, including 5 proteins without any detectable expression in the ancestor. The other 31 parallel changes indicate reduced expression in the evolved clones, including 7 proteins with no measurable expression in either evolved clone. Two protein spots that differ between the ancestor and the evolved clones confirm previous mutations identified using insertion sequence elements as molecular markers in these long-term populations (Papadopoulos et al. 1999). In particular, IS-associated deletions of the rbs operon, including its promoter region, were found in all 12 populations (Cooper et al. 2001), and the pykF gene was inactivated by an IS transposition in population Ara − 1 (Schneider et al. 2000). Here, we observed the parallel loss of RbsB, the D-ribose-binding periplasmic protein, in both focal populations, consistent with these known deletions. By contrast, PykF (pyruvate kinase I) disappeared only in population Ara − 1, with no significant change in expression in population Ara + 1 (Table 1). These same mutations also explain why parallel changes at the transcription level were seen in these same clones for the rbs operon, but not for pykF (Cooper et al. 2003).
Figure 1.—
Two-dimensional protein gel electrophoresis of the ancestor and an evolved clone. Protein extracts were prepared from cultures grown in DM250 medium for 24 hr at 37°. Laboratory-made dry strips (18 cm, pH range from 4 to 8) were hydrated in 500-μg protein samples from the ancestral strain (A) and from an evolved clone isolated at generation 20,000 from population Ara + 1 (B). The steady-state isoelectrofocalization patterns were analyzed by 12% acrylamide-bis SDS–PAGE and Coomassie blue staining. Protein spots numbered M1 and M2 did not show any variation among the three genotypes analyzed but are shown as internal standards (□) (see Table 1). (○) Protein spots modified in both evolved clones. Spots decreasing or disappearing in both evolved clones are marked on the ancestral protein pattern (A); spots increasing or appearing in both evolved clones are marked on the evolved protein pattern (B). (▵) Protein spots modified only in the Ara + 1 evolved clone. (▿) Protein spots showing decreased intensity only in the Ara − 1 evolved clone. The insets show enlarged parts of each gel with the maltoporin LamB and the maltose-binding protein MalE (open arrowheads). Mw, molecular weight; pI, isoelectric point.
TABLE 1.
Identity of protein spots that differ between the ancestor and evolved clones and two protein spots shown as internal standards for visualization
| Protein spot no.a | Protein (gene)b | Functional groupc | Expression lower in:d | Quantitatione |
|---|---|---|---|---|
| M1 | AhpC (ahpC) | Cell processes, protection | 1.060 ± 0.065 | |
| 0.991 ± 0.034 | ||||
| M2 | GapA (gapA) | Metabolism, energy metabolism | 0.999 ± 0.011 | |
| 0.993 ± 0.079 | ||||
| 1 | CirA (cirA)* | Transport, outer membrane | Evol | 0.656 ± 0.011 |
| 0.678 ± 0.052 | ||||
| 2 | FadL (fadL)* | Transport, outer membrane | Anc | 1.956 ± 0.080 |
| 2.800 ± 0.195 | ||||
| 3 | LamB (lamB)* | Transport, outer membrane | Evol(0) | OFF |
| OFF | ||||
| 4 | ArtJ (artJ) | Transport, periplasmic space | Evol | 0.504 ± 0.038 |
| 0.536 ± 0.037 | ||||
| 5 | GlnH (glnH) | Transport, periplasmic space | Evol(0) | OFF |
| OFF | ||||
| 6–7 | MalE (malE)* | Transport, periplasmic space | Evol(0) | OFF |
| OFF | ||||
| 8–9 | MetQ (metQ) | Transport, periplasmic space | Evol(0) | OFF |
| OFF | ||||
| 10 | MglB (mglB)* | Transport, periplasmic space | Evol | 0.736 ± 0.007 |
| 0.554 ± 0.007 | ||||
| 11 | MglB (mglB)* | Transport, periplasmic space | Evol | 0.599 ± 0.036 |
| 0.451 ± 0.010 | ||||
| 12 | ModA (modA)* | Transport, periplasmic space | Evol | 0.433 ± 0.008 |
| 0.408 ± 0.034 | ||||
| 13 | RbsB (rbsB)* | Transport, periplasmic space | Evol(0) | OFF |
| OFF | ||||
| 14 | TolB (tolB) | Transport, periplasmic space | Evol | 0.165 ± 0.020 |
| 0.139 ± 0.031 | ||||
| 15 | TolB (tolB) | Transport, periplasmic space | Evol | 0.206 ± 0.008 |
| 0.258 ± 0.044 | ||||
| 16 | ZnuA (znuA) | Transport, periplasmic space | Anc(0) | ON |
| ON | ||||
| 17 | AroG (aroG) | Metabolism, amino-acid biosynthesis | Evol | 0.127 ± 0.008 |
| 0.203 ± 0.014 | ||||
| 18 | IlvB (ilvB)** | Metabolism, amino-acid biosynthesis | Anc(0) | ON |
| ON | ||||
| 19 | LeuA (leuA)* | Metabolism, amino-acid biosynthesis | Anc(0) | ON |
| ON | ||||
| 20 | LeuA (leuA)* | Metabolism, amino-acid biosynthesis | Anc(0) | ON |
| ON | ||||
| 21 | SerC (serC)* | Metabolism, amino-acid biosynthesis | Evol | 0.320 ± 0.008 |
| 0.380 ± 0.012 | ||||
| 22 | AceB (aceB) | Metabolism, central intermediary metabolism | Anc | 4.959 ± 0.186 |
| 6.017 ± 0.285 | ||||
| 23 | GpmA (gpmA) | Metabolism, central intermediary metabolism | Evol | 0.403 ± 0.071 |
| 0.344 ± 0.013 | ||||
| 24 | NfnB (nfnB) | Metabolism, central intermediary metabolism | Anc(0) | ON |
| ON | ||||
| 25 | ATPF (atpF) | Metabolism, energy metabolism | Evol | 0.461 ± 0.002 |
| 0.524 ± 0.025 | ||||
| 26 | ATPF (atpF) | Metabolism, energy metabolism | Evol | 0.409 ± 0.064 |
| 0.492 ± 0.014 | ||||
| 27 | Mdh (mdh)* | Metabolism, energy metabolism | Evol | 0.532 ± 0.047 |
| 0.545 ± 0.024 | ||||
| 28 | SucC (sucC)* | Metabolism, energy metabolism | Evol | 0.631 ± 0.005 |
| 0.742 ± 0.022 | ||||
| 29 | SucD (sucD)* | Metabolism, energy metabolism | Evol | 0.463 ± 0.014 |
| 0.484 ± 0.029 | ||||
| 30–31 | PflB (pflB)* | Metabolism, energy metabolism | Evol(0) | OFF |
| OFF | ||||
| 32 | FabI (fabI) | Metabolism, fatty acid biosynthesis | Evol | 0.497 ± 0.012 |
| 0.424 ± 0.026 | ||||
| 33 | FklB (fklB) | Information transfer, chaperone, folding | Evol | 0.374 ± 0.017 |
| 0.333 ± 0.012 | ||||
| 34 | GroEL (groL)** | Information transfer, chaperone, folding | Evol | 0.300 ± 0.037 |
| 0.399 ± 0.040 | ||||
| 35 | HtpG (htpG) | Information transfer, chaperone, folding | Evol | 0.447 ± 0.026 |
| 0.454 ± 0.044 | ||||
| 36 | Aat (aat) | Information transfer, protein degradation | Evol | 0.555 ± 0.038 |
| 0.564 ± 0.026 | ||||
| 37 | HslU (hslU) | Information transfer, protein degradation | Evol | 0.468 ± 0.063 |
| 0.414 ± 0.020 | ||||
| 38 | Fur (fur)* | Information transfer, transcription | Evol(0) | OFF |
| OFF | ||||
| 39 | AsnS (asnS)** | Information transfer, translation | Evol | 0.556 ± 0.043 |
| 0.653 ± 0.019 | ||||
| 40 | AsnS (asnS)** | Information transfer, translation | Evol | 0.505 ± 0.032 |
| 0.379 ± 0.029 | ||||
| 41 | GlyS (glyS) | Information transfer, translation | Evol | 0.216 ± 0.039 |
| 0.238 ± 0.024 | ||||
| 42 | GlyS (glyS) | Information transfer, translation | Evol | 0.200 ± 0.019 |
| 0.157 ± 0.021 | ||||
| 43 | Mannosyl-transferasef | Cell processes | Anc(0) | ON |
| ON | ||||
| 44 | MinD (minD) | Cell processes, cell division | Evol | 0.218 ± 0.029 |
| 0.292 ± 0.017 | ||||
| 45 | MinD (minD) | Cell processes, cell division | Evol | 0.272 ± 0.028 |
| 0.255 ± 0.019 | ||||
| 46 | GTP-dependent nucleic acid- binding proteinf | GTP-binding protein | Evol | 0.739 ± 0.043 |
| 0.731 ± 0.061 | ||||
| 47 | BipA (bipA) | GTP-binding protein, elongation factor | Evol | 0.543 ± 0.029 |
| 0.442 ± 0.031 | ||||
| 48 | BipA (bipA) | GTP-binding protein, elongation factor | Evol | 0.542 ± 0.044 |
| 0.583 ± 0.040 | ||||
| 49 | YcdO (ycdO) | Hypothetical, unknown | Evol | 0.559 ± 0.068 |
| 0.633 ± 0.027 | ||||
| 50 | OppA (oppA) | Transport, periplasmic space | Ara + 1 | 0.315 ± 0.006 |
| 0.975 ± 0.090 | ||||
| 51 | ArgT (argT) | Metabolism, amino-acid biosynthesis | Ancg | 2.204 ± 0.322 |
| 1.054 ± 0.037 | ||||
| 52 | MetE (metE) | Metabolism, amino-acid biosynthesis | Ara + 1 | 0.544 ± 0.048 |
| 0.983 ± 0.055 | ||||
| 53 | MetE (metE) | Metabolism, amino-acid biosynthesis | Ara + 1 | 0.409 ± 0.040 |
| 0.950 ± 0.039 | ||||
| 54 | GlpK (glpK)* | Metabolism, carbon compound utilization | Ara − 1 | 1.066 ± 0.137 |
| 0.256 ± 0.027 | ||||
| 55 | TktA (tktA) | Metabolism, central intermediary metabolism | Ara + 1 | 0.559 ± 0.049 |
| 0.996 ± 0.045 | ||||
| 56 | PykF (pykF) | Metabolism, energy metabolism | Ara − 1(0) | 1.112 ± 0.041 |
| OFF | ||||
| 57 | GuaA (guaA)* | Metabolism, nucleotide biosynthesis | Ara − 1 | 0.996 ± 0.096 |
| 0.274 ± 0.015 | ||||
| 58 | PurH (purH) | Metabolism, nucleotide biosynthesis | Ara + 1 | 0.615 ± 0.060 |
| 0.964 ± 0.120 | ||||
| 59 | EF-Tu (tufAB)** | Information transfer, translation | Ancg | 2.476 ± 0.163 |
| —h | ||||
| 60 | SodB (sodB)* | Cell processes, adaptation to stress | Ara + 1 | 0.370 ± 0.030 |
| 1.000 ± 0.039 |
The protein spots M1 and M2 show similar intensities across all genotypes and serve as internal standards for visualization only (Figure 1). Protein spots 1–49 show significant and parallel changes in the two independently evolved clones compared to their ancestor (○ in Figure 1). Protein spots 50–60 show significant changes in only one of the two evolved clones (▵ and ▿ in Figure 1). Some proteins were identified in two different spots; this pattern corresponds to post-translational modifications or to degradation products (in each case, both protein spots showed the same changes in the evolved clones).
Regulation by cAMP–CRP and ppGpp is indicated by “*” and “**,” respectively, for the functionally characterized genes (Cashel et al. 1996 and Salgado et al. 2004 for the table; Man et al. 1997 for serC; Zhang et al. 2005 for sodB; Zheng et al. 2004 for modA, kbl, and leuA; Traxler et al. 2006 for asnS).
Functional groupings of the proteins are shown according to the data from GenProtEC (http://genprotec.mbl.edu).
Anc, expression lower in the ancestor clone; Anc(0), proteins absent from the ancestor but appearing in both evolved clones; Evol, expression lower in both evolved clones; Evol(0), proteins disappearing in both evolved clones; Ara + 1, expression lower than in the ancestor only in the Ara + 1 evolved clone; Ara − 1, expression lower than in the ancestor only in the Ara − 1 evolved clone; Ara − 1(0), protein spot disappearing in the Ara-1 evolved clone.
Quantitative gel analysis was performed using the Melanie II software (Genebio). For every spot, the protein expression of each evolved clone was standardized using the ancestral strain as a reference, after first standardizing for total protein volume over all spots (see materials and methods). For example, a value of 2.0 indicates that the relative abundance of a particular protein spot is twice as high in an evolved clone as in the ancestor, whereas a value of 0.5 indicates that the relative abundance of a protein in the evolved clone is only half that measured in the ancestor. The upper and lower entries for each protein are relative abundances measured in the Ara + 1 and Ara − 1 evolved clones, respectively. Mean values are shown along with standard deviations based on the three independent sets of gels. OFF, protein spots absent in the evolved clones, but present in the ancestor. ON, protein spots absent in the ancestor, but present in the evolved clones.
Based on protein sequence comparisons. A gene name could not be assigned.
ArgT and EF-Tu are significantly higher only in the Ara + 1 evolved clone.
The Melanie II software was unable to quantify the EF-Tu protein spot in the Ara − 1 evolved clone because the spot was fuzzy and precise delineation was impossible for two of the three replicate gels.
The 38 proteins that evolved parallel changes in expression were grouped into broad functional categories according to GenProtEC (http://genprotec.mbl.edu/). Interestingly, most of these proteins belong to just two functional groups. Twelve of them function in transport, especially periplasmic-binding proteins involved in sugar, amino-acid, and ion transfers. Two of these transport proteins, FadL and ZnuA, are among the 7 proteins that showed parallel increases in their expression during evolution. Thirteen of them function in metabolism, especially energy metabolism, central metabolism, and amino-acid biosynthesis. Four metabolic proteins—IlvB, LeuA, AceB, and NfnB—are among the 7 that evolved parallel increases in their expression levels. Genes that encode transport and metabolic functions represent ∼10 and 18% of the E. coli genome, respectively; hence, these two categories appear to be substantially overrepresented among those found to have undergone parallel evolutionary changes in protein expression.
Comparison of protein and transcription profiles:
The same set of clones used in this study were previously analyzed using macroarrays to measure changes in their global transcription profiles (Cooper et al. 2003), allowing us to compare the changes found by the two methodologies. To the best of our knowledge, this is the first such comparison between mRNA and protein levels in an evolutionary context, where the magnitude of expression changes is often quite subtle. We should also emphasize, however, that the culture conditions and physiological state of cells were somewhat different in the two studies: the global mRNA levels were measured in the exact same medium as in the evolution experiment (DM25) and during exponential growth, whereas we prepared protein extracts from stationary-phase cells grown in DM250. This methodological difference reflected the need for larger quantities of protein to perform two-dimensional protein electrophoresis and to quantify a reasonable number of proteins. Nevertheless, the proteomic data support the transcription profiling in four important respects.
Several dozen genes exhibit altered expression levels in both of the independently evolved focal populations. The macroarrays revealed 59 genes that had altered mRNA levels in both populations of the entire set of E. coli genes, while protein profiles showed that 38 spots had changed in both evolved populations of the several hundred that were visualized.
In each data set, all changes that affected both populations were parallel. In other words, the changes relative to the common ancestor were in the same direction in both independently evolved clones. Under the null hypothesis that differences reflect random and unpredictable effects during either evolution or sample preparation, one would not expect to see such striking directional correspondence. Therefore, the differences revealed by these approaches are almost certainly physiologically relevant and caused by beneficial mutations that were substituted during the 20,000 generations of experimental evolution.
According to both methods, most but not all of the parallel changes involved decreased expression.
Both approaches implicate parallel changes in expression levels in the same functional categories of genes, including especially transport and binding proteins as well as those involved in metabolism.
Moreover, when we examine in greater detail the lists of genes with parallel changes, important commonalities emerge between mRNA and protein expression at two regulatory levels. First, although the genes that were identified by parallel expression changes using the two methodologies were different, most in fact belong to the same global regulatory network. In particular, approximately half (17/35) of the parallel expression changes for functionally characterized proteins detected by protein profiling (Table 1) correspond to genes regulated by guanosine tetraphosphate (ppGpp) and cAMP–cAMP receptor protein (cAMP–CRP), which is itself under the control of ppGpp (Johansson et al. 2000). In the macroarray study, also about half of the functionally characterized genes with parallel expression changes were regulated by ppGpp and cAMP–CRP (Cooper et al. 2003). This pattern led to the discovery of beneficial mutations in the spoT gene, which encodes a protein involved in synthesis and degradation of ppGpp, the molecular effector of the stringent response, which is the global regulatory network involved in adaptation to nutritional stresses (Cashel et al. 1996). Thus, despite little strict identity in the gene targets identified using macroarrays and proteomics, strong correspondence was revealed by the shared regulatory network overlying many of the parallel expression changes seen after 20,000 generations of evolution. Therefore, both parallel changes observed after 20,000 generations of experimental evolution using mRNA and protein profiling methodologies, although measured under slightly different environmental conditions, reflect the same underlying mutational basis of adaptation. However, this evolved change in the regulation of the stringent response evidently caused different subsets of genes in this network to be affected during exponential growth in DM25 medium vs. stationary phase in DM250.
Second, at the lower level of genes and operons, two parallel expression changes were detected using both methodologies. Specifically, expression of both the ribose operon and the maltose regulon decreased after 20,000 generations of experimental evolution (Table 1; see Cooper et al. 2003 for mRNA data). These changes may therefore reflect beneficial mutations in these regulons. Indeed, deletions of the rbs operon were found previously in all 12 of the evolved populations, thus explaining the loss of expression of those genes, and these mutations were also shown to be beneficial by competitions between otherwise isogenic strains (Cooper et al. 2001). Given the existence of this information for the ribose operon, we therefore further investigated the maltose regulon in both focal populations (see Parallel substitutions in malT, the transcriptional activator of the maltose regulon).
Also, six parallel changes observed in the protein expression profiles corresponded to significant transcriptional changes in only one of the two focal populations. In all cases, the changes in protein and transcriptional expression levels were in the same direction compared to the ancestral values. These differences might, in principle, indicate mutational changes in the coding sequence or post-transcriptional regulation of the relevant genes in the population where no transcriptional differences were observed. Alternatively, occasional discrepancies might reflect the vagaries of statistical analyses. In support of the latter explanation, we note that the transcription level differed significantly between the two evolved populations in only one of these six instances (nfnB: t = 3.0483, d.f. = 6, two-tailed P = 0.0226 using data from http://myxo.css.msu.edu/ecoli/arrays).
Parallel substitutions in malT, the transcriptional activator of the maltose regulon:
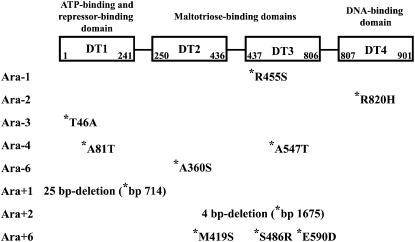
Sequencing the malT gene in both evolved clones from 20,000 generations revealed a 25-bp internal deletion in population Ara + 1, leading to a truncated protein and a point mutation in population Ara − 1 (Figure 2). To investigate further the extent of genetic parallelism, we sequenced the malT gene in one evolved clone isolated at 20,000 generations from each of the other 10 long-term populations. Nine more mutations were found in this gene in 6 of the other populations. Therefore, including the 2 focal populations, a total of 11 mutations were found in malT spread among 8 of the 12 populations (Figure 2). Such strong parallelism suggests that these mutations have a beneficial effect on fitness in the glucose-limited environment of the evolution experiment, and this hypothesis will be tested in a later section. Two mutations were found in population Ara − 4, 3 in population Ara + 6, and 1 in each of six other populations. Four of the 12 populations became mutators during the evolution experiment (Sniegowski et al. 1997; Cooper and Lenski 2000), including both populations that substituted multiple mutations in malT. Populations Ara + 1 and Ara + 2 acquired small internal deletions, while all other substitutions in malT were nonsynonymous point mutations. The MalT protein contains four domains, and the various point mutations affect all four domains in different evolved populations: 2 in DT1, 2 in DT2, 4 in DT3, and 1 in DT4 (Figure 2). The diversity of affected domains implies that the hypothesized beneficial affect does not depend on the precise location of the mutation, and it is also consistent with mutations that reduce MalT activity. Because MalT is a transcriptional activator, we also predict that the populations that evolved mutations in malT have reduced growth on maltose, which we test in the next section.
Figure 2.—
Schematic structure of the MalT protein and location of evolved mutant alleles. The protein is composed of four domains (Danot 2001; Richet et al. 2005): DT1 (residues 1–241), DT2 (residues 250–436), DT3 (residues 437–806), and DT4 (residues 807–901). DT1 binds ATP and contains surface determinants involved in the binding of repressor proteins, including Aes, MalK, and MalY. DT2 and DT3 are involved in maltotriose binding and represent putative multimerization domains. DT4 corresponds to the DNA-binding domain and presumably recruits RNA polymerase. The malT gene was sequenced in one clone isolated at 20,000 generations from each of the 12 populations designated Ara − 1–Ara − 6 and Ara + 1–Ara + 6. Amino-acid substitutions deduced from the mutations identified in the evolved clones are indicated using the one-letter code, and the residue number refers to its position in the MalT polypeptide. In both the Ara + 1 and Ara + 2 populations, small internal deletions were found in malT (their size and position relative to the malT translational start codon are shown).
Phenotypic effects of the evolved malT alleles:
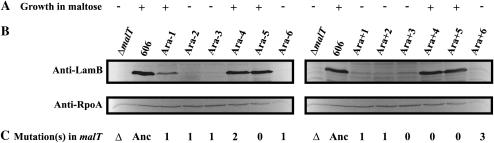
Isogenic strains with the ancestral malT allele, the Ara − 1 evolved allele, or the Ara + 1 evolved allele were constructed as described in materials and methods. Two phenotypic tests were performed for these three isogenic strains and for all 12 evolved clones from which the malT gene was sequenced; a third test was performed using only the three isogenic strains. First, we examined the growth ability of the entire set of strains in minimal medium with maltose as the sole carbon source (Figure 3A). Second, we assayed the amount of the membrane maltoporin LamB (Figure 3B), which is encoded by lamB, one of the targets of the MalT transcriptional activator (Szmelcman and Hofnung 1975). Protein extracts were prepared from all strains after growth in either glucose or maltose minimal medium. These extracts were analyzed by Western blotting with polyclonal antibodies raised against LamB (A. Meinke, Intercell AG) and with monoclonal antibodies against RpoA (M. Cashel, National Institutes of Health) as an internal standard. Third, transcriptional activities of the lamB promoter were investigated in the three ancestral strains that are isogenic except for the malT allele (Table 2). A lamB∷lacZ operon fusion was introduced by transduction (see materials and methods) in the three strains and β-galactosidase activities were measured during growth in either glucose or maltose minimal medium.
Figure 3.—
Phenotypic tests of MalT activity. The ancestral strain is labeled 606. One evolved clone was isolated at 20,000 generations from each of the 12 populations designated Ara − 1–Ara-6 and Ara + 1–Ara + 6. A malT-deleted strain (ΔmalT) served as a control. (A) Growth ability in DM250 maltose at 37°. +, Growth after 8 hr; −, no growth even after 24 hr. (B) Relative level of LamB protein. Cellular extracts were prepared from all strains after overnight culture in DM250 medium. Each sample contained 20 μg of total protein and was subjected to 12% SDS–PAGE and immunoblot analysis with polyclonal antibodies against LamB (anti-LamB) or RpoA (anti-RpoA) as a control. (C) The number of mutations identified in malT in each population is indicated. Δ, deletion allele; Anc, ancestral state.
TABLE 2.
Expression of a transcriptional fusion lamB–lacZ
| Genotype | DM250 glucose | DM250 maltose |
|---|---|---|
| 606 | 100.00 | 219.24 ± 3.55 |
| 606 malT Ara−1 | 12.99 ± 0.78 | 214.33 ± 2.36 |
| 606 malT Ara+1 | 1.22 ± 0.61 | NA |
β-Galactosidase activities of ancestral strains with the evolved malT alleles from population Ara − 1 (606 malT Ara−1) and population Ara + 1 (606 malT Ara+1) are expressed relative to the specific activity measured for the isogenic strain carrying the ancestral malT allele (606), which is arbitrarily set to the value of 100 for growth in DM250 glucose. The strains were grown overnight in DM250 containing glucose or maltose as the sole carbon source. NA, not applicable because this strain did not grow in DM250 maltose.
On the whole, these experiments showed reductions in the activity of the maltose regulon associated with the malT mutations (Figure 3), consistent with the predicted loss of function of the MalT transcriptional activator. In particular, most malT mutations were associated with the absence of growth in DM250 maltose and the loss of the LamB protein, confirming the two-dimensional protein gels. These changes occurred at the transcriptional level, at least for both focal populations, because the transcriptional activity of the lamB promoter was very low in the isogenic ancestral strains carrying each of the evolved malT alleles during growth in glucose (Table 2). The effects of the evolved alleles were not identical, however, because the activity remaining for the Ara − 1 malT allele was higher than for the Ara + 1 allele (see below).
However, there were three exceptions to the overall pattern summarized above. First, the evolved clone from population Ara − 1, as well as the ancestral strain with the corresponding evolved malT allele, showed the ancestral phenotype when grown in DM250 maltose (Figure 3A and Table 2). However, a strong decrease of both the LamB protein level of the evolved clone (Figure 3B) and the lamB promoter activity (Table 2) was detected in glucose minimal medium. Therefore, this particular evolved mutation appears to affect the basal activity of the MalT transcriptional activator during growth on glucose, leaving unchanged the induction level during growth on maltose. Second, the evolved clone sampled from population Ara − 4 grew perfectly well in maltose minimal medium and had ancestral levels of LamB in both glucose and maltose minimal media, despite having two nonsynonymous point mutations in malT. These mutations evidently do not affect the activity of MalT. Third, the evolved clone from population Ara + 3 was unable to grow on maltose and revealed a complete loss of the LamB protein (Figure 3B). However, this clone has no mutation anywhere in malT, and we also found no mutations in lamB, suggesting the presence of some (as yet unidentified) mutation in the promoter region of the malK–lamB–malM operon or in another regulator of the maltose regulon.
These few exceptions notwithstanding, the data demonstrate that the parallel genetic changes in malT were linked to parallel phenotypic losses of expression of the maltose regulon during the 20,000 generations of evolution in a glucose-limiting medium.
Beneficial fitness effects of the malT mutations:
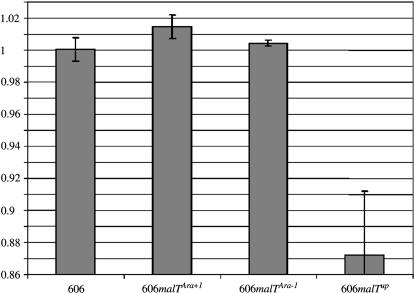
The isogenic ancestral strains with the Ara + 1 or Ara − 1 evolved malT allele were each competed against the ancestor carrying the wild-type malT allele under the same culture conditions that prevailed during the long-term evolution experiment (Figure 4). In each case, the genotype of interest competed against a variant of the ancestor bearing a neutral marker (arabinose utilization) that allowed the strains to be distinguished. As shown previously (Lenski et al. 1991), this marker had no discernible effect on fitness (H0 = 1, n = 6, ts = 0.2470, P = 0.8147). However, each evolved allele was beneficial, with fitness relative to the ancestral malT allele of 1.0146 for the Ara + 1 mutation (H0 = 1, n = 6, ts = 5.5603, P = 0.0026) and 1.0044 for the Ara − 1 mutation (H0 = 1, n = 6, ts = 5.8080, P = 0.0021). Although these fitness increments are small, both are highly significant when measured over several days in replicated experiments. Moreover, these gains were eliminated when each evolved allele was replaced again by the ancestral allele (data not shown), confirming that the gains were caused by the malT mutations rather than by some hypothetical secondary mutations that might have accidentally occurred during the strain constructions. Also, when assayed in an environment where glucose was replaced by maltose, the Ara − 1 malT evolved allele was neutral (data not shown). The Ara + 1 evolved allele could not be assayed in this second environment because it confers a Mal− phenotype and, therefore, its fitness is effectively zero.
Figure 4.—
Fitness effects of two evolved malT mutations and a mutation with upregulated expression of malT, all moved into the ancestral genetic background. Competitions were performed in the same medium used in the long-term evolution experiment. Error bars are 95% confidence intervals based on six replicate competition assays for each genotype. From left to right, the four genotypes are as follows: 606 is the ancestral strain with the ancestral malT allele; malTAra+1 and malTAra−1 are alleles that evolved in the focal populations Ara + 1 and Ara − 1, respectively, which were then moved into the ancestral chromosome; and malTup is an upregulated allele also moved into the ancestral chromosome.
To examine further whether the beneficial effects conferred by the evolved malT alleles were caused by the decreased activity of MalT, we introduced an upregulated mutant allele of malT into the ancestral strain. This allele was obtained from an evolved clone isolated from another evolution experiment performed in a chemostat environment (Notley-McRobb and Ferenci 1999), where it conferred a growth advantage. When introduced into our ancestral strain, it conferred high constitutive expression of the maltose regulon, as detected by high levels of LamB in glucose minimal medium (data not shown). When assayed by competitions in our serial-transfer regime, the upregulated malT allele was very detrimental (H0 = 1, n = 6, ts = 8.1517, P = 0.0005; Figure 4), confirming that low MalT activity is indeed beneficial in this environment.
Like the malT mutations, the rbs deletions also conferred only a small fitness benefit (Cooper et al. 2001). This small benefit acted in concert with an unusually high mutation rate from Rbs+ to Rbs−, leading to the rapid substitution of the rbs mutations in all 12 populations. To investigate whether a high mutation rate might also have influenced the parallel substitutions in malT, we performed a fluctuation test with 60 independent cultures to measure the ancestor's rate of mutation from Mal+ to Mal−. To analyze the data, we employed a program written by P. J. Gerrish (Los Alamos National Laboratory) that generates expected Luria–Delbrück distributions according to the method of Ma et al. (1992) and then uses maximum likelihood to refine the mutation rate estimate. This procedure gave a mutation rate of 4.2 × 10−6/cell/generation. The entire maltose regulon is ∼30,000 bp. Excluding third positions (synonymous) and other sites where nonsynonymous mutations would not cause a Mal− phenotype leaves ∼10,000 bp that could be mutated to produce a Mal− phenotype. The resulting mutation rate from Mal+ to Mal− is thus ∼4 × 10−10/bp, which is close to typical mutation rates estimated previously for E. coli (Drake 1991; Lenski et al. 2003). Therefore, locus-specific hypermutability does not contribute to the parallel substitutions observed in malT during the long-term evolution experiment. Also, given the small fitness effects of the evolved malT alleles relative to some other beneficial mutations observed in the long-term experiment, some of which are ∼10% (Lenski and Travisano 1994; Cooper et al. 2003; Crozat et al. 2005), we expect that the malT mutations were substituted either fairly late in the evolution experiment or by hitchhiking with other beneficial mutations of stronger effect. We investigate these dynamics in the next section.
Genetic and phenotypic evolutionary dynamics:
To examine in more detail the temporal dynamics of the appearance and substitution of the evolved malT alleles in populations Ara − 1 and Ara + 1, we performed PCR–RFLP experiments (see materials and methods) using 63 clones sampled from population Ara +1 and 70 clones isolated from population Ara − 1. In population Ara + 1, the malT mutation was present in 0/13 clones at 4000 generations, in 14/18 clones at 5000 generations, and in 32/32 clones from 7000 and 10,000 generations. In population Ara − 1, the evolved malT allele was present in 0/36 clones at 2500 and 3000 generations, in 15/16 clones at 4000 generations, and in 18/18 clones at 5000 generations. Therefore, the evolved malT alleles contributed to the somewhat slower fitness increase that followed the very rapid fitness gains seen over the first 2000 generations of the experiment (Lenski and Travisano 1994).
In population Ara + 1, clones isolated at 20,000 generations and carrying the identified malT mutation were unable to grow on maltose minimal medium (Figure 3A). We would therefore expect to observe two different phenotypic subpopulations during the evolution experiment: one able to grow on maltose and retaining the ancestral malT allele and the other unable to grow on maltose and carrying the evolved malT allele. In fact, on the basis of the ability to grow on maltose minimal medium, we identified three different subpopulations in population Ara + 1 that coexisted between 4000 and 7000 generations (Table 3). One type grew on maltose within 8 hr at 37° and was therefore phenotypically Mal+. A second type was unable to grow on maltose even after 24 hr, and it was therefore phenotypically Mal−. The third type showed no growth on maltose after 8 hr but strong growth after 24 hr, which we designated as MalS. We then sought to understand the molecular mechanism underlying the MalS phenotype of the third subpopulation. The malT gene of the MalS clone sampled at 5000 generations (Table 3) was sequenced. We found a nonsynonymous point mutation leading to an I138F change in domain DT1 of the MalT protein. This mutation may explain the absence of growth in the maltose medium after 8 hr. When left for 24 hr, however, this clone grew perfectly well. This culture was subsequently plated on LB agar and four isolated colonies were chosen at random. All four now showed growth on maltose minimal medium after 8 hr of growth, strongly suggesting that they had acquired suppressor mutations of the original I138F mutant allele. Sequencing malT revealed no further mutation, however, and therefore implies extragenic suppression. Therefore, this third category of evolved clones seems to have a distinct class of malT mutations, leading to a suppressible deficiency of maltose utilization. In any case, this third MalS category, together with the ancestral Mal+ category, eventually disappeared and was replaced by the nonsuppressible Mal− clones.
TABLE 3.
Phenotypic evolutionary dynamics of maltose growth ability
| Generations | Mal+ | Mal− | Mals |
|---|---|---|---|
| 2000 | 15/15 | — | — |
| 3000 | 13/15 | — | 2/15 (suppressible mutation different from I138F) |
| 4000 | 5/13 | — | 8/13 (suppressible mutation different from I138F) |
| 5000 | 1/18 | 16/18 (14 malT Ara+1a and 2 unknown) | 1/18 (I138F suppressible mutation) |
| 7000 | — | 14/14 | — |
Individual clones were isolated at several time points from population Ara + 1 and assessed for their growth capacity in DM250 maltose medium at 37°. The number of clones in each category relative to the total number of clones analyzed is given. Mal+, growth after 8 hr; Mal−, no growth after 24 hr; Mals, no growth after 8 hr but growth after 24 hr. In relevant cases, additional data on the malT genotype are given in parentheses.
malT Ara+1, malT evolved allele eventually substituted in population Ara + 1.
The transient polymorphism in population Ara + 1 was even more complex at the genetic level. At 7000 generations, all Mal− clones carried the small internal deletion that was eventually substituted (Figure 2), but at 5000 generations some Mal− clones (2 of 16) did not have that same malT deletion allele (Table 3; the Mal+ and MalS clones also did not have the deletion allele). Thus, the two other Mal− clones carried yet another nonsuppressible mal mutation. At 5000 generations, the MalS evolved clone had the I138F MalT suppressible change. The presence of the same mutation was investigated by PCR–RFLP in the 10 MalS clones sampled at 3000 and 4000 generations. They did not carry that mutation, indicating the presence of at least one more mal allele. These data demonstrate a high level of transient diversity in the maltose regulon, with at least three phenotypic and five genotypic subpopulations present between generations 3000 and 5000 in the Ara + 1 population (Table 3). A second population, Ara − 6, was also investigated for phenotypic polymorphism. A similar degree of polymorphism was detected, with Mal+, Mal−, and MalS phenotypic subpopulations coexisting between 3000 and 10,000 generations, before the nonsuppressible Mal− allele, caused by a point mutation in the DT2 domain of malT, was eventually fixed.
DISCUSSION
We describe parallel evolutionary changes in protein expression profiles that occurred in two populations of E. coli during 20,000 generations in a defined environment (Lenski et al. 1991; Lenski and Travisano 1994; Cooper and Lenski 2000; Lenski 2004). The global transcription profiles of the same two evolved clones were previously obtained (Cooper et al. 2003), allowing a comparison between transcript and protein expression for the whole E. coli genome in this evolutionary context. Despite some differences in the culture conditions used to perform the transcription and protein profiles, we detected remarkable similarities in the evolved response using both global methodologies. In particular, there were many parallel expression changes, and about half of them involved genes in the same global regulatory network: the ppGpp and cAMP–CRP regulon. Parallel expression changes in this pathway were therefore a key outcome of this evolution experiment, and both types of global analysis of gene expression revealed the effects of these regulatory changes. In addition to the parallel changes in this global regulatory network, two other common changes of a more localized nature were detected using both DNA macroarrays and protein profiling. One was loss of expression of the rbs operon, which was previously shown to be caused by beneficial mutations that were substituted in all 12 populations of the long-term evolution experiment (Cooper et al. 2001). The other common change was loss of protein expression of the maltose regulon. These parallel expression changes led us to discover a mutation in malT, the transcriptional regulator of the maltose regulon, in both focal populations. Each mutation was then moved into the ancestral chromosome, and competition experiments between each of these constructs and the malT+ ancestor revealed that both mutations were beneficial under the culture conditions in the evolution experiment. The analysis of malT was extended to all 12 independently evolved populations, and strong genetic parallelism was detected, with malT mutations found in 8 of the 12 populations. Analyzing the expression of lamB, one of the regulatory targets of MalT, by Western blots and transcriptional fusions revealed that the malT mutations caused the loss or decrease of the maltose regulon function. By using numerous clones sampled from the two focal populations at various time points, we showed that the malT mutations were not substituted during the early period of most rapid fitness increase, but instead they contributed to the somewhat slower adaptation that occurred after 2000 generations (Lenski and Travisano 1994). The maltose regulon also revealed a high level of genetic and phenotypic diversity, with several subpopulations coexisting within a single population for several thousands of generations.
As noted, slightly different growth conditions were used for the transcription (Cooper et al. 2003) and proteomic analyses of the evolved clones. In the case of the transcript study, cells were collected in exponential phase during growth in DM25 (25 μg glucose/ml), whereas the protein profiles were performed with cells in stationary phase from cultures in DM250 (250 μg glucose/ml) to obtain a sufficient yield of proteins for the two-dimensional protein electrophoresis. One might therefore be concerned that the two sets of data are not comparable. However, our comparisons emphasize the occurrence of beneficial mutations that affected two levels of gene regulation, rather than the precise identity of the affected genes. Thus, differences in the physiological state of the bacteria probably explain why only two identical changes were observed when we compared expression at the mRNA and protein levels in the same evolved clones. However, these changes allowed identification of two beneficial mutations in the focal populations: one already known that affects the rbs operon for ribose catabolism (Cooper et al. 2001) and one discovered in this study affecting the malT transcriptional regulator of the maltose operons (see below). The fact that the same changes were identified under the different culture conditions may reflect the specific effects of mutations that govern expression at the operon level.
More generally, although the two methodologies detected parallel expression changes in different genes, about half of them belong to the stringent response pathway. Therefore, both methodologies revealed the same overarching changes in a global regulatory network. These high-level changes reflect mutational events that occurred during the long-term evolution experiment in genes encoding global regulators, which control the architecture of the cell's regulatory network. In particular, changes in gene expression detected by both transcription and proteomic analyses reflect the presence of the beneficial mutations that were found in spoT (Cooper et al. 2003). Thus, parallel changes in transcription and proteomic profiles that occurred during experimental evolution reveal bacterial adaptation through modifications that occurred at multiple levels of genetic organization, ranging from the level of operons up to global regulators of genetic networks. These parallel changes in expression allowed the discovery of beneficial mutations that show a high degree of molecular parallelism, as mutations in the same three genes (rbs, malT, spoT) were found in at least 8 of the 12 independently evolved populations (Figure 2; Cooper et al. 2001, 2003). By contrast, no parallel mutations were found when randomly chosen genes were sequenced (Lenski et al. 2003). These parallel genetic changes, discovered by parallel changes in gene expression, thus highlight important targets of selection and adaptation in this experiment. Genetic parallelism has also been reported in other evolution experiments using bacteriophages (Wichman et al. 1999) and bacteria (Treves et al. 1998), although these other studies did not examine resulting changes in global regulatory networks.
Despite the similarities observed in the transcription and protein profiles, the maltose regulon shows some interesting exceptions. Transcription profiling found significant and parallel changes in three genes in this regulon (malK, malP, and malQ). However, the same transcription profiles reveal no significant changes in either lamB or malT in either focal population (four two-tailed t-tests in all, each with 6 d.f., all P > 0.1, using data from Cooper et al. 2003 available at http://myxo.css.msu.edu/ecoli/arrays). The transcriptional profiles also show no effect of an evolved spoT allele on expression of either lamB or malT (two t-tests, each with 6 d.f., both P > 0.1, using the same data source as above). Thus, the proteomic data resolved certain changes, such as the parallel reductions in LamB, that were not found by the transcriptional study; these changes in LamB expression were unequivocally confirmed by finding the responsible mutations in malT. Another difference between the two studies is that the transcriptional analysis found significant parallel changes in ∼1.5% of all genes (Cooper et al. 2003), whereas in our proteomic study ∼10% of the 300–400 proteins detected on the gels showed significant parallel changes. This difference may reflect post-transcriptional levels of gene regulation or, alternatively, it could reflect differences in the statistical power of the two studies and their methodologies.
A new set of beneficial mutations in this long-term evolution experiment was discovered that affected the malT gene. A total of 11 mutations were found in eight different populations. Measurement of the mutation rate from Mal+ to Mal− in the ancestor indicated a mutation rate typical of other estimates for E. coli (Drake 1991). Five mutations occurred in populations that had retained the low ancestral mutation rate, with 1 mutation in each of five different populations. Two of these mutations were small internal deletions with no obvious direct sequence repeats at the extremities. The 6 other mutations occurred in populations that had evolved defects in their DNA repair pathways, including 1 in population Ara − 2, 2 in Ara − 4, and 3 in Ara + 6. Populations Ara − 2 and Ara − 4 evolved defects in methyl-directed mismatch repair (Sniegowski et al. 1997; Shaver et al. 2002); all three mutations in these populations were C·G to T·A transitions, which are characteristic of this class of mutators. Population Ara + 6 evolved a mutator phenotype that was previously suggested to involve a defect in mutT on the basis of the finding that six mutations found in randomly screened genes were all A·T to C·G transversions (Lenski et al. 2003). All three malT mutations found here in Ara + 6 were of the same class, strongly supporting the previous suggestion of a mutT defect.
The mutations found in malT in the various populations were located throughout the gene, affecting the several domains of the MalT activator protein. The MalT protein belongs to a family of bacterial transactivators characterized by high molecular mass, usually >90 kDa (Danot 2001). The MalT protein has four domains (Figure 2): DT1 (residues 1–241), DT2 (residues 250–436), DT3 (residues 437–806), and DT4 (residues 807–901) (Danot 2001; Richet et al. 2005). Numerous signals control the activity of the MalT transcriptional activator. Activation of MalT and its subsequent binding to target promoters requires the formation of a high-order MalT oligomer, induced by the maltotriose inducer and ATP. By contrast, the monomeric form of MalT, which is inactive, is stabilized by three different proteins: MalK, the ABC subunit of the maltose transporter; MalY, a cytoplasmic protein of unknown function that has a βC-S lyase activity; and Aes, a protein with sequence similarities to lipases that has esterase activity. The DT1 domain binds ATP (with a Walker A motif from residues 42 to 47) and the repressor proteins Aes and MalK, while DT1 and DT2 are involved in recognition of the repressor protein MalY; DT2 and DT3 are required together for strong maltotriose binding, while DT3 alone binds it with low affinity; and DT4 carries the DNA-binding site (Danot 2001; Joly et al. 2002; Schlegel et al. 2002).
The malT mutations in the eight evolved populations produced a Mal− phenotype in six cases (Ara + 1, Ara + 2, Ara + 6, Ara − 2, Ara − 3, and Ara − 6). In four cases, a likely mechanism can be found: populations Ara + 1 and Ara + 2 have malT deletions leading to truncated proteins without the DT2–DT4 domains and without domain DT4, respectively; the mutation in population Ara − 3 eliminates the threonine residue 46, which lies in the Walker A motif and is involved in ATP binding; and the mutation in population Ara − 2 affects the DT4 domain and may interfere with DNA-binding activity, the interaction with RNA polymerase, or both. In the two other cases, possible explanations are more speculative. The mutation in population Ara − 6 affects the DT2 domain and might interfere with the transition from the monomeric to the oligomeric forms or, alternatively, with the overall conformation of MalT. A similar effect could also apply to population Ara + 6, which has three mutations, one in DT2 and two in DT3. In two populations (Ara − 1 and Ara − 4), the malT mutations did not lead to a Mal− phenotype. Despite having a Mal+ phenotype that is indistinguishable from the ancestor, population Ara − 4 has two mutations in malT, one in DT1 and the other in DT3. Population Ara − 1 has one mutation in the DT3 domain leading to reduced activity of MalT in minimal glucose medium, although the induction level is fully conserved in minimal maltose medium. The modified residue (R455) may therefore be involved in the affinity of maltotriose for the MalT protein. The crystal structure of DT3 has been solved and reveals eight copies of a two-helix bundle motif arranged in a right-hand superhelical fold encompassing ∼80% of DT3 (Steegborn et al. 2001). The structure also indicates interaction between the DT3 domains, which may explain the MalT oligomerization. Two residues are characteristic for the DT3 motif, G541 and A547, and they are well conserved in the different helices of the structure. One of the mutations found in population Ara − 4 affects the A547 residue, although the MalT activity is not measurably affected. More specific studies of these mutations of the complex DT3 domain might give more information about its functioning.
Most of the observed mutations in malT caused loss of the ability to grow on maltose as a sole carbon source, a trend consistent with a previously reported tendency toward ecological specialization in these populations (Cooper and Lenski 2000). Such specialization can occur by two population-genetic processes: mutation accumulation, whereby neutral mutations are substituted in genes that are not maintained by selection, and antagonistic pleiotropy, where mutations substituted because they are beneficial in the selective environment have detrimental side effects in other environments (Futuyma and Moreno 1988; Rose 1991; Cooper and Lenski 2000). We constructed isogenic strains with the ancestral and evolved malT alleles, and we performed competitions that show that the evolved malT alleles are beneficial in the glucose medium in which they evolved. This result provides a direct demonstration of the role of antagonistic pleiotropy in the ecological specialization of these populations, as did previous experiments on the parallel losses of the ribose–catabolic function encoded by the rbs operon (Cooper et al. 2001). The benefit associated with the loss of MalT activity may reflect, at least in part, the elimination of unneeded functions costly to fitness in the glucose medium (Graña and Acerenza 2001; see also Dekel and Alon 2005). Alternatively, the benefit caused by the loss of MalT activity might be more complex, involving, for example, regulatory feedbacks or metabolic intermediates (e.g., Dykhuizen 1978). Using the Melanie II software, we estimate that the LamB spot represents ∼0.13% of the total protein on the ancestral gels, while the two MalE spots together represent ∼0.48%. However, these are only 2 of the 14 proteins in the maltose operons; the others were not detected on our gels, nor were hundreds of other proteins. Therefore, we are not in a position to say whether the 1.5% fitness gain that resulted from the complete loss of expression of the maltose operons in population Ara + 1 is fully explained by energetic savings. Moreover, it has been shown that MalT is a pivotal component of an elaborate regulatory network that connects several metabolic pathways (Boos and Böhm 2000). As noted before, the DT1 and DT3 domains of MalT interact directly with proteins including MalK, MalY, and Aes, whose functions are not fully understood (Richet et al. 2005). In any case, the benefit of the malT mutations seems to be related to reduced expression of the maltose regulon; and the deliberate introduction of a malT constitutive mutation, which caused high expression of the maltose regulon even in glucose medium, was detrimental in the glucose-limited environment.
Mutations in malT were also found in another evolution experiment performed in a very different environment, with E. coli propagated for several hundred generations in glucose-limited chemostats (Notley-McRobb and Ferenci 1999). In contrast to the serial transfer, or seasonal, environment in our experiments (with lag, exponential, and stationary phases each day), bacteria in chemostats experience constant physiological conditions with very low glucose concentration and high cell density. Under the chemostat conditions, malT mutations evolved that caused constitutive expression of the maltose regulon, which enhanced fitness in the severely glucose-limited environment (Notley-McRobb and Ferenci 1999). These mutations upregulated the lamB gene, leading to increased expression of this high-affinity glucose transport pathway through the outer membrane maltoporin. Increased expression of LamB was also found by proteomic profiling of the ancestor and evolved clones in another chemostat evolution experiment using E. coli, although the responsible mutation was not identified (Kurlandzka et al. 1991). By contrast, the loss of maltose regulon expression was shown to be beneficial under the seasonal conditions in our evolution experiment. As a further confirmation of this difference, introduction of a chemostat-evolved malT constitutive mutation into our ancestral strain strongly reduced fitness in the seasonal environment used in our experiments. This difference also indicates that, under these conditions, glucose uptake involves porins other than LamB. The best candidate is OmpF because the ancestral E. coli B strain lacks OmpC (Schneider et al. 2002), although other porins like OmpN might also be involved (Prilipov et al. 1998). In the chemostat environment, increased expression of the OmpC and OmpF porins also evolved along with upregulation of LamB (Kurlandzka et al. 1991); by contrast, we found no significant changes in OmpF expression after 20,000 generations in the seasonal environment (data not shown). The following factors might explain the opposing selection on the malT gene and the LamB expression in these two environments. In chemostats, cells can draw down the concentration of limiting glucose to very low levels, at which point porins may limit glucose uptake. Although the LamB porin is specifically identified as a maltose porin, it also allows glucose molecules to diffuse across the outer membrane because glucose is smaller than maltose (Luckey and Nikaido 1980). By contrast, the serial-transfer regime produces boom–bust cycles of glucose availability rather than perpetual scarcity, and so precise regulation may be more important than maximizing expression of total porins. The malT gene regulates lamB, and different mutations prevailed in malT that led to increased or reduced expression of LamB in the chemostat and the serial-transfer regimes, respectively.
The dynamics of variation in the Mal phenotype were examined more closely in three of the long-term populations in our study. In two of them, extensive phenotypic polymorphism was detected with at least three subpopulations coexisting for several thousand generations: one Mal+, one Mal−, and one suppressible MalS. In one population, we showed that at least five different malT alleles were present. This finding implies clonal interference, in which subpopulations that carry different beneficial mutations compete with one another until one eventually prevails (Gerrish and Lenski 1998); this phenomenon was previously reported in the long-term populations on the basis of evidence at other loci (Shaver et al. 2002; Crozat et al. 2005). A high level of genotypic diversity was also previously detected in these populations by using IS elements as genetic markers (Papadopoulos et al. 1999), although it was not clear from those earlier data whether the polymorphisms were important for fitness. Here, we showed that substantial allelic diversity was transiently maintained in precisely those genes that were accumulating beneficial mutations, as was also seen in the chemostat experiments described above (Notley-McRobb and Ferenci 1999). These results indicate that evolving asexual populations do not necessarily experience discrete selective sweeps involving single beneficial mutations, as supposed under the original model of periodic selection (Atwood et al. 1951). Instead, such populations may harbor multiple subpopulations, each with different beneficial alleles, which can coexist for long periods even as they compete with one another. In that case, the eventual winner is a subpopulation that accumulates one or more additional beneficial mutations that allow it to prevail (Yedid and Bell 2001; Hegreness et al. 2006).
Finally, a total of five different genes or operons have been shown thus far to be targets of selection in the long-term evolution experiment based on competition assays performed with isogenic strains: rbs (Cooper et al. 2001), spoT (Cooper et al. 2003), topA and fis (Crozat et al. 2005), and malT (this study). Assuming that their beneficial effects were additive, mutations in these genes would account for a fitness increase of ∼30%, which represents less than half of the 70% gain measured after 20,000 generations (Cooper and Lenski 2000). Four other loci were recently identified as targets of selection on the basis of parallel substitutions in many or all of the populations (Woods et al. 2006), although isogenic constructs were not made and thus their fitness effects have not been measured to date. In any case, it is clear that the malT mutations account for only a small fraction of the total fitness increase. Thus, genetic adaptation has involved many loci, and the same is likely also true for the expression changes that mediate the phenotypic changes on which selection acts. Again using isogenic constructs, transcriptional profiling showed that the spoT mutation could account for only some of the many changes found by using that methodology (Cooper et al. 2003), and the same inference probably holds for the many changes in protein expression found here (Table 1). Indeed, we examined the proteomic profiles of ancestral constructs bearing two of the evolved malT evolved alleles, and we found that these mutations were responsible for the disappearance of the LamB and MalE proteins (data not shown). Our proteomic data also revealed the loss of the RbsB protein, which is obviously linked to the rbs deletion mutations that were previously found in these populations. Also, a recent study identified genes whose expression was sensitive to the level of DNA supercoiling (Peter et al. 2004), which is yet another phenotype that has evolved in the long-term populations owing to mutations in topA and fis (Crozat et al. 2005). Seven of these supercoiling-sensitive genes encode proteins that evolved parallel changes in expression in these populations (tolB, ilvB, gpmA, nfnB, mdh, asnS, and minD: Table 1). There are few data available on the Fis regulon, and some relate to Salmonella (Gonzalez-Gil et al. 1996; Yoon et al. 2003; Kelly et al. 2004), but three parallel changes in protein profiles might be related to Fis (GlnH, RbsB, and AceB: Table 1) as might three other changes observed in only one of the evolved clones (GlpK, EF-TU, and SodB: Table 1). To relate all the parallel and divergent expression changes to the beneficial mutations found in this experiment, it will be necessary to perform a comprehensive study of changes in global expression profiles using otherwise isogenic strains carrying all of these mutations, alone and in various combinations, in the ancestral genetic background. Such an analysis will not be easy, of course, but it will provide a unique perspective on the evolution of gene regulatory networks, including both global and local controls.
Acknowledgments
We thank Michael Cashel (National Institutes of Health) and Andreas Meinke (Intercell AG, Vienna) for their generous gifts of anti-RpoA and anti-LamB antibodies, respectively. We thank Tom Ferenci (University of Sydney) and Evelyne Richet (Institut Pasteur, Paris) for sharing strains BW3216 and pop7164, respectively. We thank Phil Gerrish (Los Alamos National Laboratory) for mutation rate calculations. We are indebted to Thierry Rabilloud [Commissariat à l'Energie Atomique (CEA), Grenoble, France] for his valuable assistance with the Melanie II software used to analyze the two-dimensional protein gels. This work was supported by grants from the French Centre National de Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM), the Rhône-Alpes Region and Genopole, the Université Joseph Fourier, the Life Science Division of the CEA, and the National Science Foundation. L. Kühn and J. Garin acknowledge Fondation Rhône-Alpes Futur for funding. L. Pelosi thanks CNRS for a postdoctoral fellowship.
References
- Abou-Sabé, M., J. Pilla, D. Hazuda and A. Ninfa, 1982. Evolution of the D-ribose operon of Escherichia coli B/r. J. Bacteriol. 150: 762–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood, K. C., L. K. Schneider and F. J. Ryan, 1951. Periodic selection in Escherichia coli. Proc. Natl. Acad. Sci. USA 37: 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos, W., and A. Böhm, 2000. Learning new tricks from an old dog: MalT of the Escherichia coli maltose system is part of a complex regulatory network. Trends Genet. 16: 404–409. [DOI] [PubMed] [Google Scholar]
- Cashel, M., V. J. Gentry, V. J. Hernandez and D. Vinella, 1996. The stringent response, pp. 1458–1496 in Escherichia coli and Salmonella: Cellular and Molecular Biology, edited by F. C. Neidhardt. American Society of Microbiology, Washington, DC.
- Colosimo, P. F., K. E. Hosemann, S. Balabhadra, G. Villarreal, Jr., M. Dickson et al., 2005. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307: 1928–1933. [DOI] [PubMed] [Google Scholar]
- Cooper, T. F., D. E. Rozen and R. E. Lenski, 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. USA 100: 1072–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, V. S., and R. E. Lenski, 2000. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407: 736–739. [DOI] [PubMed] [Google Scholar]
- Cooper, V. S., D. Schneider, M. Blot and R. E. Lenski, 2001. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J. Bacteriol. 183: 2834–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crandall, K. A., C. R. Kelsey, H. Imamichi, H. C. Lane and N. P. Salzman, 1999. Parallel evolution of drug resistance in HIV: failure of nonsynonymous/synonymous substitution rate ratio to detect selection. Mol. Biol. Evol. 16: 372–382. [DOI] [PubMed] [Google Scholar]
- Crozat, E., N. Philippe, R. E. Lenski, J. Geiselmann and D. Schneider, 2005. Long-term experimental evolution in Escherichia coli. XII. DNA topology as a key target of selection. Genetics 169: 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn, P. J., J. L. Yen, M. R. Bogwitz, G. Le Goff, E. Feil et al., 2002. A single P450 allele associated with insecticide resistance in Drosophila. Science 297: 2253–2256. [DOI] [PubMed] [Google Scholar]
- Danot, O., 2001. A complex signalling module governs the activity of MalT, the prototype of an emerging transactivator family. Proc. Natl. Acad. Sci. USA 98: 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel, E., and U. Alon, 2005. Optimality and evolutionary tuning of the expression level of a protein. Nature 436: 588–592. [DOI] [PubMed] [Google Scholar]
- DeRisi, J. L., V. R. Iyer and P. O. Brown, 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686. [DOI] [PubMed] [Google Scholar]
- Dorman, C. J., and P. Deighan, 2003. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 13: 179–184. [DOI] [PubMed] [Google Scholar]
- Drake, J. W., 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88: 7160–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen, D., 1978. Selection for tryptophan auxotrophs of Escherichia coli in glucose-limited chemostats as a test of the energy conservation hypothesis of evolution. Evolution 32: 125–150. [DOI] [PubMed] [Google Scholar]
- Elena, S. F., and R. E. Lenski, 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4: 457–469. [DOI] [PubMed] [Google Scholar]
- Ferea, T. L., D. Botstein, P. O. Brown and R. F. Rosenzweig, 1999. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc. Natl. Acad. Sci. USA 96: 9721–9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futuyma, D. J., 1986. Evolutionary Biology, Ed. 2. Sinauer, Sunderland, MA.
- Futuyma, D. J., and G. Moreno, 1988. The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 19: 207–233. [Google Scholar]
- Gerrish, P. J., and R. E. Lenski, 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102/103: 127–144. [PubMed] [Google Scholar]
- Gonzalez-Gil, G., P. Bringmann and R. Kahmann, 1996. FIS is a regulator of metabolism in Escherichia coli. Mol. Microbiol. 22: 21–29. [DOI] [PubMed] [Google Scholar]
- Graña, M., and L. Acerenza, 2001. A model combining cell physiology and population genetics to explain Escherichia coli laboratory evolution. BMC Evol. Biol. 1: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegreness, M., N. Shoresh, D. Hartl and R. Kishony, 2006. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311: 1615–1617. [DOI] [PubMed] [Google Scholar]
- Huey, R. B., G. W. Gilchrist, M. L. Carlson, D. Berrigan and L. Serra, 2000. Rapid evolution of a geographic cline in size in an introduced fly. Science 287: 308–309. [DOI] [PubMed] [Google Scholar]
- Johansson, J., C. Balsalobre, S. Y. Wang, J. Urbonaviciene, D. J. Jin et al., 2000. Nucleoid proteins stimulate stringently controlled bacterial promoters: a link between the cAMP-CRP and the (p)ppGpp regulons in Escherichia coli. Cell 102: 475–485. [DOI] [PubMed] [Google Scholar]
- Joly, N., O. Danot, A. Schlegel, W. Boos and E. Richet, 2002. The Aes protein directly controls the activity of MalT, the central transcriptional activator of the Escherichia coli maltose regulon. J. Biol. Chem. 277: 16606–16613. [DOI] [PubMed] [Google Scholar]
- Kelly, A., M. D. Goldberg, R. K. Carroll, V. Danino, J. C. D. Hinton et al., 2004. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology 150: 2037–2053. [DOI] [PubMed] [Google Scholar]
- Kurlandzka, A., R. F. Rosenzweig and J. Adams, 1991. Identification of adaptive changes in an evolving population of Escherichia coli: the role of changes with regulatory and highly pleiotropic effects. Mol. Biol. Evol. 8: 261–281. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K., 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Lea, D. E., and C. A. Coulson, 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49: 264–285. [DOI] [PubMed] [Google Scholar]
- Lederberg, S., 1966. Genetics of host-controlled restriction and modification of deoxyribonucleic acid in Escherichia coli. J. Bacteriol. 91: 1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, P. S., and K. H. Lee, 2003. Escherichia coli—a model system that benefits from and contributes to the evolution of proteomics. Biotechnol. Bioeng. 84: 801–814. [DOI] [PubMed] [Google Scholar]
- Lelong, C., and T. Rabilloud, 2003. Prokaryotic proteomics, pp. 157–171 in Methods and Tools in Biosciences and Medicine: Prokaryotic Genomics, edited by M. Blot. Birkhäuser Verlag, Basel, Switzerland/Boston/Berlin.
- Lenski, R. E., 2004. Phenotypic and genomic evolution during a 20,000-generation experiment with the bacterium Escherichia coli. Plant Breed. Rev. 24: 225–265. [Google Scholar]
- Lenski, R. E., and M. Travisano, 1994. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl. Acad. Sci. USA 91: 6808–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski, R. E., M. R. Rose, S. C. Simpson and S. C. Tadler, 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138: 1315–1341. [Google Scholar]
- Lenski, R. E., J. A. Mongold, P. D. Sniegowski, M. Travisano, F. Vasi et al., 1998. Evolution of competitive fitness in experimental populations of E. coli: what makes one genotype a better competitor than another? Antonie van Leeuwenhoek 73: 35–47. [DOI] [PubMed] [Google Scholar]
- Lenski, R. E., C. L. Winkworth and M. A. Riley, 2003. Rates of DNA sequence evolution in experimental populations of Escherichia coli during 20,000 generations. J. Mol. Evol. 56: 498–508. [DOI] [PubMed] [Google Scholar]
- Lewontin, R. C., 1974. The Genetic Basis of Evolutionary Change. Columbia University Press, New York.
- Link, A. J., D. Phillips and G. M. Church, 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179: 6228–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos, J. B., T. R. Jackman, A. Larson, K. de Queiroz and L. Rodriguez-Schettino, 1998. Contingency and determinism in replicated adaptive radiations of island lizards. Science 279: 2115–2118. [DOI] [PubMed] [Google Scholar]
- Luckey, M., and H. Nikaido, 1980. Specificity of diffusion channels produced by λ phage receptor protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 77: 167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria, S. E., and M. Delbrück, 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W. T., G. H. Sandri and S. Sarkar, 1992. Analysis of the Luria-Delbrück distribution using discrete convolution powers. J. Appl. Probab. 29: 254–267. [Google Scholar]
- Man, T-K., A. J. Pease and M. E. Winkler, 1997. Maximization of transcription of the serC (pdxF)-aroA multifunctional operon by antagonistic effects of the cyclic AMP (cAMP) receptor protein-cAMP complex and Lrp global regulators of Escherichia coli K-12. J. Bacteriol. 179: 3458–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. H., 1992. A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Nachman, M. W., H. E. Hoekstra and S. L. D'Agostino, 2003. The genetic basis of adaptive melanism in pocket mice. Proc. Natl. Acad. Sci. USA 100: 5268–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil, P., B. J. Crespi and C. P. Sandoval, 2002. Host-plant adaptation drives the parallel evolution of reproductive isolation. Nature 417: 440–443. [DOI] [PubMed] [Google Scholar]
- Notley, L., and T. Ferenci, 1995. Differential expression of mal genes under cAMP and endogenous inducer control in nutrient stressed Escherichia coli. Mol. Microbiol. 16: 121–129. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb, L., and T. Ferenci, 1999. The generation of multiple co-existing mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ. Microbiol. 1: 45–52. [DOI] [PubMed] [Google Scholar]
- Papadopoulos, D., D. Schneider, J. Meier-Eiss, W. Arber, R. E. Lenski et al., 1999. Genomic evolution during a 10,000-generation experiment with bacteria. Proc. Natl. Acad. Sci. USA 96: 3807–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, B. J., J. Arsuaga, A. M. Breier, A. B. Khodursky, P. O. Brown et al., 2004. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 5: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prilipov, A., P. S. Phale, R. Koebnik, C. Widmer and J. P. Rosenbush, 1998. Identification and characterization of two quiescent porin genes, nmpC and ompN, in Escherichia coli BE. J. Bacteriol. 180: 3388–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey, P., and M. Travisano, 1998. Adaptive radiation in a heterogeneous environment. Nature 394: 69–72. [DOI] [PubMed] [Google Scholar]
- Richet, E., N. Joly and O. Danot, 2005. Two domains of MalT, the activator of the Escherichia coli maltose regulon, bear determinants essential for anti-activation by MalK. J. Mol. Biol. 347: 1–10. [DOI] [PubMed] [Google Scholar]
- Rose, M. R., 1991. Evolutionary Biology of Aging. Oxford University Press, Oxford.
- Salgado, H., S. Gama-Castro, A. Martinez-Antonio, E. Diaz-Peredo, F. Sanchez-Solano et al., 2004. RegulonDB (version 4.0): transcriptional regulation, operon organization and growth conditions in Escherichia coli K-12. Nucleic Acids Res. 32: D303–D306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schat, H., R. Vooijs and E. Kuiper, 1996. Identical major gene loci for heavy metal tolerances that have independently evolved in different local populations and subspecies of Silene vulgaris. Evolution 50: 1888–1895. [DOI] [PubMed] [Google Scholar]
- Schlegel, A., O. Danot, E. Richet, T. Ferenci and W. Boos, 2002. The N terminus of the Escherichia coli transcription activator MalT is the domain of interaction with MalY. J. Bacteriol. 184: 3069–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter, D., 2004. Parallel evolution and inheritance of quantitative traits. Am. Nat. 163: 809–822. [DOI] [PubMed] [Google Scholar]
- Schneider, D., E. Duperchy, E. Coursange, R. E. Lenski and M. Blot, 2000. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics 156: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, D., E. Duperchy, J. Depeyrot, E. Coursange, R. E. Lenski et al., 2002. Genomic comparisons among Escherichia coli strains B, K-12, and O157:H7 using IS elements as molecular markers. BMC Microbiol. 2: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber, V., C. Steegborn, T. Clausen, W. Boos and E. Richet, 2000. A new mechanism for the control of a prokaryotic transcriptional regulator: antagonistic binding of positive and negative effectors. Mol. Microbiol. 35: 765–776. [DOI] [PubMed] [Google Scholar]
- Shaver, A. C., P. G. Dombrowski, J. Y. Sweeney, T. Treis, R. M. Zappala et al., 2002. Fitness evolution and the rise of mutator alleles in experimental Escherichia coli populations. Genetics 162: 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski, P. D., P. J. Gerrish and R. E. Lenski, 1997. Evolution of high mutation rates in experimental populations of Escherichia coli. Nature 387: 703–705. [DOI] [PubMed] [Google Scholar]
- Steegborn, C., O. Danot, R. Huber and T. Clausen, 2001. Crystal structure of transcription factor MalT domain III: a novel helix repeat fold implicated in regulated oligomerization. Structure 9: 1051–1060. [DOI] [PubMed] [Google Scholar]
- Szmelcman, S., and M. Hofnung, 1975. Maltose transport in Escherichia coli K-12: involvement of the bacteriophage lambda receptor. J. Bacteriol. 124: 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler, M. F., D-E. Chang and T. Conway, 2006. Guanosine 3′5′-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc. Natl. Acad. Sci. USA 103: 2374–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treves, D. S., S. Manning and J. Adams, 1998. Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol. Biol. Evol. 15: 789–797. [DOI] [PubMed] [Google Scholar]
- Vasi, F., M. Travisano and R. E. Lenski, 1994. Long-term experimental evolution in Escherichia coli. II. Changes in life-history traits during adaptation to a seasonal environment. Am. Nat. 144: 432–456. [Google Scholar]
- Wang, J. C., 1971. Interaction between DNA and an Escherichia coli protein omega. J. Mol. Biol. 55: 523–533. [DOI] [PubMed] [Google Scholar]
- Wichman, H. A., M. R. Badgett, L. A. Scott, C. M. Boulianne and J. J. Bull, 1999. Different trajectories of parallel evolution during viral adaptation. Science 285: 422–424. [DOI] [PubMed] [Google Scholar]
- Woods, R., D. Schneider, C. L. Winkworth, M. A. Riley and R. E. Lenski, 2006. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc. Natl. Acad. Sci. USA 103: 9107–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron, C., J. Vieira and J. Messing, 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33: 103–119. [DOI] [PubMed] [Google Scholar]
- Yedid, G., and G. Bell, 2001. Microevolution in an electronic microcosm. Am. Nat. 157: 465–487. [DOI] [PubMed] [Google Scholar]
- Yoon, H., S. Lim, S. Heu, S. Choi and S. Ryu, 2003. Proteome analysis of Salmonella enterica serovar Typhimurium fis mutant. FEMS Microbiol. Lett. 226: 391–396. [DOI] [PubMed] [Google Scholar]
- Zhang, J., 2003. Parallel functional changes in the digestive RNases of ruminants and colobines by divergent amino acid substitutions. Mol. Biol. Evol. 20: 1310–1317. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., G. Gosset, R. Barabote, C. S. Gonzalez, W. A. Cuevas et al., 2005. Functional interactions between the carbon and iron utilization regulators, Crp and Fur, in Escherichia coli. J. Bacteriol. 187: 980–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, D., C. Constantinidou, J. L. Hobman and S. D. Minchin, 2004. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 32: 5874–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]