Abstract
In early mammalian development, one of the two X chromosomes is silenced in each female cell as a result of X chromosome inactivation, the mammalian dosage compensation mechanism. In the mouse epiblast, the choice of which chromosome is inactivated is essentially random, but can be biased by alleles at the X-linked X controlling element (Xce). Although this locus was first described nearly four decades ago, the identity and precise genomic localization of Xce remains elusive. Within the X inactivation center region of the X chromosome, previous linkage disequilibrium studies comparing strains of known Xce genotypes have suggested that Xce is physically distinct from Xist, although this has not yet been established by genetic mapping or progeny testing. In this report, we used quantitative trait locus (QTL) mapping strategies to define the minimal Xce candidate interval. Subsequent analysis of recombinant chromosomes allowed for the establishment of a maximum 1.85-Mb candidate region for the Xce locus. Finally, we use QTL approaches in an effort to identify additional modifiers of the X chromosome choice, as we have previously demonstrated that choice in Xce heterozygous females is significantly influenced by genetic variation present on autosomes (Chadwick and Willard 2005). We did not identify any autosomal loci with significant associations and thus show conclusively that Xce is the only major locus to influence X inactivation patterns in the crosses analyzed. This study provides a foundation for future analyses into the genetic control of X chromosome inactivation and defines a 1.85-Mb interval encompassing all the major elements of the Xce locus.
IN mammals, X chromosome inactivation serves to equalize X-linked gene expression between the sexes. Early in female development, each somatic cell inactivates one of its two X chromosomes. This choice is then faithfully transmitted to all daughter cells through mitosis, such that the adult female is a mosaic of two different cell lineages (Lyon 1961). Two forms of X inactivation that differ in their mechanism of choice take place in the mouse embryo. The extraembryonic tissues undergo imprinted X inactivation, where the choice is dictated by parental origin. This results in nonrandom inactivation of the paternally inherited chromosome (Takagi and Sasaki 1975; Huynh and Lee 2001; Sado et al. 2001; Wang et al. 2001; Sado and Ferguson-Smith 2005). In contrast, embryonic cells undergo random inactivation, and either X chromosome can be chosen for silencing (Lyon 1961; Krietsch et al. 1986).
Although theoretically the two X chromosomes in a somatic cell have an equal chance of being inactivated, the X-linked locus Xce (Xcontrolling element) can significantly bias this choice in mice (Cattanach and Isaacson 1967; Cattanach and Williams 1972). Previous experiments have shown that Xce exerts a primary effect on choice, as skewed X inactivation patterns are observed even in embryos isolated soon after X inactivation occurs (Rastan 1982) and because the effect persists even in the face of a selective advantage for one chromosome over the other (Drews et al. 1974). Three alleles of Xce have been defined in inbred mouse strains on the basis of their influence on the X inactivation pattern in genetic crosses: a weak allele Xcea (C3H/HeJ, 101/H, A/J, CBA/J and BALB/cByJ), an intermediate allele Xceb (C57BL/6J, DBA/2J and JU/Ct), and a strong allele Xcec (CAST/Ei), although additional alleles are thought to exist in other strains (Cattanach et al. 1969; West and Chapman 1978; Johnston and Cattanach 1981; Simmler et al. 1993). In Xce heterozygotes, the chromosome carrying the weaker of the two alleles is more likely to be inactivated. The degree of skewing can be quite profound; in Xcea/Xcec heterozygotes, the mean X inactivation pattern is ∼25:75, whereby the chromosome carrying the Xcea allele is active in only one-quarter of cells (Plenge et al. 2000; de La Casa-Esperon et al. 2002). In contrast, choice in Xce homozygotes is largely unbiased (Krietsch et al. 1986; Plenge et al. 2000).
In earlier studies, the Xce locus was mapped to a region between the ectodysplasin-A (Eda, at position 94.5 Mb in Mm Build 34) and the phosphoglycerate kinase (Pgk1, 100.7 Mb) genes, a region that encompasses the X inactivation center (Xic) (Cattanach et al. 1970, 1982; Cattanach and Papworth 1981). The Xist locus, which encodes a noncoding RNA required in cis to initiate X inactivation, is located in the Xic (Borsani et al. 1991; Brockdorff et al. 1991; Lee et al. 1999), along with its antisense counterpart Tsix (Lee et al. 1999). However, an ancestral recombination event identified in the well-characterized strain JU/Ct excluded these as positional candidates for Xce (Simmler et al. 1993). Although this analysis suggested that the distal boundary of the Xce candidate region was located between DXPas29 and DXPas28 (98 and 97.9 Mb, respectively), the proximal boundary of the candidate interval has not yet been refined.
Although the nature of the Xce locus and its molecular mode of action has not yet been identified, most models of X inactivation (Lyon 1971; Brown and Chandra 1973; Russell and Cacheiro 1978; Rastan 1983) hypothesize that it serves as a binding site for trans-acting factors that in turn regulate the expression of other loci in the Xic, such as Xist or Tsix. The various Xce alleles are thus predicted to have differential binding affinities for this factor (or factors), leading to a bias in the choice between chromosomes. While no such factors have been identified to date, mutagenesis has uncovered three candidate loci, all of which are autosomally encoded (Xiaf1–3) (Percec et al. 2002, 2003). We have shown previously that naturally occurring genetic variation between inbred mouse strains can also influence X chromosome choice (Chadwick and Willard 2005), suggesting that it may be possible to identify additional modifiers of choice in inbred strains using standard quantitative trait (QTL) mapping approaches.
As the precise location and nature of the Xce locus is not known, genetic studies of X inactivation in mice rely upon tightly linked markers to infer Xce genotype. Previously, the markers DXMit18 and DXMit171 were used for this purpose (Percec et al. 2002, 2003). However, we found that progeny testing did not always support this interval (data not shown), suggesting that Xce in fact may lie proximal to these markers. In this study, we used QTL mapping techniques to define the Xce candidate interval to a maximum 1.85-Mb region of the mouse X chromosome and provide progeny test data to support this localization. We then used a similar approach to search for additional naturally occurring modifiers of X inactivation patterns.
MATERIALS AND METHODS
Mice and mouse crosses:
C57BL/6J (B6), BALB/cByJ (BALB), and CAST/Ei (CAST) mice used in these crosses were purchased from The Jackson Laboratory. Mice were housed in accordance with Institutional Animal Use and Care Committee guidelines. F1 crosses were carried out using B6 or BALB females and CAST males. F1 progeny were then intercrossed to generate F2 females. In addition, we backcrossed B6CASTF1 females to B6 males or BALBCASTF1 females to BALB males for two to eight generations, selecting for the presence of CAST X chromosome alleles at each backcross generation. After the first backcross generation, both males and females were used for backcrossing. Ear biopsies were collected at weaning for later RNA isolation.
RNA isolation and cDNA synthesis:
RNA was isolated from ear biopsies and whole-mouse embryos using the RNeasy miniprep kit (QIAGEN, Chatsworth, CA), according to the manufacturer's recommendations. For ear RNA, the modified fibrous tissue protocol was used. cDNA synthesis was carried out as described (Percec et al. 2003), using random primers.
Allele-specific expression assays:
The Pctk1 expression assay was carried out as described previously (Plenge et al. 2000), except that products were separated and analyzed on an ABI 3100 capillary sequencer. We designed an assay similar to Idh3g for use with animals that were not informative at Pctk1. This Idh3g assay was performed as the Pctk1 assay with the following differences: the primers used were 5′-AACTATGGCCATGTGTATGC-3′ and 5′-CTCCAATATCTGGGGTATGC-3′, and the products were digested with TaqαI.
Genotyping:
All genotyping was carried out by PCR amplification of microsatellite markers, and PCR products were separated on an ABI 3100 capillary sequencer. Genetic location of markers and primer sequences were taken from the Mouse Genome Database (Blake et al. 2003).
Embryo dissections:
Embryos were collected at 10.5 days post-coitum (dpc) according to established protocols (Hogan et al. 1994). Mating was ascertained by checking for vaginal plugs, with the first day after mating designated as 0.5 dpc. The sex of embryos was determined by PCR amplification of the Smcx and Smcy genes, as described previously (Mroz et al. 1999).
QTL mapping:
QTL mapping was performed by standard interval mapping (Lander and Botstein 1989), using the software R/qtl (Broman et al. 2003), an add-on package to the general statistical software, R (Ihaka and Gentleman 1996). Statistical significance with adjustment for a genomewide scan was determined by a permutation test (Churchill and Doerge 1994); 1000 permutation replicates were used.
SNP discovery:
To identify SNPs that differentiated the CAST haplotype from BALB and B6 haplotypes, genomic DNA from each strain was PCR amplified and sequenced using the BigDye Terminator v 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). Polymorphic sites were identified by directly comparing sequences from the three strains. Sequenced regions, primers used, and SNP genotypes are listed in Table 1.
TABLE 1.
SNPs identified and tested in this study
| SNP | Primer A | Primer B | No. of SNPs in amplicon | B6/BALB allele | CAST allele |
|---|---|---|---|---|---|
| SNP-846 | 5′-GGCTAAGCCATCACTTATCC-3′ | 5′-AAGATTCCTAACTGTTCTGGG-3′ | 3 (all in dbSNP) | GCT | TTC |
| SNP-828 | 5′-CCATGGAGATGATGACAAGC-3′ | 5′-GGCCTACAAAGCGACTTCC-3′ | 1 novel | A | G |
| LeeSNPG-Ia | 5′-GCTTGGTTCGTCTATCTTGTGGG-3′ | 5′-CCAGAGTCTGATGTAACGGAGG-3′ | 1 | Not digested by ScrFI | Digested by ScrFI |
| SNP-843 | 5′-TGTTTCCATGCCTCAGAAGC-3′ | 5′-GAACCACACTGCTTAACTAGC-3′ | 3 novel 2 in dbSNP | TACCA | ACTAG |
SNPs were genotyped by PCR amplification of genomic DNA using the primers indicated followed by sequencing.
This assay was published in Stavropoulos et al. (2001). To genotype this SNP, genomic DNA was PCR amplified with the primers indicated and digested with ScrFI, and products were separated by gel electrophoresis.
RESULTS
Identification of major genetic influences on X inactivation patterns:
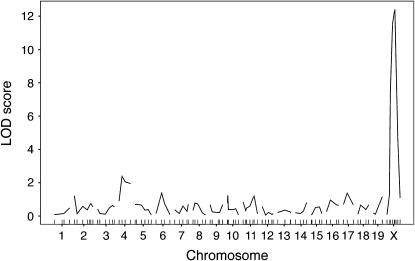
We have demonstrated previously that genetic background differences segregating in the B6CASTF2 cross resulted in significant effects on early events that determine the X inactivation pattern (the proportion of cells that have one or the other chromosome active) in Xce heterozygous mice (Chadwick and Willard 2005). X inactivation can be considered a quantitative trait, where the X inactivation pattern is treated as a continuous quantitative phenotype. To determine whether these effects represented a global influence on X chromosome choice (regardless of Xce genotype), we used a QTL mapping strategy in a population of B6CASTF2 Xce homozygotes and Xce heterozygotes. We genotyped 72 B6CASTF2 Xce heterozygous and 28 B6CASTF2 Xce homozygous females using a panel of microsatellite markers with ∼20 cM average spacing based on one developed previously (Iakoubova et al. 2000). We identified a single major QTL (Figure 1) in this cross. As this locus was X linked, it seemed likely that this association was due to Xce. Thus, we conclude that Xce is the major locus in the genome that influences X chromosome choice.
Figure 1.—
LOD curves from the B6CASTF2 Xce heterozygotes/Xce homozygotes whole-genome scan. LOD score is on the y-axis; chromosomes are on the x-axis. Genetic location of markers tested are indicated by tick marks on the x-axis. The genomewide level of significance as determined by permutation tests is LOD = 3.5. Significance was reached only on the X chromosome, indicating that Xce is the only major locus influencing X inactivation patterns in this cross.
Defining the Xce candidate region:
Although the Xce locus was first described nearly 40 years ago (Cattanach and Isaacson 1967), its genomic localization on the X chromosome has not been rigorously defined and encompasses a large genetic interval (Cattanach et al. 1970, 1982; Cattanach and Papworth 1981; Simmler et al. 1993). We sought to use QTL approaches in an effort to further define the Xce candidate interval.
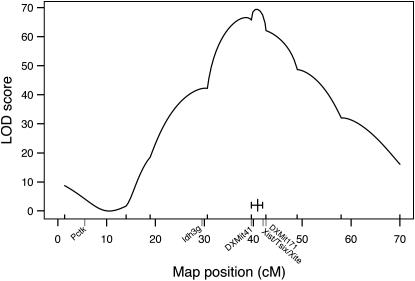
For this analysis, we used a sample of B6CAST (n = 528) and BALBCAST (n = 127) female mice collected from a variety of crosses (intercrosses and backcrosses) that had previously been bred for other purposes. This sample was not predicated on the Xce genotype, and analysis included mice both heterozygous (Xceb/Xcec or Xcea/Xcec) and homozygous (Xceb/Xceb or Xcea/Xcea) at Xce. However, each sample was heterozygous for at least one of the marker genes used to determine the X inactivation pattern, Pctk1 (5.5 cM) or Idh3g (29.5 cM). For each individual, we determined the X inactivation pattern and the X chromosome genotypes for markers spanning the X chromosome (DXMit55: 1.4 cM; DXMit165: 14 cM; DXMit75: 18.9 cM; DXMit42: 30.6 cM; DXMit41: 39.6 cM; DXMit97: 49 cM; DXMit151: 58 cM; and DXMit156: 70 cM), including one marker thought to be tightly linked to the Xce locus (DXMit171: 42.6 cM). Interval mapping with this data set (Figure 2) indicated that the Xce locus was located within a 2.3-cM region (1.5-LOD confidence interval) spanning from 39.6 cM (DXMit41) to 41.9 cM (slightly proximal of DXMit171), consistent with and refining previous mapping studies (Cattanach et al. 1970, 1982; Cattanach and Papworth 1981; Simmler et al. 1993).
Figure 2.—
X chromosome LOD curve from extended Xce heterozygote/Xce homozygote analysis (n = 655, from B6CAST and BALBCAST intercrosses and backcrosses). LOD scores are on the y-axis; genetic positions of markers tested (from MGD) are on the x-axis. The genetic locations of Pctk1, Idh3g, Xist/Tsix/Xite, DXMit41, and DXMit171 are indicated. The 1.5-LOD confidence interval is indicated by a short, solid horizontal line, intersected by a vertical line indicating the peak. This suggests that the Xce locus is located between DXMit41 and DXMit171.
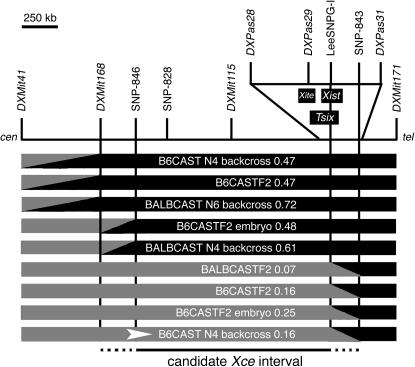
To refine further the candidate region, we identified nine animals (six from B6CAST crosses, three from BALBCAST crosses) with crossovers between DXMit41 and DXMit171 and genotyped them at intervening markers: DXMit168, DXMit115, DXPas28, DXPas29, and DXPas31 (used by Simmler et al. 1993). The haplotypes and X inactivation patterns for these individuals are shown in Figure 3. It is important to note that the apparent nonrandom representation of alleles in these females (all carrying the CAST allele at DXMit41 while carrying B6 or BALB alleles at DXMit171) reflects the requirement of our approach: the recombinant chromosomes also need to carry CAST alleles at either Pctk or Idh3g (both located proximal to this region) such that X inactivation assays can be carried out in female progeny. To classify these animals into phenotypic classes (Xce homozygous like and Xce heterozygous like), we determined the probability that each individual's X inactivation pattern fell into a normal distribution with a mean of 0.5 and SD of 0.1 (predicted for Xce homozygotes given a random choice; Plenge et al. 2000) or with a mean of 0.29 and SD of 0.13 (Chadwick and Willard 2005). As a result of this analysis, we identified five individuals predicted to be Xce homozygotes (X inactivation patterns 0.47–0.72, 76–96% probability of being homozygous at Xce) and four mice predicted to be Xce heterozygotes (X inactivation patterns 0.07–0.25, 94–99% probability of being heterozygous at Xce). Three of the so-defined Xce homozygous females had heterozygous genotypes at DXMit168 but homozygous genotypes at DXMit115, thus defining the proximal boundary of the candidate interval. In addition, we also identified four Xce heterozygous females that had heterozygous genotypes at DXPas29 but homozygous genotypes at DXPas31, indicating that the distal boundary of the candidate interval lay between these two markers. As no other published microsatellite markers were available, we sequenced selected genomic segments in the relevant strains and identified additional SNPs in these regions (between DXMit168 and DXMit115 and between DXPas29 and DXPas31) that distinguished the CAST haplotype from that of the two classical inbred strains used in our crosses (Table 1). Both individuals with recombinant haplotypes between DXMit168 and DXMit115 had crossover events close to DXMit168; while they had heterozygous genotypes at DXMit168, both had B6 genotypes at SNP-846. All four individuals with recombinant haplotypes between DXPas29 and DXPas31 had crossover events that occurred within a 165-kb region between LeeSNPG-I (a SNP located within the Xist locus; Stavropoulos et al. 2001) and SNP-843. Thus, by analyzing recombinant chromosomes, we have established that the Xce candidate interval spans a minimum of 1.5 Mb and a maximum of 1.85 Mb from DXMit168 (96.3 Mb) to SNP-843 (98.1 Mb), as shown in Figure 3.
Figure 3.—
Haplotype map of females with crossovers between DXMit41 and DXMit171. Heterozygous genotypes (B6/CAST or BALB/CAST) are indicated as shaded segments; homozygous genotypes (B6/B6 or BALB/BALB) are indicated as solid segments. Diagonal regions between markers indicate the location of crossover events. The locations of Xite, Tsix, and Xist are indicated. The genetic background and X inactivation pattern of each individual are also indicated. Animals with homozygous genotypes between SNP-846 and DXMit171 had Xce homozygous-like X inactivation patterns, suggesting that the proximal boundary of the Xce interval lay between DXMit168 and SNP-846. Conversely, animals with heterozygous genotypes between DXMit41 and SNP-843 had Xce heterozygous-like X inactivation patterns, suggesting that the distal boundary of the candidate interval lay between LeeSNPG-I and SNP-843. The B6CAST N4 backcross individual selected for further progeny testing (Figure 4) is indicated by an arrowhead.
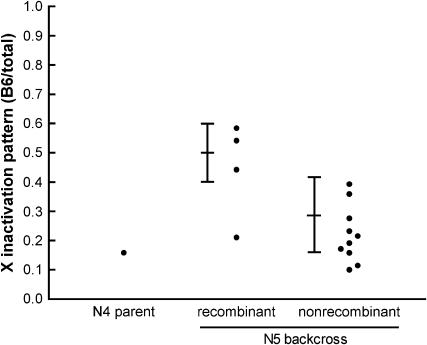
To confirm these results, we progeny tested a B6CAST N4 backcross female that had a recombinant haplotype between LeeSNPG-I and SNP-843 (Figure 4; the individual tested is indicated in Figure 3). The X inactivation patterns of the N5 backcross progeny confirmed our placement of the distal boundary of the Xce candidate region; female progeny that had inherited the CAST haplotype (and thus that were heterozygous throughout the Xce candidate region) had skewed, Xce heterozygous-like X inactivation patterns, while progeny that had inherited the B6 haplotype in this region generally had X inactivation patterns in the Xce homozygous-like range. This confirmed our placement of the distal Xce boundary between LeeSNPG-I and SNP-843.
Figure 4.—
Progeny testing to confirm the distal boundary of the Xce candidate region. A B6CAST N4 backcross female with a crossover between LeeSNPG-I and SNP-843 was crossed to a B6 male and the X inactivation patterns of the N5 progeny were determined. The predicted range of X inactivation patterns for Xce homozygotes (mean = 0.5, SD = 0.1, as suggested by Plenge et al. 2000) and Xce heterozygotes arising from this cross (mean = 0.29, SD = 0.13, from Chadwick and Willard 2005) are indicated. “Recombinant” animals do not carry the heterozygous haplotype of the parent in the DXMit41–DXMit171 candidate region; “nonrecombinant” animals have the same haplotype as the N4 parent. As expected, N5 females with the recombinant haplotype have X inactivation patterns that are generally consistent with an Xce homozygous genotype, while N5 females with the nonrecombinant haplotype have X inactivation patterns consistent with an Xce heterozygous genotype.
Nonrandom distribution of crossovers in the DXMit41–DXMit171 interval:
While identifying recombination breakpoints in the nine females that were recombinant between DXMit41 and DXMit171, we observed a nonrandom distribution of crossovers in these individuals. Although we found two independent crossover events occurring in the 258-kb region between DXMit168 and SNP-846 and four independent crossover events in the 165-kb region between LeeSNPG-I and SNP-843, we did not observe a single crossover in the intervening ∼1.5 Mb (Figure 3). To determine whether this apparent suppression of recombination was unusual for an area of this physical size, we sought to calculate the relationship between genetic and physical distances in the DXMit41–DXMit171 interval, compared to another region of the X chromosome.
We collected genotype information from individuals, both male and female, arising from B6CAST crosses in which the mother carried a fully nonrecombinant CAST X chromosome opposite a nonrecombinant B6 X chromosome. This situation was true in both intercrosses (n = 921) and in some backcrosses (n = 612). We selected individuals for which we had genotype information at both DXMit41 (39.6 cM) and DXMit171 (42.6 cM) (n = 967), and individuals with genotype information at both DXMit165 (14 cM) and DXMit75 (18.9 cM) (n = 1297). We observed 11 crossovers between DXMit41 and DXMit171 in 967 individuals, a genetic distance of 1.1 cM. The actual physical distance between the two was 2.74 Mb, and the calculated recombination rate was thus 0.40 cM/Mb in this interval. In the DXMit165–DXMit75 interval, we observed 93 crossovers in 1297 meioses, a genetic distance of 7 cM over a 17.8-Mb region, or 0.39 cM/Mb, which is similar to that observed in the DXMit41–DXMit171 interval and to the genomewide average (0.5 cM/Mb) (Buchner et al. 2003; Kelmenson et al. 2005). If recombination rates were consistent across the entire length of the chromosome, we would have expected to identify three or four individuals with crossovers between SNP-846 and LeeSNPG-I. Conversely, the 165-kb segment between LeeSNPG-I and SNP-843 appears to contain a recombination hotspot with a recombination frequency of 3.6 cM/Mb, more than sevenfold higher than the genome average.
QTL mapping to identify autosomal modifiers of Xce:
Because QTL mapping approaches proved quite useful in refining the Xce candidate region, we used a similar strategy to search for additional loci that influenced X inactivation patterns (Figure 5). To control for the effect of the major X-linked QTL (i.e., Xce), we used only Xce heterozygous females, who would be predicted to have a similar range of X inactivation patterns. We genotyped 72 Xce heterozygous B6CASTF2 females at loci across the genome (20 cM average spacing) with the panel of microsatellite markers used previously. Interval mapping identified one region on chromosome 4 that showed suggestive association with differences in the X inactivation pattern, although this fell slightly below the genomewide level of significance determined by permutation testing (LOD = 3.49, P = 0.08). We added additional markers in this region, but this did not increase the level of significance.
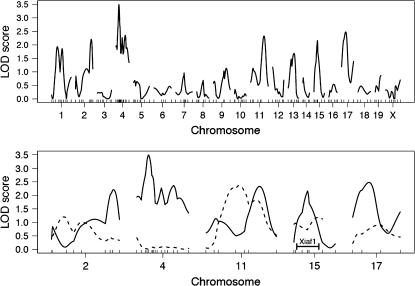
Figure 5.—
LOD curves from B6CASTF2 Xce heterozygote whole-genome scan. (Top) The LOD scores from the whole-genome analysis of B6CASTF2 Xce heterozygous adult samples. The genomewide significance level as determined by permutation tests is LOD = 3.5. (Bottom) The LOD scores of adults (solid lines) and embryos (dashed lines) for the five most significant chromosomes from the adult analysis. The location of Xiaf1 on chromosome 15 is indicated (Percec et al. 2002).
To confirm or exclude this region as a candidate locus, we sought to reduce variation in the phenotype by using whole-embryo RNA rather than RNA obtained from adult ear biopsies (Chadwick and Willard 2005), in principle making it easier to detect potential QTL. Thus, we genotyped Xceb/Xcec B6CASTF2 embryos (n = 79) on the chromosomes with the highest LOD scores in the initial scan: chromosomes 2, 4, 11, 15, and 17. None of these chromosomes showed a significant association with the phenotype in whole embryos, including chromosome 4, which was most significant in the initial screen (Figure 5), and chromosome 15, which harbors Xiaf1, an ENU-induced mutation known to influence X chromosome choice (Percec et al. 2002, 2003). Thus, while we have demonstrated that genetic background plays a role in influencing X inactivation patterns (Chadwick and Willard 2005), we were not able to identify any significantly associated autosomal loci with the available data.
DISCUSSION
A genetically defined candidate region for Xce:
In this study we have used a combination of QTL mapping, analysis of recombinant chromosomes, and progeny testing to demonstrate conclusively that Xce is the only major locus that influences X inactivation patterns (and likely, therefore, X chromosome choice) in the mouse crosses analyzed. Furthermore, we defined a 1.85-Mb region of the mouse X chromosome that contains most (if not all) elements of the Xce locus, as this interval accounts fully for the known effects of Xce on X inactivation patterns. As the nature of and the underlying molecular mechanisms of Xce remain elusive, this has implications for future study of X chromosome choice.
Previously, Simmler et al. (1993) attempted to map the Xce locus by examining microsatellite marker haplotypes of mouse strains with well-characterized Xce alleles. They observed discordant alleles at DXPas29 and DXPas31 in the JU/Ct strain relative to other Xceb strains, indicating that the Xce locus lay proximal to DXPas29 and thus was distinct from Xist. Combined with the results of our study, the Xce candidate interval could be even smaller than the region identified here. However, a potential limitation of the Simmler et al. study is its reliance upon microsatellite repeat polymorphisms, which are known to have somewhat higher mutation rates than other sequences (Hastbacka et al. 1992). Thus, in the absence of additional recombinants, we feel it is prudent to consider the larger interval as a guide to further study. Our data establish a relatively similar distal boundary for the Xce candidate interval (on the basis of genetic mapping and progeny testing), but do not allow us to exclude Xist, Tsix, or Xite as Xce candidate loci. This allows for the possibility that polymorphisms located either within these transcripts or in their regulatory regions may contribute to the bias in X chromosome choice observed in Xce heterozygous females. In fact, in humans a rare variant found within the XIST promoter has been shown to cause significant skewing of the X inactivation pattern (Plenge et al. 1997). This bias in determining the X inactivation pattern is thought to be the result of a dramatic increase in CTCF binding to the XIST promoter on the variant allele (Pugacheva et al. 2005).
In addition to clarifying the relationship between Xce candidate sequences and known genes within the Xic, we have defined ∼1.7 Mb of additional candidate sequence that can now be tested. Although it is tempting to postulate that all X chromosome choice elements are located in the immediate vicinity of Xist and Tsix, it will be important to consider the remainder of the candidate interval. Subsequent studies may further define critical sequences by identifying additional recombinants, by establishing congenic lines carrying overlapping introgressed segments, or by using a sequence substitution approach in female embryonic stem cells.
It is interesting to note the nonrandom distribution of crossover events that we observed within the Xce candidate region. While we observed no recombination in the 1.5-Mb region between SNP-846 and LeeSNPG-I, recombination in the adjacent region (LeeSNPG-I–SNP-843) appeared to be over sevenfold higher than the genomewide average (Buchner et al. 2003; Kelmenson et al. 2005). Several recombination hotspots have been defined in mammals, the most well characterized being at the major histocompatibility complexes of humans and mice (Shiroishi et al. 1990; Jeffreys et al. 2001). In yeast, recombination hotspots are relatively common and distributed throughout the genome (Gerton et al. 2000); recent evidence suggests that this may also be true in mammalian genomes (Crawford et al. 2004; McVean et al. 2004). If this is indeed the case, the “granular” distribution of crossovers that we observed in the Xce region may not be unusual. Alternatively, the Xce locus may be marked by genomic rearrangements (such as inversions) between strains, as these have been shown to bias recombination rates (Singleton 1964), even in mice (Zheng et al. 1999; Klysik et al. 2004). Such genomic differences could either ablate binding sites for trans-acting factors entirely or bring these sites closer to (or farther from) other enhancer elements or the Tsix promoter. Future genomic studies in strains with well-characterized Xce alleles may address this question.
An approach for identifying trans-acting factors influencing X chromosome choice:
Using QTL mapping techniques, we were able to define the Xce candidate interval on the mouse X chromosome. However, we were unable to identify autosomal modifier loci using a similar strategy in our initial data set of 72 Xce heterozygous F2 females. Nonetheless, we believe that this approach may have great potential for identifying trans-acting factors, although a significantly larger sample size would likely be required. Another possibility is to investigate other crosses, particularly with additional wild-derived strains. Wild-derived inbred strains are highly polymorphic relative to classical inbred strains, and many such polymorphisms could have functional consequences (Ideraabdullah et al. 2004) that would obviate the need for extensive mutagenesis screens.
Although our initial screen did identify one suggestive autosomal QTL, we were unable to confirm this locus in F2 embryos. We can imagine several scenarios that could explain this. First, as is true in any linkage mapping experiment, the initial identification of a locus on chromosome 4 could have been a false positive. Alternatively, the QTL could exert a subtle effect that we were able to identify in the initial experiment, and for which we had low power to detect in the second, smaller experiment. A further possibility is that the chromosome 4 QTL affected an aspect of X inactivation, such as clone size, that would be irrelevant or undetectable in whole embryos. A QTL that affected clone size, perhaps by altering the timing of X inactivation, would not be detectable in embryos, where we determined X inactivation patterns in whole animals. Aside from this limitation, whole embryos would appear to be a promising system with which to further investigate the genetic regulation of X inactivation. The significant reduction in phenotypic variation relative to adult populations makes them well suited both for QTL analyses and for further study of X inactivation choice in general.
Acknowledgments
The authors are grateful to Stephanie Merrett for assistance with mouse husbandry and to Petko Petkov and Joanne Thorvaldsen for helpful discussions. This work was funded in part by research grants GM45441 (to H.F.W.) and GM74768 (to M.S.B.) from the National Institutes of Health.
References
- Blake, J. A., J. E. Richardson, C. J. Bult, J. A. Kadin and J. T. Eppig, 2003. MGD: the Mouse Genome Database. Nucleic Acids Res. 31: 193–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani, G., R. Tonlorenzi, M. C. Simmler, L. Dandolo, D. Arnaud et al., 1991. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351: 325–329. [DOI] [PubMed] [Google Scholar]
- Brockdorff, N., A. Ashworth, G. F. Kay, P. Cooper, S. Smith et al., 1991. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351: 329–331. [DOI] [PubMed] [Google Scholar]
- Broman, K. W., H. Wu, S. Sen and G. A. Churchill, 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Brown, S. W., and H. S. Chandra, 1973. Inactivation system of the mammalian X chromosome. Proc. Natl. Acad. Sci. USA 70: 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner, D. A., M. Trudeau, A. L. George, Jr., L. K. Sprunger and M. H. Meisler, 2003. High-resolution mapping of the sodium channel modifier Scnm1 on mouse chromosome 3 and identification of a 1.3-kb recombination hot spot. Genomics 82: 452–459. [DOI] [PubMed] [Google Scholar]
- Cattanach, B. M., and J. H. Isaacson, 1967. Controlling elements in the mouse X chromosome. Genetics 57: 331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattanach, B. M., and D. Papworth, 1981. Controlling elements in the mouse. V. Linkage tests with X-linked genes. Genet. Res. 38: 57–70. [DOI] [PubMed] [Google Scholar]
- Cattanach, B. M., and C. E. Williams, 1972. Evidence of non-random X chromosome activity in the mouse. Genet. Res. 19: 229–240. [DOI] [PubMed] [Google Scholar]
- Cattanach, B. M., C. E. Pollard and J. N. Perez, 1969. Controlling elements in the mouse X-chromosome. I. Interaction with the X-linked genes. Genet. Res. 14: 223–235. [DOI] [PubMed] [Google Scholar]
- Cattanach, B. M., J. N. Perez and C. E. Pollard, 1970. Controlling elements in the mouse X-chromosome. II. Location in the linkage map. Genet. Res. 15: 183–195. [DOI] [PubMed] [Google Scholar]
- Cattanach, B. M., T. H. Bucher and S. J. Andrews, 1982. Location of Xce in the mouse X chromosome and effects on PGK-1 expression. Genet. Res. 40: 103–104. [Google Scholar]
- Chadwick, L. H., and H. F. Willard, 2005. Genetic and parent-of-origin influences on X chromosome choice in Xce heterozygous mice. Mamm. Genome 16: 691–699. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, D. C., T. Bhangale, N. Li, G. Hellenthal, M. J. Rieder et al., 2004. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36: 700–706. [DOI] [PubMed] [Google Scholar]
- de La Casa-Esperon, E., J. C. Loredo-Osti, F. Pardo-Manuel de Villena, T. L. Briscoe, J. M. Malette et al., 2002. X chromosome effect on maternal recombination and meiotic drive in the mouse. Genetics 161: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, U., S. R. Blecher, D. A. Owen and S. Ohno, 1974. Genetically directed preferential X-activation seen in mice. Cell 1: 4–8. [Google Scholar]
- Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown et al., 2000. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastbacka, J., A. de la Chapelle, I. Kaitila, P. Sistonen, A. Weaver et al., 1992. Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nat. Genet. 2: 204–211. [DOI] [PubMed] [Google Scholar]
- Hogan, B., R. Beddington, F. Costantini and E. Lacey, 1994. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Huynh, K. D., and J. T. Lee, 2001. Imprinted X inactivation in eutherians: a model of gametic execution and zygotic relaxation. Curr. Opin. Cell Biol. 13: 690–697. [DOI] [PubMed] [Google Scholar]
- Iakoubova, O. A., C. L. Olsson, K. M. Dains, J. Choi, I. Kalcheva et al., 2000. Microsatellite marker panels for use in high-throughput genotyping of mouse crosses. Physiol. Genomics 3: 145–148. [DOI] [PubMed] [Google Scholar]
- Ideraabdullah, F. Y., E. de la Casa-Esperon, T. A. Bell, D. A. Detwiler, T. Magnuson et al., 2004. Genetic and haplotype diversity among wild-derived mouse inbred strains. Genome Res. 14: 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka, R., and R. Gentleman, 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5: 299–314. [Google Scholar]
- Jeffreys, A. J., L. Kauppi and R. Neumann, 2001. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 29: 217–222. [DOI] [PubMed] [Google Scholar]
- Johnston, P. G., and B. M. Cattanach, 1981. Controlling elements in the mouse. IV. Evidence of non-random X-inactivation. Genet. Res. 37: 151–160. [DOI] [PubMed] [Google Scholar]
- Kelmenson, P. M., P. Petkov, X. Wang, D. C. Higgins, B. J. Paigen et al., 2005. A torrid zone on mouse chromosome 1 containing a cluster of recombinational hotspots. Genetics 169: 833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klysik, J., C. Dinh and A. Bradley, 2004. Two new mouse chromosome 11 balancers. Genomics 83: 303–310. [DOI] [PubMed] [Google Scholar]
- Krietsch, W. K., M. Fehlau, P. Renner, T. Bucher and R. Fundele, 1986. Expression of X-linked phosphoglycerate kinase in early mouse embryos homozygous at the Xce locus. Differentiation 31: 50–54. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. T., L. S. Davidow and D. Warshawsky, 1999. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 21: 400–404. [DOI] [PubMed] [Google Scholar]
- Lyon, M. F., 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373. [DOI] [PubMed] [Google Scholar]
- Lyon, M. F., 1971. Possible mechanisms of X chromosome inactivation. Nat. New Biol. 232: 229–232. [DOI] [PubMed] [Google Scholar]
- McVean, G. A., S. R. Myers, S. Hunt, P. Deloukas, D. R. Bentley et al., 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304: 581–584. [DOI] [PubMed] [Google Scholar]
- Mroz, K., L. Carrel and P. A. Hunt, 1999. Germ cell development in the XXY mouse: evidence that X chromosome reactivation is independent of sexual differentiation. Dev. Biol. 207: 229–238. [DOI] [PubMed] [Google Scholar]
- Percec, I., R. M. Plenge, J. H. Nadeau, M. S. Bartolomei and H. F. Willard, 2002. Autosomal dominant mutations affecting X inactivation choice in the mouse. Science 296: 1136–1139. [DOI] [PubMed] [Google Scholar]
- Percec, I., J. L. Thorvaldsen, R. M. Plenge, C. J. Krapp, J. H. Nadeau et al., 2003. An N-ethyl-N-nitrosourea mutagenesis screen for epigenetic mutations in the mouse. Genetics 164: 1481–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenge, R. M., B. D. Hendrich, C. Schwartz, J. F. Arena, A. Naumova et al., 1997. A promoter mutation in the XIST gene in two unrelated families with skewed X-chromosome inactivation. Nat. Genet. 17: 353–356. [DOI] [PubMed] [Google Scholar]
- Plenge, R. M., I. Percec, J. H. Nadeau and H. F. Willard, 2000. Expression-based assay of an X-linked gene to examine effects of the X-controlling element (Xce) locus. Mamm. Genome 11: 405–408. [DOI] [PubMed] [Google Scholar]
- Pugacheva, E. M., V. K. Tiwari, Z. Abdullaev, A. A. Vostrov, P. T. Flanagan et al., 2005. Familial cases of point mutations in the XIST promoter reveal a correlation between CTCF binding and pre-emptive choices of X chromosome inactivation. Hum. Mol. Genet. 14: 953–965. [DOI] [PubMed] [Google Scholar]
- Rastan, S., 1982. Primary non-random X-inactivation caused by controlling elements in the mouse demonstrated at the cellular level. Genet. Res. 40: 139–147. [DOI] [PubMed] [Google Scholar]
- Rastan, S., 1983. Non-random X-chromosome inactivation in mouse X-autosome translocation embryos: location of the inactivation centre. J. Embryol. Exp. Morphol. 78: 1–22. [PubMed] [Google Scholar]
- Russell, L. B., and N. L. Cacheiro, 1978. The use of mouse X-autosome translocations in the study of X-inactivation pathways and nonrandomness. Basic Life Sci. 12: 393–416. [DOI] [PubMed] [Google Scholar]
- Sado, T., and A. C. Ferguson-Smith, 2005. Imprinted X inactivation and reprogramming in the preimplantation mouse embryo. Hum. Mol. Genet. 14(Suppl. 1): R59–R64. [DOI] [PubMed] [Google Scholar]
- Sado, T., Z. Wang, H. Sasaki and E. Li, 2001. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128: 1275–1286. [DOI] [PubMed] [Google Scholar]
- Shiroishi, T., N. Hanzawa, T. Sagai, M. Ishiura, T. Gojobori et al., 1990. Recombinational hotspot specific to female meiosis in the mouse major histocompatibility complex. Immunogenetics 31: 79–88. [DOI] [PubMed] [Google Scholar]
- Simmler, M. C., B. M. Cattanach, C. Rasberry, C. Rougeulle and P. Avner, 1993. Mapping the murine Xce locus with (CA)n repeats. Mamm. Genome 4: 523–530. [DOI] [PubMed] [Google Scholar]
- Singleton, J. R., 1964. A mechanism intrinsic to heterozygous inversions affecting observed recombination frequencies in adjacent regions. Genetics 49: 541–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos, N., N. Lu and J. T. Lee, 2001. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc. Natl. Acad. Sci. USA 98: 10232–10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi, N., and M. Sasaki, 1975. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256: 640–642. [DOI] [PubMed] [Google Scholar]
- Wang, J., J. Mager, Y. Chen, E. Schneider, J. C. Cross et al., 2001. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat. Genet. 28: 371–375. [DOI] [PubMed] [Google Scholar]
- West, J. D., and V. M. Chapman, 1978. Variation for X chromosome expression in mice detected by electrophoresis of phosphoglycerate kinase. Genet. Res. 32: 91–102. [DOI] [PubMed] [Google Scholar]
- Zheng, B., M. Sage, W. W. Cai, D. M. Thompson, B. C. Tavsanli et al., 1999. Engineering a mouse balancer chromosome. Nat. Genet. 22: 375–378. [DOI] [PubMed] [Google Scholar]