Abstract
A detailed genetic linkage map of Brassica rapa has been constructed containing 545 sequence-tagged loci covering 1287 cM, with an average mapping interval of 2.4 cM. The loci were identified using a combination of 520 RFLP and 25 PCR-based markers. RFLP probes were derived from 359 B. rapa EST clones and amplification products of 11 B. rapa and 26 Arabidopsis. Including 21 SSR markers provided anchors to previously published linkage maps for B. rapa and B. napus and is followed as the referenced mapping of R1–R10. The sequence-tagged markers allowed interpretation of the pattern of chromosome duplications within the B. rapa genome and comparison with Arabidopsis. A total of 62 EST markers showing a single RFLP band were mapped through 10 linkage groups, indicating that these can be valuable anchoring markers for chromosome-based genome sequencing of B. rapa. Other RFLP probes gave rise to 2–5 loci, inferring that B. rapa genome duplication is a general phenomenon through 10 chromosomes. The map includes five loci of FLC paralogues, which represent the previously reported BrFLC-1, -2, -3, and -5 and additionally identified BrFLC3 paralogues derived from local segmental duplication on R3.
THE genus Brassica includes oilseed, vegetable, fodder, and condiment crops. Brassica rapa (syn. campestris; A genome), B. napus (AC genome), B. juncea (AB), and B. carinata (BC) contribute ∼12% of the global supply of edible vegetable oil (Labana and Gupta 1993). B. rapa and B. oleracea (C genome) provide many vegetables that contribute to a healthy human diet, being a valuable source of dietary fiber, vitamin C, and other health-enhancing factors such as anticancer compounds (Fahey and Talalay 1995). The Brassica A genome therefore has worldwide importance in agriculture, with the quality and economic value of derived products such as processed oils and kimchi being dependent upon appropriate combinations of alleles. B. rapa includes a variety of vegetables crops such as Chinese cabbage, Pakchoi, turnip, and broccoletto as well as oilseed crops such as turnip rape and sarson (Gomez-Campo 1999).
The high degree of neutral DNA polymorphisms of most Brassica species (Figdore et al. 1988) has facilitated the development of molecular linkage maps, with at least 15 described to date for B. oleracea (Slocum et al. 1990; Kianian and Quiros 1992; Lan et al. 2000), B. rapa (Song et al. 1991; Chyi et al. 1992; Teutenico and Osborn 1994), B. nigra (Lagercrantz and Lydiate 1996), B. Juncea (Cheung et al. 1997; Pradhan et al. 2003), and B. napus (Landry et al. 1991; Uzunova et al. 1995). Where common sets of DNA markers and/or parental genotypes have been used, it has been possible to designate linkage groups according to a common nomenclature (Parkin et al. 1995, 2005; Butruille et al. 1999; Sebastian et al. 2000). Thus for B. napus linkage groups N1–N10 representing the A genome correspond to B. rapa R1–R10, and linkage groups N11–N19 representing the C genome correspond to B. oleracea O1–O9. Bohuon et al. (1996) demonstrated that marker order and linkage group structure had been conserved between the diploid (B. oleracea) and amphidiploid (B. napus) C genomes. In this study, we generated a detailed linkage map using sequenced EST clones derived from tissue-specific libraries of B. rapa. To establish the identity of linkage groups corresponding to R1–R10, we used SSR markers from Suwabe et al. (2002) and Lowe et al. (2004).
The Brassica genomes are closely related to the model plant Arabidopsis thaliana, diverging ∼20 MYA (Koch et al. 2001), and remain collinear. Comparative mapping of RFLP probes among the three diploid species B. rapa (n = 10), B. oleracea (n = 9), and B. nigra (n = 8) has suggested that genomes of the Brassica species are composed of three rearranged variants of an ancestral genome and descended from a common hexaploid ancestor (Lagercrantz and Lydiate 1996). All comparative studies of Arabidopsis and Brassica to date have revealed extensive duplications, with Arabidopsis segments being conserved an average of three times within the diploid Brassica genomes (Truco et al. 1996; Lan et al. 2000; Lukens et al. 2003; Parkin et al. 2005). Fiber–FISH mapping has been used to compare a 431-kb Arabidopsis BAC contig with B. rapa mitotic chromosomes (Jackson et al. 2000). Cytogenetic study using 21 Brassicaceae species revealed that the tribe Brassiceae comprising ∼240 species descended from a common hexaploid ancestor that has a similar genome to Arabidopsis (Lysak et al. 2005). Comparative genome analysis revealed that genes are reduced by deletion in the triplication blocks in the Brassica genome (O'Neill and Bancroft 2000; Rana et al. 2004; Park et al. 2005). Recently, we showed the sequence-level indel in four BAC clones that represent a triplicated and segmentally duplicated FLC region of B. rapa and are homologous with 125 kb of Arabidopsis chromosome 5 (Yang et al. 2006).
In this study we demonstrate the conservation of genome segments within and between chromosomes, on the basis of sequence-tagged markers.
MATERIALS AND METHODS
Population development and DNA extraction:
F2:3 families (40 F3 seedling) of 134 F2 lines (“JWF3p”) were developed from Chinese cabbage F1 cultivar Jangwon (B. rapa ssp. pekinensis). These two biennial inbred parent lines were made available courtesy of the former “Seoul Seed” company (Korea). To induce flowering, all seedling plants were vernalized for 35 days in a cold room at 5° with a 16-hr photoperiod. Tissues were collected from F2:3 families after 15 days of growing in greenhouses. Genomic DNA was extracted from the lyophilized tissue following the method of Cho et al. (1994), using 1 ml saturated phenol (BM Co.) for every 1-g sample.
Gel electrophoresis and Southern blot analysis:
To screen for polymorphisms, an average of 10 μg genomic DNA from the inbred parent lines was digested with seven restriction enzymes (BamHI, DraI, EcoRI, HindIII, EcoRV, XbaI, and ScaI) and fractionated on 0.9% agarose gels. Electrophoresis and Southern blotting were conducted as described by Cho et al. (1994). BamHI, EcoRI, EcoRV, and ScaI enzymes were used for digestion of the segregating progeny populations.
EST clones used as RFLP probes:
Four different tissue-specific libraries were used as a source of RFLP probe. These were prepared from mRNA isolated from immature flowers (BIF), anthers (BAN), roots (BR), and dark-grown seedlings (BDS) of B. rapa line Jangwon (Kim et al. 1996; Lim et al. 2000). Plasmid DNA preparation and nucleotide sequencing were conducted as described by Lim et al. (2000). We have used less redundant cDNA clones on the basis of their sequence and BLASTN search in GenBank (National Center for Biotechnology Information). Insert DNA was amplified by PCR using T7 and T3 primers and eluted by QIAGEN (Valencia, CA) gel extraction kits. Probe labeling was conducted by random hexamer labeled with 32P-dCTP (Feinberg and Vogelstein 1983). Hybridization followed the method described by Cho et al. (1994). Hybridized filters were washed with three stringency steps (2×, 1×, and 0.5× SSC with 0.5, 0.1, and 0.1% SDS), respectively, and exposed to X-ray film (Fuji, Stamford, CT) for 2–3 days.
Genome sequence tag markers used as RFLP probes:
Genome sequence tags (GSTs) representing 24 genes from Arabidopsis chromosomes 4 and 5 were generated by PCR amplication using Arabidopsis ecotype Columbia genomic DNA. The DNA were cloned and sequenced prior to use as RFLP probes. The 10 GSTs derived from Arabidopsis chromosome (chr)4 were At4RPP5 (At4g16860), At4ML1 (At4g21750), At4TR1 (At4g24520), At4CBF2a (At4g25480), At4PRHA (At4g29940), At4CPK5 (At4g35310), At4FAH1 (At4g36220), AtAP2 (At4g36920), At4HLS1 (At4g37580), and At4CESA2 (At4g39350). The other 14 GSTs derived from Arabidopsis chr5, At5HAT2 (At5g4730), At5COR78 (At5g52310), At5PDC2 (At5g54960), At5ILL1 (At5g56650), At5MSI1 (At5g58230), At5NPH3 (At5g64330), AtMYB68 (At5g6579), At5LCY (U50738), and 6 R-EST genes containing a cluster of NBS-LRR resistance recognition motif were used. Two flowering-time genes, AtFCA (At4g16280) and AtLFY (At5g61850) of Arabidopsis, were developed as probes. As for B. rapa genes, 6 flowering-time genes of B. rapa BrFLC (AY273164), BrAGL20 (AY345237), BrCO (AY356370), BrGI (AY356369), BrSVP (AY356366), and BrFLC5 gene-specific PCR product (forward primer, 5′-TTACCGCCTCTTTTATCCTTCTC-3′; reverse primer, 5′-CATATAACAACAAAAACCCCAATC-3′) were used in this genetic map. The 5 function genes of B. rapa, BrGST, BrMyrosinase, BrSAM, BrSLP, and BrDFRI, were surveyed. Characteristics of genetic markers are summarized in Table 1.
TABLE 1.
Characteristics of genetic markers in this linkage map
| Probe | Marker type | Source | No. of probes | No. of loci | Status | Reference |
|---|---|---|---|---|---|---|
| RFLP | EST_BIF | Immature flower library of B. rapa cv. Jangwon | 114 | 157 | pBluescript | Kim et al. (1996); Lim et al. (2000) |
| EST_BAN | Anther library of B. rapa cv. Jangwon | 176 | 235 | pBluescript | ||
| EST_BDS | Dark-grown seeding library of B. rapa cv. Jangwon | 45 | 55 | pBluescript | ||
| EST_BR | Root library of B. rapa cv. Jangwon | 24 | 27 | pBluescript | ||
| At4GTS | Chromosome 4-located genes of Arabidopsis cv. Colombia | 10 | 10 | pGemT | GenBank | |
| At5GTS | Chromosome 5-located genes of Arabidopsis cv. Colombia | 14 | 17 | pGemT | GenBank | |
| AtGene | Full length of expressed gene from A. thaliana | 2 | 2 | pGemT | GenBank | |
| BrGene | Full length of expressed gene from B.rapa | 11 | 17 | pGemT | BrFLC_AY273164; BrAGL20_AY345237, etc. | |
| PCR | SSR | R1–R10-located SSR markers | 11 | 11 | Primer | Lowe et al. (2004) |
| SSR | R1–R10-located SSR markers | 10 | 10 | Primer | Suwabe et al. (2002) | |
| BrH80A08_FLC1 | 5′-ttcccaagcttgctggtact-3′ 5′-gagatttccctcgcttgatg-3′ | 1 | 1 | Primer | KBrH80A08_AC155344 | |
| BrH04D11_FLC2 | 5′-gcgccaattataaatttgattttc-3′ 5′-tcctcctgaacctggtcttg-3′ | 1 | 1 | Primer | KBrH04D11_AC155341 | |
| BrH80C09_MAF | 5′-cagtgaagttcaaccgcagta-3′ 5′-catgagtgaacataaaacagtgaaa-3′ | 1 | 1 | Primer | KBrH80C09_A166741 | |
| Random primer | Operon primer S14 | 1 | 1 | Primer | BM commercial product | |
| Total | 421 | 545 |
PCR-based genetic markers:
Twenty-one previously developed SSR marker assays were selected on the basis of their ability to identify known A genome linkage groups in B. napus and B. rapa (Suwabe et al. 2002; Lowe et al. 2004). Two SSR markers were developed from BACs containing BrFLC sequences and one SSR marker was derived from BACs containing the BrMAF gene. One RAPD marker (operon primer S14) was included since it was codominant in this population.
Linkage analysis:
Linkage analysis and map construction were carried out using JoinMap 3.0 (van Ooijen and Voorrips 2001). Segregating data were sorted according to locus order for each linkage group using MSExcel. This facilitated detection of errors associated with putative “double-recombinant” events and guided visual checking of original autoradiographs and revision of data points where these had been misscored or typed. All editing operations were recorded and are traceable. Linked loci were grouped on the basis of pairwise LOD values between 5 and 8, and centimorgan distances were estimated with the Kosambi mapping function (Kosambi 1944). Locus order within the LOD grouping was decided through an optimized algorithm using three rounds of linked markers. Multiple segregating loci detected by a probe were indicated by the addition of a suffix (-a, -b, -c, -d) to the locus names. Linkage maps were visualized using MapChart (Voorrips 2002) and PowerPoint.
RESULTS
EST marker characteristics:
A total of 551 cDNA clones from four B. rapa tissue-specific EST libraries were screened to obtain informative RFLP markers. Of these 440 were polymorphic between the parents. A high degree of polymorphism with such markers has previously been reported within Brassica subspecies (Song et al. 1988). To obtain segregating genotypes among the progeny, four restriction enzymes were used, with EcoRV found to detect the highest level of polymorphism (Table 2).
TABLE 2.
Summary of the polymorphism detected by EST clones
| Source of restriction endonuclease | BIF | BAN | BDS | BR | Total (%) |
|---|---|---|---|---|---|
| EcoRI | 32 | 45 | 9 | 6 | 92 (25.6) |
| EcoRV | 37 | 65 | 14 | 8 | 124 (34.6) |
| BamHI | 21 | 29 | 10 | 5 | 65 (18.1) |
| ScaI | 24 | 37 | 12 | 5 | 78 (21.7) |
| Total | 114 | 176 | 45 | 24 | 359 (100) |
The JWF3 B.rapa linkage map:
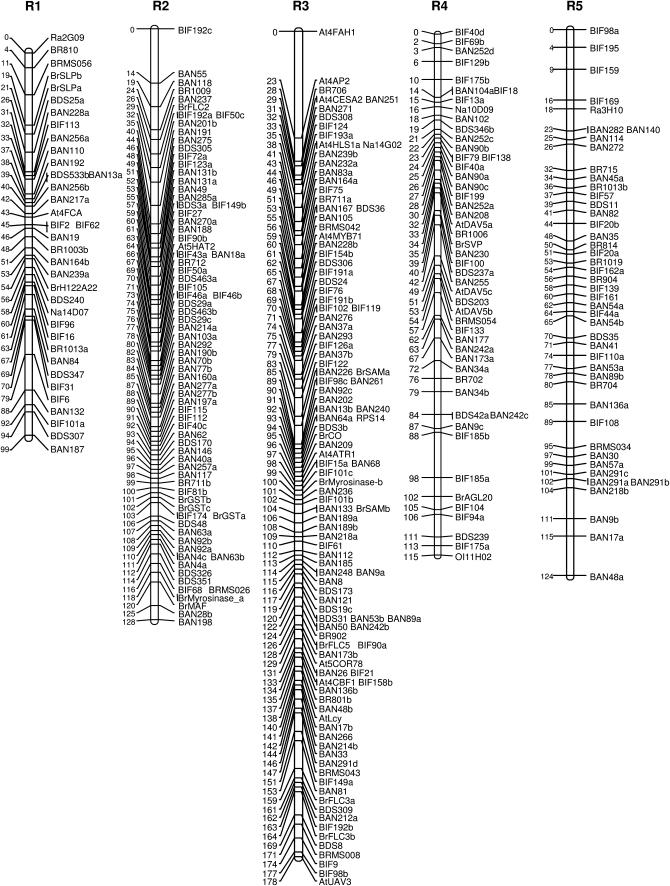
This genetic map of B. rapa was generated on the basis of 545 markers, 520 RFLPs and 25 PCR-based markers assigned to 10 linkage groups covering a total map length of 1287 cM with an average 2.4-cM interval. The 10 linkage groups are most likely to correspond to the 10 chromosomes of B. rapa (Figure 1). It was possible to assign each linkage group to a previously determined classification (R1–R10) on the basis of evidence from the location of previously published SSR markers designated to A genome linkage groups within the context of the B. napus (Parkin et al. 1995; Lowe et al. 2004) and B. rapa (Suwabe et al. 2002) genetic maps. From a total of 75 mapped A genome SSR markers screened against the parents, 21 displayed polymorphism. Each of the 10 linkage groups had at least one SSR marker that provides an anchor to existing published maps.
Figure 1.—
Composite detailed genetic linkage map of Brassica rapa.
The two longest linkage groups R3 (178 cM) and R9 (193 cM) correspond to the two longest groups N3 and N9 of B. napus (Lowe et al. 2004; Udall et al. 2005). Within the Korean Brassica Genome Project (KBGP, http://www.brassica.rapa.org) it had previously been reported that R9 corresponds to cytogenetic chromosome 1 and that R3 corresponds to cytogenetic chromosome 2 (Lim et al. 2005). We compared the relative lengths of each linkage group with the length of the corresponding cytogenetic chromosome identified by Lim et al. There was good agreement, with a calculated correlation coefficient of 0.87. When the same cytogenetic chromosome lengths were compared with the length of corresponding A genome linkage groups reported for recent genetic maps of B. napus, correlation coefficients of 0.66 (Parkin et al. 2005) and 0.78 (Udall et al. 2005) were obtained.
Sequence homology:
B. rapa probe sequences used to establish marker loci were compared against all sequences in GenBank release using BLASTn. The supplemental table (http://www.genetics.org/supplemental/) lists the B. rapa loci for which nucleotide sequence homology was determined with a cutoff of 1E-12, together with the matching GenBank database accession. The similarity data indicated that 422 (77%) of all loci corresponded to genes of known sequence, of which 89 aligned with Arabidopsis expressed, putative, or hypothetical protein-coding sequences. A study of the hit sequence based on their organisms showed that 317 of 422 (75%) had highest sequence homology to the Arabidopsis genome and 97 of 422 (23%) were matched on four species of Brassica, which were B. rapa (39/97), B. napus (32/97), B. oleracea (19/97), and B. juncea (7/97). Only 2% (8 of 422) showed the sequence homology on other organisms, including on rice. The probe sequences used to generate the marker loci appeared to represent a wide range of gene classes, including regulatory factors and structural genes involved in membrane transport, signal transduction, cell cycle regulation, carbon metabolism, stress response, DNA synthesis, and fatty acid metabolism.
Single-locus genes:
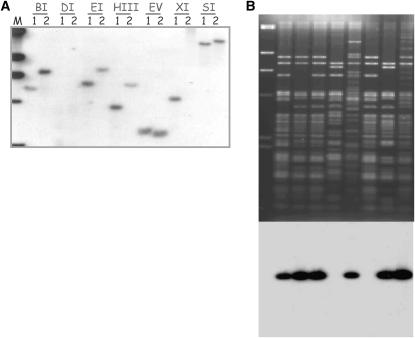
From the screening of RFLP probes against parental lines using seven restriction enzymes, 12% of clones gave single hybridization fragments, and 62 of these were incorporated into the linkage map. Probe BAN235 (Figure 2A) mapped to a single locus on R9, and this was confirmed by screening the probe to an 11× genome coverage HindIII BAC library (Park et al. 2005). Positive hybridization signals were detected for 14 BAC clones, and the isolated DNA was digested with HindIII enzyme. The resultant fingerprints were consistent with the BACs forming a single contig by Southern analysis using the same probe, BAN235 (Figure 2B).
Figure 2.—
Examples of single-locus genes of a parent survey, colony hybridization, and confirmation by Southern blotting to positive BAC fingerprints using BAN235 that it is located on R9 (61 cM). (A) RFLP autoradiography of parent genomic DNA digested with seven restriction enzymes using a BAN 235 single-locus probe. (B) Autoradiography of HindIII fingerprint of positive BAC clones using a BAN235 probe.
The genetic map location of the 62 single-locus markers and their corresponding BLAST comparison data are shown with an asterisk (*) next to their locus name in the supplemental table (http://www.genetics.org/supplemental/) . Of these, 26% (17/62) had no sequence similarity to any sequence in GenBank, indicating that these may be Brassica unique genes, and 25% (16/62) displayed highest similarity to sequences corresponding to uncharacterized gene models within the Arabidopsis genome (expressed protein, unknown protein, or full-length cDNA). Single-locus-specific probes are distributed across all B. rapa chromosomes with no apparent clustering.
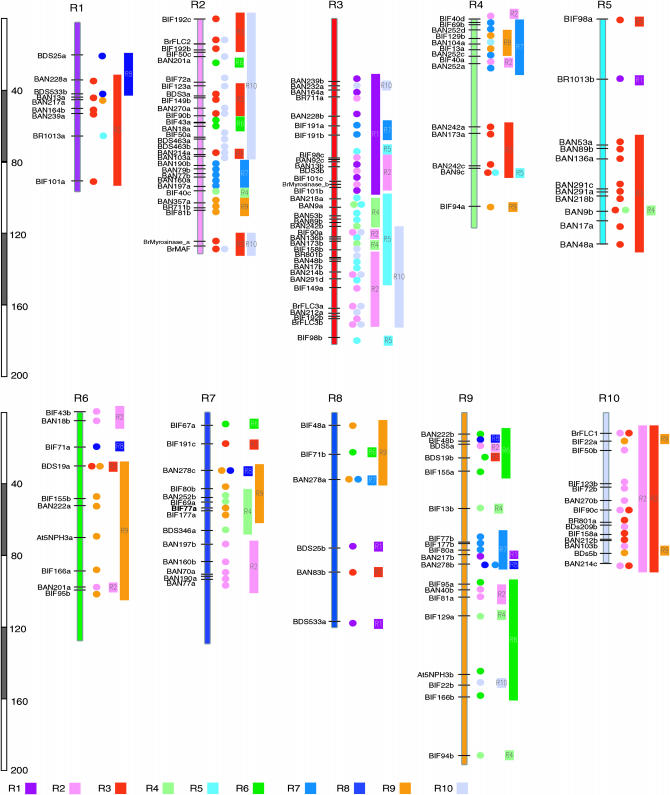
Duplicated marker and homologous linkage groups:
The remainder of the RFLP probes detected more than one segregating locus, with an average of 1.31 loci per probe (520/396). A total of 102 of the 396 mapped probes gave rise to multiple loci (229), with an average of 2.25 loci per probe. Eighty-one detected 2 loci, 18 detected 3, 2 detected 4, and a single probe detected 5 loci. Of these, 72 probes revealed locus duplication (164 loci) of two or three copies on different linkage groups. The pattern of duplications within the B. rapa genome was revealed by comparing ordered clusters of loci derived from common gene probes. At the top of the largest linkage group R3 with five loci duplicated R1 and the middle region of R3 corresponds to sections of R4 and R5, whereas the lower region corresponds to sections represented by 10 loci on R2 and 7 loci on R10. Four marker loci, spanning 19 cM at the top of R4, are also duplicated within R7 and R9 where the four duplicated loci span 17 and 11 cM, respectively. Most chromosomal parts show two or three duplication blocks (Figure 3). These relationships between homeologous chromosome segments provide good evidence for a series of historical segmental duplication events in this genome. However, since all genetic mapping experiments are based on polymorphism of genetic markers, the fine detailed pattern of duplications or triplication is incomplete due to the presence of monomorphic or dimorphic markers.
Figure 3.—
Interchromosomal duplication of Brassica rapa. Circles show duplicated loci of the same markers. Each linkage group is assigned 10 different colors.
Comparison of flowering-time-related genes:
Six genes involved in regulating flowering time isolated from B. rapa (FLC, GI, CO, SVP, AGL20, and VRN1) and two from Arabidopsis (LFY and FCA) were used as RFLP probes. Most probes hybridized to two or three major bands, and some of them were polymorphic between the parents. It was not possible to map any paralogous loci of VRN1 due to the presence of three major monomorphic bands when tested with seven restriction enzymes. The BrFLC (AY273164) isolated from B. rapa cv. Maeryuk (F1) detected two polymorphic segregating bands and an additional three monomorphic bands between the two parents. Two distinct polymorphic loci BrFLC-a and BrFLC-b were mapped 5 cM apart on the long-arm telomere region of R3. BrFLC-a was found to correspond to BrFLC3 sequences within the BAC clone KBrH117M18 (AC146875) and BrFLC-b to KBrH52O08 (AC155342). This was determined by Southern hybridization to BAC HindIII fingerprints. Two SSR markers, BrH80A08_FLC1 (KBrH080A08, AC155344) and BrH04D11_FLC2 (KBrH004D11, AC155341) were derived from two individual BAC sequences. They were classified on the basis of results of colony hybridization, HindIII fingerprints, and hybridization pattern using the BrFLC gene as probe. BrH80A08_FLC1 was located on the short arm of R10 and BrH04D11 was assigned to the short arm of R2, substantiating the positions determined by Schranz et al. (2002). A synthetic GST probe, designated from the second exon to the fifth exon of BrFLC5, showed a single polymorphic band and was mapped to a position 33 cM away from BrFLC3a. All of the duplicated BrFLC genes are located near telomeres (of R2, R3, and R10), with linked markers in the distal regions usually showing skewed segregation, mostly toward the maternal genotype. Data are consistent with the map location reported in a previous study (Schranz et al. 2002), although we report here one more BrFLC3 paralogue derived from recent segmental duplication and the reverse map orientation. An SSR marker derived from BrH80C09_MAF (KBrH80C09, AC166741) was mapped to the long arm of R2 and appears to correspond to the VFR1 locus on R2 (Schranz et al. 2002).
DISCUSSION
Genetic linkage map:
A detailed genetic linkage map of the Brassica A genome has been constructed, on the basis of 134 B. rapa F2:3 families. It contains 545 loci and most of them were detected by RFLP analysis using sequenced EST probes from four different tissue-specific libraries (474 loci/359 probes). Additional gene-specific probes were derived from Arabidopsis chromosome 4 (10 loci/10 probes) and chromosome 5 (17 loci/14 probes). Several flowering-time genes from Arabidopsis (2 loci/2 probes), B. rapa (7 loci/6 probes), and BrMyrosinase, BrDFRI, BrGST, BrSAM, and BrSLP (10 loci/5 probes) functional genes were mapped on this linkage map. The detection of 1.31 loci per RFLP probe (520/396) closely matches that reported (1.27 and 1.34 from 220 and 269 probes, respectively) for previous B. rapa maps (Song et al. 1991; Chyi et al. 1992). In contrast, Udall et al. (2005) observed a higher level of polymorphism with codominant and dominant segregation in the amphidiploid B. napus, where there are twice as many potential loci.
The accumulated set of sequence-tagged genetic markers provides a valuable source of information for study and navigation of the Brassica A genome, not only in B. rapa but also in the context of B. napus and B. juncea. Since the model dicotyledonous plant, A. thaliana, is closely related to Brassica the genus and share, on average, 87% sequence identity (Cavell et al. 1998), there is an expectation that understanding the genetic control of basic biological processes in Arabidopsis can be transferable to other species (Lagercrantz 1998). However, Brassica EST markers that do not correspond to genes derived from other species are of additional value, as they provide insight into the identity and location of novel gene functions, which may be related to the well-characterized adaptability and plasticity of this crop genus.
We have obtained an average marker density of 2.4 cM. B. rapa has the smallest diploid Brassica genome, estimated at 529 Mb (Johnston et al. 2005). Thus we calculate that the current map provides a genetic marker on average at least every 1 Mb. This information may be exploited in at least two ways. First, within the ongoing B. rapa genome-sequencing project (Yang et al. 2005) 62 single-locus gene markers are now available that will assist in the isolation and confirmation of “seed” BACs, as well as provide anchored markers to span between adjacent BAC contigs to integrate the physical map. Second, there is the prospect of benefiting from the rich source of biological information and genetic resources from Arabidopsis functional genomics research to benefit Brassica crop plants.
Multiple duplicated FLC genes of B.rapa:
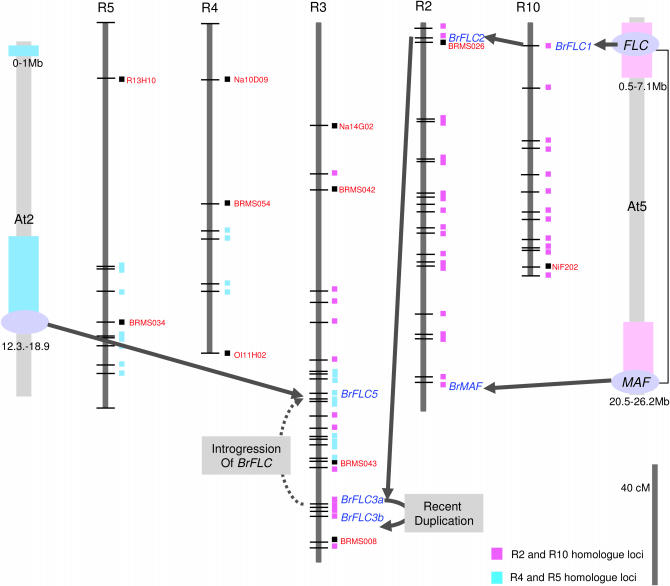
The MADS-box flowering-time regulator FLC, located at the top of chromosome 5 of Arabidopsis, has a repressive function role on flowering time (Michaels and Amasino 1999). There are some differences on the number of orthologous or paralogous BrFLC loci with that of Schranz et al. (2002). Using the BrFLC gene as an RFLP probe, we obtained two polymorphic and three monomorphic bands. These two loci were located at the telomere of the long arm of R3, whereas Schranz et al. were able to assign only one locus to R3. This difference appears to result from the use of a backcross population by Schranz et al., where heterozygote genotypes are not detected. In contrast, the JWF3 is an F3 pooled population (40 seedlings per line) that is able to represent F2 segregation. We were able to detect eight recombinant genotypes in the population between two alleles of BrFLC3-a and -b, where one was homozygous for one parent line (genotype A), and the other represented the heterozygote (H), resulting in a map interval of 5 cM on R3. Other monomorphic BrFLC fragments were detected using SSR markers derived from B. rapa BACs that contain BrFLC genes. Sequences of four BACs, KBrH080A08, KBrH004D11, KBrH117M18, and KBrH52O08 containing BrFLC1, -2, -3a, and -3b, respectively, are collinear to the FLC region at 3.0–3.35 Mb of Arabidopsis chromosome 5 with indels (Yang et al. 2006). This genetic map surrounding BrFLC1, -2, -3a, and -3b shows synteny between each linkage group and with the 3-Mb region of Arabidopsis chromosome 5 (Figure 4). Meanwhile BrFLC5 is identified in a BAC clone KBrH038M21 (not submitted yet) that is collinear with 12.7–12.91 Mb of Arabidopsis chromosome 2 and the genetic position was determined 33 cM away from BrFLC3a on the long arm of R3. From this we infer that this genomic segment was replicated by an insertion within the homeologous region of R3 and this region and mostly shows the homeologous blocks to R4 and R5. Another BAC clone KrH80C09 corresponds to the MAF gene locus (At565050) within the 25.8- to 26.2-Mb region of Arabidopsis chromosome 5 (Yang et al. 2006).
Figure 4.—
Comparative mapping of B. rapa linkage groups containing replicated FLC genes to the Arabidopsis genome. Vertical lines represent homeologous linkage groups in B. rapa, two loci of BrFLC3a and FLC3b are the polymorphic segregating RFLP genotype data using the BrFLC gene (AY273164), and the other three, BrFLC1, BrFLC2, and BrMAF, are the segregated genotype data using developed SSR markers from BACs containing FLC and MAF genes. R2, R3, and R10 show the sequenced homology to Arabidopsis chr5 (0.5–7 Mb and 20.5–26.2 Mb, respectively). Originated FLC of Arabidopsis is triplicated at the top of R10 and R2 and at the bottom of R3, and a local duplication event has happened at the bottom of R3. The BrFLC5 gene mapped region shows R4 and -5 homeologues and corresponds to Arabidopsis chr3 (0–1 Mb and 12.3–18.9 Mb, respectively).
Comparison of flowering-time genes of B. rapa, B. napus, and A. thaliana and QTL of B. rapa have been reported (Osborn et al. 1997; Kole et al. 2001). A major QTL, “VFR2” has been shown to correspond to BrFLC1, while the QTL “FR1” corresponded to BrFLC2 (Kole et al. 2001). An additional QTL “FR2” corresponds to BrFLC5, although another vernalization response QTL1 “VFR1” was not accounted for by a corresponding flowering-time-related gene (Schranz et al. 2002). A BAC clone KBrH80C09, containing a tandem array of three MAF genes (Yang et al. 2006) and mapped on the long arm of R2, suggests that this gene may play a role for QTL of VFR1.
The genomes of Brassica species have triplicated counterparts to corresponding homeologous segments of Arabidopsis (O'neill and Bancroft 2000; Rana et al. 2004; Lysak et al. 2005). Almost 88% of triplicated genes near the FLC regions returned to a single-copy or a two-copy state by deletion (Yang et al. 2006). Because of this reason, hybridization data using a single EST probe might have limitations for inferring genome duplication. But, overall distributions of duplicate or triplicate regions are detected from the hybridization data of multiloci EST markers (Figure 3), suggesting that the genome-level triplication might have happened in the ancestor of Brassica.
The KBGP is currently underway and is aiming to generate the first complete Brassica chromosome sequence of R9 (cytogenetic chromosome 1) (www.brassic.rapa.org). We have selected nine seed BACs through BAC library screening using single-locus EST markers. FISH and sequence information generally coincided with our expectations. The complete set of 62 locus-specific single-copy EST markers will be valuable markers for the primary anchoring of “seed” BACs for each linkage group.
Acknowledgments
This work was supported by a grant from the BioGreen 21 Program and by the National Institute of Agricultural Biotechnology, Rural Development Administration. G.J.K. is supported by the United Kingdom Biotechnology & Biological Sciences Research Council.
References
- Bohuon, E. J., D. J. Keith, I. A. P. Parkin, A. G. Sharpe and D. J. Lydiate, 1996. Alignment of the conserved C genomes of Brassica oleracea and Brassica napus. Theor. Appl. Genet. 93: 833–839. [DOI] [PubMed] [Google Scholar]
- Butruille, D. V., R. P. Guries and T. C. Osborn, 1999. Linkage analysis of molecular markers and quantitative trait loci in populations of inbred backcross lines of Brassica napus L. Genetics 153: 949–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavell, A. C., D. Lydiate, I. A. P. Parkin, C. Dean and M. Trick, 1998. Collinearity between a 30-centimorgan segment in Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome 41: 62–69. [PubMed] [Google Scholar]
- Cheung, W. Y., G. Champange, N. Hubert and B. S. Landry, 1997. Comparison of the genetic map of Brassica napus and Brassica oleracea. Theor. Appl. Genet. 94: 569–582. [Google Scholar]
- Cho, Y. G., M. Y. Eun, S. R. Mccouch and Y. A. Chae, 1994. The semidwarf gene, sd-1, of rice (Oryza sativa L.). II. Molecular mapping and marker-assisted selection. Theor. Appl. Genet. 89: 54–59. [DOI] [PubMed] [Google Scholar]
- Chyi, Y. S., M. E. Honeck and J. L. Sernyk, 1992. A genetic linkage map of restriction fragment length polymorphism loci for Brassica rapa (syn. campestris). Genome 35: 746–757. [Google Scholar]
- Fahey, J. W., and P. Talalay, 1995. The role of Crucifers in cancer chemoprotection, pp. 87–93 in Phytochemicals and Health, edited by D. L. Gustine and H. E. Flores. American Society of Plant Physiologists, Rockville, MD.
- Feinberg, A. P., and B. Vogelstein, 1983. A technique for radiolabeling DNA restriction fragments to a high specific activity. Ann. Biochem. 132: 6–13. [DOI] [PubMed] [Google Scholar]
- Figdore, S. S., W. C. Kenard, K. M. Song, M. K. Slocum and T. C. Osborn, 1988. Assessment of the degree of restriction fragment length polymorphism in Brassica. Theor. Appl. Genet. 75: 833–840. [Google Scholar]
- Gomez-Campo, C., 1999. Biology of Brassica Coenospecies. Elsevier, Amsterdam/New York.
- Jackson, S. A., Z. Cheng, M. L. Wang, H. M. Goodman and J. Jiang, 2000. Comparative fluorescence in situ hybridization mapping of a 431-kb Arabidopsis thaliana bacterial artificial chromosome contig reveals the role of chromosomal duplications in the expansion of the Brassica rapa genome. Genetics 156: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, J. S., A. E. Pepper, A. E. Hall, Z. J. Chen, G. Hodnett et al., 2005. Evolution of genome size in Brassicaceae. Ann. Bot. 95: 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kianian, S. F., and C. F. Quiros, 1992. Generation of a Brassica oleracea composite RFLP map: linkage arrangements among various populations and evolutionary implications. Theor. Appl. Genet. 84: 544–554. [DOI] [PubMed] [Google Scholar]
- Kim, H. U., B. S. Park, T. Y. Chung and S. K. Kang, 1996. Isolation of anther-preferentially expressed genes from Chinese cabbage. Mol. Cells 6: 666–672. [Google Scholar]
- Koch, M., B. Haubold and T Mitchell-Olds, 2001. Molecular systematics of the Brassicaceae: evidence from coding plastidic MATK and nuclear CHS sequences. Am. J. Bot. 88: 534–544. [PubMed] [Google Scholar]
- Kole, C., P. Quijada, S. D. Michaels, R. M. Amasino and T. C. Osborn, 2001. Evidence for homology of flowering-time genes VFR2 from Brassica rapa and FLC from Arabidopsis thaliana. Theor. Appl. Genet. 102: 425–430. [Google Scholar]
- Kosambi, D. D., 1944. The estimation of map distance from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Labana, K. S., and M. L. Gupta, 1993. Importance and origin, pp. 1–20 in Breeding Oilseed Brassica, edited by K. S. Labana, S. S. Banga and S. K. Banga. Springer-Verlag, Berlin.
- Lagercrantz, U., 1998. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150: 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz, U, and D. Lydiate, 1996. Comparative genome mapping in Brassica. Genetics 144: 1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, T. H., T. A. Delmonte, K. P. Reischamann, J. Hyman, S. Kowalski et al., 2000. EST-enriched comparative map of Brassica oleracea and Arabidopsis thaliana. Genome Res. 10: 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, B. S., N. Hubert, T. Etoh, J. J. Harada and S. E. Lincoln, 1991. A genetic map for Brassica napus based on restriction fragment length polymorphisms detected with expressed DNA sequences. Genome 34: 543–552. [Google Scholar]
- Lim, J. Y., C. S. Shin, E. Chung, J. S. Kim, H. U. Kim et al., 2000. Analysis of expressed sequence tags form Brassica rapa L. ssp. pekinensis. Mol. Cells 10: 399–404. [PubMed] [Google Scholar]
- Lim, K. B., H. De Jong, T. J. Yang, J. Y. Park, S. J. Kwon et al., 2005. Characterization of rDNAs and tandem repeats in heterochromatin of Brassica rapa. Mol. Cells 19: 436–444. [PubMed] [Google Scholar]
- Lowe, A. J., C. Moule, M. Trick and K. J. Edwards, 2004. Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theor. Appl. Genet. 108: 1103–1112. [DOI] [PubMed] [Google Scholar]
- Lukens, L., F. Zou, D. Lydiate, I. A. Parkin and T. Osborn, 2003. Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics 164: 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak, M. A., M. A. Koch, A. Pecinka and I. Schubert, 2005. Chromosome triplication found across the tribe Brassiceae. Genome Res. 15: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S. D., and R. M. Amasino, 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, C. M., and I. Bancroft, 2000. Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant J. 23: 233–243. [DOI] [PubMed] [Google Scholar]
- Park, J. Y., D. H. Koo, C. P. Hong, S. J. Lee, J. W. Jeon et al., 2005. Physical mapping and microsynteny of Brassica rapa ssp. pekinensis genome corresponding to a 222 kb gene-rich region of Arabidopsis chromosome 4 and partially duplicated on chromosome 5. Mol. Gen. Genet. 274: 579–588. [DOI] [PubMed] [Google Scholar]
- Parkin, I. A. P., A. G. Sharpre, D. J. Keith and D. J. Lydiate, 1995. Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38: 1122–1131. [DOI] [PubMed] [Google Scholar]
- Parkin, I. A. P., J. M. Gulden, A. G. Sharpe, L. Lukens, M. Trick et al., 2005. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan, A. K., V. Gupta, A. Mukhopadhyay, N. Arumugam, Y. S. Sodhi et al., 2003. A high-density linkage map in Brassica juncea (Indian mustard) using AFLP and RFLP markers. Theor. Appl. Genet. 106: 607–614. [DOI] [PubMed] [Google Scholar]
- Rana, D., T. van den Boogaart, C. M. O'Neill, L. Hynes, E. Bent et al., 2004. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 40: 725–733. [DOI] [PubMed] [Google Scholar]
- Sebastian, R. L., E. C. Howell, G. J. King, D. F. Marshall and M. J. Kearsey, 2000. An integrated AFLP and RFLP Brassica oleracea linkage map from two morphologically distinct doubled-haploid mapping populations. Theor. Appl. Genet. 100: 75–81. [Google Scholar]
- Schranz, M. E., P. Quijada, S. B. Sung, L. Lukens, R. Amasino et al., 2002. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocum, M. K., S. S. Figdore, W. C. Kennard, J. Y. Suzuri and T. C. Osborn, 1990. Linkage arrangements of restriction fragment length polymorphism loci in Brassica oleracea. Theor. Appl. Genet. 80: 57–64. [DOI] [PubMed] [Google Scholar]
- Song, K. M., T. C. Osborn and P. H. Williams, 1988. Brassica taxonomy based on nuclear restriction fragment length polymorphism (RFLPs). A genome evolution of diploid and amphidiploid species. Theor. Appl. Genet. 75: 784–794. [Google Scholar]
- Song, K. M., J. Y. Susuki and M. K. Slocum, 1991. A linkage map of Brassica rapa (syn. B. campestris) based on restriction fragment length polymorphism loci. Theor. Appl. Genet. 82: 296–304. [DOI] [PubMed] [Google Scholar]
- Suwabe, K., H. Iketani, T. Nunome, T. Kage and M. Hirai, 2002. Isolation and characterization of microsatellites in Brassica rapa L. Theor. Appl. Genet. 104: 1092–1098. [DOI] [PubMed] [Google Scholar]
- Teutenico, R. A., and T. C. Osborn, 1994. Mapping of RFLP and qualitative trait loci in Brassica rapa, and comparison to linkage maps of B. napus, B. oleracea, and Arabidopsis thaliana. Theor. Appl. Genet. 89: 885–894. [DOI] [PubMed] [Google Scholar]
- Truco, M. J., J. Hu, J. Sadowski and C. F. Quiros, 1996. Inter- and intra-genomic homology of the Brassica genomes: implications for their origin and evolution. Theor. Appl. Genet. 93: 1225–1233. [DOI] [PubMed] [Google Scholar]
- Udall, J. A., P. A. Quijada and T. C. Osborn, 2005. Detection of chromosomal rearrangements derived from homeologous recombination in four mapping populations of Brassica napus L. Genetics 169: 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzunova, M., W. Ecke, K. Weissleder and G. Robbelen, 1995. Mapping the genome of rapeseed (Brassica napus L.) I. Construction of an RFLP linkage map and localization of QTLs for seed glucosinate content. Theor. Appl. Genet. 90: 194–204. [DOI] [PubMed] [Google Scholar]
- Van Ooijen, J. W., and R. E. Voorrips, 2001. JoinMap Version 3.0: Software for the Calculation of Genetic Linkage Maps. Plant Research International, Wageningen, The Netherlands.
- Voorrips, R. E., 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Yang, T. J., J. S. Kim, K. B. Lim, S. J. Kwon, J. A. Kim et al., 2005. The Korea Brassica Genome Project: a glimpse of the Brassica genome based on comparative genome analysis with Arabidopsis. Comp. Funct. Genomics 6: 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, T. J., J. S. Kim, S. J. Kwon, K. B. Lim, B. S. Choi et al., 2006. Sequence-level analysis of the diploidization process in the triplicated FLC region of Brassica rapa. Plant Cell 18: 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]