Abstract
Chromosome architecture undergoes extensive, programmed changes as cells enter meiosis. A highly conserved change is the clustering of telomeres at the nuclear periphery to form the “bouquet” configuration. In the fission yeast Schizosaccharomyces pombe the bouquet and associated nuclear movement facilitate initial interactions between homologs. We show that Bqt2, a meiosis-specific protein required for bouquet formation, is required for wild-type levels of homolog pairing and meiotic allelic recombination. Both gene conversion and crossing over are reduced and exhibit negative interference in bqt2Δ mutants, reflecting reduced homolog pairing. While both the bouquet and nuclear movement promote pairing, only the bouquet restricts ectopic recombination (that between dispersed repetitive DNA). We discuss mechanisms by which the bouquet may prevent deleterious translocations by restricting ectopic recombination.
MEIOSIS, the specialized form of nuclear division that reduces the diploid number of chromosomes by half, consists of one round of DNA replication followed by two successive nuclear divisions. At the first meiotic division (MI) homologous chromosomes (homologs), each consisting of two sister chromatids, are segregated to opposite poles, reducing the chromosome number by half. The second meiotic division (MII), like mitosis, segregates sister chromosomes to opposite poles, producing four haploid nuclei.
The elevated rate of recombination characteristic of meiosis is due to a programmed set of meiosis-specific events that includes the formation of DNA double-strand breaks (DSBs) by Spo11, called Rec12 in Schizosaccharomyces pombe (Keeney et al. 1997; Cervantes et al. 2000). The DSBs are then repaired via interaction with an intact duplex, giving rise to gene conversions and crossovers (reviewed in Roeder 1997; Keeney 2001). At least one crossover per homolog pair is required to promote the proper attachment of each homolog in a pair to spindle microtubules from opposite poles, ensuring segregation of homologs at MI (reviewed in Page and Hawley 2003; Petronczki et al. 2003). In addition to its critical role in segregation, recombination generates diversity in subsequent generations by creating new combinations of alleles.
An elevated frequency of recombination is not sufficient to promote proper meiotic chromosome segregation; recombination must involve homologs rather than sister chromatids and must frequently produce a crossover. In meiosis, unlike mitosis, gene conversion is frequently associated with crossing over (Grimm et al. 1994; Virgin et al. 2001; Cromie et al. 2005 and references therein). Consequently, the recombination events that occasionally occur between dispersed repetitive DNA, such as transposons, genes for tRNAs, subtelomeric sequences, and multigene families, may frequently involve crossovers. Such crossovers can produce deleterious chromosomal rearrangements (reviewed in Shaffer and Lupski 2000). However, in both S. pombe and the budding yeast Saccharomyces cerevisiae, recombination between dispersed repetitive DNA is significantly lower than allelic recombination (Munz et al. 1982; Kohli et al. 1984; Kupiec and Petes 1988a,b; Goldman and Lichten 1996, 2000; Virgin and Bailey 1998; Schlecht et al. 2004). The restriction of ectopic recombination may, in some cases, result from an insufficient length of sequence identity. The restriction of ectopic recombination may also reflect the recombination-independent propensity of chromosomes to align, in register, along their entire length. In the latter case, if homologs were unable to align, ectopic recombination would not be restricted and dispersed repetitive elements might recombine as efficiently as allelic sequences.
Despite the elevated frequency of recombination in meiosis, in many organisms the number of crossovers per genome is not much larger than the number of chromosomes (reviewed in Hillers 2004). If these crossovers were placed randomly throughout the genome, a large fraction of meioses would contain at least one homolog pair with no crossovers and therefore be prone to missegregation at MI. However, in many organisms the placement of crossovers is regulated; for instance, the presence of one crossover reduces the likelihood of a second nearby crossover (reviewed in van Veen and Hawley 2003; Hillers 2004). This phenomenon, crossover interference, is manifest as a frequency of double crossovers lower than that expected for two independent events. In S. pombe, which does not have crossover interference, each of its three chromosomes receives 10–20 randomly placed crossovers, resulting in a very low frequency of chromosomes without a crossover (Munz 1994).
Before recombination can take place, two DNA molecules must first be in close proximity. Thus, a critical step in meiotic recombination is the juxtaposition of homologous chromosomes. Homologs are brought together in a stepwise process (reviewed in Gerton and Hawley 2005). In this process we define homologs as being “aligned” when they are in register along their entire length. Homologs are subsequently defined as being “paired” when they are intimately associated along their entire length. Clustering of telomeres at the nuclear periphery, the bouquet configuration, is a conserved feature of meiosis that is thought to facilitate the alignment of homologs (reviewed in Scherthan 2001; Yamamoto and Hiraoka 2001). In S. pombe the bouquet consists of a tight cluster of all telomeres at the spindle-pole body (SPB). This clustering requires the telomere-binding protein Taz1, the Taz1-binding protein Rap1, and the heterochromatin protein Rik1 (Cooper et al. 1997, 1998; Nimmo et al. 1998; Chikashige and Hiraoka 2001; Kanoh and Ishikawa 2001; Tuzon et al. 2004). Telomere- and SPB-led oscillatory nuclear movement (“horsetail” movement) occurs throughout meiotic prophase (Chikashige et al. 1994) and depends on Dhc1, the microtubule motor protein dynein (Ding et al. 1998; Yamamoto et al. 1999). Perturbing either the bouquet or horsetail movement reduces pairing and meiotic recombination (Shimanuki et al. 1997; Cooper et al. 1998; Nimmo et al. 1998; Yamamoto et al. 1999; Niwa et al. 2000; Miki et al. 2002; Ding et al. 2004; Saito et al. 2005). By providing a physical linkage between chromosomes, recombination stabilizes their initial alignment and pairing promoted by the bouquet and horsetail movement (Nabeshima et al. 2001; Ding et al. 2004). Thus, the initial alignment and pairing of homologs are required for wild-type levels of meiotic recombination, which in turn is required for stable homolog pairing.
The Bqt2 protein of S. pombe is a meiosis-specific SPB component and is required for telomere clustering (Martin-Castellanos et al. 2005; Chikashige et al. 2006). In bqt2Δ mutant meioses SPB movement still occurs but telomeres are dispersed throughout the nucleus and chromosome movement is diminished. Here we show that, as expected, pairing of homologs in meiotic prophase and allelic recombination are reduced in bqt2Δ mutants. Furthermore, recombination events in both bqt2Δ and dhc1Δ mutants display negative interference: the presence of one recombination event increases the likelihood of a second nearby event. Bqt2, but not Dhc1, restricts ectopic recombination. We suggest a model in which ectopic recombination is restricted by the position of the two repetitive DNA elements relative to their nearest telomere, rather than pairing per se.
MATERIALS AND METHODS
Yeast strains, media, and culture conditions:
Solid media were YEA + 4S, YEA + 5S, YEAG, or supplemented EMM2 used at 32° as described previously (Davis and Smith 2003). Liquid cultures were grown at 30° in YEL + 5S. Sporulation was at 25° on supplemented SPA (Gutz et al. 1974) for 2–4 days. The yeast strains and mutant alleles used are described below or in references in supplemental Table S1 at http://www.genetics.org/supplemental/.
Genetic screen for meiotic segregation mutants:
In the absence of recombination (e.g., in rec12Δ mutants), S. pombe possesses a residual ability to segregate homologs at MI (Davis and Smith 2003 and references therein). Missegregation of homologs at MI results in an elevated frequency of heterozygous diploid spores. Random segregation at MI is expected to produce ∼10-fold more heterozygous diploid spores than that observed in rec12 mutant meioses (Davis and Smith 2003, 2005). Heterozygous diploid spore formation in rec12 mutants is thus expected to be increased by loss of the recombination-independent MI homolog segregation ability. To identify gene products required for this process, we enriched for mutants of strain GP2640 (h90 ade6-52 leu1-32 ura4-294 his3-D1 fus1∷LEU2 rec12-152∷LEU2) as follows. Haploid fus1 mutant cells are unable to mate and therefore cannot sporulate; however, fus1 mutant diploids, if heterozygous at the mating-type locus, are able to properly complete meiosis and sporulation (Petersen et al. 1995). By coupling the ability of fus1Δ to prevent sporulation of haploid cells with the ability to selectively kill nonsporulated cells using glusulase treatment (Ponticelli and Smith 1989), the meiotic progeny of diploid cells can be efficiently selected.
Strain GP2640 carrying plasmid pDW220 (ura4+ fus1+; Petersen et al. 1995) was mutagenized by random integration of linearized plasmid pAF1 (his3+; Ohi et al. 1996) into the genome. Pools of mutagenized cells were sporulated, and the vegetative cells were killed and spores liberated from asci by treatment with glusulase. The spore suspensions were allowed to germinate in EMM2 medium, and cells that lost pDW220 were selected on plates containing 5-fluoroorotic acid. The remaining cells, a mixture of haploids and diploids, were phenotypically Fus1−, and therefore only the diploids could sporulate. Each subsequent round of sporulation of these diploid spores coupled with killing of haploid cells results in a theoretical 10-fold enrichment for mutants with random segregation at MI. Nine pools of His+ transformants (totaling ∼10,000 individual colonies) were carried through two rounds of enrichment. The resulting mutants were screened by testing ∼25 individual colonies from each pool for those that produced an elevated level of diploid spores (i.e., I2-staining spore colonies on EMM2 medium), and two mutants were identified. One mutant contained an insertion within the klp6 (SPBC649.01C) coding sequence. Klp6 is a kinesin-like protein that belongs to the kinesin-8 family of microtubule-destabilizing proteins (Lawrence et al. 2004; Miki et al. 2005). The role of Klp6 in mitosis and meiosis has been described by others (West et al. 2001, 2002; Garcia et al. 2002a,b; Li and Chang 2003; Sanchez-Perez et al. 2005). The other mutant contained an insertion within the coding sequence of SPAC1002.06C. This gene, recently named bqt2, is required for meiotic bouquet formation (Martin-Castellanos et al. 2005; Chikashige et al. 2006) and is the subject of this study.
Microscopy:
To assay pairing, strains were used in which lacO was integrated near the centromere of chromosome I (ChrI) (Nabeshima et al. 1998) and bound by a variant of the green fluorescent protein-LacI-nuclear localization signal fusion (GFP13-LacI12-NLS; Straight et al. 1998), adapted for S. pombe (Davis and Smith 2003). Approximately 107 cells were mated on supplemented SPA and collected after 16–24 hr. Live zygotes, arrested in prophase by the mei4 mutation (Shimoda et al. 1985; Hiraoka et al. 2000), were examined by fluorescence microscopy performed on a Nikon Eclipse 600 microscope using a Nikon 60× 1.40 NA Plan Apo objective (Nikon, Melville, NY). Images were captured using MetaMorph software (Molecular Devices, Sunnyvale, CA) and a Cascade 512B CCD camera (Photometrics, Tucson, AZ). GFP foci were counted in two experiments and statistical significance was calculated for each experiment independently using a χ2-test.
Deletion constructs:
A complete replacement of the bqt2 coding sequence with 3HA-6His-kanMX6 was constructed using the method of Bahler et al. (1998). A PCR was performed using as template plasmid pFA6a-3HA-6His-kanMX6 (Davis and Smith 2003). The forward and reverse primers in this reaction contained nucleotides corresponding to the 5′ and 3′ ends of bqt2+ (nucleotides 10,147–10,226 and 9555–9634, respectively, of cosmid SPAC1002; GenBank accession no. AL136078). The resulting PCR product was used to transform S. pombe strain GP363 (h+ ade6-M26 ura4-294 arg3-124) to G418 resistance, conferred by kanMX. Deletion of bqt2 (bqt2-168∷kanMX) was confirmed by a PCR.
Recombinant frequencies:
Intergenic recombinant frequencies among Ade+ viable spores were determined by plating spore suspensions on YEAG to select Ade+ spores and, after 3–5 days, colonies were toothpicked to grids on YEAG. After growth overnight, the segregants were replicated to the appropriate test media. Otherwise, recombinant frequencies were determined as previously described (Young et al. 2002). Statistical significance of Ade+ recombinant frequencies was calculated using Student's t-test. χ2-tests were used for statistical analysis of genetic interference data. For crossover interference in the ura4-aim–tps16–arg1 intervals we determined whether or not the observed frequency of double crossovers was greater than that expected for two independent events. For interference between a conversion and a crossover we determined whether or not the frequency of crossovers was greater among the Ade+ spores than among total spores.
Unequal sister-chromatid exchange (SCE) frequencies were determined as follows. Appropriately diluted mitotic cultures of the ade6-Dup-containing strain were plated on YEA + 4S to determine the total number of viable cells and on YEAG to determine the frequency of Ade+ recombinants. The ade6-Dup strain and the appropriate ade6-D19 strain were then mated on supplemented SPA. Spores were harvested and spore suspensions were plated on YEA + 4S to determine the total number of viable cells and on YEAG to determine the frequency of Ade+ recombinants. The mitotic frequency, which was typically 5–10% of the meiotic frequency and never >27%, was subtracted from the meiotic frequency to give the final meiotic SCE frequency. Ten crosses were performed for each genotype and the statistical significance was calculated using Student's t-test.
RESULTS
Bqt2 is required for homolog pairing in meiotic prophase:
We isolated a mutation in the S. pombe gene SPAC1002.06c in a screen for mutations that increase MI chromosome missegregation in a rec12 background (see materials and methods for details). SPAC1002.06c was subsequently named bqt2 and reported to encode a meiosis-specific spindle-pole body protein required for telomere clustering and wild-type levels of meiotic recombination (Martin-Castellanos et al. 2005; Chikashige et al. 2006). To determine whether, as expected, the telomere-clustering defect of bqt2Δ mutants resulted in defective homolog pairing, we examined both wild-type and bqt2Δ mutants marked with a tandem array of lacO DNA near the centromere of ChrI (Nabeshima et al. 1998). As a control, we also examined pairing in the absence of Dhc1, the heavy chain of the microtubule motor dynein, which is required for meiotic horsetail movement and efficient homolog pairing (Yamamoto et al. 1999; Ding et al. 2004). Pairing was visualized by fluorescence microscopy of the GFP-LacI-NLS fusion protein, which binds to the lacO array.
Homolog pairing in S. pombe is a dynamic process (Ding et al. 2004). To aid the analysis, we used the mei4 mutation, which arrests cells in meiotic prophase, after horsetail nuclear movement, with paired homologs (Yokobayashi and Watanabe 2005). To the best of our knowledge, the precise position of the mei4 arrest point relative to the dynamics of pairing is unknown. A single-GFP focus indicates pairing of the lacO array, while unpaired arrays generate two GFP foci in a single nucleus. Three or four GFP foci in a single nucleus indicate a defect in sister-chromatid cohesion. Two experiments, each with wild type and mutants on the same SPA plate, were performed, one ∼16 hr and the other ∼24 hr after the cells were mated. A single-GFP focus was found in 66, 39, and 43% of prophase nuclei in wild-type, bqt2Δ, and dhc1Δ cells, respectively, in the first experiment, and 96, 51, and 53% in the second (Table 1). The difference in the absolute level of pairing observed in the two experiments may be due to the degree to which the cells reached the mei4 arrest point in each experiment. The fraction of paired homologs for both bqt2Δ and dhc1Δ was statistically different from that in wild type in each experiment (P < 0.0005). This indicates that pairing of the lacO array was significantly reduced in both bqt2Δ and dhc1Δ mutants. In all of the strains examined, ≤2% prophase nuclei contained three or four GFP foci, indicating that sister-chromatid cohesion is not significantly altered in bqt2Δ and dhc1Δ mutants.
TABLE 1.
Bqt2 is required for pairing of homologous chromosomes in meiotic prophase
| Homologous chromosomes
|
Sister chromatids: | |||||
|---|---|---|---|---|---|---|
| % paired
|
% unpaired
|
% separated
|
||||
| Parental genotypes | Expt. 1 | Expt. 2 | Expt. 1 | Expt. 2 | Expt. 1 | Expt. 2 |
| mei4-B2 (GP4101) | 66 | 96 | 31 | 4 | 2 | 0 |
| bqt2Δ mei4-B2 (GP5764) | 39 | 51 | 60 | 49 | 1 | 0 |
| dhc1Δ mei4-B2 (GP5733) | 43 | 53 | 56 | 45 | 2 | 2 |
Cells were induced to mate and arrested in meiotic prophase. GFP dots, reflecting LacI-GFP bound to the lacO array at lys1, were counted in two experiments. In the first experiment 131, 99, and 129 zygotes were counted in wild-type, bqt2Δ, and dhc1Δ cells, respectively, and in the second 98, 102, and 113 were counted. One and two dots indicate paired and unpaired homologs, respectively. Three or four dots indicate separation of sister chromatids. The numbers are the percentage of zygotes in indicated classes for both experiments. In each experiment the fraction of paired homologs for both bqt2Δ and dhc1Δ was statistically different from that of wild type (P < 0.0005, contingency χ2-tests).
Telomere clustering is required for wild-type levels of homologous intergenic recombination:
In bqt2Δ mutants meiotic DSB formation and repair is nearly wild type but recombination is reduced by factors of ∼3–7 in the three intervals examined (Martin-Castellanos et al. 2005; Chikashige et al. 2006). To better understand this apparent discrepancy between reduced recombination and normal formation and repair of DSBs, which are expected to produce recombinants, we extended the analysis of meiotic recombination. We measured intergenic recombination (crossovers) in four intervals: lys3–met5 on ChrI, pat1–leu1 on ChrII, and ura4-aim–tps16–arg1 on ChrIII (Figure 1A). Recombinant frequency in the bqt2Δ mutants was reduced by a factor of 2–4 in the regions tested, a reduction similar to that seen in the dhc1Δ mutant (Tables 2 and 3). The lys3–met5 interval encompasses a region with DSB frequency and kinetics of repair that are nearly wild type in bqt2Δ mutant meioses (Martin-Castellanos et al. 2005). Significantly, recombination in this interval was reduced in the bqt2Δ mutant by a factor of ∼4 (Table 2). These data suggest that repair of meiotic DSBs in the bqt2Δ mutant, and perhaps all pairing mutants, frequently involves the use of either sister chromatids or homologous nonallelic sequences as a template.
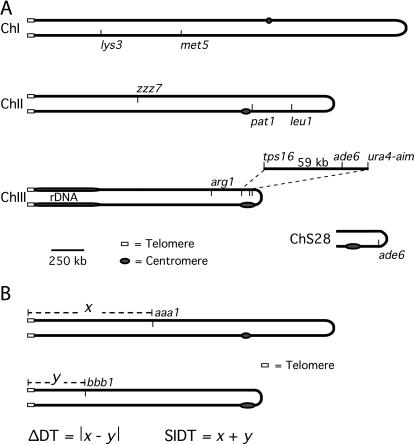
Figure 1.—
S. pombe chromosomes. (A) Centromeres, telomeres, rDNA, and relevant genetic markers are indicated. ChS28 is a deletion derivative of ChrIII (Niwa et al. 1989). Drawn to scale. (B) Graphical representation of the sum of insert distances to their nearest telomere (SIDT) and the difference between the distances of each locus from its nearest telomere (ΔDT) for two loci, aaa1 and bbb1, on heterologous chromosomes.
TABLE 2.
Pairing mutants demonstrate reduced allelic recombination and negative interference between a conversion and a crossover
| Recombinant frequency
|
||||||
|---|---|---|---|---|---|---|
| Wild type
|
bqt2Δ
|
dhc1Δ
|
||||
| Interval (Chr) | Among total | Among Ade+a | Among totalb | Among Ade+a | Among totalb | Among Ade+a |
| lys3–met5 (I) | 0.30 | 0.27 | 0.08** | 0.10 | 0.14** | 0.14 |
| Clys3–met5c | 0.92 | 1.18 | 1.02 | |||
| leu1–pat1 (II) | 0.37 | 0.43 | 0.16** | 0.22* | 0.14** | 0.21* |
| Cleu1–pat1c | 1.15 | 1.31* | 1.46*** | |||
| tps16–arg1 (III) | 0.43 | 0.40 | 0.14** | 0.31** | 0.14** | 0.34** |
| Ctps16–arg1c | 0.93 | 2.29** | 2.49** | |||
Three or more independent crosses were performed for each interval. For the lys3–met5 interval the strains crossed were: wild type, GP13 × GP5570 (four crosses); bqt2Δ, GP5478 × GP5644 (four crosses); dhc1Δ, GP5572 × GP5642 (four crosses). For the leu1–pat1 interval: wild type, GP13 × GP5585 (three crosses); bqt2Δ, GP5478 × GP5584 (three crosses); dhc1Δ, GP5571 × GP5583 (three crosses). For the tps16–arg1 interval: wild type, GP5217 × GP5224 (five crosses) and GP5216 × GP5223 (two crosses); bqt2Δ, GP5219 × GP5226 (five crosses) and GP5218 × GP5225 (two crosses); dhc1Δ, GP5221 × GP5279 (three crosses) and GP5220 × GP5280 (one cross). Each frequency is based on the cumulative number of spore colonies, >500 colonies in each case.
Statistical significance relative to frequency among totals: *P < 0.02; **P < 0.0005.
Statistical significance relative to wild type: **P < 0.0005.
The coefficient of coincidence, C, equals the observed frequency of crossovers among Ade+ spores divided by the frequency of crossovers among total spores. Statistically significant difference from C = 1: *P < 0.02; ***P < 0.0025; **P < 0.0005.
TABLE 3.
Pairing mutants demonstrate negative crossover interference
| Recombinant frequency
|
|||
|---|---|---|---|
| Interval (Chr) | Wild type | bqt2Δa | dhc1Δa |
| aim–tps16 (III) | 0.11 | 0.04** | 0.04** |
| tps16–arg1 (III) | 0.43 | 0.14** | 0.14** |
| Caim–tps16–arg1b | 1.2 | 2.5*** | 2.9 |
Three or more independent experiments were performed for each interval. Each frequency is based on >500 colonies analyzed. tps16–arg1 data are from Table 2.
Statistical significance relative to wild type: **P < 0.0005.
The coefficient of coincidence, C, equals the observed frequency of double crossovers divided by the product of the frequencies of the respective single crossovers. The numbers of double crossovers observed for wild type, bqt2Δ, and dhc1Δ were 62, 18, and 12, respectively. Statistically significant difference from C = 1: ***P < 0.05.
Recombination in pairing mutants displays negative interference:
In many organisms the presence of one crossover reduces the likelihood of a second nearby crossover (reviewed in Hillers 2004). This phenomenon, called crossover interference, is manifest as a lower-than-expected frequency of double crossovers. Interference is defined as I = 1 − C, where C (coefficient of coincidence) = 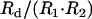 and R1, R2, and Rd are the frequencies of crossovers in interval one, interval two, and double crossovers, respectively. When I = 0, double crossovers occur at the frequency expected for two independent events; when I = 1, no double crossovers are observed. In rare instances the opposite situation has been reported whereby the presence of one crossover increases the likelihood of a second nearby crossover: I < 0, a situation called negative interference. In wild-type S. pombe there is no meiotic crossover interference, either positive or negative (Munz 1994).
and R1, R2, and Rd are the frequencies of crossovers in interval one, interval two, and double crossovers, respectively. When I = 0, double crossovers occur at the frequency expected for two independent events; when I = 1, no double crossovers are observed. In rare instances the opposite situation has been reported whereby the presence of one crossover increases the likelihood of a second nearby crossover: I < 0, a situation called negative interference. In wild-type S. pombe there is no meiotic crossover interference, either positive or negative (Munz 1994).
If recombination is limited by inefficient pairing, the presence of a crossover may select for cells in which adjacent chromosomal intervals are necessarily in close proximity and may also stabilize the interaction between homologs. This may increase the likelihood of a second event, resulting in negative interference. We calculated the coefficient of coincidence (C) for the two adjacent intervals, ura4-aim–tps16 and tps16–arg1, on ChrIII (see Figure 1A) using the recombination data from Table 3. In wild-type crosses C = 1.2, not significantly different from 1 (P > 0.3). In bqt2Δ mutant crosses C = 2.5, significantly >1 (P < 0.05). Similarly, in dhc1Δ mutant crosses C = 2.9. Although 2.9 was not significantly >1 (0.05 < P < 0.1), we suspect that this reflects the limited number of observed double crossovers (12). These data indicate that bqt2Δ, and perhaps dhc1Δ, mutants exhibit negative crossover interference.
The negative interference described above was between two crossovers. Similarly, positive interference is typically observed between two crossovers; in both S. cerevisiae and the filamentous fungus Neurospora crassa a gene conversion without an associated crossover does not exhibit interference with an adjacent interval (Fogel and Hurst 1967; Stadler and Towe 1968; Malkova et al. 2004). However, if one allelic interaction promotes the interaction of nearby chromosomal regions, negative interference might be observed between a conversion and a crossover. The crosses used to measure homologous intergenic recombination, above, also contained heteroalleles of ade6, allowing us to address this possibility. We measured intergenic recombination among selected ade6+ gene convertants in these crosses and calculated C. Here 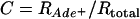 , where
, where 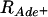 and
and 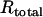 are the frequencies of crossovers among ade6+ convertants and among total spores, respectively. For gene convertants at ade6 and crossovers in the tps16–arg1 interval, which is ∼60–300 kb from ade6 on ChrIII (Figure 1A), C = 0.93 in wild-type crosses, not significantly different from 1 (P > 0.25; Table 2). In bqt2Δ and dhc1Δ mutant crosses C = 2.29 and 2.49, respectively, and is significantly >1 (P < 0.0005; Table 2). The data in Table 2 include ade6+ gene convertants with and without an associated crossover between ura4+-aim and tps16. When only those convertants without an associated crossover are considered, C = 2.45 and 2.51 in bqt2Δ and dhc1Δ mutant crosses, respectively, and is significantly >1 (P < 0.0005). These data indicate that both bqt2Δ and dhc1Δ mutants exhibit negative interference between a conversion and a crossover.
are the frequencies of crossovers among ade6+ convertants and among total spores, respectively. For gene convertants at ade6 and crossovers in the tps16–arg1 interval, which is ∼60–300 kb from ade6 on ChrIII (Figure 1A), C = 0.93 in wild-type crosses, not significantly different from 1 (P > 0.25; Table 2). In bqt2Δ and dhc1Δ mutant crosses C = 2.29 and 2.49, respectively, and is significantly >1 (P < 0.0005; Table 2). The data in Table 2 include ade6+ gene convertants with and without an associated crossover between ura4+-aim and tps16. When only those convertants without an associated crossover are considered, C = 2.45 and 2.51 in bqt2Δ and dhc1Δ mutant crosses, respectively, and is significantly >1 (P < 0.0005). These data indicate that both bqt2Δ and dhc1Δ mutants exhibit negative interference between a conversion and a crossover.
An alternative explanation for apparent negative interference is a subpopulation of “hot” meiotic cells that are recombinationally more competent than the bulk population. To address this possibility, we determined the recombinant frequency in the lys3–met5 and pat1–leu1 intervals, on ChrI and -II, respectively, among ade6+ (ChrIII) convertants. If there is a significant subpopulation of hot cells, then the recombinant frequency in both intervals would be higher among ade6+ convertants than among total cells (C > 1), despite the intervals being on different chromosomes. The recombinant frequency in the lys3–met5 interval was not significantly higher among ade6+ convertants than among total cells in wild type, bqt2Δ, or dhc1Δ (C = 0.92, P > 0.3; C = 1.18, P > 0.3; C = 1.02, P > 0.9, respectively; Table 2). These data argue against a subpopulation of hot cells.
In the pat1–leu1 interval the recombinant frequency was modestly higher among ade6+ convertants than among total spores in wild type, bqt2Δ, and dhc1Δ (Table 2). In wild-type crosses C = 1.15, not significantly different from 1 (P > 0.05), but in bqt2Δ and dhc1Δ mutant crosses C = 1.31 and 1.46, respectively, and was significantly >1 (P < 0.02 and P < 0.0025, respectively). To determine if the increase in recombinant frequency among ade6+ convertants observed in bqt2Δ and dhc1Δ mutants was significantly different from the increase observed in wild type, we determined whether or not the observed frequency of crossovers was greater than that expected if C = 1.15, the wild-type value. The increase in recombinant frequency among ade6+ convertants observed in the bqt2Δ mutant was not significantly different from the increase observed in wild type (P > 0.25), but that in the dhc1Δ mutant was significantly different (P < 0.05). These results indicate that the observed increase of recombinant frequencies among ade6+ convertants in the pat1–leu1 interval (ChrII) is independent of the bqt2Δ mutation but partially dependent on the dhc1Δ mutation. The reason for this result is unknown, but in light of the results with the lys3–met5 interval (ChrI), it does not indicate that the negative interference observed at the ura4-aim–ade6–tps16–arg1 region of ChrIII in bqt2Δ and dhc1Δ mutants is due to hot cells specifically present in the mutant population.
Telomere clustering, but not horsetail movement, restricts ectopic recombination:
To determine whether reduced pairing resulted in excess ectopic (nonallelic) recombination, we first determined the recombinant frequency between the ade6-M26 allele on ChrIII and the ade6-M210 allele on an artificial minichromosome (ChS28; Niwa et al. 1989 and see Figure 1A). In this assay, the frequency of Ade+ recombinants was increased, relative to that in wild type, by a factor of 18 in bqt2Δ mutants but was not affected in dhc1Δ mutants (Table 4). We infer that the minichromosome is released from its position near the telomeres of ChrI, -II, and -III in the bqt2Δ mutant and can more readily come into proximity to the ade6 locus on ChrIII and therefore recombine with it.
TABLE 4.
Telomere clustering, but not horsetail movement, limits ectopic recombination
| Recombinant frequency (× 105)
|
||||
|---|---|---|---|---|
| Locia | ΔDTb | wt | dhc1Δ | bqt2Δ |
| ade6 (allelic) | 0 | 530 ± 55 | 140 ± 8 | 120 ± 15 |
| ade6 × ChS28 | ∼1.34 Mb | 2.2 ± 0.5 | 2.4 ± 0.4 | 40 ± 6.2 |
| ade6 × zzz7 | ∼0.85 Mb | 8.9 ± 0.8 | 13 ± 1.5 | 32 ± 3.3 |
| ade6-Dup (SCE) | ∼5 kb | 1250 ± 160 | 1230 ± 120 | 1910 ± 160 |
Recombinant frequencies are the mean ± SEM for at least four experiments. wt, wild type.
The ade6-M26 and ade6-M210 alleles were used in the ade6 (allelic) and ade6 × ChS28 experiments. The ade6-M26 and ade6-469 alleles were used in the ade6 × zzz7 and ade6-Dup (SCE) experiments. The M210 and 469 alleles are 3 bp apart (Szankasi et al. 1988; G. Freyer, personal communication).
ΔDT is the difference between the distances of each locus from its nearest telomere (Figure 1B).
We next determined the recombinant frequency between the ade6-M26 allele on ChrIII and the ade6-469 allele ectopically transplaced on ChrII (zzz7; Virgin and Bailey 1998 and see Figure 1A). In this assay, the frequency of Ade+ recombinants was increased, relative to that in wild type, by a factor of 3.5 in bqt2Δ mutants but was not significantly affected in dhc1Δ mutants (P > 0.05; Table 4). In bqt2Δ mutants, recombinant frequencies increased in both assays of ectopic recombination but decreased by a factor of 4.4 for allelic recombination (at the endogenous locus) between the ade6-M26 and ade6-M210 alleles (Table 4). These data indicate that Bqt2, but not Dhc1, restricts the interaction of ectopic sequences.
The normal frequency of DSB formation and repair (Martin-Castellanos et al. 2005), but reduced homolog recombination (Tables 2 and 3), suggested that the bqt2Δ mutation might increase the frequency of meiotic sister-chromatid exchange. To address this possibility, we determined the frequency of recombination between tandemly duplicated copies of the ade6 gene, one marked with the M26 allele and the other with the 469 allele, flanking ura4+ at the endogenous ade6 locus (Schuchert and Kohli 1988). In this assay, recombination must use the sister as a template, since ade6 is deleted from the homologous chromosome. In bqt2Δ mutant meioses the Ade+ recombinant frequency was modestly but significantly greater than that in wild type (Table 4; P = 0.009). The Ade+ recombinant frequency in dhc1Δ mutant meioses was not significantly different from that in wild type (Table 4; P > 0.9). These data indicate that Bqt2, but not Dhc1, restricts unequal sister-chromatid exchange.
DISCUSSION
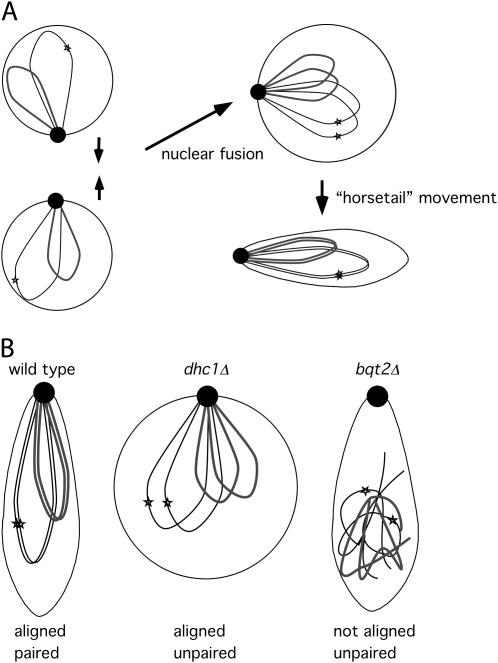
In S. pombe, telomere clustering and horsetail nuclear movement promote the pairing of homologous chromosomes during meiotic prophase (reviewed in Burgess 2004), perhaps by limiting the space that must be searched for a homologous sequence. Recombination requires homolog proximity but also promotes pairing, perhaps by stabilizing the initial alignment of homologs. To understand the role that each of these processes plays in homolog pairing, we have analyzed mutations that specifically abolish telomere clustering (bqt2Δ; Martin-Castellanos et al. 2005; Chikashige et al. 2006) and horsetail nuclear movement (dhc1Δ; Yamamoto et al. 1999). Bqt2 is a meiosis-specific SPB component that, together with Bqt1, tethers the telomere protein Rap1 to the SPB protein Sad1 (Martin-Castellanos et al. 2005; Chikashige et al. 2006). Dhc1, the heavy chain of the microtubule motor dynein, is required for horsetail nuclear movement and, although expression of Dhc1 is not meiosis specific, it has no detected mitotic phenotype (Yamamoto et al. 1999). We first showed that Bqt2, as predicted, is required for efficient pairing of homologs in meiotic prophase (Table 1). We then extended the analysis of meiotic recombinant frequencies in bqt2Δ and dhc1Δ mutants. In light of our results, we propose that (1) the bouquet promotes the alignment of homologs but not their full pairing, (2) horsetail movement facilitates the transition from alignment to full pairing (Figure 2A), and (3) ectopic recombination is restricted by the alignment of homologs, rather than pairing per se.
Figure 2.—
The role of telomere clustering and “horsetail” nuclear movement in meiotic homolog pairing. (A) Prior to nuclear fusion, telomeres cluster at the SPB (solid circle). In the prophase nucleus, the bouquet promotes alignment of homologs, and horsetail movement facilitates the transition to full pairing. (B) Proposed chromosome configuration in wild-type, dhc1Δ, and bqt2Δ mutant prophase nuclei. Only two of the three chromosome pairs are shown. The stars represent homologous sites on a pair of homologs.
Recombination in homolog-pairing mutants displays negative interference:
Positive crossover interference is widespread in meiotic recombination (reviewed in Hillers 2004). Negative crossover interference (higher than expected frequency of double crossovers) is less common and has been associated with special chromosomal regions—centromeres in Drosophila melonogaster and several species of plants (Green 1975; Sinclair 1975; Denell and Keppy 1979; Peng et al. 2000; Boyko et al. 2002; Esch and Weber 2002; Esch 2005) and heterozygous translocations in the mosquito Aedes aegypti and several species of plants (Auger and Sheridan 2001, and references therein). Several proposals have been put forth to explain these cases of negative interference. Apparent negative crossover interference has been attributed to gene conversion of the central marker, i.e., one, not two, events (Green 1975). Denell and Keppy (1979) suggested that negative interference may be characteristic of chromosomal regions, such as centromeres, that have a low density of recombination events (per unit physical length). Auger and Sheridan (2001) suggested that negative interference is a result of reduced competence for crossover formation near translocation breakpoints. Just as hot cells result in negative interference (Grossenbacher-Grunder 1985), so would hot regions of chromosomes—the chromosomal regions that do pair in a pairing-deficient mutant. After submission of this article, negative crossover interference was reported in a zip4Δ mutant of S. cerevisiae using a single-interval assay (nonparental ditype ratio; Tsubouchi et al. 2006). Our results suggest that inefficient homolog pairing leads to negative interference in S. pombe.
bqt2Δ and dhc1Δ mutants display negative interference, both between two crossovers and between a conversion and a crossover (Tables 2 and 3). This is in contrast to wild-type S. pombe where there is no interference (Tables 2 and 3; Kohli and Bahler 1994; Munz 1994). The negative crossover interference in the ura4-aim–tps16–arg1 intervals is unlikely to result from gene conversion of the central marker for the following reason. The frequency of conversion at tps16 would have to be 0.9%, the frequency of apparent double crossovers in excess of the expected frequency, in the ura4-aim–tps16–arg1 intervals in bqt2Δ and dhc1Δ mutants. This is more than twice the frequency of conversion observed in wild type (0.4%; Zahn-Zabal et al. 1995). Given that both mutations reduce all allelic recombination examined by more than a factor of 2 (Tables 2–4), it is unlikely that conversion at tps16 could explain these results. Additionally, to explain the negative interference between ade6+ convertants and tps16–arg1 crossovers (Table 2), the frequency of conversion at tps16 would have to be at least 17%, but only among ade6+ spores.
Gene conversion in S. pombe meiosis is frequently associated with crossing over between flanking markers (Grimm et al. 1994; Cromie et al. 2005). Negative interference between a conversion and a crossover could be explained if the conversion and crossover were not separate events. However, to explain the negative interference between ade6+ convertants and apparent tps16–arg1 crossovers, a single recombination event would frequently have to cover more than the 59 kb between ade6 and tps16. While we cannot formally rule out this possibility, it seems unlikely that the frequency of this type of event would be increased in bqt2Δ and dhc1Δ mutants.
We propose that negative interference in bqt2Δ and dhc1Δ mutants is the consequence of inefficient homolog pairing and an otherwise wild-type ability to repair meiotic DSBs and reflects a propensity for localized proximity of homologs to extend to larger regions—at least 59 kb, the distance between ade6 and tps1 (Figure 1A). The propensity for extended proximity may be related to recombination in two distinct ways. First, an initial recombination event may stabilize the interaction between homologs. This would limit the space that an adjacent chromosomal interval must search for a homologous sequence with which to recombine, thus increasing the likelihood of a second nearby recombination event. In this model, recombination is required to extend localized proximity. Second, when, by chance, one locus is close enough to its homolog to recombine, adjacent chromosomal intervals are necessarily in close proximity, thus increasing the likelihood of a second nearby recombination event. In this model, the first recombination event does not cause local proximity to be extended; rather, extended proximity reflects simply the physical properties of the chromosome. Our genetic data do not differentiate between these models, although a cytological assay for local alignment in a Rec− mutant may be able to do so.
Telomere clustering, but not horsetail movement, restricts ectopic recombination:
Crossovers between dispersed repetitive DNA such as transposons, genes for tRNA, subtelomeric sequences, and multigene families can produce deleterious chromosomal rearrangements. Endogenous repetitive sequences are unlikely to be identical. At least in S. cerevisiae, the mismatch repair (MMR) pathway, which can detect regions of sequence divergence, restricts recombination between diverged sequences (reviewed in Borts et al. 2000). Despite this restriction, ectopic recombination (that between nonallelic sequences) does occur. In humans, a significant number of diseases and syndromes are due to chromosomal translocations, duplications, or deletions generated by meiotic recombination between repetitive DNA (reviewed in Stankiewicz and Lupski 2002). This emphasizes the importance of restricting ectopic recombination. In fact, processes other than MMR must restrict ectopic recombination since the frequency of recombination between nearly identical repeats is significantly lower than that of allelic recombination in both S. cerevisiae and S. pombe (Goldman and Lichten 1996, 2000; Virgin and Bailey 1998; Schlecht et al. 2004). The mechanisms that limit ectopic recombination have not been well characterized. Our data indicate that the meiotic bouquet plays a critical role in S. pombe, perhaps by promoting the alignment of chromosomes, in register, along their entire length.
We have shown that, in S. pombe, ectopic recombination is predominantly constrained by telomere clustering, not homolog pairing per se. Both bqt2Δ and dhc1Δ mutants reduce pairing and allelic recombination, although the meiotic DSB frequency is nearly wild type (Tables 1–4; Yamamoto et al. 1999; Ding et al. 2004; Martin-Castellanos et al. 2005; Chikashige et al. 2006; C. Ellermeier and G. R. Smith, unpublished data). The high viable spore yield in bqt2Δ and dhc1Δ mutants (∼50% of wild type; our unpublished data) indicates that the meiotic DSBs are repaired. We had initially inferred that DSB repair in both mutants frequently involves the use of either sister chromatids or homologous nonallelic sequences as a template. Instead, we found that unequal sister-chromatid exchange and ectopic recombination were unaffected in the dhc1Δ mutant (Table 4). Perhaps in dhc1Δ mutant meioses DSBs are repaired by equal sister-chromatid exchange or by nonhomologous end joining. In bqt2Δ mutant meioses unequal sister-chromatid exchange was elevated 1.5-fold, relative to wild type (Table 4). If sister-chromatid exchange is more frequent than exchange between homologs in wild-type S. pombe, a reduced ability to repair DSBs using the homolog as template (e.g., in pairing mutants) would result in only a small increase in SCE. Ectopic recombination, measured in two different assays, was elevated 3.5- and 18-fold in bqt2Δ mutant meioses (Table 4). Both bqt2Δ and dhc1Δ mutants reduce homolog pairing and allelic recombination to approximately the same extent (Tables 1–4) while the restriction of meiotic ectopic recombination is eased in bqt2Δ mutants and maintained in dhc1Δ mutants. This indicates that the restriction is not dependent on homolog pairing per se. Instead, we suggest that ectopic recombination is restricted predominantly by chromosomal position.
We propose that ectopic recombination between two dispersed repetitive sequences is restricted by their relative positions in the prophase nucleus. Because of telomere clustering, the distance, in base pairs, from the nearest telomere determines the position of a locus within the nucleus (Ding et al. 2004). As a result, the frequency of ectopic recombination is inversely proportional to the difference between the distances of each locus from its nearest telomere (ΔDT, Figure 1B). This proposal is supported by the following data. First, in wild type, the frequency of recombinants increases as ΔDT decreases: the frequency of recombinants is highest when ΔDT = 0 (i.e., in allelic recombination) and decreases as ΔDT increases (Table 4). When ΔDT is the highest, the ratio of allelic to ectopic recombinant frequencies is 240 (ade6 × ChS28, Table 4). Second, in bqt2Δ mutant meioses the effect of ΔDT was nearly eliminated: the ratio of allelic to ectopic recombinant frequencies was never >4 (Table 4). Together, these data suggest that the effect of ΔDT on ectopic recombination reflects the role of telomere clustering in restricting ectopic recombination. Determination of recombinant frequencies at additional pairs of loci is required to establish the generality of the effect of ΔDT on ectopic recombination.
We interpret the different effects of bqt2Δ and dhc1Δ mutants on ectopic recombination as follows. Alignment and subsequent pairing of homologs along their entire length are required to ensure wild-type levels of meiotic recombination. Bqt2 (telomere clustering) and Dhc1 (horsetail movement) contribute to this in different ways (Figure 2). Prior to mating, chromosomes in wild-type S. pombe are in the “Rabl” orientation; i.e., the centromeres are clustered at the SPB and telomeres are dispersed (Funabiki et al. 1993). When mating is induced, centromeres are released and telomeres move to the SPB (Chikashige et al. 1994, 1997). After nuclear fusion, the bouquet promotes alignment but not full pairing of homologs. The Dhc1-dependent horsetail movement facilitates the transition to full pairing but is dependent on telomere clustering for this effect. In a dhc1Δ mutant meiosis telomeres still cluster at the SPB and the bouquet is sufficient to promote alignment of homologs but not full pairing (Figure 2B). In the absence of telomere clustering (bqt2Δ), chromosomes are not able to align (Figure 2B). In this view, both bqt2Δ and dhc1Δ mutants are scored as pairing defective, measured cytologically at a single locus, but have distinctly different configurations of homologous chromosomes. One prediction of this model is that meiotic ectopic recombination should be elevated to the same extent in the bqt2Δ dhc1Δ double mutant as in bqt2Δ.
We expect that like bqt2Δ, other mutations that disrupt telomere clustering such as bqt1Δ, taz1Δ, rap1Δ, and rik1Δ would result in elevated meiotic ectopic recombination. Additionally, we expect that mutations that disrupt horsetail nuclear movement without disrupting telomere clustering would have no effect on meiotic ectopic recombination. Like Bqt2, Mcp6 is a meiosis-specific SPB protein in S. pombe. In mcp6Δ mutant meioses telomere clustering appears normal but horsetail nuclear movement is reduced (Saito et al. 2005; Tanaka et al. 2005). In contrast to our view above, the frequency of ectopic recombinants is increased modestly in mcp6Δ mutant meioses (Saito et al. 2005). Additional experiments may determine whether telomere clustering is fully wild type in mcp6Δ mutants.
In an extensive analysis of ectopic recombination in S. cerevisiae, the authors concluded that for loci on heterologous chromosomes the efficiency of ectopic recombination is negatively correlated with the sum of insert distances to their nearest telomeres (SIDT) (see Figure 1B; Goldman and Lichten 1996; Schlecht et al. 2004). In S. cerevisiae, Ndj1 is required for bouquet formation and pairing (Conrad et al. 1997; Trelles-Sticken et al. 2000). The negative correlation of ectopic recombination efficiency with SIDT does not depend on Ndj1 (Schlecht et al. 2004). Additionally, ectopic recombination efficiencies are only modestly increased in ndj1Δ mutants (Schlecht et al. 2004). In S. cerevisiae, unlike in S. pombe, the bouquet does not play a major role in restricting ectopic recombination. Perhaps in S. cerevisiae the bouquet is not required for alignment, and the synaptonemal complex promotes alignment and restricts ectopic recombination.
Pairing functions and the distribution of meiotic recombination events:
Our data have several important implications. First, we suggest that by promoting homolog pairing, the bouquet and horsetail movement prevent negative interference in S. pombe. Similarly, in S. cerevisiae, Ndj1 and presumably the bouquet contribute to positive interference (Chua and Roeder 1997). In both of these highly diverged species, the bouquet affects the distribution of recombination events. Second, the bouquet functions to restrict ectopic recombination in S. pombe. This function is critical for successful completion of meiosis. Ectopic recombination not only results in deleterious chromosomal rearrangements (reviewed in Stankiewicz and Lupski 2002), but also disturbs meiotic chromosome segregation in S. cerevisiae (Jinks-Robertson et al. 1997). Understanding the mechanism by which the bouquet functions in S. pombe should contribute to our understanding of how chromosome rearrangements are formed in humans.
Acknowledgments
We are grateful to Yasushi Hiraoka for providing strains and sharing unpublished data; Chad Ellermeier and Greg Freyer for unpublished data; Olaf Nielsen for strains and plasmids; and Sue Amundsen, Gareth Cromie, Joseph Farah, and Alastair S. H. Goldman for helpful comments on the manuscript. This work was supported by National Institutes of Health research grant GM32194 to G.R.S. and postdoctoral fellowship F32-GM20125 to L.D.
References
- Auger, D. L., and W. F. Sheridan, 2001. Negative crossover interference in maize translocation heterozygotes. Genetics 159: 1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie, 3rd et al., 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. [DOI] [PubMed] [Google Scholar]
- Borts, R. H., S. R. Chambers and M. F. Abdullah, 2000. The many faces of mismatch repair in meiosis. Mutat. Res. 451: 129–150. [DOI] [PubMed] [Google Scholar]
- Boyko, E., R. Kalendar, V. Korzun, J. Fellers, A. Korol et al., 2002. A high-density cytogenetic map of the Aegilops tauschii genome incorporating retrotransposons and defense-related genes: insights into cereal chromosome structure and function. Plant Mol. Biol. 48: 767–790. [DOI] [PubMed] [Google Scholar]
- Burgess, S. M., 2004. Homolog pairing in S. pombe: the ends are the means. Mol. Cell 13: 766–768. [DOI] [PubMed] [Google Scholar]
- Cervantes, M. D., J. A. Farah and G. R. Smith, 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5: 883–888. [DOI] [PubMed] [Google Scholar]
- Chikashige, Y., and Y. Hiraoka, 2001. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr. Biol. 11: 1618–1623. [DOI] [PubMed] [Google Scholar]
- Chikashige, Y., D. Q. Ding, H. Funabiki, T. Haraguchi, S. Mashiko et al., 1994. Telomere-led premeiotic chromosome movement in fission yeast. Science 264: 270–273. [DOI] [PubMed] [Google Scholar]
- Chikashige, Y., D. Q. Ding, Y. Imai, M. Yamamoto, T. Haraguchi et al., 1997. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J. 16: 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige, Y., C. Tsutsumi, M. Yamane, K. Okamasa, T. Haraguchi et al., 2006. Meiotic proteins Bqt1 and Bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125: 59–69. [DOI] [PubMed] [Google Scholar]
- Chua, P. R., and G. S. Roeder, 1997. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 11: 1786–1800. [DOI] [PubMed] [Google Scholar]
- Conrad, M. N., A. M. Dominguez and M. E. Dresser, 1997. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science 276: 1252–1255. [DOI] [PubMed] [Google Scholar]
- Cooper, J. P., E. R. Nimmo, R. C. Allshire and T. R. Cech, 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747. [DOI] [PubMed] [Google Scholar]
- Cooper, J. P., Y. Watanabe and P. Nurse, 1998. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392: 828–831. [DOI] [PubMed] [Google Scholar]
- Cromie, G. A., C. A. Rubio, R. W. Hyppa and G. R. Smith, 2005. A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over. Genetics 169: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, L., and G. R. Smith, 2003. Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics 163: 857–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, L., and G. R. Smith, 2005. Dynein promotes achiasmate segregation in Schizosaccharomyces pombe. Genetics 170: 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denell, R. E., and D. O. Keppy, 1979. The nature of genetic recombination near the third chromosome centromere of Drosophila melanogaster. Genetics 93: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, D. Q., Y. Chikashige, T. Haraguchi and Y. Hiraoka, 1998. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 111: 701–712. [DOI] [PubMed] [Google Scholar]
- Ding, D. Q., A. Yamamoto, T. Haraguchi and Y. Hiraoka, 2004. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell 6: 329–341. [DOI] [PubMed] [Google Scholar]
- Esch, E., 2005. Estimation of gametic frequencies from F2 populations using the EM algorithm and its application in the analysis of crossover interference in rice. Theor. Appl. Genet. 111: 100–109. [DOI] [PubMed] [Google Scholar]
- Esch, E., and E. Weber, 2002. Investigation of crossover interference in barley (Hordeum vulgare L.) using the coefficient of coincidence. Theor. Appl. Genet. 104: 786–796. [DOI] [PubMed] [Google Scholar]
- Fogel, S., and D. D. Hurst, 1967. Meiotic gene conversion in yeast tetrads and the theory of recombination. Genetics 57: 455–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki, H., I. Hagan, S. Uzawa and M. Yanagida, 1993. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121: 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, M. A., N. Koonrugsa and T. Toda, 2002. a Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 21: 6015–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, M. A., N. Koonrugsa and T. Toda, 2002. b Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol. 12: 610–621. [DOI] [PubMed] [Google Scholar]
- Gerton, J. L., and R. S. Hawley, 2005. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat. Rev. Genet. 6: 477–487. [DOI] [PubMed] [Google Scholar]
- Goldman, A. S., and M. Lichten, 1996. The efficiency of meiotic recombination between dispersed sequences in Saccharomyces cerevisiae depends upon their chromosomal location. Genetics 144: 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, A. S., and M. Lichten, 2000. Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proc. Natl. Acad. Sci. USA 97: 9537–9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, M. M., 1975. Conversion as a possible mechanism of high coincidence values in the centromere region of Drosophila. Mol. Gen. Genet. 139: 57–66. [DOI] [PubMed] [Google Scholar]
- Grimm, C., J. Bahler and J. Kohli, 1994. M26 recombinational hotspot and physical conversion tract analysis in the ade6 gene of Schizosaccharomyces pombe. Genetics 136: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossenbacher-Grunder, A.-M., 1985. Spontaneous mitotic recombination in Schizosaccharomyces pombe. Curr. Genet. 10: 95–101. [DOI] [PubMed] [Google Scholar]
- Gutz, H., H. Heslot, U. Leupold and N. Loprieno, 1974. Schizosaccharomyces pombe, pp. 395–446 in Handbook of Genetics, edited by R. C. King. Plenum, New York.
- Hillers, K. J., 2004. Crossover interference. Curr. Biol. 14: R1036–R1037. [DOI] [PubMed] [Google Scholar]
- Hiraoka, Y., D. Q. Ding, A. Yamamoto, C. Tsutsumi and Y. Chikashige, 2000. Characterization of fission yeast meiotic mutants based on live observation of meiotic prophase nuclear movement. Chromosoma 109: 103–109. [DOI] [PubMed] [Google Scholar]
- Jinks-Robertson, S., S. Sayeed and T. Murphy, 1997. Meiotic crossing over between nonhomologous chromosomes affects chromosome segregation in yeast. Genetics 146: 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh, J., and F. Ishikawa, 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 11: 1624–1630. [DOI] [PubMed] [Google Scholar]
- Keeney, S., 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53. [DOI] [PubMed] [Google Scholar]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kohli, J., and J. Bahler, 1994. Homologous recombination in fission yeast: absence of crossover interference and synaptonemal complex. Experientia 50: 295–306. [DOI] [PubMed] [Google Scholar]
- Kohli, J., P. Munz, R. Aebi, H. Amstutz, C. Gysler et al., 1984. Interallelic and intergenic conversion in three serine tRNA genes of Schizosaccharomyces pombe. Cold Spring Harbor Symp. Quant. Biol. 49: 31–40. [DOI] [PubMed] [Google Scholar]
- Kupiec, M., and T. D. Petes, 1988. a Allelic and ectopic recombination between Ty elements in yeast. Genetics 119: 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec, M., and T. D. Petes, 1988. b Meiotic recombination between repeated transposable elements in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 2942–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C. J., R. K. Dawe, K. R. Christie, D. W. Cleveland, S. C. Dawson et al., 2004. A standardized kinesin nomenclature. J. Cell Biol. 167: 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., and E. C. Chang, 2003. Schizosaccharomyces pombe Ras1 effector, Scd1, interacts with Klp5 and Klp6 kinesins to mediate cytokinesis. Genetics 165: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova, A., J. Swanson, M. German, J. H. McCusker, E. A. Housworth et al., 2004. Gene conversion and crossing over along the 405-kb left arm of Saccharomyces cerevisiae chromosome VII. Genetics 168: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Castellanos, C., M. Blanco, A. E. Rozalen, L. Perez-Hidalgo, A. I. Garcia et al., 2005. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr. Biol. 15: 2056–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, F., K. Okazaki, M. Shimanuki, A. Yamamoto, Y. Hiraoka et al., 2002. The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol. Biol. Cell 13: 930–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, H., Y. Okada and N. Hirokawa, 2005. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 15: 467–476. [DOI] [PubMed] [Google Scholar]
- Munz, P., 1994. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics 137: 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz, P., H. Amstutz, J. Kohli and U. Leupold, 1982. Recombination between dispersed serine tRNA genes in Schizosaccharomyces pombe. Nature 300: 225–231. [DOI] [PubMed] [Google Scholar]
- Nabeshima, K., T. Nakagawa, A. F. Straight, A. Murray, Y. Chikashige et al., 1998. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell 9: 3211–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima, K., Y. Kakihara, Y. Hiraoka and H. Nojima, 2001. A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J. 20: 3871–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo, E. R., A. L. Pidoux, P. E. Perry and R. C. Allshire, 1998. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392: 825–828. [DOI] [PubMed] [Google Scholar]
- Niwa, O., T. Matsumoto, Y. Chikashige and M. Yanagida, 1989. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J. 8: 3045–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, O., M. Shimanuki and F. Miki, 2000. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 19: 3831–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi, R., A. Feoktistova and K. L. Gould, 1996. Construction of vectors and a genomic library for use with the his3-deficient strains of Schizosaccharomyces pombe. Gene 174: 315–318. [DOI] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2003. Chromosome choreography: the meiotic ballet. Science 301: 785–789. [DOI] [PubMed] [Google Scholar]
- Peng, J., A. B. Korol, T. Fahima, M. S. Roder, Y. I. Ronin et al., 2000. Molecular genetic maps in wild emmer wheat, Triticum dicoccoides: genome-wide coverage, massive negative interference, and putative quasi-linkage. Genome Res. 10: 1509–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, J., D. Weilguny, R. Egel and O. Nielsen, 1995. Characterization of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol. Cell. Biol. 15: 3697–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki, M., M. F. Siomos and K. Nasmyth, 2003. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112: 423–440. [DOI] [PubMed] [Google Scholar]
- Ponticelli, A. S., and G. R. Smith, 1989. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics 123: 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, G. S., 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11: 2600–2621. [DOI] [PubMed] [Google Scholar]
- Saito, T. T., T. Tougan, D. Okuzaki, T. Kasama and H. Nojima, 2005. Mcp6, a meiosis-specific coiled-coil protein of Schizosaccharomyces pombe, localizes to the spindle pole body and is required for horsetail movement and recombination. J. Cell Sci. 118: 447–459. [DOI] [PubMed] [Google Scholar]
- Sanchez-Perez, I., S. J. Renwick, K. Crawley, I. Karig, V. Buck et al., 2005. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 24: 2931–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan, H., 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell. Biol. 2: 621–627. [DOI] [PubMed] [Google Scholar]
- Schlecht, H. B., M. Lichten and A. S. Goldman, 2004. Compartmentalization of the yeast meiotic nucleus revealed by analysis of ectopic recombination. Genetics 168: 1189–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchert, P., and J. Kohli, 1988. The ade6–M26 mutation of Schizosaccharomyces pombe increases the frequency of crossing over. Genetics 119: 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer, L. G., and J. R. Lupski, 2000. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu. Rev. Genet. 34: 297–329. [DOI] [PubMed] [Google Scholar]
- Shimanuki, M., F. Miki, D. Q. Ding, Y. Chikashige, Y. Hiraoka et al., 1997. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol. Gen. Genet. 254: 238–249. [DOI] [PubMed] [Google Scholar]
- Shimoda, C., A. Hirata, M. Kishida, T. Hashida and K. Tanaka, 1985. Characterization of meiosis-deficient mutants by electron microscopy and mapping of four essential genes in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 200: 252–257. [DOI] [PubMed] [Google Scholar]
- Sinclair, D. A., 1975. Crossing over between closely linked markers spanning the centromere of chromosome 3 in Drosophila melanogaster. Genet. Res. 26: 173–185. [DOI] [PubMed] [Google Scholar]
- Stadler, D. R., and A. M. Towe, 1968. A test of coincident recombination in closely linked genes of Neurospora. Genetics 58: 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz, P., and J. R. Lupski, 2002. Genome architecture, rearrangements and genomic disorders. Trends Genet. 18: 74–82. [DOI] [PubMed] [Google Scholar]
- Straight, A. F., J. W. Sedat and A. W. Murray, 1998. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 143: 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi, P., W. D. Heyer, P. Schuchert and J. Kohli, 1988. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination hot spot allele ade6–M26. J. Mol. Biol. 204: 917–925. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., T. Kohda, A. Yamashita, N. Nonaka and M. Yamamoto, 2005. Hrs1p/Mcp6p on the meiotic SPB organizes astral microtubule arrays for oscillatory nuclear movement. Curr. Biol. 15: 1479–1486. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken, E., M. E. Dresser and H. Scherthan, 2000. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J. Cell Biol. 151: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi, T., H. Zhao and G. S. Roeder, 2006. The meiosis-specific Zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with Zip2. Dev. Cell 10: 809–819. [DOI] [PubMed] [Google Scholar]
- Tuzon, C. T., B. Borgstrom, D. Weilguny, R. Egel, J. P. Cooper et al., 2004. The fission yeast heterochromatin protein Rik1 is required for telomere clustering during meiosis. J. Cell Biol. 165: 759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen, J. E., and R. S. Hawley, 2003. Meiosis: when even two is a crowd. Curr. Biol. 13: R831–R833. [DOI] [PubMed] [Google Scholar]
- Virgin, J. B., and J. P. Bailey, 1998. The M26 hotspot of Schizosaccharomyces pombe stimulates meiotic ectopic recombination and chromosomal rearrangements. Genetics 149: 1191–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin, J. B., J. P. Bailey, F. Hasteh, J. Neville, A. Cole et al., 2001. Crossing over is rarely associated with mitotic intragenic recombination in Schizosaccharomyces pombe. Genetics 157: 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, R. R., T. Malmstrom, C. L. Troxell and J. R. McIntosh, 2001. Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol. Biol. Cell 12: 3919–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, R. R., T. Malmstrom and J. R. McIntosh, 2002. Kinesins klp5+ and klp6+ are required for normal chromosome movement in mitosis. J. Cell Sci. 115: 931–940. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., and Y. Hiraoka, 2001. How do meiotic chromosomes meet their homologous partners?: lessons from fission yeast. BioEssays 23: 526–533. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., R. R. West, J. R. McIntosh and Y. Hiraoka, 1999. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 145: 1233–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi, S., and Y. Watanabe, 2005. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell 123: 803–817. [DOI] [PubMed] [Google Scholar]
- Young, J. A., R. W. Schreckhise, W. W. Steiner and G. R. Smith, 2002. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell 9: 253–263. [DOI] [PubMed] [Google Scholar]
- Zahn-Zabal, M., E. Lehmann and J. Kohli, 1995. Hot spots of recombination in fission yeast: inactivation of the M26 hot spot by deletion of the ade6 promoter and the novel hotspot ura4-aim. Genetics 140: 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]