Abstract
It is standard genetic practice to determine whether or not two independently obtained mutants define the same or different genes by performing the complementation test. While the complementation test is highly effective and accurate in most cases, there are a number of instances in which the complementation test provides misleading answers, either as a result of the failure of two mutations that are located in different genes to complement each other or by exhibiting complementation between two mutations that lie within the same gene. We are primarily concerned here with those cases in which two mutations lie in different genes, but nonetheless fail to complement each other. This phenomenon is often referred to as second-site noncomplementation (SSNC). The discovery of SSNC led to a large number of screens designed to search for genes that encode interacting proteins. However, screens for dominant enhancer mutations of semidominant alleles of a given gene have proved far more effective at identifying interacting genes whose products interact physically or functionally with the initial gene of interest than have SSNC-based screens.
A BRIEF HISTORY OF THE COMPLEMENTATION TEST
The early fly geneticists were certainly aware of a small number of genes that appeared to be defined by multiple alleles, in the sense that these mutations were recombinationally inseparable and produced the same or similar phenotypes (Muller 1932; Stone 1935; Stern and Schaeffer 1943). Moreover, these geneticists did indeed carefully analyze the phenotypes of various double heterozygotes in an attempt to understand gene function. Among other things these studies led to the discovery both of intragenic recombination (Green and Green 1949, 1956; Lewis 1951; Green 1990) and of complex interactions between various allelic combinations (cf. Stern and Schaeffer 1943; Green 1961). But given the small number of genes defined by multiple alleles, it is not clear that these workers saw the construction of double heterozygotes as a tool for addressing the single question that underlies the complementation test: namely, among a set of independently arising mutations causing the same or similar phenotypes, which of those mutations define the same gene(s)?
The first experimental approach designed to address this question was referred to as the cis–trans test (Pontecorvo 1958). According to Pontecorvo, the cis–trans test was first proposed by Ed Lewis (Lewis 1951) to describe those cases where heterozygotes for two tightly linked mutations exhibit a phenotype when the mutations are arranged as a trans-heterozygote but not as a cis-heterozygote. To quote Pontecorvo (1958, p. 37), “if a heterozygote having two different allelic recessives (arisen by independent mutations), one on one chromosome and the other on the homologous chromosome (trans or repulsion arrangement), has a recessive phenotype, or a more nearly recessive phenotype than the corresponding double heterozygote with both recessives on one chromosome and a normal homolog,” then the two mutations define the same gene. Lewis (1951) refers to this as “position pseudoallelism,” while Pontecorvo refers to it simply as the “Lewis effect.” Despite Pontecorvo's attribution of the idea to Lewis, Lewis himself attributes the origin of this idea to Seymour Benzer, who both coined the term “cis–trans test” in the course of his studies of the T4 rIIa and rIIb genes and popularized its usage (Benzer 1955, 1962; Lewis 1967). It is unclear, at least to us, which of these two scientists truly deserves credit for devising the cis–trans test, and so we suggest that credit be shared equally2.
The power of the classical cis–trans test is that performing the cis component of the test (m1 m2/+ +) as well as the trans component (m1 +/+ m2) effectively rules out a potentially serious error that might occur in the case of two tightly linked genes with related functions (such as the rIIa and rIIb genes in phage T4). One could imagine a case where heterozygosity at both loci created combined haplo-insufficiency, misleading the investigator into believing that the mutations were allelic. But if this were the case, then the same mutant phenotype would be observed in cis- as well as trans-heterozygotes. Unfortunately, the cis component of the cis–trans test is rarely done in higher eukaryotes both because it is critical only in the case of very tightly linked and functionally related loci and because when two mutations are tightly linked it is often difficult, if not impossible, to obtain the necessary doubly mutant chromosomes by recombination3.
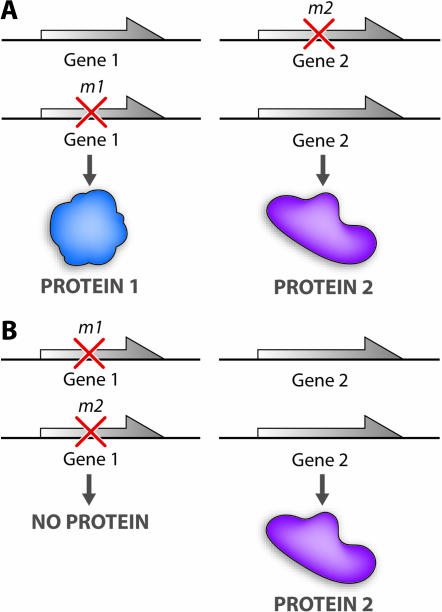
The modern version of the complementation test was first described in detail by Giles (1958) and Fincham (1958) in articles published in the Proceedings of the 10th International Congress of Genetics. The test rapidly became a mainstay of genetic analysis in fungi (cf. Case and Giles 1960) and in higher organisms, such as Drosophila (cf. Green 1961). The basic form of this test is described in Figure 1. Consider two independently isolated mutations, m1 and m2. Both of these mutations are fully recessive, and m1/m1 and m2/m2 homozygotes manifest similar mutant phenotypes (for example, a change in wing structure in flies or auxotrophy for histidine in yeast). As shown in Figure 1A, if m1 and m2 are not in the same gene then the wild-type (+) alleles of both genes are still present in the double heterozygote and fully functional forms of both proteins are produced. In this instance, the two mutations are said to “complement each other” and to define different genes. However, Figure 1B displays the case where both mutations m1 and m2 lie within the same gene. In this case the double heterozygotes (m1/m2) possess only mutant copies of this gene and will thus manifest only the mutant phenotype4. In this case, we say that m1 and m2 to “fail to complement each other” and that they define the same gene.
Figure 1.—
The trans-component of the complementation test in its simplest form.
The value of this test cannot be overrated. For example, it allows the mutants obtained in a given screen to be quickly sorted into complementation groups. To consider a small-scale screen, our laboratory used a germline clone assay to screen 50,000 EMS-treated chromosomes for a meiotic defect. Among the 12 mutants recovered, complementation testing revealed that fully a third were alleles of a single gene (ald), two more were alleles of a previously identified meiotic gene [c(2)M], while the remaining six appear to define novel genes. For the most dramatic example of the utility of this test, consider the case of the classic screen by Nusslein-Volhard et al. (1984), where the use of the complementation test allowed the authors to sort >250 new zygotic lethal mutants into a tractable number of complementation groups. Similarly, the classic screen by Hartwell et al. (1973) for mutants that control the progression of the cell through the cell cycle yielded 149 mutants that were sorted into a much smaller number of genes (32), or complementation groups, solely by the complementation test. It is this test that makes the analysis of the outcome of mutant screens, both large and small, a tractable process.
That said, there are two ways in which the complementation test can “lie to you.” First, as detailed below, it is possible for mutations at two different loci to interact in such a way as to produce a mutant phenotype in m1 +/+ m2 heterozygotes, even though m1 and m2 define separate genes. This phenomenon is discussed in detail below. Second, it is also possible for two mutants in different domains of the same gene to fully complement each other, provided that they define separate functional elements of either the gene or its protein product. This phenomenon is referred to as intragenic complementation5.
However, despite these possible errors, we would be at a loss without the complementation test. Moreover, a variant of the complementation test, transformation rescue, is considered to be the gold standard for proving that one has indeed identified, at the DNA sequence level, a gene previously defined only by genetic mutations. This type of complementation test asks the question: Can a wild-type copy of gene X rescue the function of the mutant allele that is believed to define gene X? If you can show that a homozygote for the mutant of interest can be “rescued” by the addition of a wild-type copy of your candidate gene, and you have identified one or more mutant alleles at the level of DNA sequence alterations, then you have cloned the gene defined by that mutant.
CAVEATS FOR THE USE OF THE COMPLEMENTATION TEST
The complementation test is straightforward, but there are three simple caveats for its use. First, as originally conceived the complementation test should be done only when both loss-of-function mutations are fully recessive. Although the test has also been used in cases in which one of the mutations exhibits a dominant or semidominant phenotype due solely to haplo-insufficiency, there is a serious concern that a failure to see complementation with a second mutation might not reflect allelism, but rather simply the ability of a mutation in a second gene to enhance the phenotypic effects of the first mutation (see below). In those cases where one wishes to determine whether or not two truly anti- or neomorphic mutations are allelic or whether one such mutation is allelic to loss-of-function mutations that map in the same interval, the only alternative is to “revert” the dominant in hopes of creating a testable loss-of-function, and thus recessive, mutation (cf. Rasooly et al. 1991).
Second, the complementation test does not require that the two mutants have the same phenotype. As we have noted previously, different mutations in the same gene can produce rather different phenotypes. Sometimes a mutation that alters, but does not destroy, function will have a weaker effect on the organism's phenotype than does a null or “knockout” mutation (for example, one might have alleles of a gene that result in lethality when homozygous and weaker alleles that cause a visible phenotype). In these cases the double heterozygote usually exhibits the phenotype associated with the weaker of the two alleles.
Third, it is important to note that the complementation test is only a test of gene function and provides no information regarding the nature or position of the mutations. Two mutations that alter the same base pair in the same gene will fail to complement, just as will two loss-of-function mutations at different sites in the same gene. Moreover, any nearby mutation or aberration that acts to inactivate a given gene will fail to complement other alleles of that gene even if the mutation or the breakpoint is some distance away.
A particularly illustrative example of this phenomenon arose in our lab. As noted above, a recent mutagenesis in our lab recovered four mutants that fell into a single complementation group. All four mutations were found to map to a small interval that included the ald gene, which encodes the fly mps1 homolog (Gilliland et al. 2005). Mutations in the ald gene cause defects in both meiosis and mitosis, and all four new ald alleles failed to complement the original ald allele in terms of the meiotic defect. All four mutants were also rescued by a single copy of our ald+ transgene construct. However, there were clear differences among the four mutants. Two mutants displayed both the meiotic and mitotic phenotypes and had DNA sequence changes in the ald gene (in one case an early stop codon and in the other a nine-codon deletion). However, although the other two mutants exhibited obvious meiotic defects, they had no mitotic defects, nor could we find DNA sequence changes associated with these two mutations either in the ald gene itself or in the region delimited by the rescue construct.
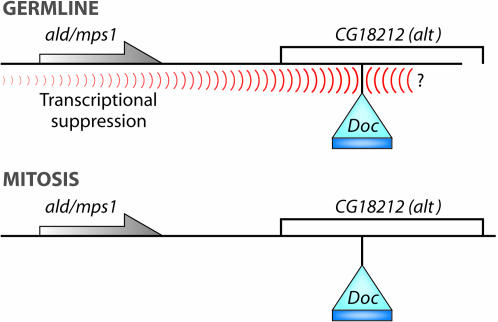
The answer to this paradox came when we discovered that both of these unusual mutants contained a new insertion of a transposable element known as a Doc in the next gene downstream of ald, a gene called CG18212. These insertions were 1638 or 2042 nucleotides downstream from the end of the ald transcript and were thus well outside of the region included in our fully functional ald+ transgene. We had considered CG18212 as a candidate gene when we were cloning the ald1 mutation and had ruled it out by creating a P-element excision that deleted virtually all of the gene, demonstrating that this mutation fully complemented the original ald mutation. As shown in Figure 2, it turns out that the answer to this riddle lay in our discovery that these Doc elements can suppress the function of at least one nearby gene (ald) in cis in the germline, but not in the soma6,7. As a result of this cis-inactivation in the germline, the Doc insertions in CG18212 fail to complement canonical ald mutations in terms of the meiotic, but the not the mitotic defect. As novel as this might seem, it is really no different from the inactivation of genes by moving them close to a block of heterochromatin, a phenomenon known as position-effect variegation (PEV). By moving the gene near the heterochromatin one can inactivate its function without damaging the gene—and that strongly variegating (inactivated) gene will fail to complement a loss-of-function allele of the same gene borne by its homolog, despite the fact that the gene itself is structurally intact.
Figure 2.—
Germline-specific inactivation of the wild-type copy of the ald+ gene causes it to fail to complement a mutant allele of ald carried by the homologous chromosome, but only in the germline.
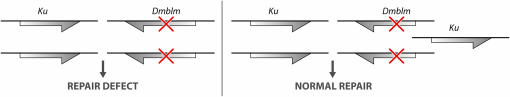
There is a similar cautionary tale regarding interpreting transformation rescue experiments. For example, repair-deficient mutations in the Drosophila mus309 gene were mapped to a small genetic region that contained the fly homolog of the Ku gene, which encodes a well-characterized protein involved in DNA repair. Moreover, these mutants were rescued by a single-copy insertion of a gene encoding the DNA repair protein Ku (Beall and Rio 1996). It thus seemed at the time a reasonable inference that the mus309 gene encoded the fly Ku protein. The only missing piece of data was the sequencing of mus309 alleles to identify the expected sequence lesion. Unfortunately, as shown in Figure 3, it turns out that mus309 mutations are not alleles of the Ku gene. Rather, they were shown to be alleles of the adjacent Dmblm locus (Kusano et al. 2001). Dmblm is a homolog of the human Bloom syndrome gene, which encodes a helicase of the RECQ family. Both genes function in the repair of double-strand breaks and increasing the dose of Ku, even by 50%, can suppress the effects of homozygosity for loss-of-function mutations at the Dmblm gene.
Figure 3.—
An extra copy of the nearby Ku gene rescues the phenotypic defects caused by homozygosity for mutation at the Dmblm gene.
If there is a lesson to be learned from either story, it is the need to identify the structural lesions responsible for the individual mutations that define any gene or complementation group. To do otherwise might both allow one to be seriously misled and, more importantly, preclude the discovery of a valuable novelty.
SECOND-SITE NONCOMPLEMENTATION
In rare cases, two mutations in unlinked genes, which by themselves are fully recessive, can create a mutant phenotype in double heterozygotes. In other words, although individuals heterozygous for either m1 or m2 alone (m1/+ and m2/+) are wild type, doubly heterozygous individuals (m1/+; m2/+) exhibit a mutant phenotype. This phenomenon is referred to as nonallelic noncomplementation or second-site noncomplementation (SSNC). Since the term SSNC seems to have the wider usage, we use it here. Hawley and Walker (2003) have divided SSNC into three separate categories (types 1–3) that differ in terms of allele specificity at the two loci.
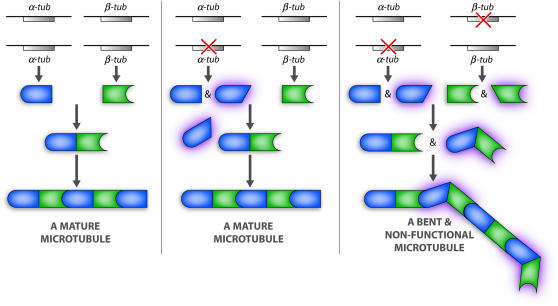
In type 1 SSNC, the interaction is restricted to specific missense alleles at both loci, and neither allele can be a null. The double-mutant combination may be thought of as a “synthetic antimorph” because only the combination of the two specific mutant proteins produces a poisonous gene product (see Figure 4). As intriguing as this phenomenon can be, to the best of our knowledge it has been demonstrated in only two cases (see below).
Figure 4.—
Two missense alleles of the α- and β-tubulin genes encode proteins that combine to create a poisonous tubulin monomer.
In type 2 SSNC, allele specificity is required at only one locus (gene A), but the mutation at the other gene (B) can be any loss-of-function allele, including a deletion. This phenomenon is thought to identify physical interactors by multiple mechanisms. Two of these cases, which are documented below, include examples in which the mutant form of protein A sequesters some of a limited quantity of protein B into inactive complexes or in which normal levels of protein A are required to allow a mutant form of protein B to form the homotetramer that is required for function. In addition to identifying interactions between protein-encoding genes, there is no reason to preclude examples of this type of SSNC in which one mutation occurs in a gene encoding an RNA product (although no such examples of this phenomenon have yet been found).
In type 3 SSNC, also known as “combined haplo-insufficiency,” the interaction is allele independent at both loci. The phenomenon can be, and often is, observed when both mutations are nulls. This type of SSNC neither requires nor implies the physical interaction of the two proteins. (Moreover, as in the case for type 2 SSNC, there is no reason that these types of interactions need be limited to genes that encode protein rather than RNA products.) Rather, it suggests only that reducing the dosage of the product of gene A does not create a phenotype, unless the cell or organism is further crippled by a reduction in the dosage of gene B. Although this type of SSNC is probably the most common, and sometimes the least interesting, it has uncovered important examples in which the proteins encoded by the noncomplementing genes do indeed act in functionally related processes.
Type 1 SSNC (poisonous interactions)—the interaction is allele specific at both loci:
Type 1 SSNC is both the rarest and the most interesting of these phenomena. By definition it requires allele specificity at both genes, and neither allele can be a null mutation. Although this type of interaction can be explained by asserting that the two mutant proteins physically interact to produce a poisonous protein dimer or complex (see Figure 4), as shown below it need not always imply such an interaction. For illustration, we describe two cases in which type I SSNC did indeed reflect physical interaction and a third case in which it most certainly did not.
The first case of SSNC that did confirm a physical interaction was the discovery of dual allele-specific SSNC by mutations in the yeast α- and β-tubulin genes. In 1988 Tim Stearns and David Botstein (Stearns and Botstein 1988) screened for mutants that displayed cold-sensitive SSNC in the presence of a recessive cold-sensitive allele of β-tubulin. Over 20,000 mutagenized cells were screened, and only one second-site noncomplementer was found. This new mutant indeed turned out to carry a mutation in the α-tubulin gene that fails to complement one of the β-tubulin mutants in an allele-specific fashion. The failure of complementation presumably reflects the combination of the two mutant proteins to form a defective and poisonous tubulin subunit whose incorporation into a growing microtubule leads to severe defects (see Figure 4). The Stearns and Botstein (1988) article is widely viewed as the hallmark on SSNC research, primarily because the screen did identify a mutation in a gene whose product, α-tubulin, physically interacted with β-tubulin8.
A second case in which dual allele-specific noncomplementation appears to reflect the physical interaction of two proteins is provided by the work of Berg and his collaborators on the genes encoding the yeast proteins RFA (the large subunit of the heterotrimeric single-stranded DNA-binding protein RPA) and the DSB repair protein Rad52 (Firmenich et al. 1995). These authors identified a missense allele of the RFA gene (rfa1-44) that displayed a number of unusual interactions with the RAD52 gene, including the failure to complement a specific missense allele of RAD52 (rad52-34). Hays et al. (1998) went on to demonstrate that the RFA and RPA proteins did indeed physically interact, at least as assayed by the yeast two-hybrid system. Moreover, the rad52-34 mutation lies in the region of RAD52 required to bind RFA, and this mutation blocks the binding of mutant Rad52 protein to wild-type RFA. (Unfortunately, the coexpression of both mutant proteins was toxic to yeast, and thus it was not possible to ascertain whether or not the protein encoded by the rfa1-44 mutation does indeed bind to the mutant Rad52 protein to create a poisonous complex, but this seems a likely possibility.)
Unfortunately, one can also observe dual allele-specific SSNC even in cases where the two proteins do not physically interact. The classic example of this phenomenon stems from the efforts of David Drubin and his collaborators, who set out to identify actin-interacting proteins by screening for second-site mutations that failed to complement either of two temperature-sensitive alleles of the yeast actin gene (Vinh et al. 1993; Welch et al. 1993). The screening of >55,000 mutagenized colonies yielded a total of 14 extragenic noncomplementing mutants. Although one of these mutations (anc1) caused several properties that made its product likely to be an actin interactor (such as defects in actin filament formation on its own), the Anc1 protein turns out not to be a physical partner of actin. Rather, the Anc1 protein is a yeast-specific subunit of the transcription factor TAF14, a component of the mediator complex (Cairns et al. 1996). Deletion of ANC1/TFG3 results in diminished transcription. Thus anc1 mutants appear to interact with actin mutants not by physical interaction of mutant gene products, but instead by global effects on the transcription process.
Thus we are aware of but two true examples in which dual-allelic-specific second-site noncomplementers did indeed identify proteins that physically interact. To our knowledge, no other bona fide examples of this phenomenon exist in the literature and screens. For this reason, and because of the type of interactions observed by Drubin and his collaborators, screening for dual-allelic-specific SSNC as a means for identifying physically interacting proteins is rarely done and indeed has largely, and appropriately, been replaced by far more powerful proteomics-based methodologies.
Type 2 SSNC–the interaction is allele specific at one locus:
In this second type of second-site noncomplementation, we observe allele specificity only for one of the two genes. The mutation at the other gene needs only to be a loss-of-function allele; indeed, a deficiency of the second gene usually will work just fine. There are a variety of interactions that can produce this result. Although we discuss but two examples, there may be many more possibilities. In the first case, which involves the interaction of mutations in the α- and β-tubulin genes in Drosophila, the failed complementation resulted from the ability of the mutant α-tubulin protein to sequester β-tubulin subunits into inactive and nonfunctional complexes. This case is considered in detail below. In a second case, digenic retinitis pigmentosa in humans and mice, a mutant protein (RDS1) can form only functional homotetramers in the presence of normal levels of a second protein called ROM. In the presence of reduced levels of ROM the mutant RDS1 protein fails to form the requisite heterotetramers, and retinal degeneration is observed (Kajiwara et al. 1994; Goldberg and Molday 1996; Kedzierski et al. 2001). In both of these cases, type 2 SSNC does indeed imply physical or functional interaction.
The classic example of SSNC by sequestration of functional protein A into inactive complexes by mutant protein B is provided by Fuller and her collaborators, who recovered a mutation in a Drosophila α-tubulin gene that failed to complement mutations in a spermatogenesis-specific β-tubulin gene in flies (Hays et al. 1989). Flies heterozygous for the mutant α-tubulin gene and for various recessive alleles of the β-tubulin gene, including a known null, are sterile (Hays et al. 1989). In other words, although heterozygotes for mutations at either gene are fertile, doubly heterozygous flies are sterile. The interaction of this specific α-tubulin mutation with β-tubulin mutations is not mimicked by a deficiency for the α-tubulin gene, but rather requires a specific allele of the α-tubulin gene. Fuller and her colleagues advanced the hypothesis that the α-tubulin protein acts to sequester the normal β-tubulin subunits into inactive dimers. As a result, the total number of functional tubulin dimers is reduced below some critical threshold. Approximately half of normal levels, as would be observed in the presence of heterozygosity for the β-tubulin null mutant alone, is apparently good enough. But reducing that concentration further, to one-quarter of normal levels, impairs spermatogenesis.
The observed interaction between α- and β-tubulin mutations provided an obvious basis for a screen for mutations in other genes whose protein products might interact with either α- or β-tubulin. Aware that many such interactions might be spurious, these workers wisely focused their attention on those interactions that met two further quite stringent criteria. First, the interaction had to be allele specific for at least one of the two loci, and second, the noncomplementing (nc) mutation had to cause a defect in male fertility on its own (in the absence of the tubulin mutation). Although mutations meeting these criteria were found (cf. Regan and Fuller 1988; Green et al. 1990), as detailed below it is not clear that any of the proteins encoded by these genes has a functional, much less a physical, relationship to β-tubulin.
We first consider the noncomplementing mutation that defined the whirligig gene (Green et al. 1990). Although this allele of whirligig (denoted wrlnc4) was initially identified on the basis of its failure to complement a deletion that included the α-tubulin gene, it was subsequently shown to fail to complement β-tubulin mutations as well. (In fact, it also fails to complement one of the other tubulin noncomplementing mutations, haywire, which is described below). The interaction of whirligig mutations with β-tubulin null mutations is allele specific in the sense that the wrlnc4 mutation fails to complement some β-tubulin mutations and not others. Moreover, whirligig mutants display multiple anomalies in microtubule organization, including defects in spermatid differentiation, disordered spermatid components, disrupted flagellar axonemes, and axonemes lacking one or both central pair microtubules (Green 1990). Strengthening the case for a functional interaction between the Whirligig protein and tubulin, although a deletion for whirligig is a dominant male sterile, that sterility is rescued by heterozygosity for the β-tubulin null mutation. Thus, sterility can result from any combination of mutations that alters the relative concentrations of these two proteins, and fertility can be restored by making the concentration of these proteins more equal. Unfortunately, attempts to clone whirligig have been complicated by the observation that the recessive male sterility and the β-tubulin noncomplementing phenotypes are recombinationally separable, suggesting that the two phenotypes may be caused by two distinct mutations. (J. A. Brill, personal communication). Thus, it is not clear that the gene defined by the B2t noncomplementing mutation plays a direct role in spermatogenesis, nor is there yet any compelling evidence that its product directly interacts with tubulin.
A similarly frustrating story evolved from the study of one of the best characterized of these second-site noncomplementing genes, the haywire gene. Again, the interaction of haywire mutations with β-tubulin null mutations is allele specific (Regan and Fuller 1988), and males homozygous for these haywire mutations have defects in several major microtubule-based processes. Nonetheless, when the haywire gene was cloned, it was shown to encode a general RNA polymerase II transcription factor that is also essential for nucleotide excision repair (Mounkes et al. 1992; Mounkes and Fuller 1999). Presumably, as was the case for the actin noncomplementer described in the previous section, the aberrant haywire gene product results in the impaired expression of some gene or set of genes in a fashion that cripples meiosis and spermatogenesis in a cell with reduced levels of β-tubulin.
The reason that this type of screen for interactors pulls up proteins involved in gene expression is fairly obvious. Once the normal level of a tubulin subunit has been reduced by a mutation in that gene, any second mutation that further diminishes the expression of tubulin or tubulin-interacting genes is likely to create a phenotype. For example, many, if not most, yeast screens for mitotic mutants have identified splicing genes. This observation was explained by Biggins et al. (2001), who noted that the yeast α-tubulin gene contains an intron; it is thus one of the few yeast genes that must be spliced. Therefore mutants with defects in the splicing machinery might well be expected to have mitotic defects. Indeed, when Biggins et al. (2001) made an intronless TUB1 gene, the splicing mutants no longer had mitotic defects.
Taken together, these observations suggest that sequestration-based examples of SSNC are relatively rare. They also provide a warning that screens for second-site noncomplementers of null alleles are unlikely to identify genes encoding proteins whose products interact functionally, much less physically, but rather are far more likely to identify genes whose products are required for the expression of the gene defined by the original null allele.
Type 3 SSNC (combined haplo-insufficiency)—the interaction is not allele specific at either locus:
This type of SSNC neither requires nor implies the physical interaction of the two proteins. Rather, it suggests only that reducing the dosage of the product of gene A does not create a phenotype unless the cell or organism is further crippled by a reduction in the dosage of gene B. Combined haplo-insufficiency does not require allele specificity at either gene, and it is sometimes created by using null alleles or deficiencies at one or both genes. This type of SSNC is probably the most common and sometimes the least interesting. One could imagine, for example, a case where the mutation in gene B simply depressed the rate of transcription of gene A, thus decreasing the level of A protein production below some threshold of function.
However, there are also important examples in which the observation of combined haplo-insufficiency does indicate that two genes do act in functionally related processes. Consider, for example, a case involving Hoxb-5 and Hoxb-6 genes that has been reported by Rancourt et al. (1995). Hoxb-5 and Hoxb-6 are adjacent genes in the mouse HOXB locus and are members of the homeotic transcription factor complex that governs establishment of the mammalian body plan. Although loss-of-function mutations at the Hoxb-5 and Hoxb-6 genes are fully recessive, hoxb-5, hoxb-6 trans-heterozygotes (Hoxb-5−, Hoxb-6+/Hoxb-5+, Hoxb-6−) display a mutant phenotype, presumably as a result of combined haplo-insufficiency. There are in fact many other examples of SSNC in HOX clusters besides these two genes (R. Krumlauf, personal communication). For examples in both mice and humans of severe phenotypic abnormalities in limb development that result from combined haplo-insufficiency involving mutations at the Hoxd-13 and Hoxa-13 genes see Debeer et al. (2002), Fromental-Ramain et al. (1996), Goodman and Scambler (2001), and Davis and Capecchi (1996).
A second such example can be found in the analysis of genes required for spindle pole body function in yeast, namely MPS1 and SPC42. The SPC42 gene encodes a core component of the spindle pole body whose function is directly regulated via phosphorylation by the Cdc28 kinase. However, in addition to this direct form of regulation by Cdc28, Cdc28 also regulates Spc42 function indirectly by a pathway that involves phosphorylation of Mps1 (Jaspersen et al. 2001). Neither of these two independent pathways is essential on its own for cell viability, but disruption of both pathways can lead to defects in spindle pole body duplication. While mutations in the MPS1 or SPC42 gene that block Cdc28-dependent phosphorylation are viable in either haploids or diploids, diploids that are doubly heterozygous for both mutations are not viable (Jaspersen et al. 2001). Apparently, as a consequence of the changes in spindle pole body size that accompany the shift from a haploid to a diploid cell, the combined haplo-insufficiency for Cdc-28 phosphorylatable forms of these two proteins becomes a lethal event (S. Jaspersen, personal communication).
Such examples aside, anyone devising a screen for functionally related genes on the basis of searching for mutants that exhibit combined haplo-insufficiency is likely to be frustrated by the rather large number of nonspecific modifiers described in the first paragraph of this section. Only when such a screen is coupled with a powerful secondary screen, such as demanding that the new mutations cause relevant phenotypes as homozygotes in the absence of a mutation at the original locus, is there a good likelihood of identifying interesting new loci. However, the recent development of techniques designed by Boeke and his collaborators (cf. Ye et al. 2005) may allow the development in yeast of high-throughput means to identify examples of SSNC that can be sorted through by exactly such secondary screens.
DOMINANT ENHANCEMENT: THE TECHNIQUE THAT ACTUALLY WORKS!
Although screens for second-site noncomplementers that demand that both mutations be fully recessive have not proven terribly successful in identifying proteins that physically interact or even in identifying proteins that function in the same or parallel pathways, real successes have been obtained from screens for dominant enhancer mutations of semidominant mutations. These screens are by definition not screens for noncomplementation at all, because they begin with a semidominant or weakly poisonous antimorphic mutation and then screen for dominant enhancers of that mutation. (Recall that a true complementation test requires that both mutations be fully recessive!) In addition, one could expand the types of “bait” mutants employed in such screens to include the sorts of overexpression constructs created in Drosophila by Rorth and her colleagues (Rorth et al. 1998) or conversely to screen a collection of overexpressing lines for their ability to enhance a given “threshold” mutation.
For example, in Caenorhabditis elegans Yook et al. (2001) observed strong interactions between heterozygous mutations in the genes encoding the physically interacting synaptic proteins UNC-13 and syntaxin/UNC-64, but only when at least one mutation encoded a partially functional, but weakly poisonous, gene product. In this instance, dominant enhancement was observed only if the altered gene products impair the protein complexes with which they are normally associated (Yook et al. 2001). These authors then tested mutant alleles of other genes, whose protein products were also known to interact with syntaxin/UNC-64, for their ability to interact with a poisonous allele of this gene. Although the strongest effects were observed between loci encoding gene products that bind to one another, interactions were also observed between proteins that do not directly interact but are members of the same complex. Yook et al. (2001) also observed strong dominant enhancement between genes that function at distant points in the same pathway, implying that physical interactions are not required for this process. However, mutations in genes that function in different processes such as neurotransmitter synthesis or synaptic development did complement one another. Thus, this genetic interaction was specific for genes acting in the process of synaptic vesicle trafficking.
A similar approach was taken by Dan Kiehart and his colleagues who used dominant enhancement to identify a group of genes whose protein products interact with nonmuscle myosin in Drosophila (Halsell and Kiehart 1998; Halsell et al. 2000). In Drosophila, the zipper (zip) gene encodes the nonmuscle myosin protein. Mutations in this gene produce malformed legs in the adult. Indeed, a missense allele (zip[Ebr]) proved useful as bait in a screen for dosage-sensitive enhancers or suppressors. Kiehart and colleagues scanned through 70% of the Drosophila genome by testing a large collection of deficiencies for their ability to modify the expression of a semidominant zip[Ebr] and identified three chromosomal deficiencies that strongly enhanced the phenotype associated with the zip[Ebr] mutation. More importantly, the interaction of those deletions with zipper can be explained as the result of reducing the dose of specific genes located within the regions defined by these deficiencies. These include genes encoding cytoplasmic myosin, collagen IV, and the signal transduction protein RhoA.
Similarly, Fehon and his collaborators identified a number of interactors for the Drosophila homolog of the human neurofibromatosis type 2 protein (NF2), a gene known as Merlin, by screening for dominant enhancers of semidominant effects on head, eye, wing, and leg morphology, of a dominant-negative allele of this gene (La Jeunesse et al. 2001). From a screen of 100,000 mutagenized chromosomes they recovered 29 dominant enhancers of Mer3, 23 of which fell into five complementation groups. In many cases these mutations also modified the expression of a hypomorphic Mer3 mutation as well. Finally, mutations in two of these complementation groups were shown to modify the cellular localization of wild-type Merlin protein and thus clearly do define genes whose products interact with Merlin.
But perhaps the most impressive use of this method has been the work by Rubin and his collaborators to dissect the role of the Ras signaling pathway in eye development in Drosophila. For example, Simon et al. (1991) performed an elegant screen for mutations in genes whose products interacted with the protein encoded by the sevenless gene in Drosophila. The product of the sevenless gene specifies a membrane receptor tyrosine kinase whose function is required for the proper differentiation of one type of photoreceptor cell (R7) that is present in each of the 800 repeating units that compose the fly's compound eye. Simon et al. (1991) used site-directed mutagenesis to screen for dominant enhancers of a temperature-sensitive sevenless allele. Twenty such enhancers, defining seven lethal complementation groups, were recovered from >30,000 treated chromosomes. The genes defined by these mutations encode critical components of a Ras-dependent signaling process that acts downstream of the Sevenless protein to mediate R7 differentiation.
Of course, not all such interactions will define interacting functions. Consider, for example, the case of Minute mutations in Drosophila, which define the genes that encode ribosomal proteins. There are numerous examples of mutations that would be recessive in a non-Minute background but cause a phenotype, even as a heterozygote, in the presence of the Minute mutation. Obviously, screens based on dominant enhancement, just like those based on canonical SSNC, must include rigorous secondary screens to filter out those genes that just alter the expression of the bait gene. Moreover, as we noted in the preceding section, the development of high-throughput screens, such as the RNAi-based screen for dominant enhancers recently carried out in C. elegans by Lehner et al. (2006), may provide a rapid method for identifying such mutations and facilitate their rapid characterization.
SUMMARY AND CONCLUSIONS
To quote Lewis (1967, p. 20), “In many situations the cis–trans test works well.” Indeed, we would go a bit further and argue that in the vast majority of cases the assignment of new mutations to individual genes by use of the complementation test works well. Nonetheless, misleading results can be obtained quite easily, as a result of the failure of mutations in different genes to complement. The existence of several fascinating examples of SSNC has led various investigators to perform a number of screens designed to identify functionally related genes on the basis of second-site noncomplementation. Unfortunately, such screens have not been terribly successful or at least not as successful as screens for dominant enhancers of weakly dominant alleles of the gene of interest.
Acknowledgments
The authors thank Michael Ashburner, Doug Bishop, Julie Brill, Kim Collins, James Crow, Susan Dutcher, Mel Green, Susan Flynn, Robb Krumlauf, Cathy Lake, Sue Jaspersen, Beth Jones, and Julia Richards for reading parts of this manuscript and/or for valuable discussions. We also thank Nina Kolich for help in preparing this manuscript. R.S.H. is an American Cancer Society (ACS) research professor and W.D.G. is supported by a postdoctoral fellowship from ACS. Both R.S.H. and W.D.G. thank Scott Page and other members of the Hawley lab for performing the mutant screen described here.
Footnotes
2However, we note that the online encyclopedia Wikipedia has decided the issue in favor of Ed Lewis (http://en.wikipedia.org/wiki/Complementation). That must surely settle the issue.
3However, it might well still be worth the effort of doing the cis component of the test in at least some cases—see the discussion below of cases of combined haplo-insufficiency involving tightly linked genes in the mouse.
4There are several well-described instances in which the phenotype of a double heterozygote for two mutations within a given gene is considerably more extreme than that of either of the two homozygotes. This phenomenon is referred to as “negative complementation” (Fincham 1966). Negative complementation presumably reflects the ability of the two abnormal protein products to form a dimer or multimer that is not only nonfunctional (as are homodimers of the two mutant proteins) but poisonous as well. For a modern example of this phenomenon in Drosophila see Bickel et al. (1997).
5The phenomenon of intragenic complementation is usually used to describe a case in which the protein of interest has two or more different and separable functional domains, such that a gene with a loss-of-function missense mutation in domain 1 might complement a missense mutation in domain 2. This can occur if the protein of interest has two or more separable functional domains. A loss-of-function missense mutation affecting domain 1 may fully complement a missense mutation affecting domain 2 since both domains are present in fully functional form in the doubly heterozygous diploid. A second form of intragenic complementation may occur even in proteins with single functional domains, so long as the protein contains two or more identical subunits. In the double heterozygote, the two differently mutant subunits are thought to mutually ameliorate their respective defects by entering into an oligomeric complex whose structure is more nearly normal than that found in either homozygote. Numerous examples of this phenomenon in lower eukaryotes have been described by Schlesinger and Levinthal (1965) and by Zabin and Villarejo (1975), but for a more recent consideration see Hehir et al. (2000). Examples of this phenomenon in eukaryotes have been described by Hawley and Walker (2003). However, a very different type of intragenic complementation is exhibited in cases in which one of the two mutations lies in an upstream regulatory element and the other lies within the coding region. These effects appear to reflect the ability of various enhancer-like elements to function in trans in an organism like Drosophila, which has ubiquitous somatic pairing. In Drosophila this type of intragenic complementation can be disrupted by heterozygosity for chromosomal rearrangements, a phenomenon referred to as transvection (for review see Morris et al. 1999 and Duncan 2002). Similar effects have been noted in a variety of organisms (Tartof and Henikoff 1991; Matzke 2001).
6This interpretation was confirmed once antibodies against Ald/Mps1 protein were available. Western blotting demonstrated that ovaries from mutant homozygous females have far lower amounts of ald protein than wild type, yet larval brains have normal amounts of protein (W. Gilliland and R. S. Hawley, unpublished data).
7As a postscript to this story, we decided to assign a name to CG18212. As this gene is uninvolved in anything of any importance to female meiosis, but two deceitful interlopers (the two Doc elements) attempted to mislead us into thinking that it was critically important, we decided to name this gene alt, which stands for aluminum tubes. Because when someone presents you with pictures of aluminum tubes and tells you that they are intended for nuclear uses, you must be very careful about the inferences you make.
8We feel compelled to point out that despite the heuristic value of this article in terms of its genetic elegance, the finding that α- and β-tubulin physically interact did not exactly come as a huge surprise to our colleagues in cell biology. Nonetheless, the finding of the α-tubulin mutant turned out to be of real value to yeast geneticists because, prior to this study, no viable mutant alleles of the α-tubulin gene were available. Moreover, Stearns and Botstein continued their efforts by screening for second-site noncomplementers of the α-tubulin mutation. This screen also yielded both a new cold-sensitive allele of the original β-tubulin gene, and, more critically, the first recovered point mutation in the second α-tubulin gene (TUB3). The only previous existing alleles of TUB3 were null mutations generated by gene disruption.
References
- Beall, E. L., and D. C. Rio, 1996. Drosophila IRBP/Ku p70 corresponds to the mutagen-sensitive mus309 gene and is involved in P-element excision in vivo. Genes Dev. 10: 921–933. [DOI] [PubMed] [Google Scholar]
- Benzer, S., 1955. Fine structure of a bacteriophage. Proc. Natl. Acad. Sci. USA 41: 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer, S., 1962. The fine structure of the gene. Sci. Am. 206: 70–84. [DOI] [PubMed] [Google Scholar]
- Biggins, S., N. Bhalla, A. Chang, D. L. Smith and A. W. Murray, 2001. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics 159: 453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel, S. E., D. W. Wyman and T. L. Orr-Weaver, 1997. Mutational analysis of the Drosophila sister-chromatid cohesion protein ORD and its role in the maintenance of centromeric cohesion. Genetics 146: 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns, B. R., N. L. Henry and R. D. Kornberg, 1996. TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol. Cell. Biol. 16: 3308–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case, M. E., and N. H. Giles, 1960. Comparative complementation and genetics maps of the pan-2 locus in Neurospora crassa. Proc. Natl. Acad. Sci. USA 46: 659–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, A. P., and M. R. Capecchi, 1996. A mutational analysis of the 5′ HoxD genes: dissection of genetic interactions during limb development in the mouse. Development 122: 1175–1185. [DOI] [PubMed] [Google Scholar]
- Debeer, P., C. Bacchelli, P. J. Scambler, L. De Smet, J. P. Fryns et al., 2002. Severe digital abnormalities in a patient heterozygous for both a novel missense mutation in HOXD13 and a polyalanine tract expansion in HOXA13. J. Med. Genet. 39(11): 852–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, I. W., 2002. Transvection effects in Drosophila. Annu. Rev. Genet. 36: 521–556. [DOI] [PubMed] [Google Scholar]
- Fincham, J., 1966. Genetic Complementation. Benjamin, New York.
- Fincham, J. R. S., 1958. The role of chromosomal loci in enzyme formation. Proceedings of the 10th International Congress of Genetics, pp. 355–363, Montreal.
- Firmenich, A. A., M. Elias-Arnanz and P. Berg, 1995. A novel allele of Saccharomyces cerevisiae RFA1 that is deficient in recombination and repair and suppressible by RAD52. Mol. Cell. Biol. 15(3): 1620–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromental-Ramain, C., X. Warot, N. Messadecq, M. LeMeur, P. Dollé et al., 1996. Hoxa13 and Hoxd13 play a crucial role in the patterning of the limb autopod. Development 122: 2997–3011. [DOI] [PubMed] [Google Scholar]
- Giles, N. H., 1958. Mutations at specific loci in Neurospora. Proceedings of the 10th International Congress of Genetics, pp. 261–279, Montreal.
- Gilliland, W. D., S. M. Wayson and R. S. Hawley, 2005. The meiotic defects of mutants in the Drosophila mps1 gene reveal a critical role of Mps1 in the segregation of achiasmate homologs. Curr. Biol. 15(7): 672–677. [DOI] [PubMed] [Google Scholar]
- Goldberg, A. F., and R. S. Molday, 1996. Defective subunit assembly underlies a digenic form of retinitis pigmentosa linked to mutations in peripherin/rds and rom-1. Proc. Natl. Acad. Sci. USA 93: 13726–13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, F. R., and P. J. Scambler, 2001. Human HOX gene mutations. Clin. Genet. 59: 1–11. [DOI] [PubMed] [Google Scholar]
- Green, L. L., N. Wolf, K. L. McDonald and M. T. Fuller, 1990. Two types of genetic interaction implicate the whirligig gene of Drosophila melanogaster in microtubule organization in the flagellar axoneme. Genetics 126: 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, M. M., 1961. Complementation at the yellow locus in Drosophila melanogaster. Genetics 46: 1385–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, M. M., 1990. The foundation of genetic fine structure: a perspective from memory. Genetics 124: 793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, M. M., and K. C. Green, 1949. Crossing over between alleles of the lozenge locus in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 35: 586–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, M. M., and K. C. Green, 1956. A cytogenetic analysis of the lozenge pseudoalleles in Drosophila melanogaster. Genet. Res. 1: 452–461. [DOI] [PubMed] [Google Scholar]
- Halsell, S. R., and D. P. Kiehart, 1998. Second-site noncomplementation identifies genomic regions required for Drosophila nonmuscle myosin function during morphogenesis. Genetics 148: 1845–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsell, S. R., B. I. Chu and D. P. Kiehart, 2000. Genetic analysis demonstrates a direct link between rho signaling and nonmuscle myosin function during Drosophila morphogenesis. Genetics 155: 1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L. H., R. K. Mortimer, J. Culotti and M. Culotti, 1973. Genetic control of cell division in yeast: V. Genetic analysis of cdc mutants. Genetics 74: 267–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley, R. S., and M. Y. Walker, 2003. Advanced Genetic Analysis. Blackwell Science, Malden, MA.
- Hays, S. L., A. A. Firmenich, P. Massey, R. Banerjee and P. Berg, 1998. Studies of the interaction between Rad52 protein and the yeast single-stranded DNA binding protein RPA. Mol. Cell. Biol. 18(7): 4400–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays, T. S., R. Deuring, B. Robertson, M. Prout and M. T. Fuller, 1989. Interacting proteins identified by genetic interactions: a missense mutation in alpha-tubulin fails to complement alleles of the testis-specific beta-tubulin gene of Drosophila melanogaster. Mol. Cell. Biol. 9: 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehir, M. J., J. E. Murphy and E. R. Kantrowitz, 2000. Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic complementation. J. Mol. Biol. 304(4): 645–656. [DOI] [PubMed] [Google Scholar]
- Jaspersen, S. L., B. J. Huneycutt, T. H. Giddings, K. A. Resing, N. G. Ahn et al., 2001. Cdc28/Cdk1 regulates spindle pole body duplication through phosphorylation of Spc42 and Mps1. Dev Cell. 7(2): 263–274. [DOI] [PubMed] [Google Scholar]
- Kajiwara, K., E. L. Berson and T. P. Dryja, 1994. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science 264: 1604–1608. [DOI] [PubMed] [Google Scholar]
- Kedzierski, W., S. Nusinowitz, D. Birch, G. Clarke, R. R. McInnes et al., 2001. Deficiency of rds/peripherin causes photoreceptor death in mouse models of digenic and dominant retinitis pigmentosa. Proc. Natl. Acad. Sci. USA. 98(14): 7718–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano, K., D. M. Johnson-Schlitz and W. R. Engels, 2001. Sterility of Drosophila with mutations in the Bloom syndrome gene—complementation by Ku70. Science 291: 2600–2602. [DOI] [PubMed] [Google Scholar]
- LaJeunesse, D. R., B. M. McCartney and R. G. Fehon, 2001. A systematic screen for dominant second-site modifiers of Merlin/NF2 phenotypes reveals an interaction with blistered/DSRF and scribbler. Genetics 158: 667–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner, B., C. Crombie, J. Tischler, A. Fortunato and A. G. Fraser, 2006. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 18: 896–903. [DOI] [PubMed] [Google Scholar]
- Lewis, E. B., 1951. Pseudoallelism and gene evolution. Cold Spring Harbor Symp. Quant. Biol. 15: 151–172. [DOI] [PubMed] [Google Scholar]
- Lewis, E. B., 1967. Genes and gene complexes, pp. 17–47 in Heritage From Mendel, edited by E. A. Brink. University of Wisconsin Press, Madison, WI.
- Matzke, A. J., 2001. A test for transvection in plants: DNA pairing may lead to trans-activation or silencing of complex heteroalleles in tobacco. Genetics 158: 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J. R., P. K. Geyer and C. T. Wu, 1999. Core promoter elements can regulate transcription on a separate chromosome in trans. Genes Dev. 13: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounkes, L. C., and M. T. Fuller, 1999. Molecular characterization of mutant alleles of the DNA repair/basal transcription factor haywire/ERCC3 in Drosophila. Genetics 152: 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounkes, L. C., R. S. Jones, B. C. Liang, W. Gelbart and M. T. Fuller, 1992. A Drosophila model for xeroderma pigmentosum and Cockayne's syndrome: haywire encodes the fly homolog of ERCC3, a human excision repair gene. Cell 71: 925–937. [DOI] [PubMed] [Google Scholar]
- Muller, H. J., 1932. Further studies on the nature and causes of gene mutations. Proceedings of the 6th International Congress of Genetics, Cornell University, Ithaca, NY, Vol. I, pp. 213–255.
- Nusslein-Volhard, C., E. Wieschaus and H. Kluding, 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Roux's Arch. Dev. Biol. 193: 267–282. [DOI] [PubMed] [Google Scholar]
- Pontecorvo, G., 1958. Trends in Genetic Analysis. Columbia University Press, New York.
- Rancourt, D. E., T. Tsuzuki, and M. R. Capecchi, 1995. Genetic interaction between hoxb-5 and hoxb-6 is revealed by nonallelic noncomplementation. Genes Dev. 9: 108–122. [DOI] [PubMed] [Google Scholar]
- Rasooly, R. S., C. M. New, P. Zhang, R. S. Hawley and B. S. Baker, 1991. The lethal(1)TW-6cs mutation of Drosophila melanogaster is a dominant antimorphic allele of nod and is associated with a single base change in the putative ATP-binding domain. Genetics 129: 409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan, C. L., and M. T. Fuller, 1988. Interacting genes that affect microtubule function: the nc2 allele of the haywire locus fails to complement mutations in the testis-specific beta-tubulin gene of Drosophila. Genes Dev. 2: 82–92. [DOI] [PubMed] [Google Scholar]
- Rorth, P., K. Szabo, A. Bailey, T. Laverty, J. Rehm et al., 1998. Systematic gain-of-function genetics in Drosophila. Development 125: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Schlesinger, M. J., and C. Levinthal, 1965. Complementation at the molecular level of enzyme interaction. Annu. Rev. Microbiol. 19: 267–284. [DOI] [PubMed] [Google Scholar]
- Simon, M. A., D. D. Bowtell, G. S. Dodson, T. R. Laverty and G. M. Rubin, 1991. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell 67: 701–716. [DOI] [PubMed] [Google Scholar]
- Stern, C., and E. W. Schaeffer, 1943. On primary attributes of alleles in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 29: 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns, T., and D. Botstein, 1988. Unlinked noncomplementation: isolation of new conditional-lethal mutations in each of the tubulin genes of Saccharomyces cerevisiae. Genetics 119: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, W. S., 1935. Allelomorphic phenomena. Dros. Inf. Serv. 4: 62–63. [Google Scholar]
- Tartof, K. D., and S. Henikoff, 1991. Trans-sensing effects from Drosophila to humans. Cell 65: 201–203. [DOI] [PubMed] [Google Scholar]
- Vinh, D. B., M. D. Welch, A. K. Corsi, K. F. Wertman and D. G. Drubin, 1993. Genetic evidence for functional interactions between actin noncomplementing (Anc) gene products and actin cytoskeletal proteins in Saccharomyces cerevisiae. Genetics 135: 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, M. D., D. B. Vinh, H. H. Okamura and D. G. Drubin, 1993. Screens for extragenic mutations that fail to complement act1 alleles identify genes that are important for actin function in Saccharomyces cerevisiae. Genetics 135: 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, P., B. D. Peyser, X. Pan, J. D. Boeke, F. A. Spencer et al., 2005. Gene function prediction from congruent synthetic lethal interactions in yeast. Mol. Syst. Biol. 1: 2005.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook, K. J., S. R. Proulx and E. M. Jorgensen, 2001. Rules of nonallelic noncomplementation at the synapse in Caenorhabditis elegans. Genetics 158: 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabin, I., and M. R. Villarejo, 1975. Protein complementation. Annu. Rev. Biochem. 44: 295–313. [DOI] [PubMed] [Google Scholar]