Abstract
To explore the potential involvement of aberrant Notch1 signaling in breast cancer pathogenesis, we have used a transgenic mouse model. In these animals, mouse mammary tumor virus LTR-driven expression of the constitutively active intracellular domain of the Notch1 receptor (N1IC) causes development of lactation-dependent mammary tumors that regress upon gland involution but progress to nonregressing, invasive adenocarcinomas in subsequent pregnancies. Up-regulation of Myc in these tumors prompted a genetic investigation of a potential Notch1/Myc functional relationship in breast carcinogenesis. Conditional ablation of Myc in the mammary epithelium prevented the induction of regressing N1IC neoplasms and also reduced the incidence of nonregressing carcinomas, which developed with significantly increased latency. Molecular analyses revealed that both the mouse and human Myc genes are direct transcriptional targets of N1IC acting through its downstream Cbf1 transcriptional effector. Consistent with this mechanistic link, Notch1 and Myc expression is positively correlated by immunostaining in 38% of examined human breast carcinomas.
Keywords: breast cancer, mouse model
The Notch signaling pathway controls cell fates through interactions between neighboring cells by positively or negatively affecting, and in a context-dependent fashion, processes of proliferation, differentiation, and apoptosis (1, 2). In mammals, each of four Notch receptors (Notch1–4) is synthesized as a precursor that is proteolytically processed to a cell membrane heterodimer. Two additional cleavages in response to ligand interaction release the Notch intracellular domain (NIC), which translocates to the nucleus and modulates the expression of target genes predominantly by binding and converting the ubiquitous Cbf1 repressor (also known as Csl, Rbp-Jk, Rbpsuh, etc.) to a transcriptional activator (3).
Deregulation of Notch signaling has been implicated in the development of lymphoid neoplasms, neuroblastomas, and various epithelial cancers, including mammary tumors (4, 5). However, in contrast to some human malignancies, such as leukemia/lymphoma, in which an involvement of aberrant Notch function has been established (6), evidence suggesting a role of this pathway in breast cancer is only now emerging. Thus, appreciable expression of Notch1 has been detected in the majority of examined ductal carcinoma in situ cases (see ref. 5) and in ductal carcinomas (7) but not in normal breast tissue. Interestingly, high Notch1 expression in breast cancers correlated significantly with poor survival of patients (8). The potential involvement of Notch1 in breast cancer pathogenesis was strengthened by the observation that the Notch1 antagonist Numb was greatly reduced or absent in more than half of examined carcinomas, and this correlated with increased Notch1 signaling (9). Moreover, the proliferative potential of cells derived from such tumors was dramatically suppressed by pharmacological inhibition of Notch1 (9). In addition to these correlations, compelling evidence for tumorigenic action of Notch in the mammary epithelium has been derived from animal studies (10–12).
Previously, we generated a mouse model in which all mammary glands of transgenic females expressing a constitutively active Notch1IC (N1IC) transgene driven by the mouse mammary tumor virus (MMTV) LTR develop lactation-dependent papillary tumors that are noninvasive and regress upon gland involution (12). After additional pregnancies, however, multifocal invasive (nonregressing) tumors also appear, apparently evolving from remnants of N1IC-induced in situ neoplasms through the occurrence of secondary tumorigenic events.
By using a combination of genetic and molecular analyses, we started investigating the mechanisms involved in the pathogenesis of N1IC-induced tumors and report here that one of the events mediating the development of premalignant neoplasms and later collaborating in the evolution of nonregressing invasive carcinomas is the direct transcriptional activation of Myc by Notch1.
Results
Up-Regulation of Myc Expression in N1IC-Induced Mammary Tumors.
By using microarray analysis, we compared a set of expression profiles of murine mammary tumors. In addition to N1IC-induced regressing and nonregressing neoplasms, we examined adenocarcinomas caused either by expression of MMTV LTR-driven oncogenes, including polyomavirus middle T antigen (13), Myc (14), Hras1 (15), and activated Erbb2 (16), or by ablation of the tumor-suppressors p53 and Brca2 (17). Our results (data not shown) revealed that the N1IC tumors exhibited a high degree of profile similarity only with the carcinomas induced by Myc overexpression. Moreover, differentially expressed transcripts corresponding to transcriptional target genes known to be activated or repressed by Myc were commonly detected in the profiles of Myc- and N1IC-induced tumors (see Fig. 6 and Tables 3 and 4, which are published as supporting information on the PNAS web site). Notably, in addition to MMTV-Myc-induced cancers, Myc expression was found to be increased in regressing and nonregressing N1IC neoplasms but not in the other examined tumor types. These observations were confirmed by Northern blot analysis (Fig. 1A). The data showed that the expected high level of N1IC transgene expression in regressing and nonregressing tumors resulted in up-regulation of known Notch1 transcriptional targets, such as Hes1 and Hey1 (18), and was paralleled by high Myc transcript levels (Fig. 1A). However, the amount of Myc transcripts was not as massive as that observed in the case of MMTV-Myc-induced tumors (Fig. 1B). The results of Western blot analyses were in agreement with these observations (data not shown).
Fig. 1.
Molecular characterization of N1IC-induced mammary tumors. (A) Northern blot analysis of total cell RNA from WT mammary glands of virgin (V), pregnant (P), lactating (L), and postinvolutional (PI) female mice and from regressing (R) and nonregressing (NR) N1IC-induced mammary tumors (two specimens of each class). The same membrane was sequentially hybridized (without stripping), with specific probes detecting the indicated RNA species. The relative Myc levels in the controls were approximately V 1.0, P 2.0, L 1.0, and PI 1.5. There was an ≈7-fold increase in Myc expression in regressing tumors relative to the amount in normal lactating glands. A similar Myc increase (≈7.5-fold on average) was detected in nonregressing carcinomas compared with corresponding control postinvolutional glands. Expression of the transgenic N1IC resulted in up-regulation of the endogenous Notch1 mRNA (row mN1, lanes 5–8). The level of mN1 transcripts in the control specimens was below detection limits by Northern blot analysis under our conditions. Hybridization to 18S rRNA (loading control) was performed to normalize the data for quantitation by using a PhosphorImager (Molecular Dynamics). (B) Northern blot analysis shows that the level of Myc mRNA expression in MMTV-Myc-induced carcinomas (≈45-fold greater than normal) is approximately six times higher than that found in nonregressing N1IC tumors. The transcript derived from the MMTV-Myc transgenic construct (14) is longer than the endogenous Myc mRNA. Lanes 1 and 2 are the same as Myc lanes 7 and 8 in A (different exposure times). (C) Southern blot analysis of EcoRI-digested DNA from a lactating mammary gland of a female mouse with an MMTV-N1IC/Mycfl/fl/Wapcre genotype after a second pregnancy, to assess the level of Cre-mediated DNA excision (lane 2) from the “floxed” (fl) Myc locus (generation of Δ allele). The control DNA (floxed allele; lane 1) was prepared from a nonregressing tumor of an animal with the same genotype. (D) Comparative Northern blot analysis of Myc mRNA expression levels at 2 weeks postpartum between N1IC-induced regressing tumors developed in females carrying MMTV-N1IC in functional Myc background (R; lanes 1 and 2; same as Myc lanes 5 and 6 in A) and tumor-free lactating glands of MMTV-N1IC/Mycfl/fl/Wapcre mice, in which Cre-mediated recombination (r) at the Myc locus had occurred. RNA was extracted from glands after the first pregnancy (lanes 3 and 4) or after a second pregnancy (lane 5).
Conditional Ablation of Myc in Mammary Glands Prevents Development of Palpable N1IC-Induced Regressing Tumors.
To examine whether the detected Myc up-regulation in N1IC neoplasms is functionally significant, we used cre/loxP-based mutagenesis with homozygous Myc conditional mutants (Mycfl/fl; ref. 19) and Wapcre mice (Wapcre/+ or Wapcre/cre; ref. 17), in which cre is transcribed specifically in alveolar and ductal mammary epithelial cells during late pregnancy and lactation by the Wap gene regulatory elements (20). Thus, our experimental mice (MMTV-N1IC/MycΔfl/fl/Wapcre or MMTV-N1IC/Mycfl/fl/Wapcre; n = 13) would acquire, upon pregnancy/lactation, a mammary epithelium deficient in Myc expression (MycΔfl/Δfl). As controls (n = 32), we used females carrying the MMTV-N1IC transgene and possessing an Myc+/+, Mycfl/+, or Mycfl/fl genotype but lacking Wapcre. We note that the mammary glands of MycΔfl/Δfl mice lacking the N1IC transgene were devoid of phenotypic manifestations.
By their third pregnancy, all control mice developed lactation-dependent regressing tumors that were readily detectable by visual inspection or palpation. In contrast, palpable masses were absent from the experimental animals, with a single exception (regressing tumor frequency 1/13 in experimental animals vs. 32/32 in controls; P < 10−6; χ2 test).
Southern blot analysis of lactating gland DNA indicated that the level of Cre-mediated excision was ≈40% (Fig. 1C), but this is an underestimate, because the tissue includes elements (stromal cells, adipocytes, etc.) in which the Wap promoter is inactive. Moreover, the pattern of Wap expression is normally mosaic (20, 21). Consistent with these considerations and in agreement with previous reports (22, 23), Northern blot analysis indicated that, after Cre-mediated recombination, the level of residual Myc expression in tumor-free lactating glands (“rescued” by Myc ablation) did not exceed ≈20% of the amount detected in regressing neoplasms developed in WT Myc background. (Fig. 1D).
We conclude that the development of N1IC-induced palpable regressing tumors depends on unperturbed Myc activity and that residual Myc expression in a small fraction of the epithelial cell population is inadequate for manifestation of a gross phenotype. However, histopathological analysis revealed the presence of microscopic neoplastic lesions that increased in number and size with parity round. Thus, the lactating glands of primiparous experimental females (2 weeks postpartum) contained rare and miniscule solid foci of noninvasive neoplastic lesions, in contrast to WT controls (Fig. 2Aa). After a second pregnancy, several multifocal noninvasive neoplasms were detected both in alveoli and ducts (Fig. 2Ab). However, these neoplastic foci were fewer and, on average, 3.7 times smaller than those seen in regressing neoplasms developing in MMTV-N1IC animals possessing WT Myc (Fig. 2Ac). Apparently, the detected microlesions originate from a small minority of cells that escape the Wap-cre action and retain unrecombined, expressible Myc. This conclusion is supported by the following three immunostaining patterns of Notch1 and Myc. In control lactating glands, there is a positive Notch1 signal (Fig. 2Ba) but no immunodetectable Myc (Fig. 2Bd). In contrast, in regressing tumors induced by N1IC in a WT Myc background, the neoplastic areas and the adjacent normal alveoli are positive for both Notch1 and Myc (data not shown). After widespread Myc deletion, however, only the noninvasive microlesions in second-lactation mammary specimens from experimental mice exhibit nuclear staining for Myc, whereas no signal is detectable in adjacent nonneoplastic alveoli (Fig. 2Be). As expected, the Notch1-positive pattern remains unchanged (Fig. 2Bb). The presence of microlesions correlated with a high level of proliferation, as assessed by proliferating cell nuclear antigen immunostaining, (50% positivity vs. 10% in adjacent nonneoplastic tissue; data not shown). We note that Myc heterozygosity in the mammary epithelium did not affect the development of palpable regressing neoplasms (n = 13).
Fig. 2.
Histopathological analysis and immunophenotyping of mammary tumors. (A) Comparison of sections of lactating mammary glands (hematoxylin/eosin staining). (Magnification: ×20.) The specimens were from animals carrying the Notch1IC transgene, in which the Myc gene was either conditionally ablated (a and b) or functional (control) (c). The glands were isolated at 2 weeks postpartum after a first (a) or a second (b and c) pregnancy. After a first pregnancy, only miniscule and rare solid intraalveolar lesions occupying <1% of the surface area were observed in experimental animals (Inset). (Magnification: ×400.) After a second pregnancy, however, several small papillary lesions were detected (b, outlined). Although they are morphologically the same as those found in controls (c, outlined), the lesions of experimental mice were fewer and occupied collectively 4.2% of the alveolar surface area vs. 14.5% in controls (area of each lesion 0.21 ± 0.16 and 0.78 ± 0.45 mm2, respectively; P < 10−6). (B) Immunohistochemical staining (brown reaction product) for Notch1IC and Myc of WT lactating glands, and regressing and nonregressing tumors from mice with an MMTV-N1IC/Mycfl/fl/Wapcre genotype. (Magnification: ×400.) In contrast to the positive Notch1 immunoreactivity, normal lactating glands lack Myc immunostaining. However, there is highly positive Myc immunodetection in nonneoplastic epithelial cells of transgenic mice expressing N1IC (d Inset, same genotype as in Ac). In the regressing tumor (same as in Ab) there is strong nuclear staining for Notch1 in the neoplasm (b, arrow) and also in the nonneoplastic tissue (b, arrowheads). In contrast to a mosaic pattern of Myc immunoreactivity in the regressing tumor (e, arrows), the adjacent nonneoplastic tissue is Myc-negative (e, arrowheads). The Notch1IC-induced nonregressing carcinoma shows strong labeling for both Notch1 (c) and Myc (f). (C) Morphological patterns of nonregressing tumors developing in transgenic animals carrying MMTV LTR-driven Notch1IC (a) or Myc (b) or in bitransgenic mice expressing both oncogenes (c). (Magnification: ×400.) The histological architecture of Notch1IC-induced tumors is mostly papillary (a, arrows), whereas the Myc tumors are composed of small glandular elements and nests (b, arrowheads). The carcinomas of bitransgenic animals exhibit a hybrid architecture of interspersed small glandular elements (c, arrowhead) and large solid papillary structures (c, arrow). At the cellular level (Insets), the Myc tumors consist of large cells with large irregular nuclei and exhibit many apoptotic bodies (b Inset, arrow), whereas the Notch1 tumors have smaller cells with uniform nuclei and no apoptotic bodies (a Inset). (Magnification: ×1,000.) The cellular features of the cancers in bitransgenic mice resemble those of the Myc tumors, including the presence of apoptotic bodies (c Inset, arrow). (D) An example of a human breast cancer (serial sections) exhibiting strong nuclear immunoreactivity for both NotchIC (a) and Myc (b). (Magnification: ×400.) Histologically, this carcinoma has a growth pattern of small, solid, irregular nests (c, hematoxylin/eosin staining). (Scale bars: A, 1,000 μm; Aa Inset, B, C, and D, 50 μm; C Insets, 20 μm.)
Reduced Incidence and Increased Latency in the Development of N1IC-Induced Nonregressing Mammary Tumors upon Conditional Ablation of Myc.
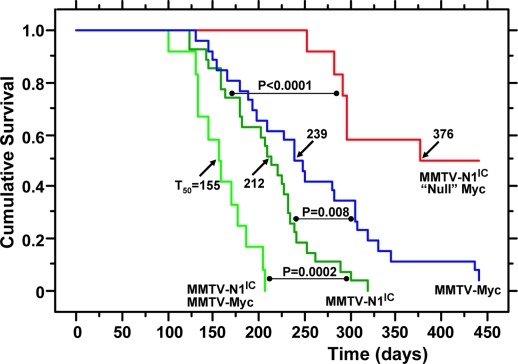
To examine the impact of Myc absence on the development of nonregressing tumors, we monitored, over a period of 15 months, the described three cohorts of N1IC transgenic animals that differed in Myc genotype. Control mice (n = 27) possessed functional Myc alleles, whereas, in experimental animals, the Myc alleles were either conditionally ablated (n = 12) or present in the heterozygous state (n = 13) in mammary glands. The results of this genetic analysis are summarized in Fig. 3.
Fig. 3.
Kaplan–Meier tumor-free mouse survival curves. The survival of mice (from the date of birth until detection of palpable N1IC-induced nonregressing tumors) is compared by a standard rank test between controls carrying MMTV-N1IC and possessing functional Myc (dark green) and females with MMTV-N1IC/MycDfl/Dfl/Wapcre genotype (red) in which the majority of the mammary epithelium has become “null” for Myc expression by Cre-mediated recombination. Also shown for further comparisons are survival curves of MMTV-Myc transgenic (blue) and MMTV-N1IC/MMTV-Myc bitransgenic (light green) animals.
Nonregressing tumors appeared in controls with a half-time for tumor-free survival (T50) of 7 months (Fig. 3), and Myc heterozygosity did not change this latency (data not shown). In contrast, only half (6/12) of the animals with conditionally ablated Myc developed nonregressing carcinomas after a much longer latency period (Fig. 3; T50 = 12.5 months; P < 0.0001, log-rank test). Moreover, these tumors were focal and rarely involved more than one breast, whereas the invasive carcinomas in control animals were multifocal and involved several mammary glands, as reported in ref. 12.
Histopathological analysis and immunostaining showed that the invasive nonregressing tumors had apparently emerged from precancerous lesions originating from Myc-retaining cells. Thus, although all cancer cell nuclei were strongly positive for N1IC as expected (Fig. 2Bc), a significant fraction of them also exhibited positive Myc immunostaining (Fig. 2Bf). In fact, we were unable to detect histologically identifiable invasive lesions totally lacking immunodetectable Myc. Moreover, a MycΔflox allele was undetectable by Southern blot analysis of DNA extracted from nonregressing tumors of animals possessing an MMTV-N1IC/Mycfl/fl/Wapcre genotype (Fig. 1C). As expected, the cells of the nonregressing tumors were highly proliferative (≈50% of the cells were positive for proliferating cell nuclear antigen immunostaining; data not shown). We also used immunophenotyping to examine whether deregulation of proliferation and inhibition of apoptosis could be correlated with differences between N1IC-induced tumors and controls in the status of important markers for these processes (see Fig. 7, which is published as supporting information on the PNAS web site).
Our genetic results clearly demonstrate that Myc is indispensable for the development of N1IC nonregressing mammary carcinomas appearing with a latency (T50 = 7 months) significantly shorter than that of tumors developing in MMTV-Myc transgenic mice (T50 = 8 months; P = 0.008; n = 26; Fig. 3). Thus, the N1IC oncogenic process is faster, despite a level of up-regulated Myc that is six times less than that observed in MMTV-Myc animals (Fig. 1B).
To examine the consequences of very high Myc expression in the context of nonregressing tumor development induced by N1IC, we also monitored MMTV-N1IC/MMTV-Myc bitransgenic females (n = 12) for the development of carcinomas and observed that they appeared with a T50 of 5 months (Fig. 3). This acceleration in tumor progression, which can be attributed to unusually high Myc levels, apparently intensifying the actions of N1IC, revealed that the two oncogenic transgenes, in addition to their functional relationship, possess other collaborating but nonoverlapping activities. Interestingly, histopathological analysis showed that the tumors of the bitransgenic mice had a “hybrid” morphological pattern, exhibiting features characteristic of both N1IC- and Myc-induced carcinomas (Fig. 2C).
The Myc Gene Is a Direct Notch1 Transcriptional Target.
Considering the causal involvement of Myc overexpression in N1IC-induced oncogenesis revealed by our genetic analysis, we thought that Myc might be a direct transcriptional target of Notch1. To test this hypothesis, we searched ≈1.7 kb of mouse Myc 5′ flanking sequence upstream from the start site of the most frequently used promoter, P2, (24) and identified three putative Cbf1 binding sites (A–C) with a YTGGGAA motif (Fig. 4A), which only slightly deviates from the canonical RTGGGAA consensus (3).
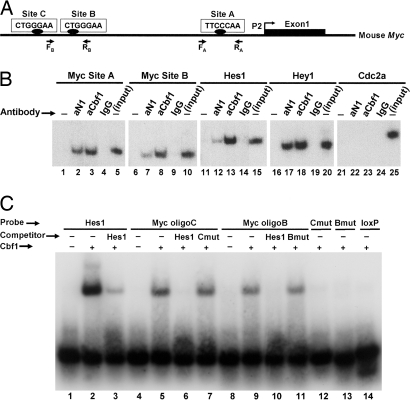
Fig. 4.
Analysis of Cbf1-binding sites in the mouse Myc promoter. (A) Shown is a diagram of the mouse Myc gene 5′ flanking region upstream from the first exon transcribed by activation of the most frequently used promoter P2. The positions of putative Cbf1-binding sites (sites A–C) and of the PCR primers used in ChIP experiments are indicated. (B) ChIP assays. In the examples shown, chromatin from a nonregressing N1IC-induced tumor was immunoprecipitated with antibodies against Notch1 (aN1) or Cbf1 (aCbf1) or with control rabbit IgG and analyzed by PCR using primers specific for the indicated promoters. The Myc promoter was analyzed for sites A and B. Hes1 and Hey1 were examined as positive controls; Cdc2a served as a negative control. “Input” corresponds to products generated by PCR (still in exponential phase; 25 cycles) by using DNA extracted from nonimmunoprecipitated chromatin (10% of the amount used in experimental samples) as a template. −, no antibody was added to the reaction mixture. (C) EMSA was performed by using a recombinant Cbf1 protein fragment and the indicated labeled oligonucleotide probes. Myc oligoB and oligoC span, respectively, the corresponding Cbf1 sites shown in A. An oligonucleotide representing two known Cbf1-binding sites of the Hes1 promoter was used as a positive control. (We attribute the shift of two bands to the presence of these two sites.) Mutated versions of Myc oligonucleotides B (Bmut) and C (Cmut) and an unrelated loxP oligonucleotide served as negative controls. In the competition assays, unlabeled Hes1, Bmut, and Cmut oligonucleotides were added to the binding mixture at a 50-fold molar excess.
To examine whether Cbf1 interacts with the Myc promoter region, we performed chromatin immunoprecipitation (ChIP) assays using anti-Notch1 and -Cbf1 antibodies. As Fig. 4B illustrates, these specific antibodies, but not a negative control IgG, were able to immunoprecipitate chromatin, which was enriched in promoter sequences of Myc and of the known Notch1 target genes Hes1 and Hey1 (positive controls), from N1IC neoplasms. In contrast, the Cdc2a promoter (negative control) was not recognized, although this gene is also overexpressed in N1IC-induced tumors (data not shown). We note that, in addition to the promoter region, there is a putative Cbf1-binding site in the middle of Myc exon 1, and two other such sites are present in intron 2 (≈200 and 300 bp from the beginning of the intron). None of these sites is conserved in the human Myc locus, and our ChIP assays did not detect enrichment for the sequences that contain them (data not shown), further emphasizing the specificity of binding of a N1IC–Cbf1 complex in the promoter region.
To establish formally the identity of the candidate Cbf1-binding sites in the Myc promoter, we performed EMSAs using, as probes or competitors, double-stranded oligonucleotides representing intact or mutated versions (3-bp substitutions) of the putative Cbf1 sites B and C (Fig. 4A) and a purified fragment of recombinant mouse Cbf1 (amino acids 203–393) that includes the DNA-binding domain (25). Fig. 4C shows that the Cbf1 protein fragment was successfully bound to WT duplexes of sites C and B (Myc oligoC and oligoB, respectively) and also to a duplex oligonucleotide containing two adjacent, experimentally verified Cbf1-binding sites of the Hes1 promoter region (positive control). In contrast, mutated versions of the C and B duplexes (Cmut and Bmut) and an unrelated double-stranded DNA fragment corresponding to a loxP site (negative control), failed to form complexes with the Cbf1 fragment. Moreover, an excess of unlabeled Hes1 oligonucleotide, which, as expected, competed successfully with the cognate Hes1-labeled probe for binding to Cbf1, was also able to out-compete the labeled C and B oligonucleotide probes, whereas the unlabeled mutated versions Cmut and Bmut used in excess were ineffective as competitors.
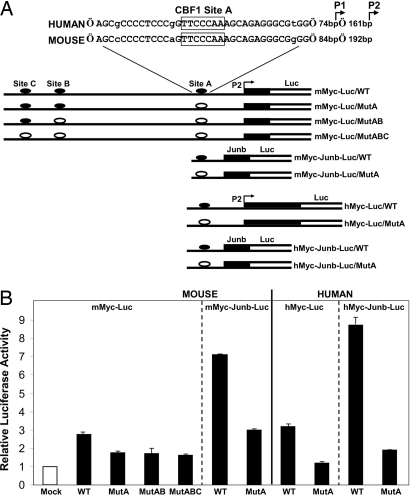
To confirm that the Cbf1 binding sites of the Myc promoter are functional, we performed luciferase reporter assays using 293T cells. In our reporter (mMyc-Luc/WT; Fig. 5A), a fragment of the mouse Myc promoter (−1,662/+181 in regard to the P2 promoter start site) was connected to the firefly luciferase gene. Reporter DNA was cotransfected with an effector gene plasmid expressing either N1IC or a chimeric Cbf1-VP16 protein (26) functioning as a transcriptional activator independently of Notch signaling. Although the reporter responded to both N1IC and Cbf-VP16, the latter effector proved to be a more potent activator. As shown in Fig. 5B, cotransfection of Cbf-VP16 with the mMyc-Luc/WT reporter resulted reproducibly to an average increase of 2.8-fold in luciferase activity compared with that attained in the absence of an effector (empty pcDNA3 vector control). The corresponding normalized activities of reporter constructs in which one, two, or three of the Cbf1-binding sites were mutated (MutA, MutAB, and MutABC; Fig. 5A) were consistently ≈40% lower (P = 0.003, Student’s t test; Fig. 5B).
Fig. 5.
Luciferase reporter assays. (A) Alignment of a region of the human and mouse Myc promoter sequences containing a recognition site for Cbf1 binding (site A). The start sites of promoters P1 and P2 are indicated. Segments of the plasmid constructs used as reporters are shown (for definition of binding sites, see Fig. 4A). In the diagrams, the putative Cbf1-binding sites (closed ovals) are intact, whereas, in the other constructs, one or more sites have been mutagenized (open ovals), as indicated. Constructs in which shorter segments of the mouse and human Myc promoters were used in association with a minimal Junb gene promoter (hatched rectangle) are also shown. (B) Results of reporter assays. 293T cells were cotransfected with Renilla luciferase plasmid and an effector plasmid expressing Cbf1-VP16 or a control plasmid (empty pcDNA3 vector; mock) in combination with one of the indicated reporter plasmids shown in A containing either intact or mutated Cbf1-binding sites. After normalization to Renilla luciferase activity, firefly luciferase activity relative to that of the control plasmid was calculated for each of the reporters. These relative values (mean ± SEM), measured in at least three independent experiments, are represented by the bars in the bar chart.
By using the same functional assay, we examined whether the human Myc is also a Notch1 target. Alignment of the mouse and human Myc promoter sequences indicated that the Cbf1-binding site A (TTCCCAA) was located in the middle of a highly conserved nucleotide stretch (33/36 base identities; Fig. 5A), whereas strong conservation in the regions of sites B and C was not evident (alignment not shown). In the reporters, the luciferase gene was driven by human Myc regulatory sequences (−389/+352) that included an intact or a mutated version of site A (hMyc-Luc/WT and hMyc-Luc/MutA, respectively). Each of these reporters was cotransfected either with the N1IC or the Cbf1-VP16 effector plasmid into 293T cells (Cbf1-VP16 again achieved higher stimulation levels). Cotransfection of hMyc-Luc/WT and Cbf-VP16 resulted in a 3.2-fold increase, on average, in luciferase activity over the level attained with the empty vector. When site A was mutagenized and the hMyc-Luc/MutA reporter was used in the assay, only a negligible response (1.2-fold relative stimulation) was observed (P = 0.001; Student’s t test; Fig. 5B). To investigate further the functional significance of binding site A while attempting to minimize the dependence of promoter activity on transcription factors other than the N1IC/Cbf1 complex, we generated additional reporters. This time, to drive luciferase gene expression, we linked to a minimal promoter of the Junb gene (−42/+136; see ref. 27) only short segments of the mouse (−366/−138) and human (−390/−133) Myc sequences containing site A (constructs mMyc-Junb-Luc/WT and hMyc-Junb-Luc/WT, respectively; Fig. 5A). Robust responses to Cbf1-VP16 were observed with both reporters (≈7-fold and ≈9-fold increase in luciferase activity for the mouse and human constructs, respectively), which were diminished upon mutation of site A (see Fig. 5B).
In summary, when considered together, our three types of molecular analysis provide unequivocal evidence that, at least in mammals, Myc is a direct Notch1 transcriptional target. Whether N1IC also exerts indirect effects contributing to Myc overexpression during tumor progression cannot be ruled out, but there is no straightforward experiment to test this possibility.
Correlation of Notch1 and Myc Expression in Human Breast Cancer.
Clearly, the potential involvement of deregulated Notch1 expression in the development of human breast cancer merits exploration. Thus, we attempted a preliminary examination of potential Notch1 and Myc coexpression in human breast carcinomas and analyzed immunohistochemically a human breast cancer tissue array by using antibodies that specifically recognize the nuclear form of either Notch1 (N1IC) or Myc. In both cases, only readily detectable nuclear staining was considered as relevant for scoring (intensity levels are defined in Methods).
First, we evaluated the presence vs. the absence of immunostaining in the carcinomas (n = 128), regardless of signal intensity (Table 1). Statistical analysis (Fisher’s exact test) indicated that the null hypothesis that the two genes are expressed independently can be overwhelmingly rejected (P < 10−6). The same conclusion can be reached by an independent test focusing only on the commonly positive cases (n = 49; Table 2) and by evaluating the frequencies of occurrence of the four possible combinations for high and low expression levels of Notch1 and Myc in the examined carcinomas. Again, the statistical analysis indicated that the observed expression levels were not independent but exhibited a high degree of correlation (P = 0.01). Overall, in >90% of the cases (20/22) exhibiting high Myc expression, Notch1 was also highly expressed (for an example, see Fig. 2D).
Table 1.
Immunophenotyping of human breast carcinomas exhibiting negative/positive staining
| Myc | Notch 1 |
|
|---|---|---|
| Negative | Positive | |
| Negative | 51 | 18 |
| Positive | 10 | 49 |
The numbers of cancer specimens in a tissue array that exhibit the indicated immunostaining intensities (defined in Materials and Methods) for Notch1IC and Myc are listed. For a statistical analysis of the Notch1/Myc immunostaining combinations, see Results. All specimens, n = 128. Double-positive specimens, n = 49.
Table 2.
Immunophenotyping of human breast carcinomas exhibiting low/high signals
| Myc | Notch 1 |
|
|---|---|---|
| Low | High | |
| Low | 12 | 15 |
| High | 2 | 20 |
The numbers of cancer specimens in a tissue array that exhibit the indicated immunostaining intensities (defined in Materials and Methods) for Notch1IC and Myc are listed. For a statistical analysis of the Notch1/Myc immunostaining combinations, see Results. All specimens, n = 128. Double-positive specimens, n = 49.
Discussion
We have shown conclusively that Myc is a direct transcriptional target of aberrant Notch1 signaling, playing an indispensable role in Notch1IC-induced murine mammary tumorigenesis. It is likely that this relationship is special, because Myc ablation in mammary glands cannot rescue the development of tumors induced by the polyomavirus middle T antigen (S. Koul, K.P., and A.E., unpublished data). Previous in vitro experiments indicated that retroviral transduction of N1IC into mammalian cells (28, 29) results in up-regulation of Myc expression, but a direct Notch1/Cbf1 effect on Myc promoter activity was not demonstrated.
In our genetic experiments, N1IC was unable to cause significant hyperproliferation of the majority of mammary epithelial cells deprived of Myc action, and development of palpable regressing tumors failed during the first pregnancy/lactation, although a minor fraction of cells had escaped Cre-mediated DNA excision. Even after a second pregnancy, only a few islands of noninvasive neoplastic structures composed of residual Myc-positive cells were encountered. Presumably, such precancerous lesions give rise to nonregressing tumors appearing with reduced incidence and increased latency, perhaps because of a diminished “target size” for the occurrence of fruitful secondary carcinogenic events. Our initial attempts to identify such events were unsuccessful. Thus, when we examined N1IC nonregressing tumors for either Kras (n = 8) or Hras1 (n = 10) potential mutations in codons 12, 13, and 61, differences from WT were not detected.
However, although necessary, Myc is not the sole downstream effector of N1IC oncogenic action, considering that lactation-dependent regressing neoplasms do not appear in MMTV-Myc transgenic mice, whereas their carcinomas develop with a latency longer than that of N1IC-induced nonregressing tumors, despite a 6-fold higher level of Myc expression. Thus, in addition to Myc, N1IC also engages other effector(s) toward attainment of a preinvasive and, eventually, a malignant state. Nevertheless, highly expressed MMTV LTR-driven Myc appears to exert more pleiotropic effects than Myc up-regulation by way of Notch1/Cbf1 action, as can be surmised from the significant reduction in latency of nonregressing tumors caused by the combined actions of MMTV-N1IC and MMTV-Myc transgenes.
It is thought that, because Notch signaling controls cell fate decisions and blocks differentiation depending on cellular context, it could promote tumorigenesis by analogous action when deregulated (4). Such a model of arrested differentiation appears consistent with the mammary oncogenic activity of constitutively expressed Notch4IC that leads to dysmorphogenesis, preventing the appearance of milk-producing lobules (30). Clearly, however, the model is not tenable for Notch1IC, because the process of mammary differentiation remains unperturbed, and the lactating females are capable of nursing their pups despite the appearance of regressing tumors. A proliferative rather than a differentiation defect is therefore a more likely working model for the development of N1IC tumors, especially in view of the crucial participation of Myc. In fact, based on the key observation that morphologically identifiable Myc-negative invasive lesions were not detected, we propose that at least the proliferative function of Myc (directly demonstrated by positive proliferating cell nuclear antigen staining) is an obligatory component of the circuitry that, in the context of N1IC-mediated events, must remain intact as a favorable precondition for the eventual manifestation of full-fledged malignancy.
Our results provide still another example of the importance of modeling cancer in mice and of the powerful combination of genetic and molecular analyses to establish causal and mechanistic relationships between concerted tumorigenic activities in aberrant signaling networks, such as those involving Notch1 and Myc. Moreover, the unraveling of a Notch1/Myc relationship by using our mouse model prompted an investigation that documented a strong association between high levels of Notch1 and Myc expression in human breast cancers.
Materials and Methods
Mice.
Mice carrying targeted Myc alleles in which exons 2 and 3 are flanked (“floxed,” fl) by directly oriented loxP sites (Mycfl; ref. 19) were kindly provided by Fred Alt (Harvard University, Boston) and crossed with MMTV-N1IC animals (12). MMTV-Myc mice (14) were obtained from Charles River Laboratories. Our Hs-cre1 (31) and Wap-cre (17) strains of mice have been described previously. Our breeding program is described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Molecular Analyses.
ChIP experiments were performed as described in ref. 32. For luciferase assays, we used pGL3 and the Dual Luciferase Assay kit (Promega). The point of reference for all mouse and human Myc sequence coordinates is the promoter P2 start site. Microarray analyses with Affymetrix (Santa Clara, CA) MGU74Av2 DNA chips were performed as described in ref. 33. Southern and Northern blot analyses and descriptions of probes, ChIP assays, EMSA, and the plasmids used for luciferase assays can be found in Supporting Materials and Methods.
Histological Analyses.
The antibodies used for immunophenotyping are listed in Supporting Materials and Methods. The results of tissue array immunostaining were scored as “negative” (absence of nuclear staining), “weakly positive” (low, <15% of the nuclei exhibiting strong immunoreactivity), and “strongly positive” (high, ≥15% of the nuclei exhibiting intense immunoreactivity).
Supplementary Material
Acknowledgments
We thank Stavros Lomvardas for invaluable help with the ChIP experiments; Fred Alt, Rhett Kovall (University of Cincinnati), Wayne Hendrickson (Columbia University, New York, NY), Jon Aster (Harvard University), Evelyne Manet (Institut National de la Santé de la Recherche Medicale, Lyon, France), and Kathryn Calame (Columbia University) for mice and reagents; Thomas Ludwig for information; Joe Terwilliger for advice on statistical analysis; Giorgio Catorretti for advice on Myc immunostaining; Ben Tycko for critical reading of the manuscript; and Guoying Chen, Thomas Kollar, Qiong Li, and Xi Sun for expert technical assistance. This work was supported by National Cancer Institute Grant 1P01 CA97403 (Project 2) (to A.E.). Additional support was provided by grants to A.E. and M.S. from the Avon Products Foundation Breast Cancer Research and Care Program through the Herbert Irving Comprehensive Cancer Center of the Columbia Medical Center. A.K. was supported by a Fellowship from the Jane Coffin Childs Memorial Fund for Medical Research.
Abbreviations
- ChIP
chromatin immunoprecipitation
- MMTV
mouse mammary tumor virus
- NIC
Notch intracellular domain
- T50
half-time of tumor-free survival.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Artavanis-Tsakonas S., Rand M. D., Lake R. J. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Lai E. C. Development (Cambridge, U.K.) 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 3.Honjo T. Genes Cells. 1996;1:1–9. doi: 10.1046/j.1365-2443.1996.10010.x. [DOI] [PubMed] [Google Scholar]
- 4.Radtke F., Raj K. Nat. Rev. Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 5.Politi K., Feirt N., Kitajewski J. Semin. Cancer Biol. 2004;14:341–347. doi: 10.1016/j.semcancer.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Pear W. S., Aster J. C. Curr. Opin. Hematol. 2004;11:426–433. doi: 10.1097/01.moh.0000143965.90813.70. [DOI] [PubMed] [Google Scholar]
- 7.Weijzen S., Rizzo P., Braid M., Vaishnav R., Jonkheer S. M., Zlobin A., Osborne B. A., Gottipati S., Aster J. C., Hahn W. C., et al. Nat. Med. 2002;8:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 8.Reedijk M., Odorcic S., Chang L., Zhang H., Miller N., McCready D. R., Lockwood G., Egan S. E. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 9.Pece S., Serresi M., Santolini E., Capra M., Hulleman E., Galimberti V., Zurrida S., Maisonneuve P., Viale G., Di Fiore P. P. J. Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dievart A., Beaulieu N., Jolicoeur P. Oncogene. 1999;18:5973–5981. doi: 10.1038/sj.onc.1202991. [DOI] [PubMed] [Google Scholar]
- 11.Callahan R., Egan S. E. J. Mammary Gland Biol. Neoplasia. 2004;9:145–163. doi: 10.1023/B:JOMG.0000037159.63644.81. [DOI] [PubMed] [Google Scholar]
- 12.Kiaris H., Politi K., Grimm L. M., Szabolcs M., Fisher P., Efstratiadis A., Artavanis-Tsakonas S. Am. J. Pathol. 2004;165:695–705. doi: 10.1016/S0002-9440(10)63333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guy C. T., Cardiff R. D., Muller W. J. Mol. Cell. Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart T. A., Pattengale P. K., Leder P. Cell. 1984;38:627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- 15.Sinn E., Muller W., Pattengale P., Tepler I., Wallace R., Leder P. Cell. 1987;49:465–475. doi: 10.1016/0092-8674(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 16.Muller W. J., Sinn E., Pattengale P. K., Wallace R., Leder P. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 17.Ludwig T., Fisher P., Murty V., Efstratiadis A. Oncogene. 2001;20:3937–3948. doi: 10.1038/sj.onc.1204512. [DOI] [PubMed] [Google Scholar]
- 18.Iso T., Kedes L., Hamamori Y. J. Cell. Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 19.de Alboran I. M., O’Hagan R. C., Gartner F., Malynn B., Davidson L., Rickert R., Rajewsky K., DePinho R. A., Alt F. W. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 20.Robinson G. W., McKnight R. A., Smith G. H., Hennighausen L. Development (Cambridge, U.K.) 1995;121:2079–2090. doi: 10.1242/dev.121.7.2079. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed F., Wyckoff J., Lin E. Y., Wang W., Wang Y., Hennighausen L., Miyazaki J., Jones J., Pollard J. W., Condeelis J. S., Segall J. E. Cancer Res. 2002;62:7166–7169. [PubMed] [Google Scholar]
- 22.Xu X., Wagner K. U., Larson D., Weaver Z., Li C., Ried T., Hennighausen L., Wynshaw-Boris A., Deng C. X. Nat. Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 23.Walton K. D., Wagner K. U., Rucker E. B., III, Shillingford J. M., Miyoshi K., Hennighausen L. Mech. Dev. 2001;109:281–293. doi: 10.1016/s0925-4773(01)00549-4. [DOI] [PubMed] [Google Scholar]
- 24.Asselin C., Nepveu A., Marcu K. B. Oncogene. 1989;4:549–558. [PubMed] [Google Scholar]
- 25.Kovall R. A., Hendrickson W. A. EMBO J. 2004;23:3441–3451. doi: 10.1038/sj.emboj.7600349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waltzer L., Bourillot P. Y., Sergeant A., Manet E. Nucleic Acids Res. 1995;23:4939–4945. doi: 10.1093/nar/23.24.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakae K., Nakajima K., Inazawa J., Kitaoka T., Hirano T. J. Biol. Chem. 1995;270:23795–23800. doi: 10.1074/jbc.270.40.23795. [DOI] [PubMed] [Google Scholar]
- 28.Rao P., Kadesch T. Mol. Cell. Biol. 2003;23:6694–6701. doi: 10.1128/MCB.23.18.6694-6701.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satoh Y., Matsumura I., Tanaka H., Ezoe S., Sugahara H., Mizuki M., Shibayama H., Ishiko E., Ishiko J., Nakajima K., Kanakura Y. J. Biol. Chem. 2004;279:24986–24993. doi: 10.1074/jbc.M400407200. [DOI] [PubMed] [Google Scholar]
- 30.Smith G. H., Gallahan D., Diella F., Jhappan C., Merlino G., Callahan R. Cell Growth Differ. 1995;6:563–577. [PubMed] [Google Scholar]
- 31.Dietrich P., Dragatsis I., Xuan S., Zeitlin S., Efstratiadis A. Mamm. Genome. 2000;11:196–205. doi: 10.1007/s003350010037. [DOI] [PubMed] [Google Scholar]
- 32.Parekh B. S., Maniatis T. Mol. Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 33.Politi K., Szabolcs M., Fisher P., Kljuic A., Ludwig T., Efstratiadis A. Am. J. Pathol. 2004;164:325–336. doi: 10.1016/S0002-9440(10)63122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.