Abstract
Plants use sophisticated strategies to balance responses to oxidative stress. Programmed cell death, including the hypersensitive response (HR) associated with successful pathogen recognition, is one cellular response regulated by reactive oxygen in various cellular contexts. The Arabidopsis basic leucine zipper (bZIP) transcription factor AtbZIP10 shuttles between the nucleus and the cytoplasm and binds consensus G- and C-box DNA sequences. Surprisingly, AtbZIP10 can be retained outside the nucleus by LSD1, a protein that protects Arabidopsis cells from death in the face of oxidative stress signals. We demonstrate that AtbZIP10 is a positive mediator of the uncontrolled cell death observed in lsd1 mutants. AtbZIP10 and LSD1 act antagonistically in both pathogen-induced HR and basal defense responses. LSD1 likely functions as a cellular hub, where its interaction with AtbZIP10 and additional, as yet unidentified, proteins contributes significantly to plant oxidative stress responses.
Keywords: basal defense, Hyaloperonospora parasitica, hypersensitive cell death, nuclear shuttling, reactive oxygen
Introduction
Rapid, regulated responses to abiotic and biotic stress are a requirement of sessile plant life. These are often mediated by signal-dependent activation of latent transcriptional regulators. The well-characterized G-box (5′-CACGTG-3′) and C-box (5′-GACGTC-3′) plant cis-elements are found in different combinatorial arrangements in promoters of genes regulated by developmental and environmental signals. Proteins that bind to these DNA elements are basic region leucine zipper (bZIP) transcription factors (TF). bZIP TFs function in the regulation of seed maturation and germination and in the integration of spatial and temporal information during floral induction (Siberil et al, 2001; Jakoby et al, 2002; Lara et al, 2003; Wigge et al, 2005). Members of the Arabidopsis ABF bZIP subfamily modulate responses to the hormone ABA and to glucose (Kim et al, 2004). Arabidopsis AtbZIP60 regulates the tunicamycin-induced endoplasmic reticulum (ER) stress response (also called the unfolded protein response). Activation of AtbZIP60 by ER stress might include translocation to the nucleus following its release from the plasma membrane (Iwata and Koizumi, 2005).
The activities of parsley bZIP TFs PcCPRF1, PcCPRF2 and PcCPRF4a are differentially regulated at both the transcriptional (Feldbrügge et al, 1994) and post-translational levels in response to light treatment (Wellmer et al, 1999, 2001). PcCPRF2 is localized to the cytoplasm of dark-adapted parsley cells, and imported into the nucleus in response to phytochrome action (Kircher et al, 1999). Nuclear uptake is preceded by rapid R/FR-dependent phosphorylation of PcCPRF2 in the cytoplasm (Wellmer et al, 1999). However, reverse genetic approaches are not feasible in parsley and thus the primary function of these bZIP TFs remains unclear.
We therefore extended our analysis to orthologous bZIP TFs in Arabidopsis. Four Arabidopsis bZIP proteins form a clade most closely related to PcCPRF2. We demonstrate that one, AtbZIP10, is actively shuttled out of the nucleus, likely via exportin. AtbZIP10, but not the other members of this clade, interacts in vivo with LSD1 (Lesions Simulating Disease resistance 1), a plant-specific zinc-finger protein. LSD1 is a negative regulator of cell death that protects plant cells from reactive oxygen-induced stress (Dietrich et al, 1994, 1997; Jabs et al, 1996; Mateo et al, 2004). The AtbZIP10–LSD1 interaction occurs in the cytoplasm, resulting in partial AtbZIP10 retention. We demonstrate genetically that AtbZIP10 is a positive regulator of pathogen-induced hypersensitive response (HR), basal defense responses and reactive oxygen-induced cell death, and that these activities are antagonized by LSD1. Our data suggest that LSD1 controls the cell death-related transcriptional activity of AtbZIP10 via altering its intracellular partitioning. Our genetic evidence demonstrates that there must be additional positive regulators of cell death that are antagonized by LSD1.
Results
Four of the 75 predicted Arabidopsis bZIP proteins define the group C (parsley CPRF2-like) clade most closely related to PcCPRF2 (AtbZIP9, AtbZIP10, AtbZIP25 and AtbZIP63; Supplementary Figure 1). Polypeptide sequence alignment of these Arabidopsis bZIPs with PcCPRF2 revealed common bZIP TF motifs: a DNA-binding basic region, an extended leucine zipper responsible for homo- and heterodimerization, bipartite nuclear localization sequences and four domains idiosyncratic for group C bZIPs (Supplementary Figure 1A). The group C Arabidopsis bZIPs, including AtbZIP10, bind G-box elements (5′-CACGTG-3′; Lara et al, 2003). We extended these findings to the related C-box element (5′-GACGTC-3′; Supplementary Figure 1). Competition experiments with excess non-labeled native or mutated G-box (5′-CACTGG-3′) or C-box (5′-GCAGTC-3′) oligonucleotides revealed that the binding of both by AtbZIP10 required intact ACGT core sequences (Supplementary Figure 1C; data not shown).
We analyzed the intracellular distribution of AtbZIP10-GFP and AtbZIP63-GFP in transiently transformed, dark-incubated parsley and Arabidopsis protoplasts. AtbZIP63-GFP was exclusively localized to the nucleus, but AtbZIP10-GFP was observed in both the nucleus and the cytoplasm (parsley protoplasts, Figure 1A; Arabidopsis protoplasts, Figure 4C, upper row). Irradiation of transformed protoplasts for 16 h with white, red, far-red or blue light did not induce any change in the intracellular distribution of AtbZIP10-GFP (C Näke and K Harter, unpublished). We concluded that signal(s) other than light control the nuclear access of AtbZIP10, or that its distribution is uncontrolled.
Figure 1.
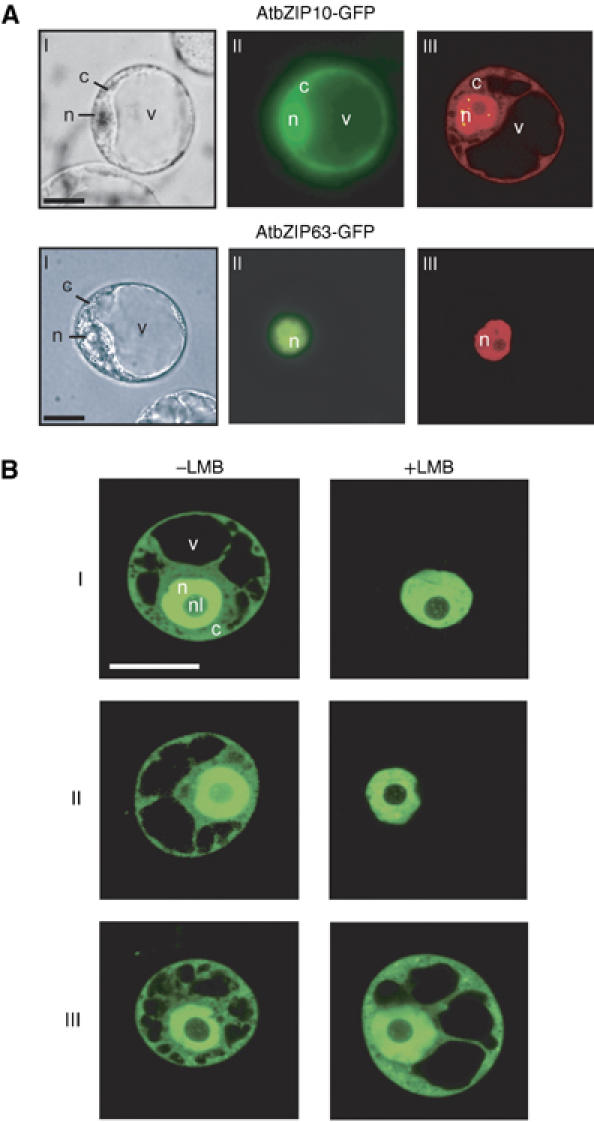
AtbZIP10 shuttles between the nucleus and the cytoplasm in plant protoplasts. (A) Bright-field (I), epifluorescence (II) and confocal laser scanning images (III) of parsley protoplasts expressing AtbZIP10-GFP or AtbZIP63-GFP. Images in I and II show identical protoplasts, and confocal images (III) are from independent cells. c, cytoplasm; n, nucleus; v, vacuole. Scale bars, 20 μm. Confocal images are false colored red. (B) Confocal laser scanning images of transiently transformed tobacco BY-2 protoplasts expressing GFP-NLS-CHS-NES (I), AtbZIP10-GFP (II) or GFP (III). Protoplasts, 16 h after transformation, were treated for 4 h with 2 μM of the nuclear export inhibitor LMB (+LMB) or mock treated (−LMB). c, cytoplasm; n, nucleus; nl, nucleolus; v, vacuole. Scale bar, 30 μm.
Figure 4.
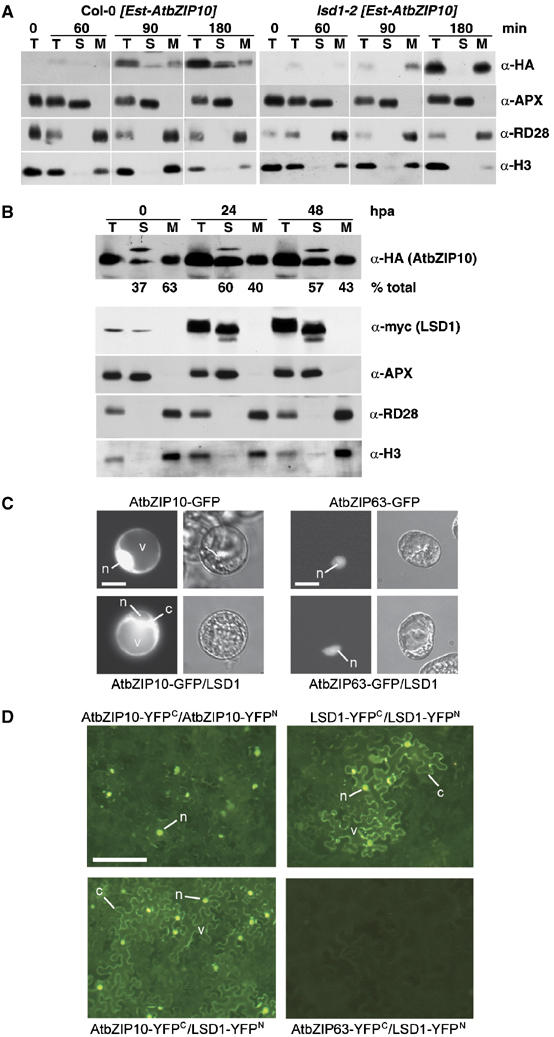
LSD1 can alter AtbZIP10 distribution and the two proteins can interact in planta. (A) F1 plants from (lsd1-2 [Est-AtbZIP10-HA] × Col-0) (left) or (lsd1-2 [Est-AtbZIP10-HA] × lsd1-2) (right) were sprayed with 20 μM estradiol. Tissue was harvested at 0, 60, 90 and 180 min after application of estradiol. Protein was extracted in sucrose buffer and fractionated. Data from one of two independent experiments are shown. (B) F1 plants from (35S-AtbZIP10-HA × Est-LSD1-myc) were sprayed with 20 μM estradiol. Tissue was harvested at 0, 24 and 48 h after application of estradiol. Protein was extracted in sucrose buffer and fractionated. Relative amounts of AtbZIP10 protein in the soluble fraction or the microsomal and nucleus-enriched fractions were determined using Image J and are presented as percent of total AtbZIP10 protein at each time point. One of three independent experiments is shown. (A, B) T, total extract; S, soluble fraction; M, microsomal and nucleus-enriched fraction; α-HA, detects HA-epitope-tagged AtbZIP10 protein; α-myc, myc-epitope-tagged LSD1 protein; α-APX, soluble ascorbate peroxidase; α-RD28, membrane-specific RD28 marker; α-H3, nuclear histone H3 protein; min, minutes after application; hpa, hours post estradiol application. (C) Arabidopsis protoplasts were transiently transformed with P35S:AtbZIP10-GFP or P35S-AtbZIP63-GFP either alone (upper row) or in combination with PLSD1:LSD1 (lower row). One day after transfection, protoplasts were analyzed by epifluorescence microscopy (see text). One of two independent experiments is shown. Representative images are shown. n, nucleus; c, cytoplasm; v, vacuole. Scale bars, 20 μm. (D) BiFC using AtbZIP10 and LSD1 in N. benthamiana epidermal leaf cells. Leaves were infiltrated with mixtures of Agrobacterium suspensions carrying plasmids encoding the indicated C-terminal (YFPC) or N-terminal (YFPN) YFP fusions of AtbZIP10, LSD1 and, as a negative control, AtbZIP63. One to two days after infiltration, the epidermal cells were analyzed by epifluorescence microscopy. An infiltrated epidermal area of 1–2 cm2 was scanned for cells showing a BiFC signal (see text). The experiments were repeated three times with similar outcomes. Representative images are shown. n, nucleus; c, cytoplasm; v, vacuole. Scale bars, 60 μm.
We anticipated that AtbZIP10 might be shuttled between the cytoplasm and the nucleus. We transiently expressed either AtbZIP10-GFP, an artificial shuttling control protein called GFP-NLS-CHS-NES (comprised of GFP, a nuclear localization signal (NLS), chalcone synthase (CHS) and a nuclear export signal (NES); Haasen et al, 1999) or GFP in protoplasts treated with the nuclear export inhibitor leptomycin B (LMB; Kudo et al, 1998). LMB limited AtbZIP10-GFP and the GFP-NLS-CHS-NES protein to the nucleus, but did not influence the localization of passively diffusing GFP (Figure 1B). Thus, AtbZIP10 is actively shuttled from the nucleus to the cytoplasm under these conditions. This conclusion is supported by our finding that AtbZIP10 interacts with Arabidopsis nuclear export factor XPO1 (Haasen et al, 1999) in the yeast two-hybrid system. The first 105 amino acids of AtbZIP10 are required for interaction with XPO1, suggesting that the NES is localized within this region (Supplementary Figure 2). We propose that an active retention mechanism might interfere with NLS-mediated nuclear import of AtbZIP10, and hence control its distribution.
LSD1, a negative regulator of plant cell death, can block AtbZIP10 nuclear function in yeast
To identify proteins that might retain AtbZIP10 in the cytoplasm, we extended the nuclear transportation trap (NTT) system for retention screening in yeast (Ueki et al, 1998; Figure 2A). Yeast cells producing LexAD-AtbZIP10 were transformed with an Arabidopsis cDNA expression library. Transformants were replica plated onto media without leucine. Clones displaying reduced growth on this media should contain plasmids encoding a cytoplasmic LexAD-AtbZIP10 retention factor (Figure 2B). From 80 000 replica-plated transformants, one clone reproducibly reduced the growth of LexAD-AtbZIP10-expressing yeast on media lacking leucine. It encoded LSD1, a negative regulator of plant cell death (Dietrich et al, 1994). Yeast expressing LexAD-AtbZIP10 exhibited reduced growth in the presence of LSD1 (Figure 2B, upper panel), but grew equally well on non-selective media (Figure 2B, lower panel). Thus, expression of LSD1 with LexAD-AtbZIP10 does not alter yeast viability. LSD1 did not alter the nuclear activity of LexAD-NLS or LexAD-AtbZIP63 (Figure 2C). We used the NTT system to define the yeast compartment where LSD1 exerts its function on AtbZIP10. Yeast expressing LexAD-LSD1 on media selective for nuclear activity did not grow (Figure 2D), indicating that LSD1 is predominantly cytoplasmic in yeast, interacts with AtbZIP10 outside of the nucleus, and retains it there.
Figure 2.
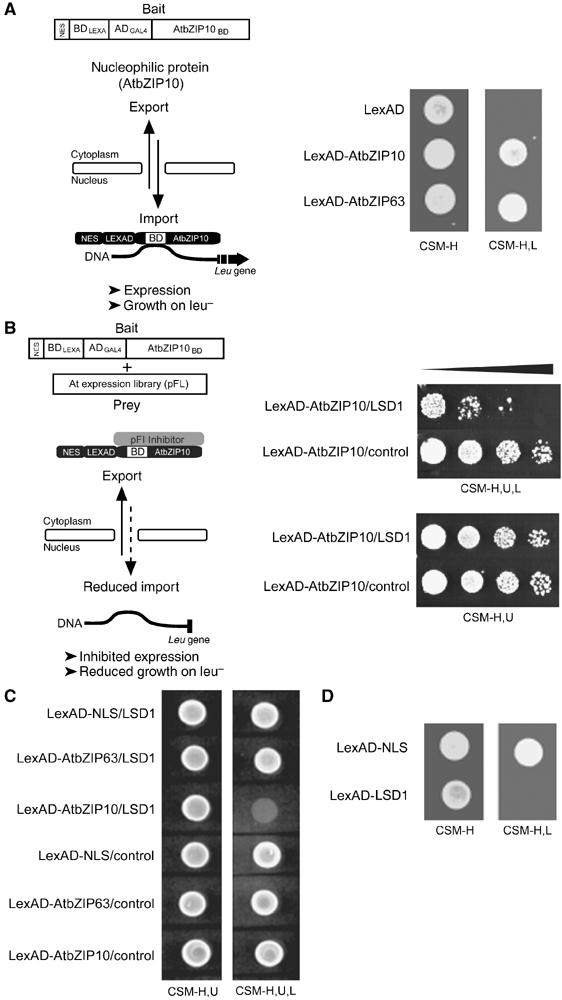
LSD1 expression prevents AtbZIP10 nuclear function in yeast. (A) AtbZIP10 and AtbZIP63 are transported into the nucleus in yeast. Schematic representation of the yeast NTT system (left panel). AtbZIP10 and AtbZIP63 were expressed in yeast (strain EGY48) as NES-BDLEXA-ADGAL4 (LexAD) fusions. Nuclear accumulation of LexAD-AtbZIP10 and LexAD-AtbZIP63, caused by the bZIP factor's NLS, induces the Leu reporter gene activity, enabling the growth of yeast cells on media without Leu (leu–, right panel). (B) Genetic screening for proteins that retain AtbZIP10 in the yeast cytosol. One of 11 colonies showed reproducibly reduced growth on CSM-H,U,L and contained a cDNA encoding full-length LSD1 (right panel). AtbZIP10/control: yeast EGY48 coexpressing LexAD-AtbZIP10 and a control protein. (C) Cytoplasmic retention activity of LSD1 in yeast is specific for AtbZIP10. Constructs encoding the indicated proteins were cotransformed into yeast strain EGY48. Nuclear localization was determined by comparative growth assay on plasmid-selective CSM-H,U media or on CSM-H,U,L media selective for nuclear accumulation (see Materials and methods). (D) LSD1 localizes to the yeast cytoplasm. The constructs encoding the indicated LexAD fusions were transformed in yeast strain EGY48. Nuclear localization was determined by comparative growth assay on CSM-H and CSM-H,L as described in (C).
LSD1 interacted with AtbZIP10 in yeast, but not with AtbZIP63, AtbZIP25 or PcCPRF2 (Figure 3A). The second and third zinc-fingers of LSD1 were required for this interaction, yet neither of these zinc-fingers alone, nor the small domain between them, was sufficient for it (Figure 3B). The C-terminal region of AtbZIP10 was sufficient for interaction with LSD1 (Figure 3C). This C-terminal domain is the least conserved region among the class C bZIP TFs (Supplementary Figure 1A), potentially explaining why AtbZIP10 is the only member of this clade that interacts with LSD1.
Figure 3.
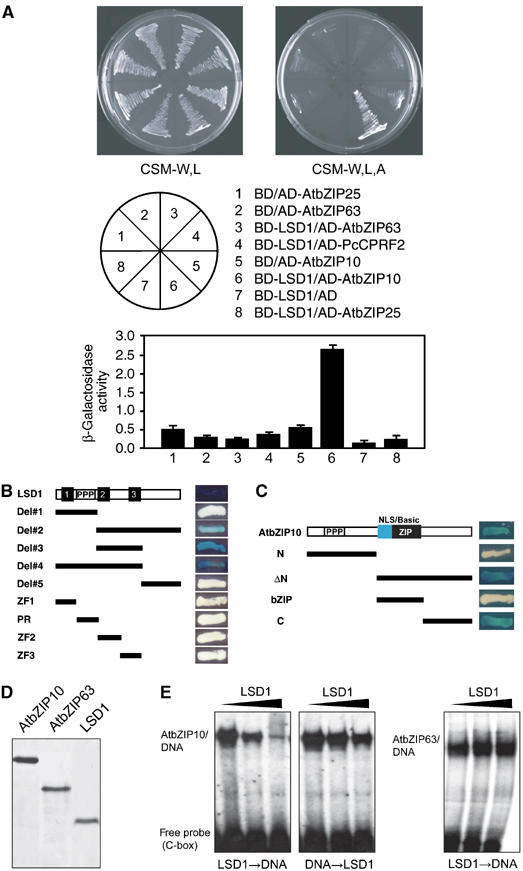
LSD1 interacts specifically with AtbZIP10, and inhibits the in vitro DNA binding capacity of AtbZIP10. (A) Specific interaction of LSD1 with AtbZIP10 tested in the yeast two-hybrid system. The indicated Gal4 DNA binding (BD) and Gal4 transactivation (AD) fusion constructs were cotransformed into PJ69-4A. Activity of reporter genes was determined either by growth on interaction selective CSM-L,W,A media or by quantitative β-galactosidase activity assay (LacZ). Error bars are s.d. (n>3). (B) Identification of the LSD1 domain required for interaction with AtbZIP10. The BD constructs encoding the indicated LSD1 polypeptides and the AD construct of full-length AtbZIP10 were cotransformed into EGY48[p2op-lacZ]. Interaction of the fusion proteins was tested using a β-galactosidase assay. 1, 2, 3 indicate the three zinc fingers, and PPP a proline-rich motif, in LSD1. (C) Identification of the AtbZIP10 domain required for interaction with LSD1. The AD constructs encoding the indicated AtbZIP10 polypeptides and the BD construct of full-length LSD1 were cotransformed and examined for protein–protein interaction as described in (B). NLS/basic indicates the NLS-containing DNA-binding domain, ZIP the leucine zipper and PPP the proline-rich transactivation domain of AtbZIP10. (D) Coomassie brilliant blue staining of SDS–PAGE-separated (His)6-tagged AtbZIP10, AtbZIP63 and LSD1 used in this study. Recombinant proteins were expressed in E. coli and purified by Ni-NTA affinity chromatography. (E) The DNA binding capacity of AtbZIP10 is inhibited by LSD1. EMSA of AtbZIP10 following coincubation with increasing amounts of LSD1 (black triangle) either before (LSD1 → DNA) or after (DNA → LSD1) the addition of radioactively labeled C-box DNA (left panels). As a control, the same experiment (LSD1 → DNA) was performed with AtbZIP63 (right panel). AtbZIP/DNA complexes are indicated.
The AtbZIP NLS sequences are near the amino terminus and overlap the basic DNA-binding domain (Supplementary Figure 1A), yet it is the AtbZIP10 C-terminus that interacts with LSD1 (Figure 3C). We tested whether LSD1 could interfere with AtbZIP10 DNA binding in vitro. Recombinant (His)6-tagged LSD1, AtbZIP10 and AtbZIP63 proteins were purified on Ni-NTA (Figure 3D). Incubation of LSD1 with AtbZIP10 before addition of labeled C-box oligonucleotide reduced AtbZIP10 DNA binding (Figure 3E, left). In contrast, only very weak inhibition of DNA binding was observed when AtbZIP10 was mixed with the C-box oligonucleotide before addition of LSD1 (Figure 3E, middle). Addition of LSD1 to C-box oligonucleotide did not block subsequent AtbZIP63 DNA binding (Figure 3E, right). BSA could not substitute for LSD1 (not shown). Thus, interaction of LSD1 with the AtbZIP10 C-terminus can hinder the NLS located within the basic DNA-binding domain. LSD1 is not able to actively remove AtbZIP10 once the latter is bound to DNA.
LSD1 can retain AtbZIP10 in the cytoplasm of plant cells
Epitope-tagged LSD1, driven by its native promoter, is soluble and complements an lsd1 null allele (Supplementary Figure 3A–D). We could not detect epitope-tagged AtbZIP10 expressed from its native promoter reliably; so we resorted to either conditional expression or constitutive expression from the viral 35S promoter of cauliflower mosaic virus. In the latter case, HA-epitope-tagged AtbZIP10 complements an atbzip10 null allele (Supplementary Figure 3E), and accumulates to approximately 10-fold lower levels than does AtbZIP10 expressed conditionally (not shown).
We conditionally overexpressed AtbZIP10 both in wild-type Col-0 and lsd1 mutant plants and studied the localization of AtbZIP10 after cell fractionation (Figure 4A). In Col-0 [Est-AtbZIP10-HA] plants, AtbZIP10 is detectable both in the soluble (S) and the microsomal-nuclear fraction (M) at 90 and 180 min after induction (Figure 4A, left). In lsd1-2 [Est-AtbZIP10] plants, AtbZIP10 localized exclusively to the nuclear-enriched microsomal fraction (Figure 4A, right), indicating that LSD1 normally slows the accumulation of newly synthesized AtbZIP10 in the nucleus.
Weak, constitutive overexpression of AtbZIP10 leads to its accumulation in both soluble and microsomal-nuclear fractions (Figure 4B, time 0). Based on the results in Figure 4A, we reasoned that conditional overexpression of LSD1 in this context would increase the relative amount of AtbZIP10 in the soluble fraction if their interaction was part of the control of AtbZIP10 localization. Figure 4B demonstrates that this was the case, despite a reproducible and modest increase in total AtbZIP10 levels over this time course.
To provide a third, independent confirmation that LSD1 can retain AtbZIP10 in the cytosol, we analyzed the intracellular distribution of constitutively overexpressed AtbZIP10-GFP (P35S:AtbZIP10-GFP) in the presence or absence of untagged LSD1 (PLSD1:LSD1) in Arabidopsis protoplasts (Figure 4C). In the absence of LSD1, 82% of transformed protoplasts exhibited mixed cytoplasmic/nuclear distribution of AtbZIP10-GFP, whereas only 18% of the cells exhibited only cytosolic AtbZIP10-GFP accumulation (n=142). In contrast, when the LSD1 construct was cotransformed, 34% of the cells displayed exclusive accumulation of AtbZIP10-GFP in the cytoplasm (n=96). Coexpression with LSD1 did not alter the nuclear localization of AtbZIP63 (Figure 4C; n=102). These data indicate that native expression levels of LSD1 are sufficient to retain a significant amount of AtbZIP10 in the cytoplasm.
LSD1 and AtbZIP10 can interact in plant cells
Retention of AtbZIP10 by LSD1 implies a direct interaction between these two proteins. We used bimolecular fluorescence complementation (BiFC; Hu et al, 2002; Walter et al, 2004) to demonstrate that LSD1 and AtbZIP10 can interact in plant cells. AtbZIP10 fused to N-terminal (YFPN) or C-terminal fragments (YFPC) of YFP and transiently overexpressed in tobacco epidermal leaf cells yielded strong YFP fluorescence either exclusively in the nucleus (83% of cells) or in both the cytoplasm and the nucleus (17% of cells; n=129), suggesting that AtbZIP10 can form homodimers under these conditions (Figure 4D, upper left). Transient overexpression of LSD1-YFP fusions showed that homodimeric LSD1 accumulated in both the cytoplasm and the nucleus in 100% of cells (n=95; Figure 4D, upper right; see below). Co-overexpression of LSD1-YFPN and AtbZIP10-YFPC reduced significantly the fraction of cells expressing exclusively nuclear fluorescence (from 83 to 31%), whereas the fraction of cells expressing fluorescence in both compartments increased significantly (from 17 to 69%) (Figure 4D, lower left; n=145). No BiFC signals were detected when AtbZIP63-YFPC was coexpressed with LSD1-YFPN, indicating that the interaction of AtbZIP10 with LSD1 was specific (Figure 4D, lower right). Although we do not know whether the BiFC constructs used are functional, and although the wild-type LSD1 subcellular fractionation is altered by overexpression, these results demonstrate that AtbZIP10 and LSD1 can interact in plant cells.
AtbZIP10 is a positive regulator of cell death
We identified a T-DNA insertion into AtbZIP10 in Col-0. We constructed a conditional overexpression AtbZIP10 allele using an estradiol-inducible promoter system (Zuo et al, 2000) in addition to the constitutive overexpression allele described above. Transgenics were generated in both Col-0 and the Col-0 lsd1-2 allele. AtbZIP10 expressed from the estradiol-inducible system accumulates to ∼10-fold higher levels than from the constitutive promoter (data not shown). To define genetic interactions between AtbZIP10 and LSD1, we generated the lsd1-2 atbzip10 double mutant.
Five-week-old Col-0, atbzip10, lsd1-2 and lsd1-2 atbzip10 plants were sprayed with 150 μM benzo(1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester (BTH), a salicylic acid analog that induces runaway cell death (rcd) in lsd1, but does not induce cell death in Col-0. We measured ion leakage of these samples over time, a readout correlated with plant cell death (Epple et al, 2003). lsd1-2 samples exhibited increased ion leakage, reaching half-maximal conductivity at ∼105 h post application (hpa), whereas Col-0 and atbzip10 samples displayed only a very slight increase in conductivity. The lsd1-2 atbzip10 double mutant displayed significant reduction of ion leakage compared to lsd1-2 (Figure 5A). Conversely, weak overexpression of AtbZIP10 in lsd1-2 (genotype 35S-AtbZIP10 lsd1-2) enhanced dramatically BTH-induced ion leakage, a phenotype so pronounced that we needed to measure conductivity at earlier time points compared to lsd1-2 (Figure 5B). Conductivity in 35S-AtbZIP10 lsd1-2 reached half-maximum by ∼40 hpa. Control Col-0 and 35S-AtbZIP10 samples showed little increase in conductivity. We also demonstrated that AtbZIP10 function is required for superoxide-induced lsd1 rcd (data not shown), indicating that AtbZIP10 is a positive regulator of rcd in lsd1 and not merely a mediator of BTH response.
Figure 5.
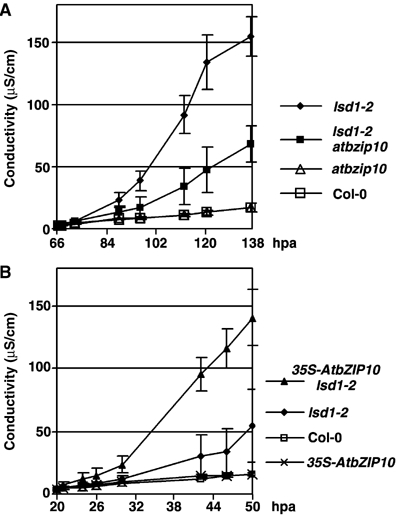
AtbZIP10 is a positive regulator of runaway cell death in lsd1. Five-week-old plants of the genotypes denoted at right were sprayed with 150 μM BTH and tissues were harvested at either 64 hpa (A) or 18 hpa (B) and processed as described in Materials and methods. Error bars represent 2 × standard error. The experiment was repeated two times. hpa, hours post application.
AtbZIP10 and LSD1 antagonistically control basal defense and cell death-mediated disease resistance against Hyaloperonospora parasitica
Hyaloperonospora parasitica (Hp) is an obligate biotrophic oomycete parasite well suited for characterization of disease resistance responses. Different RPP disease resistance genes control the recognition of different Hp isolates. Their action leads to hypersensitive cell death (HR) and cessation of pathogen growth. For weak RPP genes, the degree of HR correlates with Hp hyphal growth. In susceptible (rpp) plants, virulent Hp isolates grow and cause downy mildew, despite a plant basal defense response (Holub and Beynon, 1996). We investigated whether AtbZIP10 has a function in either RPP2-mediated HR (Sinapidou et al, 2004) or basal defense. We included the transgenic Est-AtbZIP10 line described above, and we generated a similar Est-LSD1 line for conditional estradiol-inducible overexpression of LSD1.
We probed RPP2 function by infecting 12-day-old seedlings with Hp Cala2. Sites of host cell death and Hp structures were stained with Trypan blue at 5 dpi. We evaluated microscopically the interaction sites and classified them as described in Figure 6A. A reduction in the percentage of HR sites, and corresponding increases in trailing necrosis (TN) or free hyphae (FH) interaction sites, reflects decreased RPP2 function (Holt III et al, 2002). The control genotypes, including Col-0 (Figure 6B, black labels), uniformly exhibited a high percentage of TN interactions, typical of the weak RPP2 phenotype (Sinapidou et al, 2004). lsd1-2 was completely resistant (HR at 100% of interaction sites; Aviv et al, 2002). We observed reproducible and significant decreases in RPP2 function in atbzip10 (see Supplementary Table 1 and Materials and methods for statistics). By contrast, resistance was significantly enhanced in both 35S-AtbZIP10 compared to Col-0, and in estradiol-treated Est-AtbZIP10 compared to either untreated controls or Col-0 treated with estradiol. The lsd1-2 atbzip10 double mutant was completely resistant, like lsd1. We conclude that (1) loss of AtbZIP10 function suppressed RPP2-mediated disease resistance, whereas AtbZIP10 overexpression enhanced it, and (2) loss of LSD1 enhanced RPP2-mediated disease resistance, whereas LSD1 overexpression only marginally suppressed it. Thus, antagonistic functions of LSD1 and AtbZIP10 modulate RPP2-mediated disease resistance response, presumably via their antagonistic control of host cell death. It is likely that LSD1 regulates other proteins in addition to AtbZIP10, as the lsd1-2 atbzip10 exhibits the lsd1 phenotype.
Figure 6.
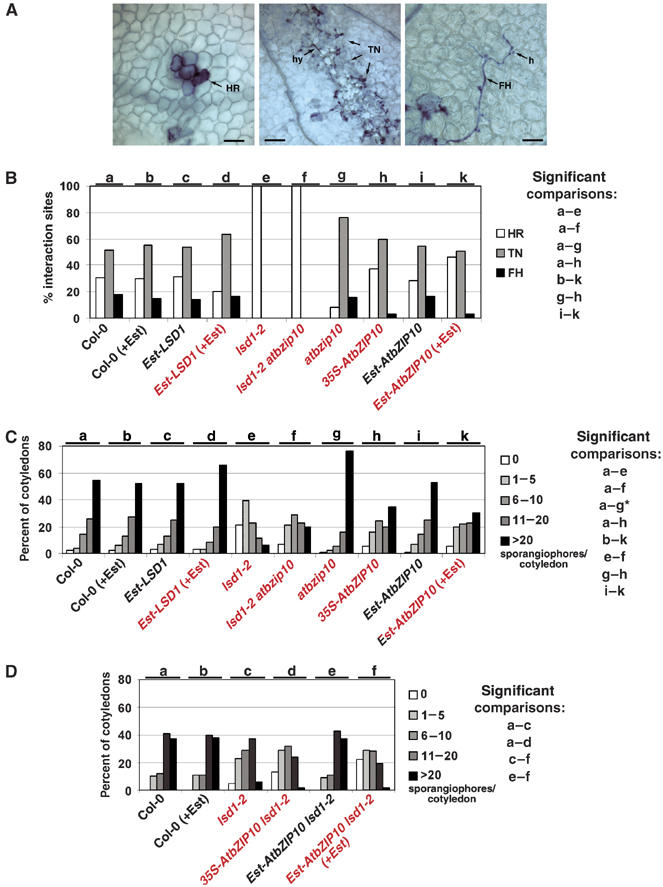
AtbZIP10 and LSD1 antagonistically control pathogen-induced cell death and disease resistance responses. (A, B) Twelve-day-old seedlings were inoculated with 5 × 104 spores/ml of the avirulent Hp isolate Cala2. Cotyledons were stained with Trypan blue at 5 dpi. (A) Presentation of HR (hypersensitive response; left panel), scale bar=100 μm; TN (trailing necrosis; middle panel), scale bar=250 μm; and FH (free hyphae; right panel), scale bar=100 μm. h denotes an Hp haustorium. (B) One randomly chosen interaction site per cotyledon from 87 to 104 interaction sites per genotype was microscopically evaluated and classified as HR, TN or FH. Control genotypes are indicated in black; (+Est) indicates treatment with 20 μM estradiol 24 h before inoculation with Hp. Three separate experiments were performed. (C, D) Twelve-day-old seedlings were inoculated with 5 × 104 spores/ml of the virulent Hp isolate Emco5. Sporangiophores per cotyledon were determined at 5 dpi (C) or 4 dpi (D). Cotyledons were classified as supporting no sporulation (0 sporangiophores), light sporulation (classes 1–5 and 6–10 sporangiophores/cotyledon), medium sporulation (class 11–20 sporangiophores/cotyledon) or heavy sporulation (>20 sporangiophores/cotyledon). Sporangiophores were counted on 100 cotyledons per genotype. Controls are indicated in black, and mutant and transgenic genotypes in red. The experiment was performed three (C) or two (D) times. For * in (C), see Materials and methods. For (B–D), we used standard contingency table analyses to make sequential pairwise comparisons between relevant genotypes designated alphabetically at top (Materials and methods; Supplementary Tables 1, 2 and 3). Significant comparisons are listed at right.
To assay basal defense, 12-day-old seedlings were inoculated with Hp Emco5. At 5 dpi, we counted sporangiophores per cotyledon (Figure 6C). Approximately 55% of the cotyledons in each control genotype (labeled black, Figure 6C) exhibited heavy sporulation. The lack of variation across these control genotypes confirmed that this assay was sensitive and robust. Sporulation was drastically reduced in lsd1-2 (Aviv et al, 2002). The atbzip10 mutant displayed a significant increase in susceptibility. By contrast, both constitutive and conditional overexpression of AtbZIP10 led to significantly decreased susceptibility compared to appropriate controls (Figure 6C and Supplementary Table 2). Finally, although both lsd1-2 and lsd1-2 atbzip10 exhibited decreased sporulation compared to Col-0, lsd1-2 was significantly more resistant than lsd1-2 atbzip10, suggesting that AtbZIP10 functions as a positive regulator of basal defense (Supplementary Table 2) We predicted that overexpression of AtbZIP10 in the lsd1 mutant background should result in added resistance to Hp Emco5. Indeed, 22% of lsd1-2 Est-AtbZIP10 cotyledons treated with estradiol did not support Hp Emco5 sporulation at 4 dpi, whereas only 5% of lsd1-2 exhibited this phenotype (Figure 6D and Supplementary Table 3).
Overexpression of AtbZIP10 did not lead to increased cell death following Hp Emco5 infection, and there is no increase in cell death in Hp Emco5-infected lsd1-2 (data not shown; Aviv et al, 2002). Thus, enhanced resistance to Hp Emco5 is due to enhanced basal defense that is apparently independent of cell death. We conclude that (1) AtbZIP10 is a positive regulator of basal defense, as its loss resulted in increased Hp sporulation and its overexpression resulted in the opposite phenotype; (2) LSD1 is a negative regulator of basal defense, as its loss led to decreased Hp sporulation; (3) LSD1 and AtbZIP10 act antagonistically in this process; and (4) LSD1 may regulate additional, unidentified positive mediators of basal defense. We infer this because lsd1-2 atbzip10, although significantly less resistant than lsd1, still more closely resembles lsd1 than atbzip10.
Discussion
Our most important findings are that AtbZIP10 is a positive mediator of plant basal defense responses and HR following pathogen attack, and that this activity is antagonized by LSD1, a negative regulator of both of these functions. We provide evidence that AtbZIP10 is shuttled out of the nucleus, perhaps in an exportin-dependent manner, and that LSD1 can participate in its cytosolic retention. We present both loss-of-function and overexpression genetic results consistent with these conclusions. We note, however, that there must be additional unidentified positive regulators of basal defense and HR that are also antagonized by LSD1. This prediction comes from our lsd1 atbzip10 double mutant analysis. We speculate that LSD1 is a hub for the regulation of transcriptional mediators of responses to various sources of oxidative stress. In support of this speculation, we identified three additional TFs as LSD1 partners in yeast two-hybrid screens (P Epple, H Kaminaka, BF Holt III and JL Dangl, data not shown). Additionally, we demonstrated that the LSD1-like protein LOL1 also antagonizes LSD1 function (Epple et al, 2003), and it interacts with the same set of TFs as LSD1 (not shown). We propose that LSD1 and LOL1 antagonize each other via competition for these TFs.
Arabidopsis class C TFs exhibit differential subcellular localization
AtbZIP10 and AtbZIP63 can form homodimers in planta (this study; Walter et al, 2004), bind to C- and G-box elements specifically in vitro and have transcriptional activity in vivo (this study; C Näke and K Harter, unpublished). AtbZIP10 and AtbZIP63 are differentially distributed in plant cells, as shown by analysis of their intracellular localization using constitutively expressed GFP and BiFC fusion proteins. AtbZIP63 is exclusively localized to the nucleus, but AtbZIP10 is detected in both the cytoplasm and the nucleus (this study; Walter et al, 2004). This was unexpected because the amino-acid composition of the bipartite NLSs is almost identical in AtbZIP10 and AtbZIP63 and the fusion of this basic region to GFP is sufficient for nuclear accumulation (Kircher et al, 1999). Our data suggest that AtbZIP10 is actively shuttling between the nuclear and the cytoplasm, probably utilizing AtXPO1. The NES required for AtXPO1-mediated nuclear export is within the N-terminal 105 amino acids of AtbZIP10. These findings suggested that retention of a significant amount of AtbZIP10 in the plant cell cytosol might regulate default re-localization into the nucleus. In contrast to previous examples (Kircher et al, 1999; Wellmer et al, 1999), AtbZIP10 was not re-localized into the nucleus following a variety of light treatments, indicating that its nuclear/cytoplasmic shuttling is modulated by signals other than light, or is unregulated.
The cytoplasmic localization of tobacco RSG, a member of a different bZIP subfamily that represses shoot growth, is likely regulated by interaction with a 14-3-3 protein (D31) and is also compromised by LMB treatment (Igarashi et al, 2001). Budding yeast Yap1 and fission yeast Pap1 are AP-1-like bZIP TFs required for oxidative stress response. Their nuclear–cytoplasmic shuttling is directly regulated by oxidation (Delaunay et al, 2000; Castillo et al, 2002) and is also mediated by CRM1/exportin1 (Toone et al, 1998; Yan et al, 1998). Importantly, AtbZIP10 partially rescues pap1, suggesting that AtbZIP10 might be a functional ortholog of Pap1, and that plants might have an oxidative stress signaling pathway analogous to that in fission yeast (Peng et al, submitted).
LSD1 contributes to cytoplasmic retention of AtbZIP10 and their antagonistic functions contribute to control of basal defense and cell death
We identified LSD1 as a protein that retained AtbZIP10 in the yeast cytoplasm. A C-terminal stretch of AtbZIP10 mediates interaction with LSD1. This domain displays the lowest degree of identity between the class C bZIP TFs (Supplementary Figure 1), and we speculate that this divergence is the molecular basis for specificity in the AtbZIP10–LSD1 interaction. The association of LSD1 with the AtbZIP10 C-terminus may cover the latter's NLS, an intrinsic part of the DNA-binding domain. As LSD1 is a cytoplasmic protein, this observation provides a likely molecular mechanism to explain the retention of AtbZIP10 in the cytoplasm. The idea that hindrance of the NLS inhibits nuclear uptake of AtbZIP10 was further supported by our observation that conditional overexpression of LSD1 leads to re-distribution of overexpressed AtbZIP10 into the soluble fraction. We could not directly assess this, as we could not reliably detect native expression levels of AtbZIP10. Nonetheless, we suggest that imbalance in AtbZIP10 distribution in lsd1 is a key contributor to the rcd phenotype of this mutant in response to reactive oxygen-dependent signals. Nevertheless, the sum of our fractionation, cell biology and genetic interaction data supports this contention.
A model of LSD1/AtbZIP10 action in Arabidopsis
Our data can be combined into an attractive, but certainly oversimplified, model: in the absence of the appropriate endogenous or environmental signal(s), LSD1 retains AtbZIP10 in the cytoplasm. We do not observe this when AtbZIP10 is overexpressed, because LSD1 retention activity is titrated. The positive regulatory effects of AtbZIP10 on HR and basal defence are inhibited by LSD1 retention activity. After perception of an appropriate reactive oxygen-derived signal, AtbZIP10 is activated, leading presumably to its dissociation from LSD1 and NLS-mediated transport into the nucleus. Inside the nucleus, AtbZIP10 induces the expression of HR- and basal defense-related target genes. Because AtbZIP10 is actively exported from the nucleus, the quantitative output in target gene expression depends on the relative intracellular amounts of LSD1 and AtbZIP10, the retention activity of LSD1 and the AtbZIP10 export rate. This mechanism enables fine-tuning of AtbZIP10-related target gene expression and, therefore, exact adjustment of the cell death response following perception of oxidative stress. Such fine-tuning is dictated by the spatial integration of these signals to determine whether cell death occurs (Torres et al, 2005). It is therefore of interest to identify immediate target genes of AtbZIP10, and to understand the biochemical mechanism by which LSD1 controls AtbZIP10 and other associated proteins.
Materials and methods
DNA constructs
(His)6-tagged AtbZIP10, AtbZIP63 and LSD1 cDNAs were cloned into pET24b (Novagen) via EcoRI/SalI (AtbZIP10, LSD1) or NheI/XhoI (AtbZIP63). For the constitutive Gal4-based yeast two-hybrid analyses, LSD1 was introduced via EcoRI/BamHI into pGADT7, AtbZIP10 via EcoRI/SalI and AtbZIP25 and AtbZIP63 via BamHI/SalI into pGBT9. For galactose-inducible yeast two-hybrid analyses, AtbZIP10 cDNA fragments were cloned into the pB42-AD vector. pGILDA-AtXPO1 is from Haasen et al (1999). The constructs for retention screening and for nuclear accumulation assays in yeast were generated by cloning AtbZIP10 and LSD1 via EcoRI/SalI and AtbZIP63 via NotI/SalI into pNS (Ueki et al, 1998). To identify interaction domains on LSD1 and AtbZIP10, full-length and deletion clones were cloned into pEG202 and pJG4-5 (Gyurius et al, 1996), respectively. For BiFC, AtbZIP10 was cloned into the binary vectors pSPYNE-35S and pSPYCE-35S; BiFC constructs of LSD1 and AtbZIP63 were as described (Walter et al, 2004). The AtbZIP10 cDNA with a C-terminal HA tag was cloned into pBAR1(35S) (Holt III et al, 2002). For conditional overexpression, AtbZIP10 and LSD1 cDNA clones (the latter with C-terminal 6.5xc-myc tag; LSD1-myc) were cloned into pER8 (Zuo et al, 2000). All clones were sequence verified.
Yeast two-hybrid, intracellular localization assays and AtbZIP10 retention screening
Small-scale transformation of yeast cells (EGY48, EGY48[p8op-lacZ], EGY48[p2op-lacZ] and PJ69-4A) for two-hybrid and intracellular localization assays was by the PEG/lithium acetate method (Lohrmann et al, 2001), or with the Frozen-EZ Yeast Transformation II Kit (Zymo Research). Reporter gene activity was determined by plating the transformants on CSM-W,L media (selection for plasmids) or CSM-W,L,A (selection for protein–protein interactions). Selection of transformants for deletion assays was carried out by plating on CSM-H,U,W. LacZ reporter gene test (β-galactosidase activity) was as described (Lohrmann et al, 2001). For intracellular localization assays based on the NTT system (Ueki et al, 1998), EGY48 yeast cell suspensions transformed with either a pNS construct alone or a pNS construct in combination with a pFL construct were plated at an identical OD600 nm on plasmid selecting media (CSM-H or CSM-H,U, respectively) and on media selecting for nuclear localization (CSM-H,L or CSM-H,U,L) and grown for 48 h at 30°C.
Large-scale transformation of the EGY48 yeast strain (Rugner et al, 2001) expressing LexAD-AtbZIP10 (His autotroph) as bait with the pFL Arabidopsis cDNA prey library (Minet et al, 1992) was used for retention screening. Transformants were plated on CSM-H,U medium and incubated at 30°C for 3 days. Colonies were replica plated onto CSM-H,U,L media using silk pads and grown for 4 days at 30°C. Transformants showing normal growth on CSM-H,U plates, but reduced growth on CSM-H,U,L plates were re-grown in liquid CSM-H,U to OD600 nm of 2.0. Suspensions were diluted in three 1:10 steps with CSM-H,U, dotted on CSM-H,U and incubated for 4 days at 30°C. Bait and prey plasmids were recovered from yeast strains with normal growth on CSM-H,U but reduced growth on CSM-H,U,L, amplified in Escherichia coli, and inserts of the pFL plasmids were sequenced. Yeast EGY48 was re-transformed with the bait and prey plasmids and the growth assay was repeated to verify differential growth.
Transient transfection of plant protoplasts, transformation of tobacco leaf cells with Agrobacterium and microscopy
Protoplasts from dark-grown parsley cell suspension cultures (Petroselinum crispum L.) were grown and transformed by electroporation (Kircher et al, 1999). Tobacco protoplasts from a dark-grown BY2 cell suspension culture were grown and transformed by PEG (Haasen et al, 1999). Protoplasts were assayed for fluorescence 12–20 h after transfection. Leaves from 2- to 4-week-old Nicotiana benthamiana plants were transiently transformed with Agrobacterium tumefaciens using the tomato bushy stunt virus p19 protein to suppress gene silencing (Voinnet et al, 2003). Co-infiltration of Agrobacterium strains containing BiFC constructs and the p19 silencing plasmid was at OD600 nm of 0.7:0.7:1.0. Epifluorescence and confocal laser scanning microscopy were performed 1–2 days after infiltration (Kircher et al, 1999).
Mutants used and transgenic lines generated
We used the Columbia (Col-0) ecotype and isogenic SALK insertion lines (Alonso et al, 2003) for atbzip10 (At4g02640; SALK_014867; insertion in codon Asp94 of exon one) and lsd1-2 (At4g20380; SALK_042687; insertion at nucleotide 57 of intron four; Arabidopsis Biological Resource Center at Ohio State University). Transgenic plants were generated by A. tumefaciens-mediated transformation (Clough and Bent, 1998).
Protein fractionation
F1 plants from crosses between lsd1-2 [Est-AtbZIP10-HA] × Col-0 and lsd1-2 [Est-AtbZIP10-HA] × lsd1-2 plants were sprayed with 20 μM estradiol at 0 and 60 min. F1 plants from a cross between estradiol-inducible LSD1-myc and 35S-AtbZIP10-HA plants were sprayed with 20 μM estradiol at 0 and 4 hpa. Leaf tissue (150–200 mg) was harvested after induction and homogenized in 750 μl of sucrose buffer (20 mM Tris–HCl pH 8, 330 mM sucrose, 1 mM EDTA, 1 mM PMSF, 5 mM DTT and 1 × protease inhibitor cocktail (Sigma)). After brief centrifugation (1000 g, 10 min, 4°C) to remove cell debris, 200 μl of the supernatant was transferred to another tube and used as ‘total protein (T)'. Another 200 μl of the supernatant was transferred to a second tube and centrifuged at 50 000 g for 45 min at 4°C. The pellet was re-suspended in 200 μl sucrose buffer and used as ‘membrane and nuclear-enriched fraction' (M), and the supernatant was used as ‘soluble fraction' (S). Equal volumes were loaded onto an SDS–PAGE gel. Western blot and protein detection were performed with anti-APX antibody (gift of Daniel Kliebenstein) at 1:10 000, anti-histone H3 antibody (Abcam, Cambridge, MA) at 1:5000, anti-RD28 (gift from Marteen Chrispeels) and standard anti-myc and anti-HA antibodies.
Conductivity assay
Conductivity assays were performed on 5-week-old plants sprayed two times (4 h interval) with 150 μM BTH (Epple et al, 2003). Leaves were harvested at 18 and 64 hpa (Figure 5A and B, respectively), leaf disks were removed with a 7-mm-diameter cork borer and floated on distilled water for 45 min. Six leaf disks per genotype were transferred to a tube containing 6 ml of distilled water. Conductivity was measured in μS/cm with an Orion (Boston) Conductivity Meter at indicated time points. Means and 2 × standard error were calculated from five replicate measurements per genotype per experiment (a total of 30 leaf disks per genotype per experiment). The experiment was repeated twice.
Pathology: Hp sporangiophore assay
Twelve-day-old seedlings were inoculated with the virulent Hp isolate Emco5 and sporangiophores counted at 4–5 dpi as described (Holt III et al, 2002).
Cell death assay
Twelve-day-old seedlings were inoculated with 5 × 104 spores/ml of avirulent Hp isolate Cala2. Cotyledons were stained with Trypan blue (Epple et al, 2003) at 5 dpi to visualize host cell death and oomycete structures. One randomly chosen interaction site on between 87 and 104 cotyledons per genotype was evaluated and classified. The experiment was repeated three times.
Statistics
We used standard contingency table analyses for sequential pairwise comparisons between relevant genetic backgrounds (Sokal and Rohlf, 1995). The null hypothesis states that the category distributions (i.e., types of interaction site for Figure 6B and numbers of sporangiophores for Figure 6C and D) for any comparison are the same. The χ2 and P-values for each pairwise comparison are presented in Supplementary Tables 1, 2 and 3. We initially designated P⩽0.05 as our nominal significance threshold. To adjust for the different number of pairwise comparisons in each data set, the threshold P-values were corrected by the Bonferroni method (Sokal and Rohlf, 1995; for Supplementary Table 1, 0.005; for Supplementary Table 2, 0.0045; for Supplementary Table 3, 0.007). For the comparison of Col-0 to atbzip10 in Figure 6C, two independent experiments suggested, but did not establish, a statistically significant difference (P-values of 0.0227 and 0.1228). In this case, we used Fisher's combined probability statistic (−2∑ln P=−2(∑−3.7854, −2.0956)=χ2 value 11.72 at 4 degrees of freedom) to reject the null hypothesis of no difference between the Col-0 and atbzip10 distributions, confirming significance.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3A–D
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplemental Materials and Methods
Acknowledgments
This work was supported by NIH (R01-GM057171) grant to JLD, JSPS overseas research fellowship to HK and DFG grants to KH (SFB388, SFB446). We thank Dr N Ueki, Yokohama Research Center, Japan, for vectors, and G Fiene and M Keinath for excellent technical support. We thank professor Corbin D Jones for expert assistance with statistics and Dr M Ellerstrøm for contributions to construction of the native promoter LSD1-myc construct.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aviv DH, Rusterucci C, Holt BF III, Dietrich RA, Parker JE, Dangl JL (2002) Runaway cell death, but not basal disease resistance, in lsd1 is SA- and NIM1/NPR1-dependent. Plant J 29: 381–391 [DOI] [PubMed] [Google Scholar]
- Castillo EA, Ayte J, Chiva C, Moldon A, Carrascal M, Abian J, Jones N, Hidalgo E (2002) Diethylmaleate activates the transcription factor Pap1 by covalent modification of critical cysteine residues. Mol Microbiol 45: 243–254 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delaunay A, Isnard AD, Toledano MB (2000) H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J 19: 5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward EJ, Ryals JA, Dangl JL (1994) Arabidopsis mutants simulating disease resistance response. Cell 77: 565–578 [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL (1997) A novel zinc-finger protein is encoded by the Arabidopsis lsd1 gene and functions as a negative regulator of plant cell death. Cell 88: 685–694 [DOI] [PubMed] [Google Scholar]
- Epple P, Mack AA, Morris VR, Dangl JL (2003) Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc Natl Acad Sci USA 100: 6831–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbrügge M, Sprenger M, Dinkelbach M, Yazaki K, Harter K, Weisshaar B (1994) Functional analysis of a light-responsive plant bZIP transcriptional regulator. Plant Cell 6: 1607–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyurius J, Golemis E, Cherkov H, Brent R (1996) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75: 791–803 [DOI] [PubMed] [Google Scholar]
- Haasen D, Kohler C, Neuhaus G, Merkle T (1999) Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J 20: 695–705 [DOI] [PubMed] [Google Scholar]
- Holt BF III, Boyes DC, Ellerstrom M, Siefers N, Wiig A, Kauffmann S, Grant MR, Dangl JL (2002) An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev Cell 2: 807–817 [DOI] [PubMed] [Google Scholar]
- Holub EB, Beynon JL (1996) Symbiology of Mouse Ear Cress (Arabidopsis thaliana) and oomycetes. Adv Bot Res 24: 228–273 [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y (2001) 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13: 2483–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N (2005) An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc Natl Acad Sci USA 102: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabs T, Dietrich RA, Dangl JL (1996) Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273: 1853–1856 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Kim S, Kang J-y, Cho D-I, Park JH, Kim SY (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40: 75–87 [DOI] [PubMed] [Google Scholar]
- Kircher S, Wellmer F, Nick P, Rugner A, Schafer E, Harter K (1999) Nuclear import of the parsley bZIP transcription factor CPRF2 is regulated by phytochrome photoreceptors. J Cell Biol 144: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M (1998) Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res 242: 540–547 [DOI] [PubMed] [Google Scholar]
- Lara P, Onate-Sanchez L, Abraham Z, Ferrandiz C, Diaz I, Carbonero P, Vicente-Carbajosa P (2003) Synergistic activation of seed storage protein gene expression by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem 278: 21003–21011 [DOI] [PubMed] [Google Scholar]
- Lohrmann J, Sweere U, Zabaleta E, Baurle I, Keitel C, Kozma-Bognar L, Brennicke A, Schafer E, Kudla J, Harter K (2001) The response regulator ARR2: a pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol Genet Genomics 265: 2–13 [DOI] [PubMed] [Google Scholar]
- Mateo A, Muhlenbock P, Rusterucci C, Chang CC, Miszalski Z, Karpinska B, Parker JE, Mullineaux PM, Karpinski S (2004) LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol 136: 2818–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2: 417–422 [DOI] [PubMed] [Google Scholar]
- Rugner A, Frohnmeyer H, Nake C, Wellmer F, Kircher S, Schafer E, Harter K (2001) Isolation and characterization of four novel parsley proteins that interact with the transcriptional regulators CPRF1 and CPRF2. Mol Genet Genomics 265: 964–976 [DOI] [PubMed] [Google Scholar]
- Siberil Y, Doireau P, Gantet P (2001) Plant bZIP G-box binding factors. Modular structure and activation mechanisms. Eur J Biochem 268: 5655–5666 [DOI] [PubMed] [Google Scholar]
- Sinapidou E, Williams K, Nott L, Bahkt S, Tor M, Crute I, Bittner-Eddy P, Beynon J (2004) Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J 38: 898–909 [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ (1995) Biometry: The Principles and Practice of Statistics in Biological Tesearch, 3rd edn. New York: Freeman [Google Scholar]
- Toone WM, Kuge S, Samuels M, Morgan BA, Toda T, Jones N (1998) Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev 12: 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL (2005) Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet 37: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Ueki N, Oda T, Kondo M, Yano K, Noguchi T, Muramatsu M (1998) Selection system for genes encoding nuclear-targeted proteins. Nat Biotechnol 16: 1338–1342 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wellmer F, Kircher S, Rugner A, Frohnmeyer H, Schafer E, Harter K (1999) Phosphorylation of the parsley bZIP transcription factor CPRF2 is regulated by light. J Biol Chem 274: 29476–29482 [DOI] [PubMed] [Google Scholar]
- Wellmer F, Schafer E, Harter K (2001) The DNA binding properties of the parsley bZIP transcription factor CPRF4a are regulated by light. J Biol Chem 276: 6274–6279 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yan C, Lee LH, Davis LI (1998) Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J 17: 7416–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3A–D
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplemental Materials and Methods


