Abstract
We have studied the molecular basis of nervous system repair in invertebrate (Hirudo medicinalis) nerve cells. Unlike in mammals, neurons in invertebrates survive injury and regrow processes to restore the connections that they held before the damage occurred. To identify genes whose expression is regulated after injury, we have used subtractive probes, constructed from regenerating and non-regenerating ganglia from the leech Hirudo medicinalis, to screen cDNA libraries made from whole leech CNS or from identified microdissected neurons. We have identified genes of known or predicted function as well as novel genes. Known genes up-regulated within hours of injury and that are widely expressed in invertebrate and mammalian cells include thioredoxin and tubulin. Other known genes, e.g. Cysteine Rich Intestinal Protein (CRIP), have previously been identified in mammalian cells though not in regenerating adult neurons. Two regulated genes identified, myohemerythrin and the novel protein ReN3 are exclusively expressed in invertebrates. Thus our approach has enabled us to identify genes, present in a neuron of known function, that are up- and down-regulated within hours of axotomy, and that may underpin the intrinsic ability of invertebrate neurons to survive damage and initiate regrowth programmes.
Keywords: axotomy, cDNA library, CRIP, dynein, myohemerythrin, novel genes, Retzius neuron, thioredoxin, tubulin, voltage-gated sodium channel
Introduction: the molecular basis of repair in the leech nervous system
We have used the nervous system of the leech Hirudo medicinalis to identify genes differentially expressed in a defined neuronal population in response to injury and during repair. The question we have asked is: when a nerve cell is damaged, which genes are turned on or off? We have approached this directly by comparing ganglia and individual nerve cells that are, and are not, regenerating. We have used a polymerase chain reaction (PCR)-based technique to construct cDNA libraries from whole ganglia as well as from individual microdissected neurons. To identify genes that are regulated after damage, we have used subtraction hybridization of normal and regenerating libraries. Because genetic information on the leech is limited, most of the sequences that we have identified in adult and regenerating neurons represent new information. Some of the sequences identified are the leech homologues of genes found in mammals including humans, and some of these have established functions. For example, the genes encoding the cytoskeletal proteins actin, tubulin and Protein 4.1, and the neuron-specific protein synapsin, are up-regulated following axotomy in leech. Such proteins might be expected to be regulated in a neuron that is rebuilding its axon and remaking synaptic connections. Other sequences that we find to be regulated by injury include Cysteine Rich Intestinal Protein (CRIP), previously shown to be expressed in developing mammalian intestinal cells (where its role is unknown) but not in adult regenerating nerve cells. Other genes regulated by injury in leech have counterparts in the mammalian genome but are ‘novel’ in having no known function. Still other regulated genes have no known homologues in vertebrate genomes, and these invertebrate-specific sequences are interesting in view of the different capacity for CNS repair in invertebrates such as the leech.
A different but related approach is to search for specific genes in leech that encode known proteins that are used during the construction of a new axon, for example genes encoding the ion channels required for electrical signalling by the axon. Accordingly, the last section describes experiments to isolate genes encoding voltage-gated sodium channels in the leech and to study their expression in regenerating nerve cells.
Why use the leech? Regeneration of nervous systems in invertebrates vs. failure of CNS repair in mammals
The invertebrates comprise at least 95% of the 15 million known species on Earth: less than 1% of species are mammals (Myers et al. 2000). Among invertebrate species there are great differences in the capacity for repair of the nervous system (Moffett, 1996), as there are in vertebrates. Wound healing and regeneration of missing body parts are widespread among invertebrates and also occur in lower vertebrates such as amphibia and reptiles. Many invertebrates can add new neurons to the adult nervous system, as can vertebrates including humans. Many cellular processes that follow injury are common to both invertebrates and mammals, for example changes in excitability following axotomy, the sealing of cut axons, or the formation of growth cones and their pathfinding. Other processes that are common in the invertebrates, for example the survival of axon segments severed from their cell bodies, are also present, though less common, in vertebrates (Gillingwater & Ribchester, 2001). A principal difference between higher vertebrates and invertebrates is in the ability to regrow injured axons following damage to the CNS.
There are two main reasons for choosing the leech nervous system to study the molecular basis of neural repair. First, unlike mammals, the leech has a demonstrated capacity to repair damage to its CNS and to restore function. Although leeches cannot regenerate lost body parts, or add new neurons to adult CNS, they are able to regrow severed axons, to re-innervate the periphery with specificity and to re-establish CNS connections. There is an established body of work at a cellular level, begun in the 1960s, that describes, in terms of individually identifiable nerve cells, the pattern of regenerative outgrowth of axons following damage; the cells and molecules that influence this outgrowth; the re-establishment of sensory and motor fields, and the specific regeneration of central connections (Nicholls, 1987). Evidently the signals exist within the leech nervous system to ensure appropriate restoration of function. We have used this nervous system with its established capacity for functional repair to study the molecular basis of the repair process.
The second advantage of the leech nervous system is that techniques have been developed for isolating single identified neurons from the ganglia of the ventral nerve cord (Ready & Nicholls, 1979). The introduction of the PCR in 1985 (Mullis, 1990) meant that it became possible to amplify vanishingly small amounts of starting material. We have used a PCR-based technique to generate cDNA libraries from small numbers of identified microdissected nerve cells as well as from whole ganglia. We have used the cDNA libraries to screen for genes whose expression changes after injury and during the repair process.
Construction of cDNA libraries from identified serotonergic neurons
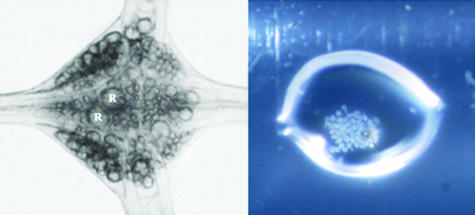
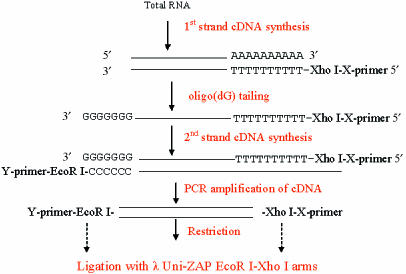
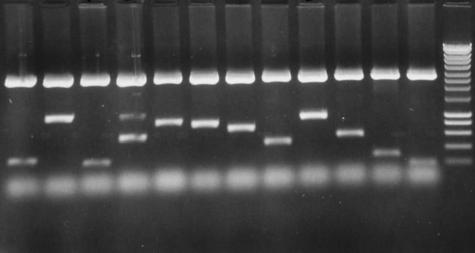
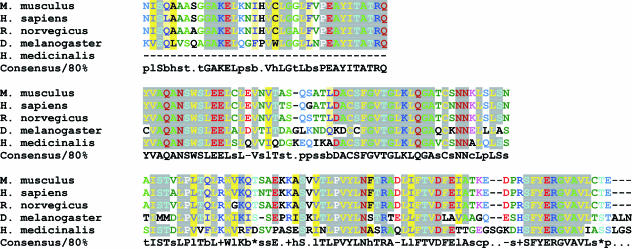
The leech nervous system contains in total around 10 000 nerve cells, organized segmentally into a series of ganglia. A typical midbody ganglion in Hirudo contains around 400 nerve cell bodies arranged as a cortical layer around the neuropile (Fig. 1). Many of these neurons have known properties, morphologies and connections. A first step was to devise a reliable and reproducible method for the construction of a neuron-specific library from a small amount of starting material, estimated equivalent to about 50 pg of mRNA (Korneev et al. 1994, 1996; Wang et al. 2002). For library construction we chose to use Retzius cells, the pair of large cells in the centre of the ganglion named after the Swedish neuroanatomist who first described them in the 1880s (Retzius, 1891). Retzius cells are among the most studied of the identified neurons in the leech. They are serotonergic modulatory neurons, influencing behaviour via serotonin release in the periphery and within the ganglion (McAdoo & Coggeshall, 1976; Willard, 1981; Henderson et al. 1983; Bruns & Jahn, 1995; Bruns et al. 2000; Trueta et al. 2003; Velazquez-Ulloa et al. 2003). Retzius neuron cell bodies were microdissected from successive ganglia and collected into a few microlitres of fluid (Fig. 1). Total RNA was extracted using TRI reagent (Helena Biosciences), and a PCR-based library constructed using poly A+ RNA as shown schematically in Fig. 2. A library constructed using around 30 microdissected Retzius cell bodies is illustrated in Fig. 3 and Table 1 (Wang et al. 2002). The library contained around 200 000 independent transformants ranging in size from 200 to 1500 bp with an average size of 600 bp. Sequence analysis performed on randomly picked clones showed that most of the clones had open reading frames and so can potentially translate a polypeptide chain. Some clones were found to code for well-characterized proteins with known functions in transport in nerve cells. Two examples are the leech homologues of α-2 tubulin, a component of microtubules in nerve cells, and cytoplasmic dynein, originally characterized as a motor protein for retrograde transport of cargo along microtubules in axons (Vallee et al. 1988; Hirokawa et al. 1990; Verhey et al. 2001). Retrograde transport plays critical roles in development and nerve regeneration by delivering neurotrophic signals from axon terminals to the cell body, and there is evidence that disruption of the axonal transport machinery causes neurodegenerative disease (Lamonte et al. 2002). The cytoplasmic dynein protein complex in mammalian cells consists of two heavy chains containing the motor domain (Lye et al. 1987), and several light and intermediate chains (Hughes et al. 1995; King et al. 1996). Dynein light chains have been shown to interact with the tubulin-binding protein gephyrin, which mediates clustering of receptors at inhibitory synapses (Fuhrmann et al. 2002). The clone isolated from the leech Retzius neuron library coded for part of cytoplasmic dynein heavy chain. Figure 4 shows the clustal alignment of amino acid sequences of this region of cytoplasmic dynein in leech, human, fly, mouse and rat.
Fig. 1.
cDNA libraries were constructed using whole ganglia from the leech ventral nerve cord or microdissected neuron cell bodies. (A) An isolated ganglion contains around 400 nerve cells including primary sensory and motor neurons, modulatory neurons and interneurons, together with macroglia and microglia, and the cells comprising the external connective tissue sheath, and the internal capsule, a structure which separates the cell bodies from the central neuropile area of the ganglion. R, Retzius cells. (B) Isolated Retzius cell bodies microdissected from successive ganglia in the ventral nerve cord, collected into a drop of fluid on the wall of an Eppendorf tube, and used to construct the Retzius neuron library.
Fig. 2.
Scheme for library construction: flow chart of the construction of amplification-based cDNA libraries from single ganglia or microdissected neurons (modified from Korneev et al. 1996).
Fig. 3.
Retzius neuron library constructed using around 30 microdissected cell bodies. (A) Twelve clones picked at random from the library are illustrated on the agarose gel. Of the 12 illustrated clones, the average insert size was about 600 bp. Most of the clones had open reading frames and some clones had polyadenylation signals.
Table 1. Sequence information for 23 clones picked at random from the Retzius neuron library. The left-hand column gives the clone notation, the central column the size of the insert in base pairs and the right-hand column the closest homology. The first seven clones listed had no homologues in existing databases and are classified as novel.
| code | length (bp) | homology |
|---|---|---|
| R5 | 284 | novel |
| R3 | 406 | novel |
| R6 | 531 | novel |
| R2 | 632 | novel |
| R33 | 653 | novel |
| R14a | 833 | novel |
| R51b/c | 860 | novel |
| R7 | 276 | hypothetical to human, mouse and fly protein |
| R49 | 491 | hypothetical protein F07C3.2 (C. elegans) |
| R39 | 595 | hypothetical protein Cj0069 (Campylobacter jejuni) |
| R18 | 434 | SMC1 gene product (Drosophila) |
| R14 | 509 | ES1 protein (zebrafish); human |
| R9 | 742 | CG5969 gene product (Drosophila); RNA binding protein (mouse) |
| R5a | 433 | zinc finger protein 265; RNA binding protein EWS |
| R6a | 502 | 26S protease-associated pad 1 homolog (mouse) |
| R17 | 589 | protein-L-isoaspartate (D-aspartate) O-methyltransferase (shorter than R8a) |
| R8a (b/c) | 622 | protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| R3a | 638 | procollagen-lysine, 2-oxogluctarate 5-dioxygenase 3; lysyl hyrdolase 2 |
| R46 | 679 | cytoplasmic dyein, heavy chain (mouse) |
| R45 | 756 | NaDH dehydrogenase (ubiquinone) Fe-S protein 1 |
| R2a | 798 | alpha-2 tubulin |
| R12 | 798 | ubiquitin-conjugating enzyme E2 variant 1, isoform b (human) |
| R8 | 1026 | s-adenoyl-L-homocysteine hydrolase 2 |
Fig. 4.
CLUSTAL alignment of the amino acid sequence of part of cytoplasmic dynein heavy chain in leech with homologous sequences from genomes of human, fly, mouse and rat.
Other randomly picked clones in the leech Retzius neuron library included a proteosome-associated protein and a ubiquitin-conjugating enzyme. Other clones showed no match in either nucleotide or protein domain databases, and were classed as novel genes. Thus the Retzius cell library is a representative collection of genes, or fragments of genes, both known and unknown, from an identified serotonergic neuron. It illustrates the open-ended nature of our approach: our aim in these experiments was not to search for one particular gene or genes, but rather to construct a representative collection of partial cDNA sequences, both known and unknown, that are representative of a known kind of nerve cell. The library is a tool to access neuron-specific genes and in particular to isolate genes whose expression changes after injury. In a wider context, because there is currently a limited amount of sequence information available in databases for Hirudo, both the single cell and the ganglion libraries that we have constructed contribute information in general on the DNA of this much studied annelid worm.
Construction of libraries from regenerating Retzius cells
To identify genes whose expression changes after injury, we have constructed libraries from regenerating neurons, in order to compare the population of genes expressed by normal and regenerating cells. As a first step we chose to use neurons whose main axons had been cut and which had been regenerating for 1 week in the animal, a relatively late stage of repair when cells are producing multiple new processes and beginning to make synapses (e.g. Baylor & Nicholls, 1971; Van Essen & Jansen, 1977; French & Muller, 1986; Bannatyne et al. 1989). To obtain regenerating ganglia, leeches were anaesthetized, a small cut was made in the ventral body wall to expose a single ganglion and the nerve roots to the body wall on both sides of the ganglion were cut; this severs the axons of all the primary sensory neurons, motoneurons and modulatory neurons, including the Retzius neurons, that project to the body wall. The leeches were then allowed to recover, and after 7 days the ganglia whose nerve roots had been cut were removed. A regenerating Retzius cell library was constructed using 15 pooled axotomized Retzius neuron cell bodies microdissected from these ganglia. Subtraction hybridization of normal and regenerating Retzius cell libraries was then performed to enrich for genes up-regulated in the regenerating neuron (Box. 1; from Korneev et al. 1997). Notable clones in the subtractive library include structural proteins 4.1 and tubulin. The level of α-tubulin appeared to double in regenerating cells. In the 7-day subtractive library we found two forms of α-tubulin, with identical coding regions but different 5′ ends (Korneev et al. 1997; Fedorov et al. 1999). Also up-regulated is the cytoskeletal protein, Protein 4.1, first isolated as a membrane-associated protein in mammalian red blood cells, and with an emerging role in synaptic plasticity in mammals by cross-linking glutamate receptors to the cytoskeleton (Shen et al. 2000; Coleman et al. 2003). The subtractive library also contained the immediate early gene CEBP-γ; the invertebrate-specific oxygen carrier myohemerythrin, a calcium regulatory protein and the neuron-specific protein synapsin, as well as many previously uncharacterized cDNAs. One novel peptide identified, which has no known homologue in the vertebrates, made up approximately 10% of clones picked at random from the subtracted library. A homologous peptide, named SPTR, was subsequently independently isolated from neurons in snail (Lymaea stagnalis) ganglia and reported as an endogenous neuropeptide with a molecular weight of 1.902 kDa (Koert et al. 2001). In snail, as in leech, this peptide is found in serotonergic neurons where it may function as a co-transmitter. Its up-regulation after axotomy in the leech suggests that it might play a role in regeneration.
Box 1. Sequence analysis of 34 randomly picked clones from the 7-day regenerating Retzius neuron subtracted library (modified from Korneev et al. 1997). Twenty sequences represent known genes. Four out of 10 novel sequences listed coded for a novel peptide designated SPTR.
| Sequence analysis of genes from the subtractive Retzius cell library |
| 20 clones represent 12 known genes |
| α-tubulin (6 clones) |
| β-tubulin (4 clones) |
| synapsin |
| C/EBPγ |
| calmodulin like |
| Protein 4.1 |
| myohaemerythrin |
| rsa 10 |
| COX I |
| ATPase inhibitor |
| ribosomal protein RS10 |
| yeast 39.4 kD protein |
| 14 clones represent 10 novel nucleotide sequences |
| 4 clones are independent isolates of a serine-rich protein |
Thus construction of a subtractive library, at a stage of repair when a neuron is extending new processes and making synapses, has identified sequences coding for the types of protein expected to be associated with axon growth and synapse formation, and validates our approach. It also identified genes with homologues in the vertebrate genome but no known function, as well as both known and unknown genes that are specific to invertebrate phyla, and which may play a role in regeneration.
Construction of libraries and probes from segmental ganglia at different stages of repair: identification of novel genes and invertebrate non-chordate genes
Having demonstrated that neuron-specific library construction coupled with subtraction hybridization is a viable way to identify regeneration-associated genes, we wished to extend our experimental approach in two ways: first to look at different times after injury, and second, to identify those genes that are down-regulated after injury, as well as those that are up-regulated. To do this we constructed cDNA libraries and cDNA probes using whole ganglia of the ventral nerve cord rather than individually microdissected nerve cells (Table 2; Emes et al. 2002). cDNA from whole ganglia contains sequences from all the different kinds of neurons in the ganglion, as well as from the macroglia and microglial cells in the ganglion, and from the cells that form the connective tissue sheath. Whole ganglion cDNA will therefore represent the genes of the different cell types, both neuronal and non-neuronal, that may contribute to the repair process. We constructed a ‘conventional’ (non-PCR-based) cDNA library using whole nerve cords from 30 animals, and also PCR-based libraries made from three regenerating ganglia at chosen times after injury. To obtain regenerating ganglia, the nerve roots on both sides of the ninth segmental ganglion were cut in anaesthetized leeches. The leech was then allowed to recover, and the regenerating ganglia removed after 1 day, 3 days or 5 days.
Table 2. The four ganglion libraries constructed using whole nerve cords (conventional, non-PCR-based library) or whole regenerating ganglia at 1, 3 or 5 days after cutting major nerve roots (PCR-based libraries). The table shows percentage recombinants, size of the libraries and average size of the inserts.
| library name | percentage recombinants | size of primary library (plaque forming units) | average insert size (bp) | |
|---|---|---|---|---|
| REHmGC | complete CNS | 91 | 3.75 × 104 | 700 |
| REHmG9r24s | subtracted 24h post-axotomy | 93 | 3.05 × 105 | 700 |
| REHmG9r72 | 72h post-axotomy | 98 | 1.36 × 105 | 600 |
| REHmG9r120 | 120h post-axotomy | 97 | 1.22 × 105 | 500 |
Table 3 illustrates a subtractive ganglion library that is enriched in sequences up-regulated at 24 h. The randomly picked clones illustrate the complexity of the library. They include a heat shock protein, 16s rRNA, subunits of cytochrome oxidase, a ubiquitin-conjugating enzyme, the structural protein actin, as well as four novel clones. One of these novel clones was the invertebrate-specific SPTR peptide that we had previously shown to be up-regulated in the Retzius cell at 7 days. Hence, in leech nervous system up-regulation of this gene in the nervous system is detectable as early as 24 h after injury. A further novel clone, designated ReN3, was also found to be specific to invertebrates. Immunohistochemistry using antibodies to a custom-made peptide showed that the peptide encoded by the ReN3 gene is widely expressed in neuronal cell bodies in Hirudo ganglia. It is also expressed in the connective tissue capsule that surrounds the ganglion, and in the internal capsule that separates the outer layer of neuron cell bodies from the inner neuropile layer of the ganglion (Fig. 5; E. J. Babington, R. Emes & S. E. Blackshaw, unpublished observations).
Table 3. Selected clones from the 24-h ganglion subtracted library (REHmG9r24S in Fig. 6). One million plaque-forming units (pfu) of amplified total CNS library (REHmGC in Fig. 6) were screened using subtracted probes (24 h vs. sham-operated control) and 105 differentially hybridizing clones were isolated. Fifty-five of these were up-regulated at 24 h post-axotomy. The individual genes identified to date are listed. The nearest homology is shown together with the accession number of the human homologue.
| Gene ID | nearest homologue§ | gene name | size (bp) | human homologue¥ accession |
|---|---|---|---|---|
| Re24s1.1.1 | Danio rerio heat shock protein hsp90beta (AF068772) | HSP90 | 612 | AF028832 |
| various clones | H. medicinalis 16s ribosomal RNA (AF325971) | 16s rRNA | 319–607 | AF011168 |
| Re24s.42 | Cyanidium caldarium cytochrome oxidase subunit II (Z48930) | COX II | 600 | |
| Re24s26.1.1 | Zenopsis nebulosus cytochrome c oxidase subunit III (AP002942) | COX III | 441 | AF346965 |
| Re24s28.1.1 | Arabidopsis thaliana ubiquitin-conjugating enzyme (X68306) | UCE | 767 | I59365 |
| Re24s3.1.1 | Bos tarus b-1,4-N-acetylglucosaminyltransferase (AB000628) | GnT | 2031 | AB024911 |
| Re24s7.1.1 | Plectus acuminatus actin (AJ012665) | actin | 1353 | S38782 |
| Re24s.19 | H. medicinalis neuron-specific protein (AAB40925) | LeSPTR | 159 | |
| Re24s4.1.1 | novel 1 | 148 | ||
| Re24s17.1.1 | novel 2 | 416 | ||
| Re24s34.2.1 | novel 3 | 417 |
blastx of nr database at NCBI http://www.ncbi.nlm.nih.gov/BLAST/
blastp of nr at NCBI using translated ORF
Fig. 5.
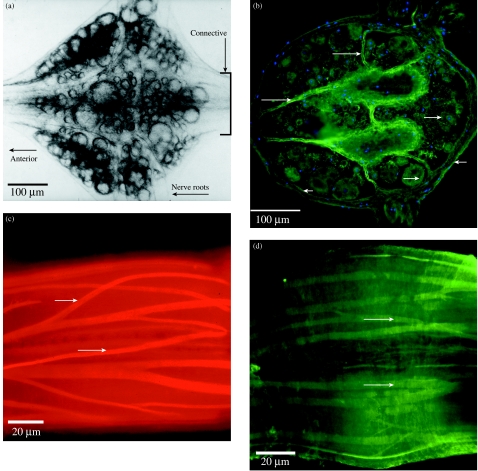
Expression of the novel peptide, designatd ReN3, in nerve cells, connective tissue and muscle of adult leech CNS. ReN3 is one of four novel clones isolated to date from the 24-h regenerating ganglion library. It has no known homologue in vertebrate genomes. (A) Isolated segmental ganglion viewed under transmitted light; ventral surface. The boxed area of the connectives is shown in C and D. (B) Horizontal section through the ganglion; orientation as in A. Immunohistochemistry using an antibody to a custom peptide shows that the encoded ReN3 peptide is widely expressed in neuron cell bodies (long arrows), in the connective tissue capsule that surrounds the ganglion (short arrows) and in the internal capsule of the ganglion (long arrows). The section is counterstained with DAPI to show nuclei. (C,D) ReN3 is also expresed in the muscle cells that extend throughout the ventral nerve cord within the connective tissue capsule. (C) A confocal section through part of the connectives that link adjacent ganglia of the ventral nerve cord (see boxed area in A), in a preparation stained with rhodamine–phalloidin to illustrate the muscle layout. (D) A confocal section through an equivalent region stained with the ReN3 antibody.
Construction of cDNA probes from whole ganglia and screening of the single neuron library for genes up- and down-regulated at 24 h post-injury
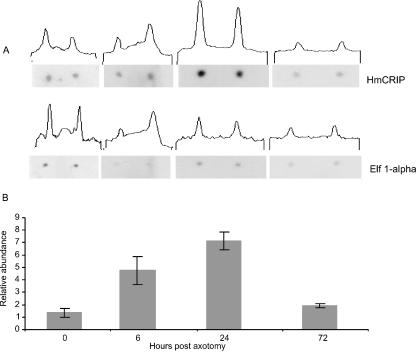
We have used cDNA from normal and regenerating ganglia to construct probes with which we have screened the single neuron library. The design of these experiments has enabled us to identify clones that are down-regulated as well as up-regulated 24 h after injury and which are also expressed in the Retzius neuron. cDNA made from intact ganglia and 24-h regenerating ganglia was used in forward and reverse subtractive hybridizations that enabled us to compare the normal and regenerating populations of cDNA, and to identify clones that are expressed in one population but not the other. The subtractive probe referred to as the control probe represents those sequences present in the normal ganglion and not present at 24 h of regeneration, i.e. clones that are down-regulated after damage. The subtractive probe referred to as the 24-h regenerating probe represents those sequences that are up-regulated after injury. We then used the ‘control’ and ‘24-h regenerating’ probes to screen differentially the Retzius neuron library (Fig. 6). This screening protocol has identified further regulated clones. One clone identified in this way is the leech homologue of CRIP, a protein previously only found in the chordates. It was first identified in developing cells of mammalian intestine and proposed to modulate the immune cell response (Birkenmeier & Gordon, 1986; Levenson et al. 1993). It was subsequently shown to be expressed in developing mammalian neurons (Shirasaki & Pfaff, 2002). Our experiments demonstrate for the first time that CRIP is expressed in adult serotonergic nerve cells, and is up-regulated after injury (Emes et al. 2003). Figure 7 illustrates CRIP expression over time in a semi-quantitative way. CRIP expression in leech CNS peaks at 24 h post-injury with levels approximately 7 times greater than at time zero, estimated relative to the control gene Elongation Factor α-1. We do not know what role CRIP plays in the injured CNS, nor whether it has an immunological function in the leech. Invertebrates are not thought to possess adaptive immune defence systems based on lymphocytes and antibodies. One possibility is that the CRIP gene in Hirudo encodes a cytokine-like molecule that is released as a result of damage and is in some way involved in the control of an inflammatory response (for discussion see Emes et al. 2003).
Fig. 6.
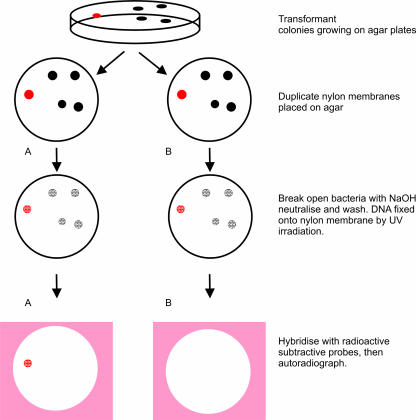
Protocol used to screen differentially the Retzius neuron library to identify clones regulated 24 h after injury and which are also expressed in this serotonergic neuron. Clones from the library were transferred in bulk to nylon membranes. Duplicate plaque lifts from 90-mm plates are shown diagrammatically. Two copies of the clones were made (A,B) to allow hybridization with the control and 24-h regenerating probes. This diagram illustrates screening of five clones: clones from the Retzius library that have hybridized with the control probe in A and with the 24-h regenerating probe in B. Comparing these autoradiographs identifies clones that are differentially expressed. For example, the red spot in A indicates a clone that is down-regulated after axotomy, denoted by the positive hybridization signal on the control film, not present on the 24-h film. The autoradiograph was then used to identify the relevant clone on the agar colony and these were excised from the agar, the DNA purified and sent for sequencing.
Fig. 7.
Semiquantitative expression of Cysteine Rich Intestinal Protein (CRIP): filters were dotted with spots of two genes, CRIP and Elongation Factor 1-α. The filters were probed with radiolabelled cDNA produced from five ganglia at chosen time points. Specific activity is measured using an NCBI image programme that determines how dark each spot was. This gives an assessment of the degree of hybridization, and hence a measure of expression level.
Genes encoding voltage-gated sodium channels in leech
In the experiments described so far we have used an open-ended approach to identify the panel of genes regulated in regenerating nerve cells, rather than a search for a particular gene or genes. A different approach is to pursue specific genes encoding for known proteins that will be required by a regenerating neuron for rebuilding its axon, for example the genes encoding ion channels required for electrical signalling by the axon. Changes in excitability after axotomy, detected using physiological techniques, are a common theme in invertebrate and mammalian neurons, although it has been more difficult to assign function to the excitability changes, and comparatively little is known about the molecular basis for the excitability changes.
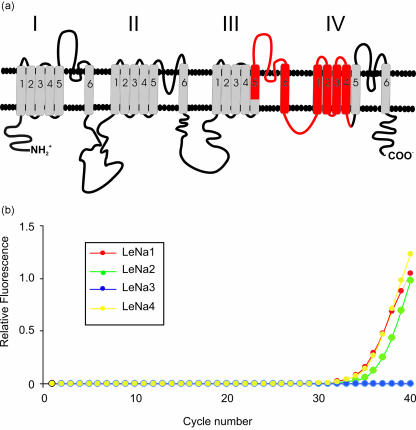
We have identified genes encoding voltage-gated sodium channels in the leech using degenerate primers and PCR (Blackshaw et al. 1999, 2003) and we have used real-time PCR to study expression of different isoforms of the channel in identified neurons in the adult ganglion. Figure 8 shows a cartoon of the α-subunit of a voltage-gated Na channel depicting the four domains I–IV, each with six probable transmembrane helices. The four domains come together in three dimensions to form the pore of the channel.
Fig. 8.
Identification of genes encoding voltage-gated sodium channel isoforms in single leech neurons. (A) Cartoon of the α-subunit of a voltage-gated Na channel representing the phospholipid bilayer; and the four domains of the channel labelled I–IV. Each of these domains has six probable transmembrane helices represented by the cylinders. The four domains come together in three dimensions to form the pore of the channel. To isolate Na-channel genes in Hirudo, we designed degnerate primers by comparing the amino acid sequence of the α-subunit of a rat brain (type IIa) Na-channel with published sequences of the putative channel in a range of different invertebrates (sea slug, fruit fly, squid, flatworm and jellyfish), and identified regions in domains III and IV of the α-subunit where the amino acid sequence is highly conserved. These primers span a segment of the α-ubunit, shown in red, 900 nucleotides long, between segment S5 of domain III and segment S4 of domain IV. This approach has isolated four homologous yet distinct isoforms of the leech Na-channel, designated LeNa 1–4. (B) Analysis of isoform expression in single Retzius neurons using real-time (quantitative) PCR shows that Retzius cells express multiple isoforms of the channel, LeNa isoforms 1, 2 and 4, but not isoform 3.
To isolate Na channel genes in Hirudo medicinalis, degenerate primers were designed by comparing the amino acid sequence of the α-subunit of a rat brain (type IIa) Na-channel with published sequences of the putative channel in a range of different invertebrates (sea slug, fruit fly, squid, flatworm and jellyfish). Regions in domains III and IV of the α-subunit where the amino acid sequence is highly conserved were identified and primers designed based on these regions. The primers span a segment of the α-subunit 900 nucleotides long, between segment S5 of domain III and segment S4 of domain IV, shown in red in Fig. 8. This part of the sequence includes a number of regions of functional importance, for example the IFM motif in the intracellular loop between domains III and IV that is critical for fast inactivation of the channel. The primers were used in PCR reactions with cDNA from nerve cord and body wall muscle, and this approach has isolated four homologous yet distinct leech sodium channel cDNAs, named LeNa 1–4 (Blackshaw et al. 1999, 2003).
We have analysed LeNa isoform expression in single leech nerve cells using isoform-specific primers and two different methods of RT-PCR, conventional and real-time. These experiments have shown that individual Retzius cells express more than one isoform of the channel – LeNa isoforms 1, 2 and 4 but not isoform 3 (Fig. 8). This is the first demonstration of multiple isoforms of voltage-gated sodium channels in an identified invertebrate neuron (Blackshaw et al. 2003). In current experiments we are studying isoform expression during regeneration (J. Malek & S. E. Blackshaw, unpublished observations).
Summary and conclusions
Questions about the molecular mechanisms of neural regeneration can be approached effectively in invertebrates because questions of specificity can be addressed using identifiable neurons. In the leech nervous system, which has a demonstrated capacity for functional repair, we have identified genes that change their pattern of expression after injury. To do this we have constructed cDNA libraries from identified and regenerating neurons dissected from leech ganglia, as well as from whole segmental ganglia. Using subtracted cDNA probes from normal and regenerating cells, we have isolated genes that change their expression after injury and which are also present in an identified 5-HT neuron of the leech. Our approach has enabled us to identify genes that are up- as well as down-regulated after injury and that are present in a defined cell type.
The sequences we have identified include both known and novel clones. Some sequences are homologues of known proteins in other species including humans. Other sequences identified are novel; and some of these sequences have homologues in mammalian expressed sequence tag (EST) databases; other sequences, such as myohemerythin and the novel peptides that we have isolated, are only found in invertebrate phyla.
We have identified genes whose expression pattern is altered at 1 week after injury – a relatively late stage of regeneration – as well as genes regulated within hours of axotomy. Successful regeneration probably depends on genes that both enhance a cell's likelihood to survive after injury, as well as genes that promote neurite extension and synapse formation. By determining the changes in gene expression that take place at different times after damage we are building a broader picture of the molecular events in a single neuron.
Throughout these experiments we have borne in mind the following questions. Are the insights to repair in the leech relevant to neural repair in humans? Are there shared molecular cues? Are there molecules and mechanisms that are unique to invertebrates? Despite the apparent diversity, studies of the development of invertebrates and vertebrates have revealed common themes at the molecular level. Similarly, the phenomenon of neural regeneration is based on molecular pathways intrinsic to neurons, and responses to a remarkably conserved chemical language.
Acknowledgments
This work was supported by the Human Frontiers Science Programme, the EP Abraham Cephalosporin Fund, the MRC and the Wellcome Trust.
References
- Bannatyne BA, Blackshaw SE, McGregor MS. New growth elicited in adult leech mechanosensory neurones by peripheral axon damage. J. Exp. Biol. 1989;143:419–434. doi: 10.1242/jeb.143.1.419. [DOI] [PubMed] [Google Scholar]
- Baylor DA, Nicholls JG. Patterns of regeneration between individual nerve cells in the central nervous system of the leech. Nature. 1971;232:268–270. doi: 10.1038/232268a0. [DOI] [PubMed] [Google Scholar]
- Birkenmeier EH, Gordon JI. Developmental regulation of a gene that encodes a cysteine-rich intestinal protein and maps near the murine immunoglobulin heavy chain locus. Proc. Natl Acad. Sci. USA. 1986;83:2516–2520. doi: 10.1073/pnas.83.8.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw SE, Gross RH, Porter DM, Henderson LP, Maue RA. Cloning of a family of putative sodium channel α subunit cDNA's from the leech. J. Physiol. 1999;518P:126. [Google Scholar]
- Blackshaw SE, Henderson LP, Malek J, Porter DM, Gross RH, Angstadt JD, et al. Single-cell analysis of a novel family of voltage-dependent sodium channel genes in the leech demonstrates cell-specific patterns of expression. J. Neurobiol. 2003;55:355–371. doi: 10.1002/neu.10214. [DOI] [PubMed] [Google Scholar]
- Bruns D, Jahn R. Real-time measure of transmitter release from single synaptic vesicles. Nature. 1995;377:62–65. doi: 10.1038/377062a0. [DOI] [PubMed] [Google Scholar]
- Bruns D, Riedel D, Klingauf J, Jahn R. Quantal release of serotonin. Neuron. 2000;28:205–220. doi: 10.1016/s0896-6273(00)00097-0. [DOI] [PubMed] [Google Scholar]
- Coleman SK, Cai C, Mottershead DG, Haapalahti J-P, Keinanen K. Surface expression of GluR-D AMPA receptor is dependent on an interaction between its C-terminal domain and a 4.1 protein. J. Neurosci. 2003;23:798–806. doi: 10.1523/JNEUROSCI.23-03-00798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emes R, Wang W-Z, Blackshaw SE. Subtracted cDNA libraries from regenerating leech ganglia identify transcripts upregulated at different times post-axotomy. J. Physiol. 2002;539P:108P. [Google Scholar]
- Emes RD, Wang W-Z, Lanary K, Blackshaw SE. HmCRIP, a cysteine-rich intestinal protein, is expressed by an identified regenerating nerve cell. FEBS Lett. 2003;533:124–128. doi: 10.1016/s0014-5793(02)03741-9. [DOI] [PubMed] [Google Scholar]
- Fedorov A, Johnston H, Korneev S, Blackshaw SE, Davies JA. The alpha-tubulin genes of the leech, Hirudo medicinalis. Gene. 1999;227:11–19. doi: 10.1016/s0378-1119(98)00603-9. [DOI] [PubMed] [Google Scholar]
- French KA, Muller KJ. Regeneration of a distinctive set of axosomatic contacts in the leech central nervous system. J. Neurosci. 1986;6:318–324. doi: 10.1523/JNEUROSCI.06-02-00318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann JC, Kins S, Rostaing P, El Far O, Kirsch J, Sheng M, et al. Gephyrin interacts with dynein light chains 1 and 2, components of motor protein complexes. J. Neurosci. 2002;22:5393–5402. doi: 10.1523/JNEUROSCI.22-13-05393.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingwater TH, Ribchester RR. Compartmental neurodegeneration and synaptic plasticity in the WldS mutant mouse. J. Physiol. 2001;534:627–639. doi: 10.1111/j.1469-7793.2001.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson LP, Kuffler DP, Nicholls JG, Zhang R. Structural and functional analysis of synaptic transmission between identified leech neurons in culture. J. Physiol. 1983;340:347–358. doi: 10.1113/jphysiol.1983.sp014766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Sato-Yoshitake R, Yoshida T, Kawashima T. Brain dynein (MAP 1C) localizes on both anterogradely and retrogradely transported membranous organelles in vivo. J. Cell Biol. 1990;111:1027–1037. doi: 10.1083/jcb.111.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SM, Vaughan KT, Herskovits JS, Vallee RB. Molecular analysis of a cytoplasmic dynein light intermediate chain reveals homology to a family of ATPase. J. Cell Sci. 1995;108:17–24. doi: 10.1242/jcs.108.1.17. [DOI] [PubMed] [Google Scholar]
- King SM, Dillman JF, III, Benashski SE, Lye RJ, Patel-King RS, Pfister KK. The mouse t-complex-encoded protein Tctex-1 is a light chain of brain cytoplasmic dynein. J. Biol. Chem. 1996;271:32281–32287. doi: 10.1074/jbc.271.50.32281. [DOI] [PubMed] [Google Scholar]
- Koert CE, Spencer GE, Van Minnen J, Li KW, Geraerts WPM, Syed NI, et al. Functional implications of neurotransmitter expression during axonal regeneration: serotonin, but not peptides, auto-regulate axon growth of an identified central neuron. J. Neurosci. 2001;21:5597–5606. doi: 10.1523/JNEUROSCI.21-15-05597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneev S, Blackshaw SE, Davies JA. cDNA libraries from a few neural cells. Prog. Neurobiol. 1994;42:339–346. doi: 10.1016/0301-0082(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Korneev S, Blackshaw SE, Kaiser K, Davies JA. cDNA libraries from identified neurons. Proc. Royal Soc. Series B. 1996;263:56–72. doi: 10.1098/rspb.1996.0010. [DOI] [PubMed] [Google Scholar]
- Korneev S, Fedorov A, Collins R, Blackshaw SE, Davies JA. A subtractive cDNA library from an identified regenerating neuron is enriched in sequences upregulated during nerve regeneration. Invertebrate Neurosci. 1997;3:185–192. doi: 10.1007/BF02480373. [DOI] [PubMed] [Google Scholar]
- Lamonte BH, Wallace KE, Holloway BA, Shelly SS, Ascano J, Tokito M, et al. Disruption of dynein/dynactin inhibits axonal transport in motor neurons causing late-onset progressive degeneration. Neuron. 2002;34:715–727. doi: 10.1016/s0896-6273(02)00696-7. [DOI] [PubMed] [Google Scholar]
- Levenson CW, Shay NF, Lee-Ambrose LM, Cousins RJ. Regulation of cysteine-rich intestinal protein by dexamthasone in the neonatal rat. Proc. Natl Acad. Sci. USA. 1993;90:712–715. doi: 10.1073/pnas.90.2.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye RJ, Porter ME, Scholey JM, McLntosh JR. Identification of a microtubule-based cytoplasmic motor in the nematode C. elegans. Cell. 1987;51:309–318. doi: 10.1016/0092-8674(87)90157-7. [DOI] [PubMed] [Google Scholar]
- McAdoo DJ, Coggeshall RE. Gas chromatographic–mass spectrometric analysis of biogenic amines in identified neurons and tissues of Hirudo medicinalis. J. Neurochem. 1976;26:163–167. [PubMed] [Google Scholar]
- Moffett SB. Nervous System Regeneration in the Invertebrates. Zoophysiology 34. Berlin: Springer-Verlag; 1996. [Google Scholar]
- Mullis KB. The unusual origin of the polymerase chain reaction. Sci. Am. 1990;262:36–43. doi: 10.1038/scientificamerican0490-56. [DOI] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, Dafonesca GAB, Kent J. Biodiversity hotspots for conservation priority. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Nicholls JG. The Search for Connections: Studies of Regeneration in the Nervous System of the Leech. Sunderland MA: Sinauer; 1987. [Google Scholar]
- Ready D, Nicholls JG. Identified neurons isolated from the leech CNS make selective connections in culture. Nature. 1979;281:67–69. doi: 10.1038/281067a0. [DOI] [PubMed] [Google Scholar]
- Retzius G. Zur Kenntniss des centralen Nervensystems des Wurmer. Biol. Untersuchsungen, Neue Folge. 1891;II:1–28. [Google Scholar]
- Shen L, Liang F, Walensky LD, Huganir RL. Regulation of AMPA receptor GluR1 subunit surface expression by a 4.1N–linked actin cytoskeleton association. J. Neurosci. 2000;20:7932–7940. doi: 10.1523/JNEUROSCI.20-21-07932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neural identity. Annu. Rev. Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Trueta C, Mendez B, De Miguel FF. Somatic exocytosis of serotonin mediated by 1-type calcium channels in cultured leech neurons. J. Physiol. 2003;547:405–416. doi: 10.1113/jphysiol.2002.030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB, Wall JS, Paschal BM, Shpetner HS. Microtubule-associated protein 1C from brain is a two-headed cytosolic dynein. Nature. 1988;332:561–563. doi: 10.1038/332561a0. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Jansen J. The specificity of re-innervation by identified sensory and motor neurons in the leech. J. Compar. Neurol. 1977;171:433–454. doi: 10.1002/cne.901710402. [DOI] [PubMed] [Google Scholar]
- Velazquez-Ulloa N, Blackshaw SE, Szczupak L, Trueta C, Garcia E, De Miguel FF. Convergence of mechanosensory inputs onto neuromodulatory serotonergic neurons in the leech. J. Neurobiol. 2003;54:604–617. doi: 10.1002/neu.10184. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Meyer D, Deehan R, Blenis J, Schnapp BJ, Rapoport TA. Cargo of kinesin identified as JIP scaffolding proteins and associated signalling molecules. J. Cell Biol. 2001;152:959–970. doi: 10.1083/jcb.152.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W-Z, Christoffers K, Emes R, Blackshaw SE. Genes regulated 24 hours after axotomy in identified 5-HT neurons in leech. J. Physiol. 2002;539P:107P. [Google Scholar]
- Willard AL. Effects of serotonin on the generation of the motor program for swimming by the medicinal leech. J. Neurosci. 1981;1:936–944. doi: 10.1523/JNEUROSCI.01-09-00936.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]