Abstract
The short interspersed repetitive element (SIRE) of Trypanosoma cruzi was first detected when comparing the sequences of loci that encode the TcP2β genes. It is present in about 1,500–3,000 copies per genome, depending on the strain, and it is distributed in all chromosomes. An initial analysis of SIRE sequences from 21 genomic fragments allowed us to derive a consensus nucleotide sequence and structure for the element, consisting of three regions (I, II, and III) each harboring distinctive features. Analysis of 158 transcribed SIREs demonstrates that the consensus is highly conserved. The sequences of 51 cDNAs show that SIRE is included in the 3′ end of several mRNAs, always transcribed from the sense strand, contributing the polyadenylation site in 63% of the cases. This study led to the characterization of VIPER (vestigial interposed retroelement), a 2,326-bp-long unusual retroelement. VIPER's 5′ end is formed by the first 182 bp of SIRE, whereas its 3′ end is formed by the last 220 bp of the element. Both SIRE moieties are connected by a 1,924-bp-long fragment that carries a unique ORF encoding a complete reverse transcriptase-RNase H gene whose 15 C-terminal amino acids derive from codons specified by SIRE's region II. The amino acid sequence of VIPER's reverse transcriptase-RNase H shares significant homology to that of long terminal repeat retrotransposons. The fact that SIRE and VIPER sequences are found only in the T. cruzi genome may be of relevance for studies concerning the evolution and the genome flexibility of this protozoan parasite.
The kinetoplastid protozoan parasite Trypanosoma cruzi is the etiological agent of Chagas disease (1). It belongs to one of the earliest extant groups of eukaryotes containing mitochondria (2). Because of their ancient lineage, they have retained unusual biological properties. T. cruzi genes are organized in polycistronic transcription units, where they are interspaced by short intergenic regions, ranging in size from 150 to 500 nt (3). Because T. cruzi seems not to regulate expression at the level of transcription initiation, and because transcription is polycistronic, modulation of gene expression is achieved initially by processing of polycistronic primary transcripts into monogenic mRNAs by means of 5′-end trans-splicing [spliced leader (SL) addition] and 3′-end polyadenylation (4–6). Both reactions occur within the intergenic regions of the polycistronic mRNAs and are coupled and governed by a common pPy tract, which is part of the SL acceptor site (7, 8). Disruption of this bifunctional signal sequence should directly affect expression of 5′ and 3′ adjacent flanking genes. The only naturally occurring situation in which such a disruption has been thoroughly documented is the insertion of a short interspersed repetitive element (SIRE) within the canonical pPy tract of the TcP2β-H1.8 gene locus, composed mainly of dTs (9). Upon insertion at this site, SIRE enlarges the distances between TcP2β and the gene upstream, destroys the original pPy signal, and presents an SL acceptor site to the TcP2β-H1.8 gene. This determines, in turn, the generation of a TcP2β-H1.8 mRNA containing, after the SL sequence, 38 bases directly transcribed from SIRE (9), and a decrease of about 40% in the efficiency of the trans-splicing reaction (10). Because of these functional properties, SIRE is unique among all previously described repetitive elements of T. cruzi and kinetoplastids in general.
Recent studies have shown that SIRE is present in about 1,500–3,000 copies per genome, depending on the strain, and that it is distributed in all chromosomes (11). Analysis of 16 genomic fragments containing SIRE sequences revealed that it frequently is linked to protein coding genes (11).
The present study was designed to evaluate the species specificity of SIRE, its conservation within the T. cruzi genome, and its relation with expressed protein coding sequences. It was expected that this analysis would disclose functional properties of this element and give some clue as to the mechanism involved in its mobility.
Materials and Methods
Parasites.
T. cruzi epimastigotes from Tulahuen 2 strain and CL-Brener clone were used in this study. Epimastigotes were grown in liver infusion tryptose (LIT) medium supplemented with 10% of FBS at 28°C (12).
Cloning Procedures.
In this study, we used a genomic T. cruzi Tulhauen 2 strain λZAPII library (11), a bloodstream trypomastigote RA strain cDNA library (13), and a normalized and a non-normalized epimastigote CL-Brener clone library, both from the T. cruzi genome project (14). High-density colony filters were constructed at Research Genetics (Huntsville, AL). Genomic fragments and cDNAs were isolated by hybridization with radioactive probes as described (11). Unless otherwise stated, sequences from cDNAs were obtained by first-pass automatic sequencing, performed as described by Vazquez et al. (11). Approximately 1 μg of plasmid template was sequenced with the Prism Ready Reaction Dye Deoxy Terminator Cycle Sequencing Kit (Applied Biosystems) by using the T7 and SIRE primers (see above) or with the Prism Ready Reaction Dye Primer (M13 forward) Cycle Sequencing Kit (Applied Biosystems) according to manufacturer's instructions. The reactions were run in ABI 373 and ABI 377 automated DNA sequencers (Applied Biosystems).
DNA and RNA Analysis.
Genomic DNA from different trypanosomatids used in Fig. 1 were kindly provided by F. Breniere (Instituto Boliviano de Biologia de la Altura ORSTOM, La Paz, Bolivia). Pulse-field gel electrophoresis, Southern blots, and dot blot hybridization were performed as described (9–11), with the exception of the Southern blot in Fig. 1, where hybridization and washing were performed at 55°C instead of the currently used 65°C. Total RNA and poly(A)+ RNA were obtained from 107 parasites and were prepared by using the RNeasy mini extraction kit (Qiagen, Chatsworth, CA) and the oligotex direct mRNA extraction kit (Qiagen), respectively. Northern and antisense probes were performed as described by Levin et al. (15).
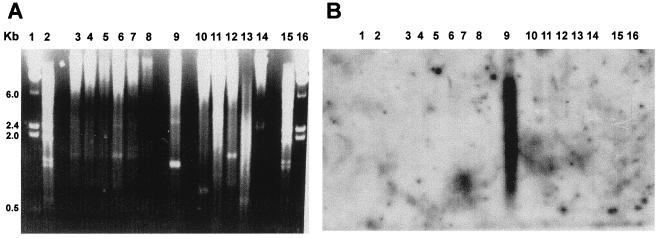
Figure 1.
Species-specificity analysis of SIRE. (A) Ethidium bromide-stained gel of EcoRI-digested genomic DNA of the following kinetoplastid species: lanes 3, Trypanosoma brucei brucei; 4, T. brucei gambiense; 5, T. brucei EAT20; 6, Trypanosoma rangeli basel; 7, T. rangeli palma; 8, T. rangeli riera; 9, T. cruzi; 10, Leptomonas collosoma; 11, Phytomonas sp. (host euphorbia characias); 12, Phytomonas sp. (hartrot disease); 13, Leishmania mexicana; 14, Crithidia fasciculata. Lanes 1 and 16, λ BsteII DNA marker; lanes 2 and 15, λ HindIII DNA marker. (B) Southern blot hybridization with a SIRE probe. Hybridization conditions were used to allow identification of low-conserved sequences.
Reverse Transcriptase (RT)-PCR Assays.
The RT-PCR was carried out as follows: 1 μg of total RNA was reverse-transcribed with 5 units of avian myeloblastosis virus reverse transcriptase by using random hexamers as primers. cDNA synthesis was performed in 50 mM KCl/0.1% Triton X-100/5 mM MgCl2/1 mM of each dNTP/20 units of human placental ribonuclease inhibitor for 40 min at 40°C. Thereafter, the cDNAs were amplified by using an oligonucleotide derived from the T. cruzi SL sequence, SL sense, 5′-AACGCTATTATTGATACAGT-3′, and one of the following antisense SIRE primers: S0, TCCTCCGGGCAGCTGGCCGGATCCTGA; S1, GGAGGACCCCAAAGTCTGCC; S2, GATGCTGGGAGAGCTGGCTA; S3, CTTACGAAGTGGCAGACTTT; HA-1, GCCAGCTCTCCCACGATCATTC. The cycling profile was as follows: 94°C for 60 sec, 52°C for 50 sec, and 72°C for 60 sec and was repeated 35 times.
Sequence Analysis.
To find expressed sequence tags (ESTs) containing SIRE, the EST database (dbEST) of T. cruzi from the National Center for Biotechnology Information (NCBI) was probed with the reference SIRE sequence by using the blastn program on the blast network service. Homologies to the reverse transcriptase of VIPER (vestigial interposed retroelement) were searched through the NCBI protein database using the blastx, blastp, and psi-blast programs (http://www.ncbi.nlm.nih.gov/BLAST/). To obtain the consensus SIRE, the sequences of SIRE derived from the genomic fragments characterized in ref. 11 and those reported under the following accession nos.: X83272, U05588, M65021, AF080220, and AF052833, were aligned by using the clustal w program.
Results
Species Specificity of SIRE.
The first SIRE sequence characterized was derived from the TcP2β-H1.8 locus (9). In this study, we used it as reference sequence for database searches and sequence comparisons, and as a probe to evaluate, among other features, the species specificity of SIRE. As shown in Fig. 1, SIRE hybridizes with EcoRI-digested genomic DNA of T. cruzi (lane 9) but not with that of other Trypanosoma species or related trypanosomatids. Taking into account that filters were washed at low stringency, this result clearly indicates that SIRE is T. cruzi specific, a result corroborated by database searches that found SIREs in DNA sequences from T. cruzi strains belonging to T. cruzi lineages 1 and 2 (16), but not in sequences from related microorganisms.
Variability, Basic Consensus Structure, and Target Integration Site of SIRE.
To evaluate the variability of SIREs in T. cruzi and to define a SIRE consensus structure, we used the SIREs derived from 16 genomic fragments of T. cruzi Tulahuen 2 strain (11) and from five different genomic sequences obtained from databases (see Materials and Methods). All SIREs could be aligned and were classified in three groups according to their homology with the reference element (9). Group A was composed by the SIREs that shared >70% homology with the reference element (14/21). Group B included copies of the element that presented only between 40% and 70% homology to SIRE TcP2β-H1.8 (2/21). The reduction of homology is partly caused by the amplification of the dinucleotide TA at the 3′ end of the element (variable part within region III, see Fig. 2A, and GenBank accession nos. X83600 and Y09145). Group C elements included five remnants of SIRE composed by short fragments of conserved SIRE sequences (GenBank accession nos. M65021, AF052833, TENU0906, and AF080220).
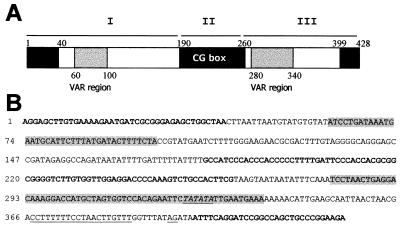
Figure 2.
(A) Schematic representation of a consensus SIRE element. Black boxes indicate the three highly conserved regions (>90%), and gray boxes indicate the two most variable regions (VAR regions, 30–50%). White boxes represent middle conserved domains (60–90%). Comparisons were made with respect to the reference SIRE (9). Horizontal bars, marked I, II, and III, identify the three regions of SIRE (see text). (B) Consensus SIRE sequence obtained from the analysis of groups A and B. Bold letters denote the highly conserved regions in the 5′ and 3′ ends and the highly conserved CG-rich region; gray-shaded letters indicate the VAR regions; the trans-splicing signal present in region III is underlined.
The consensus structure and sequence of SIRE are depicted in Fig. 2. The first 190 bp of the element (region I) start with the 5′AGGA sequence followed by a GCTTGTGA motif similar to the Chi sequence of phage λ (9). These first 12 bp and the 28 bp that follow are highly conserved. Between positions 60 and 100, sequences are variable, and region I ends at position 190, after a run of dTs with single dA insertions. The central portion of SIRE (region II) is most conserved and characterized by a high CG content. Region III contains the most variable portion of the element, easily identified by changes in the number of TA repeats. This domain is followed 40 bp downstream by a conserved DNA segment that contains the functional trans-splicing signal of SIRE. The element ends with the conserved A(A/G)GA motif.
The most probable target site of insertion was deduced from the analysis of sequences adjacent to SIRE (Table 1). The left consensus is TTTTTtNTTTTTNTT, while the pentanucleotide TTATT (Table 1) is the putative duplication of the target insertion site followed by a 3′consensus CT(G/T)PyPy.
Table 1.
Analysis of SIRE insertion target sequences
| Left (n = 23) | TDS | Right (n = 14) | |
|---|---|---|---|
| SIRE | TTCTTTTTTTTTCTT | TTATT | CTGCT |
| S-Hl8 | GTTGGCCTTTTTTTT | TTATC | TCTTT |
| S31 | TCGTGAGCTTTTATT | TTATT | CTGCT |
| S38 | ACGTGGCTTTTTATT | ND | ND |
| S10 | ATTTTTTTTTTTTTT | ND | ND |
| S12 | CTACTTTTTTTTCTT | ND | ND |
| S7 | TTTTTAATCTTATCT | ATATT | TTTAG |
| S14 | TTTTTAGTAAAATTT | TTATT | CCTTC |
| S2 | GGTTGGGTTTTTTCT | TTATT | CTCCT |
| S1 | CCTACATGGGTTCAC | TTATT | CAGCC |
| S4 | GTTTCCTGTTTTCAT | ND | ND |
| S15 | TTTTTGGGATTTTTT | TTATT | CCCTC |
| N9 | ATTTATATATTTATT | ND | ND |
| N10 | ATTTTTATATTTATT | ND | ND |
| N4 | TTTTGGGAGTCTGTT | ND | ND |
| N6 | TCTCCCCACTGTTGT | TTGTT | CTGTA |
| N3 | TCTTTTTTTTTTTCC | ND | ND |
| N7 | GCTCTCATCATGGCT | TTATT | GAACC |
| 05 | TTTTTAGTGAAATTT | ATATT | TTTTC |
| 016 | TTATTTATTTTTATT | TTATT | CGGGA |
| 018 | TTCTTTAATCTTATG | ND | ND |
| 011 | AGTAATTTATTTAAT | TTATT | CAGCC |
| 020 | TTTGTTGTTGTTGTT | TTATT | CTGTT |
| CONS. | TTTTTtNTTTTTNTT | TTATT | CT (G/T) PyPy |
Left, sequences immediately upstream of different SIRE insertions; right, sequences immediately downstream. Cons, concensus. The name of different genomic clones or cDNAs used in the analysis are indicated on the left side: SIRE, reference sequence (9); S, genomic clones; O, cDNA clones from the non-normalized library; N, cDNA clones from the normalized library. TDS, target duplication sequence; ND, not determined.
Expression of SIRE Elements.
Previous results indicated that SIRE could be expressed in the 5′ untranslated region (UTR) of the TcP2β mRNA (9), and database searches indicated that an EST with homology to the genomic marker SZ23 contained part of a SIRE in its 3′ end (11). This finding prompted us to study the representation of transcribed SIRE sequences in the mRNA population of epimastigotes. To this effect, we screened high-density colony filters containing 55,296 cDNA clones of the non-normalized and 55,296 cDNA clones of the normalized cDNA libraries of the T. cruzi genome project with the reference SIRE probe. The number of positive clones was 1,260 and 947, indicating that SIRE is present in 2.3% and 1.7% of the cloned mRNAs, respectively. Hybridization of the reference SIRE probe with a colony filter containing 5,000 trypomastigote cDNA clones showed 110 positives, confirming that SIRE is represented in 2.2% of the mRNAs. Database searches confirmed these results. The SIRE sequence hits 90 T. cruzi ESTs out of the 5,000 T. cruzi ESTs reported to the dbEST. This finding implies that SIRE is represented in 1.8% of the sequences, in accordance with results above, because most of the ESTs were obtained from the normalized T. cruzi epimastigote library.
In a first attempt to classify the expressed elements, we sequenced the SIREs of 68 cDNAs from the normalized library. These 68 SIRE sequences and 90 SIRE sequences from the dbEST were aligned with the reference SIRE. A majority of them, 51.3%, were from group A, 44.3% belonged to group B, while 4.4% were SIRE remnants, group C. The regions where the variations clustered are shown in gray in Fig. 2 A and B.
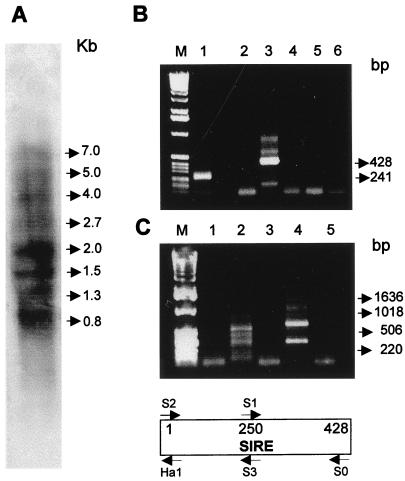
Northern hybridization of total RNA from T. cruzi epimastigotes reveals that SIRE is represented in mRNA species the size of which ranges from 800 to 7,000 b (Fig. 3A). We did not detect any transcript of 428 b, indicating that the element is not transcribed as such. However, as predicted from the previous sequence data, RT-PCR assays confirmed that mRNAs contained complete SIRE elements. Indeed, Fig. 3B shows the amplicons of the 5′ half (primers S2–S3; Fig. 3B, lane 1) and the complete element (primers S2–S0; Fig. 3B, lane 3).
Figure 3.
Transcription of SIRE elements. (A) Northern blot analysis with total T. cruzi RNA using a SIRE antisense strand-specific probe. Size of the most prominent bands are indicated in kb. (B) RT-PCR analysis of SIRE transcription. M, 1-kb ladder DNA marker (GIBCO/BRL); lane 1, amplification with primers S2–S3; lane 2, amplification with sense primers S2–S1; lane 3, amplification with primers S2–S0; lane 4, primers S2–S0, but without the addition of RT; lane 5, primers S2–S0, but without the addition of RNA during the RT reaction; lane 6, reaction without the addition of cDNA products. (C) RT-PCR analysis, orientation of SIRE transcripts. M, 1-kb ladder DNA marker; lane 1, SL primer and S2; lane 2, SL and S3; lane 3, SL and S1; lane 4, SL and Ha1; lane 5, SL and S3 without the addition of RT. The structure of SIRE with the indication of primer locations are shown below C.
Orientation of the SIRE Sequence.
Previous results have shown that the 5′-3′ SIRE sequence is oriented along the sense strand of transcription (9). That this orientation was kept at the level of mRNAs was confirmed by the fact that the SIRE hybridizing mRNAs reacted only with the antisense SIRE probe (Fig. 3A). To corroborate the orientation of the element and its linkage to the SL sequence in monocistronic T. cruzi mRNAs, we designed a series of RT-PCR assays using an SL-derived oligonucleotide as sense primer and different SIRE-derived oligonucleotides S1, S2, S3, and Ha1 as antisense primers. Amplicons were observed only when S3 (Fig. 3C, lane 2) and Ha1 (Fig. 3C, lane 3) primers were used in combination with the SL primer, confirming the strand specificity of SIRE transcription, as well as its linkage with the SL sequence.
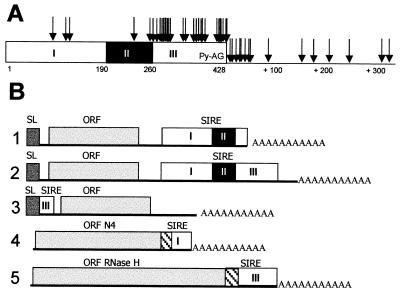
All of the results led us to the assumption that SIREs were located at the 3′ end of the mRNAs, on the sense strand of transcription. To test this hypothesis, we compared the sequences of 48 SIRE-containing cDNAs of the normalized epimastigote library and three from the trypomastigote library. It was noteworthy that in 32 out of 51 cDNAs (63%) SIRE provided the polyadenylation site (Fig. 4B, lane 1), whereas the remaining 37% of the cDNAs used polyadenylation sites located mostly within the 50 bp downstream of SIRE (Fig. 4B, lane 2). As it is shown in Fig. 4A, there is a strong tendency to polyadenylate within region III and immediately downstream, leaving, in many cases, a complete SIRE in the 3′ UTR of the mRNAs, a fact explaining the results obtained when sequencing ESTs and those observed in Fig. 3B, lane 3. In three cases, however, SIRE not only contributed the polyadenylation site but also the codons that encode the C-terminal end of two different ORFs. This was the case for the 680-bp-long cDNA N4 (GenBank accession no. Z82099) that encodes an ORF with no homology to any known protein sequence, for which the first 39 bp of region I establish the 13 C-terminal residues of the putative protein, and 43 bp downstream the polyadenylation site (Fig. 4B, lane 4). The ORFs derived from the cDNAs N5, 679 bp, and SX2, 716 bp, (sharing 87% homology at the nucleotide level) both encode for a peptide homologous to RNase H (RH). This ORF is completed by addition of 15 C-terminal amino acids derived from codons specified by the central portion of SIRE's region II, whereas the polyadenylation sites were provided by region III for SX2, and by the target duplication site TTATT for N5 (Fig. 4B, lane 5).
Figure 4.
(A) Polyadenylation sites within or near SIRE. Arrows point out poly(A) addition sites as they were identified in cDNAs containing SIRE. The black box within SIRE represents region II; in region III, py-AG indicates the position of SIRE's trans-splicing signal (9). (B) Schematic representation of five different situations where transcribed SIRE sequences are found: lanes 1, in the 3′ UTR, SIRE provides the poly(A) addition site; 2, in the 3′ UTR, poly(A) addition immediately after SIRE; 3, in the 5′ UTR when SIRE acts as a trans-splicing signal donor (9); 4 and 5, SIRE provides the codons that encode the C-terminal end of ORFs (hatched boxes). The ORFs N4 and RH are not depicted in scale. SL boxes represent SL sequences.
By analysis of the complete nucleotide sequence of cDNAs, we confirmed that the transcribed SIRE conserves its 5′-3′ orientation on the sense strand, and that it is present in the 3′ UTR of different protein coding genes, such as histone H2A, 2-hydroxyacid dehydrogenase, thimet oligopeptidase, lysosomal α-mannosidase, and an aldehyde dehydrogenase, or to several ORFs with unknown function.
SIRE Is Related to VIPER, an Unusual Retroelement.
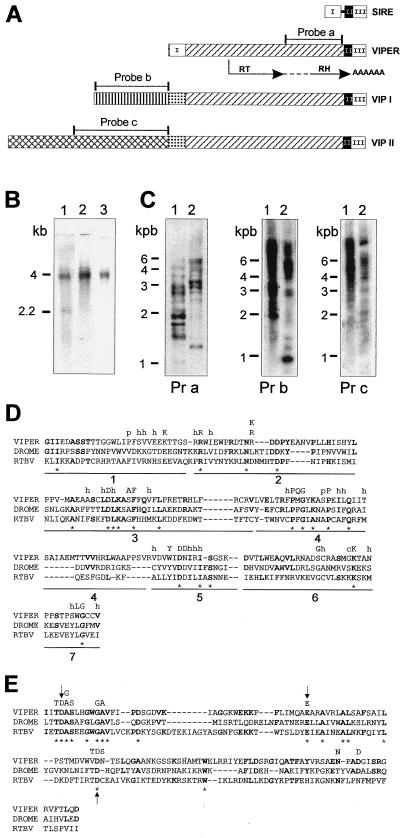
The relationship of SIRE with the coding region of a gene encoding an RH domain suggested that SIRE could be in fact associated with a RT. Screening of the genomic library with a SX2 probe led to the isolation of three genomic fragments: VIPER, 2,359 bp; VIPER-associated element (VIP) I, 3,188 bp; and VIP II, 4,395 bp (Fig. 5A). The sequence of VIPER showed that its 3′ end was formed by the same truncated form of SIRE found in SX2 and N5 cDNAs (Figs. 4 and 5), whereas its 5′ end was formed by the 5′ moiety of the element. Thus, in VIPER, the 182-bp-long 5′ and the 220-bp-long 3′ moieties of SIRE were separated by a 1,924-bp-long connecting segment (Fig. 5A). A partial reconstitution of VIPER's SIRE showed that it was >93% homologous to the reference SIRE, conserving also its typical insertion site and its corresponding duplication site TTATT. However, it was impossible to reconstitute a complete SIRE from the moieties of VIPER because the highly conserved first 30 bp of region II (Fig. 2), GCCATCCCACCCACCCCCTTGATTCCCACC, were not found, direct or inverted, inside VIPER (Fig. 5A).
Figure 5.
(A) Schematic representation of VIPER elements. Identical boxes represent highly homologous regions. SIRE is represented with its three regions as in Fig. 2A. The connecting line between region I and region II represents the 30-bp sequence of SIRE lost in VIPER. The RT and RH domains are indicated by arrows below the diagram of VIPER; the dotted line connecting the two domains indicates the 103-residue-long tether. The position of the probes used in hybridization experiments also are indicated. (B) Northern blot analysis of T. cruzi epimastigotes poly(A)+ RNA. Lane 1, probe a; lane 2, probe b; lane 3, probe c. Sizes of the bands identified are indicated on the left. (C) Southern blot analysis of T. cruzi genomic DNA. Lane 1, EcoRI digest; lane 2, BamHI digest. Pr a, probe a; Pr b, probe b; Pr c, probe c. Sizes of the molecular DNA markers are indicated on the left. (D) Alignment of VIPER RT domains with those of Drosophila melanogaster Burdock LTR-retrotransposon, DROME (18) and rice tungro bacilliform pararetrovirus, RTBV (19). The conserved subdomains proposed by Xiong and Eickbush (17) are numbered 1–7. Amino acids conserved between VIPER and the other two sequences are shown in bold, and * denotes residues conserved in all three. The highly conserved residues in the analysis of Xiong and Eickbush (17) are indicated above the comparison at the corresponding position; h, hydrophobic residue; p, small polar residue. (E) Alignment of RH domains. Highly conserved residues in the analysis of Doolittle et al. (20) are noted above the amino acid sequence. Arrows indicate the three crucial residues referred to in the text.
Comparison of the complete nucleotide sequence of VIPER with that of SX2 and N5 revealed which was its coding strand. Although the three reading frames of VIPER contained stop codons, this comparison, together with relevant EST sequences presenting homology with the central part of VIPER, allowed the reconstruction of a unique ORF for VIPER. Accordingly, VIPER would appear to be a pseudogene copy of a progenitor sequence with one or perhaps more ORFs occupying most of its length.
The reconstructed ORF starts at position 708 and stretches 487 amino acid residues downstream to a stop codon located inside the 3′ end of SIRE, as in SX2 (Fig. 5A). Typical for this ORF is the presence of RT and RH motifs separated by a 103-residue tether. Fig. 5D shows a typical alignment of RT sequences (17) derived from VIPER, the Burdock LTR-retrotransposon of Drosophila melanogaster (18) and the rice tungro bacilliform virus (RTBV) (19). Twenty of 183 residues are identical between the compared sequences (* in Fig. 5D). Individually, homologies are 45/183 between VIPER and Burdock, and 29/183 between VIPER and RTBV. It is noteworthy that the RT of VIPER does not share homology with RTs from non-LTR-retrotransposons. Fig. 5E shows alignment of the RH domains derived from those elements. Three residues (D10, E45, and D79) reported to be crucial for activity of RH (20) also are found in the three above-mentioned RHs (arrows in Fig. 5E).
A probe that spans the RH domain revealed the presence of multiple copies of VIPER in the genome (Fig. 5C, Pr a) distributed in four chromosomal bands ranging from 0.96 to 1.6 Mb (not shown). Dot blot hybridization assays indicated that there are approximately 300 copies of this or related elements in the genome of T. cruzi CL Brener clone, and Southern blot experiments demonstrated that VIPER, as SIRE, is specific of T. cruzi (not shown).
Sequence of the other two genomic fragments VIPs I and II demonstrate that they can be aligned with VIPER, because they share with it the 220-bp-long 3′ SIRE sequence and the 1,924-bp-long connecting fragment, including the ORF that encodes for the RT-RH polypeptide. Surprisingly, neither VIPI nor VIPII carries the 5′ 182-bp-long SIRE sequence; they contain instead a region of 207 bp with low similarity to the 5′ SIRE sequence (Fig. 5A). On the contrary, from this position toward the 5′ end of the fragments, the homology between VIPI and VIPII fades. In fact, sequence and Southern blot analysis show that they encode for different repetitive sequences (Fig. 5C, Pr b and Pr c). Analysis of the sequences of VIP I and VIP II also show a clustering of stop codons at the 5′ end of the coding strand; the only discernible ORF being the one homologous to VIPER's RT-RH. Accordingly, VIPI and II seem to be, as VIPER, inactive copies of progenitor sequences.
Northern blots hybridized with a probe derived from the common to VIPER, VIP I, and VIP II RH domain showed two mRNA species of about 2,200 and 4,000 bp, whereas probes derived from the unrelated 5′ ends of VIPs only hybridized mRNA species of approximately 4,000 bp (Fig. 5B, lanes 1–3).
This result can be explained assuming that VIPER is the inactive copy of a progenitor middle repetitive element that encodes a 2,200-bp mRNA species, while VIPs may be interpreted as being pseudogenes of longer elements that encode mRNAs of about 4,000 bp.
Discussion
In T. cruzi, several repeated sequences have been characterized (11, 21). Their analysis led to the observation that some were linked to the non-LTR retrotransposon L1Tc (21), closely related to the non-LTR retrotransposon Ingi of T. brucei (22), and more distantly related to the first non-LTR retrotransposon described for T. cruzi, the SL site poson CZAR (23). In turn, it has been suggested that SIRE could be related to a non-LTR retrotransposon because a truncated SIRE is adjacent to different repetitive sequences, such as C6 (24), E12, and E13 (21). Those authors found that this structural arrangement resembled that of the T. brucei Ingi element, which carries half of the mobile element RIME at either end (24). However, the substantiation of this hypothesis required a more detailed assessment of the species specificity, conservation, and functionality of SIRE.
In this report, we show that the SIRE sequence is specific of T. cruzi (Fig. 1, lane 9) and that it is highly conserved among T. cruzi strains.
The analysis of more than 150 SIRE sequences shows that the majority presents high homology with the reference SIRE (group A). Those of group B, although more different than the reference sequence, were still recognizable as complete elements, with highly conserved motifs at both ends and in the center, accounting for a third of the total SIRE sequence (Fig. 2). This conservation implies that either SIREs are successful genomic “parasites,” or that positive selection imposes genomic maintenance (25). The functional properties of SIRE speak for the latter hypothesis. In other reports, we have demonstrated that SIRE is able to donate a SL acceptor site when located immediately upstream of a protein coding gene (9), and that this insertion generates a less expressed mRNA species that includes 38 bp of the transcribed 3′ end of SIRE in its 5′ UTR (Fig. 4B, lane 3). Herein, we show that the SIRE sequence also is included in the 3′ end of mRNAs, always transcribed from the sense strand. SIRE then contributes the polyadenylation site in 63% of the studied cases (Fig. 4 A and B). In three instances, that of N4, N5, and SX2 cDNAs, it contributes in addition codons that encode the C-terminal peptides of the corresponding ORFs (Fig. 4B, lanes 4 and 5). Interestingly, the latter function is assumed by the most constant regions of SIRE, i.e., its first bases, and region II, respectively. These features of SIRE are important because in T. cruzi the polyadenylation site and surrounding intergenic sequences play a capital role for stage-specific expression of genes (26).
The analysis of cDNAs that use SIRE sequences as donor of coding regions and polyadenylation sites led to the characterization of VIPER and VIPs. We have shown that this retroelement is related to LTR-retrotransposons in T. cruzi and kinetoplastids in general, with no sequence homology to non-LTR-retrotransposons of this order (22, 23). VIPER contains only one discernible ORF with homology to RT-RH genes of LTR-retroelements (25). As VIPER's RT-RH is a pseudogene, its sequence may represent a variation of a copy of the original active VIPER element. Because LTRs that flank the coding region of LTR-retrotransposons have not been identified in VIPER, it is tempting to propose that the function of these sequences may be assumed by the 5′ and 3′ moieties of SIRE. The 5′ end of SIRE may function as an SL site donor or as a promoter region, while the 3′ moiety of SIRE functions as the poly(A) site donor. This arrangement of SIRE in a functional VIPER element probably would generate mRNA species of about 2,200 bp, as shown in Fig. 5B, lane 1.
VIPs do not present a 5′ SIRE sequence in their 5′ end, but conserve the 3′ end of SIRE with the same characteristics as in VIPER. Thus, in the active copies of VIPs, the SL acceptor site or the promoter region should be located in the 5′ end at least in VIP II, in accordance with an mRNA species of about 4,000 bp.
SIRE may have arisen by deletion of the 1,924-bp central region from a VIPER element, or VIPER may have arisen by insertion of the central region into a SIRE. Because a deletion event would have to be accompanied by a simultaneous addition of 30 bp of region II to conform a typical SIRE element, the hypothesis of an insertion within SIRE, accompanied with a concomitant loss of 30 bp seems most probable. Whether an insertion or a deletion was responsible for the generation of SIREs and VIPER, analogies may be drawn between them and other mobile elements in which a shorter element, making up the ends of a longer one, carries the sequences required in cis for transposition, and the longer element carries an encoded function required in trans (25).
LTR-retrotransposons promote a variety of recombination events in genomes, such as chromosome translocations, duplications, and deletions (25). We have identified duplication events with a particular arrangement of SIRE sequences, namely SIRE-gene-SIRE-gene-SIRE (9). SIREs have been found in the 3′ end of genes located in subtelomeric regions (11, 27), and VIPER and SIREs have been identified in a transcription strand-switch region of chromosome III (10, 11, 28). Moreover, the presence of a ribosomal pseudogene in a chromosome different from the one where the ribosomal genes normally are clustered is associated with an adjacent VIPER element (11, 29). It is thus possible that an additional role for SIRE, VIPER, and VIPs is the promotion of recombination, chromosome breaks, and perhaps even translocation of genes to subtelomeric regions.
The fact that SIRE and VIPER sequences are found only in T. cruzi may be of relevance for studies about concerted evolution of parasites and retroelements, particularly those regarding T. cruzi genome flexibility.
Acknowledgments
We thank Mariana Catalani for technical assistance. This work was supported by the T. cruzi Genome Project-Subprograma III Pragrama Ciencia y Tecnología para el Desarrollo of Iberoamerica; World Health Organization Special Programme for Research and Training in Tropical Diseases TDR; Projet Genome T. cruzi-Foundation Jean Dausset-Centre d'Étude du Polymorphisme Humain, Universidad de Buenos Aires, Ministere d'Affaires Etrangeres, France; the T. cruzi Genome Project (Centro Argentino–Brasileño de Biotecnologìa); Project IX17, University of Buenos Aires; Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina and Fondo Nicional de Ciencia y Tecnica/Proyectos de Investigación Científica y Tecnológica 01421 (Argentina). M.J.L. is a John Simon Guggenheim Foundation Fellow.
Abbreviations
- SIRE
short interspersed repetitive element
- SL
spliced leader
- UTR
untranslated region
- dbEST
database of expressed sequence tags
- RT
reverse transcriptase
- RH
RNase H
- LTR
long terminal repeat
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. Y08881, Y09442, Y09145, Y10371, Y12063, Y09742–45, Y12774, Y09115, Y09441, AJ0042, Z82099, AF096925–26, AF098062–65, and AF227564–227618).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050578397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050578397
References
- 1.Rosenbaum M B. Prog Cardiovasc Dis. 1964;7:199–225. doi: 10.1016/s0033-0620(64)80020-7. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez A P, Nelson K, Beverley S M. Proc Natl Acad Sci USA. 1993;90:11608–11612. doi: 10.1073/pnas.90.24.11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kendall G, Wilderspin A F, Ashall F, Miles M A, Kelly J M. EMBO J. 1990;9:2751–2758. doi: 10.1002/j.1460-2075.1990.tb07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang J, Van der Ploeg L H T. EMBO J. 1991;10:3877–3885. doi: 10.1002/j.1460-2075.1991.tb04957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapotas N, Bellofatto V. Nucleic Acids Res. 1993;17:4067–4072. doi: 10.1093/nar/21.17.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.López-Estranio C, Tschudi C, Ullu E. Mol Cell Biol. 1998;8:4620–4628. doi: 10.1128/mcb.18.8.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LeBowitz J, Smith H, Rusche L, Beverly S. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 8.Matthews K, Tschudi C, Ullu E. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 9.Vazquez M, Schijman A, Levin M J. Mol Biochem Parasitol. 1994;67:327–336. doi: 10.1016/0166-6851(94)00026-3. [DOI] [PubMed] [Google Scholar]
- 10.Vazquez M, Levin M J. Gene. 1999;239:217–225. doi: 10.1016/s0378-1119(99)00386-8. [DOI] [PubMed] [Google Scholar]
- 11.Vazquez M, Lorenzi H, Schijman A, Ben-Dov C, Levin M J. Gene. 1999;239:207–216. doi: 10.1016/s0378-1119(99)00387-x. [DOI] [PubMed] [Google Scholar]
- 12.Kelly J, Ward H, Miles M, Kendall G. Nucleic Acids Res. 1992;15:3963–3969. doi: 10.1093/nar/20.15.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy Yeyati P, Bonnefoy S, Mirkin G, Debrabant A, Lafon S, Panebra A, Gonzalez-Cappa E, Dedet J P, Hontebeyrie-Joskowicz M, Levin M J. Immunol Lett. 1991;31:27–34. doi: 10.1016/0165-2478(92)90006-a. [DOI] [PubMed] [Google Scholar]
- 14.Urmenyi T, Bonaldo M, Soares M, Rondinelli E. J Eukaryot Microbiol. 1999;46:542–544. doi: 10.1111/j.1550-7408.1999.tb06072.x. [DOI] [PubMed] [Google Scholar]
- 15.Levin M J, Mesri E, Benarous R, Levitus G, Schijman A, Levy-Yeyati P, Chiale P, Ruiz A, Kahn A, Rosembaum M, et al. Am J Trop Med Hyg. 1989;41:530–539. doi: 10.4269/ajtmh.1989.41.530. [DOI] [PubMed] [Google Scholar]
- 16.Briones M, Souto R, Stolf B, Zingales B. Mol Biochem Parasitol. 1999;104:219–232. doi: 10.1016/s0166-6851(99)00155-3. [DOI] [PubMed] [Google Scholar]
- 17.Xiong Y, Eickbush T. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tchurikov N A, Gerasimova T I, Johnson T K, Barbakar N I, Kenzior A L, Georgiev G P. Mol Gen Genet. 1989;219:241–248. doi: 10.1007/BF00261183. [DOI] [PubMed] [Google Scholar]
- 19.Hay J M, Jones M C, Blakerbrought M L, Dasgupta I, Davies J W, Hull R. Nucleic Acids Res. 1991;19:2615–2621. doi: 10.1093/nar/19.10.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doolittle R, Feng D, Johnson M, McClure M. Q Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 21.Martín F, Marañón C, Olivares M, Alonso C, López C. J Mol Biol. 1995;247:49–59. doi: 10.1006/jmbi.1994.0121. [DOI] [PubMed] [Google Scholar]
- 22.Kimmel B E, Ole-Moiyoi O K, Young J. Mol Cell Biol. 1987;7:1465–1475. doi: 10.1128/mcb.7.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villanueva M, Williams S, Beard C, Richards F, Aksoy S. Mol Cell Biol. 1991;11:6139–6148. doi: 10.1128/mcb.11.12.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Araya J, Cano M I, Gomes H B M, Novak E M, Requena J M, Alonso C, Levin M J, Guevara P, Ramirez J L, da Silveira F J. Parasitology. 1997;115:563–570. doi: 10.1017/s0031182097001674. [DOI] [PubMed] [Google Scholar]
- 25.Li W-H. In: Molecular Evolution. Sinauer A D, editor. Sunderland, MA: Sinauer; 1997. pp. 335–360. [Google Scholar]
- 26.Weston D, Patel B, Van Vorhis W. Mol Biochem Parasitol. 1999;98:105–116. doi: 10.1016/s0166-6851(98)00152-2. [DOI] [PubMed] [Google Scholar]
- 27.Chiurrillo M, Cano I, da Silveira J, Ramirez J. Biochem Parasitol. 1999;100:173–183. doi: 10.1016/s0166-6851(99)00047-x. [DOI] [PubMed] [Google Scholar]
- 28.Andersson B, Aslund L, Tammi M, Tran A N, Hoheisel J D, Pettersson U. Genome Res. 1998;8:809–816. doi: 10.1101/gr.8.8.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vieira de Arruda M, Reinach F, Colli W, Zingales B. Mol Biochem Parasitol. 1990;40:35–42. doi: 10.1016/0166-6851(90)90077-y. [DOI] [PubMed] [Google Scholar]