Abstract
SWAP-70 is a component of an enzyme complex that recombines Ig switch regions in vitro. We report here the cloning of the human cDNA and its B lymphocyte-specific expression. Although its sequence contains three nuclear localization signals, in small resting B cells, SWAP-70 is mainly found in the cytoplasm. On stimulation, SWAP-70 translocates to the nucleus. In activated, class-switching B cell cultures, it is associated with membrane IgG, but not IgM. The membrane Ig association requires a functional pleckstrin homology domain and is controlled by the C terminus. We suggest that SWAP-70 is involved not only in nuclear events but also in signaling in B cell activation.
The B cell receptor (BCR) is composed of the Ig heavy and light chains and of the covalently bound accessory molecules Ig-α and Ig-β. Crosslinking of the BCR by antigen stimulates the activation of intracellular protein tyrosine kinases, including src-family members. These events lead to the initiation of signaling cascades, including activation of vav, phosphatidylinositol 3-kinase, and ras (reviewed in refs. 1 and 2). How initiation of the appropriate signaling cascade is regulated is not yet understood in detail. Apart from blast formation, cell division, and differentiation into plasma or memory cells, B cell activation also leads to hypermutation of the Ig variable regions and to heavy chain class switching. In the latter process, the Ig constant region of μ is replaced by that of another class (γ, α, ɛ). Class switching is achieved by a looping out and deletion mechanism between the switch region of μ and the switch region of the isotype that is to be expressed (reviewed in ref. 3). Although the Ku70 and the Ku86 proteins of the general DNA repair machinery are necessary for completing the heavy chain class switch (4, 5), the elements that recognize specific switch regions and cause a DNA break—the putative switch recombinase—have remained elusive. Also, the biochemical link between the activated BCR on the cell surface and activation, or inactivation, of the switch recombinase in the nucleus is not known. In this paper, we explore the possibility that the SWAP-70 protein acts as a link between these two processes. SWAP-70 was isolated in the mouse as part of a complex that is able to promote recombination between two switch regions in vitro (6, 7). Here, we report the cloning of the human homolog and a study of its cellular and intracellular localization before and after B cell stimulation.
Materials and Methods
Antibodies.
Polyclonal antibody to the purified recombinant SWAP-70 protein was generated in rabbit and affinity purified (6). It was used at a concentration of 10 μg/ml. Secondary antibody for immunofluorescence was a goat anti-rabbit Alexa 488 conjugate (Molecular Probes). Antibodies to human CD2, clones 6G4 and 4B2, were a gift of Lucien Aarden (Central Laboratory of the Netherlands Red Cross Blood Transfusion Service, Amsterdam).
Screening of cDNA Library.
The human Raji Burkitt lymphoma cDNA library (CLONTECH) was screened by hybridization with two fragments from the mouse SWAP-70 cDNA sequence (accession no. AF053974) spanning nucleotides 1–256 and 442-1342, respectively. Plaques that hybridized with both fragments were used for a secondary screening. λgt11 DNA from positive plaques was prepared, and the cDNA insert was sequenced. The 5′ end of the cDNA was determined by 5′ rapid amplification of cDNA ends (CLONTECH).
Immunohistochemistry.
Immunostaining was performed on 4-μm formalin-fixed, paraffin-embedded tissue sections. Sections were deparaffinized and rehydrated, and the antigen was retrieved by using Declere (Cell Marque, Austin, TX) according to manufacturer's protocol. Slides were bar-coded for use on the Ventana Gen II automated immunostainer. Staining was performed according to the Ventana diaminobenzidine staining and detection protocol. Slides were counterstained with hematoxylin, dehydrated in ethanol, and mounted with Permount (Fisher).
Immunofluorescence.
Cytospins were fixed in 3% paraformaldehyde solution for 15 min at room temperature and rinsed in PBS. Cells were permeabilized by incubating for 5 min in staining buffer (PBS containing 5% goat serum, 0.1% Triton X-100, and 0.1% sodium azide). The first antibody was diluted to the final concentration in staining buffer, applied onto cells, and incubated for 1 h. After three washes of 5 min each in staining buffer, the secondary antibody was applied and incubated for 30 min. Cytospins were washed three times in staining buffer, one time in PBS, and a final time in tap water, and mounted in mounting medium (Fluoromount-G; Southern Biotechnology Associates).
Human Peripheral Blood Lymphocyte Activation.
Human purified peripheral blood lymphocytes were in vitro activated as previously described (8). Briefly, human peripheral blood from healthy volunteers was fractionated through Ficoll (Amersham Pharmacia), and lymphocytes in the interphase were recovered. Contaminating monocytes and natural killer cells were depleted by incubating the cells with a mixture containing anti-human CD14 and CD16 monoclonal antibodies (PharMingen), followed by immunomagnetic beads (Dynal, Great Neck, NY). Purified B and T lymphocytes were plated in activating medium [RPMI medium 1640 supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 10% FCS, 50 μM 2-mercaptoethanol, 20 μg/ml human transferrin (GIBCO), 1 μg/ml anti-CD2 antibody clone 6G4 and 1 μg/ml anti-CD2 antibody clone 4B2, 3 ng/ml IL-2 (Roche Molecular Biochemicals), and 50 ng/ml IL-4 (GIBCO)] at 5,000 cells in 200 μl per well in a round-bottom 96-well plate. Cultures were tested for class switching to IgE by an IgE ELISA at day 9 of activation. For activation of purified B lymphocytes, T cells were depleted by incubating the mononuclear cells after Ficoll fractionation with a mixture of anti-CD3, anti-CD4, and anti-CD8 antibodies, followed by immunomagnetic beads. Purified B lymphocytes were plated in activating medium (RPMI medium 1640 supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 10% FCS, 50 μM 2-mercaptoethanol, 20 μg/ml human transferrin, 1 μg/ml anti-CD40, and 50 ng/ml IL-4) at 5,000 cells in 200 μl per well in a round-bottom 96-well plate.
Cocapping.
For cocapping experiments, cells were incubated with the appropriate antibody at 37°C for 30 min, without the use of sodium azide. Cells were then washed, centrifuged on a slide, fixed in 3% paraformaldehyde, and stained for SWAP-70.
Coimmunoprecipitation.
Lipopolysaccharide B (LPS; S. typhosa 0901)-activated mouse spleen cells and cell lines WEHI-279 and 2PK3 were washed in PBS and lysed on ice for 30 min in a buffer containing 20 mM Tris⋅HCl (pH 7.4), 100 mM (NH4)2SO4, 0.1 mM EDTA, 2 mM MgCl2, 30 mM β-glycerol phosphate, 1 mM sodium orthovanadate, protease inhibitors, and 0.75% Brij 58. Antibodies used for immunoprecipitation were prebound to protein A-Sepharose. Immunoprecipitations were carried out at 4°C for 3 h.
Subcellular Fractionation.
A total of 107 cells were harvested and washed twice in PBS. The cell pellet was resuspended in 400 μl of hypotonic buffer [10 mM Hepes (pH 7.5)/5 mM KCl/2 mM EDTA/2 mM DTT/1 mM Na3VO4/protease inhibitors) and incubated on ice for 15 min to allow cells to swell. Cells were opened by passing the suspension several times through a 25-gauge needle; cell breakage was monitored under the microscope. The suspension was spun at 6,000 rpm in an Eppendorf 5415 C tabletop centrifuge for 5 min. Pelleted nuclei were washed twice in hypotonic buffer containing 0.5% Nonidet P-40 to lyse unbroken cells. Nuclei were opened with 400 μl of hypotonic buffer containing 0.5% SDS. Postnuclear supernatant was spun at 100,000 × g for 30 min to pellet the microsomal fraction. The cleared cytosolic fraction was recovered, and the membrane pellet was dissolved in 400 μl of hypotonic buffer containing 0.5% SDS.
Mutagenesis.
Green fluorescent protein (GFP) cDNA (CLONTECH) was fused in-frame upstream of wild-type SWAP-70 by conventional cloning procedures. PCR primers encoding N- or C-terminally truncated proteins were designed and used to create SWAP-70 mutants fused to GFP (including 1–525, 1–312, and 1–312 ΔNLS) and cloned into the retroviral expression vector pGfus. In the 1–312 ΔNLS construct, the NLS (291–294 KKKK) was mutated to 291–294 EMNT, the sequence in the corresponding region of β-spectrin pleckstrin homology (PH) domain (9). The point mutation W297→A, in the PH domain of full-length SWAP-70 fused to GFP, was constructed by using Quik-Change (Stratagene). Stable BJAB cell lines expressing each of the mutants were generated by using standard retroviral techniques (10). Crosslinking of the BCR in BJAB was obtained by incubating the cells with an anti-human Ig antibody for 30 min at room temperature.
Cell Culture.
Retroviral producer cells, Phoenix-A (10, 11), and human BJAB (12) cells were maintained as described. To generate recombinant retroviruses expressing GFP-SWAP and SWAP mutants, Phoenix-A producer cells were transfected by calcium phosphate procedure as described (10), and supernatants were harvested for infection. BJAB cells were infected by the spin method at 2,500 rpm, 32°C for 90 min (13).
Results and Discussion
SWAP-70 Human Homolog.
We isolated the human cDNA homolog of SWAP-70 from the Raji Burkitt lymphoma cell line. The human is 95% homologous to the mouse protein, and it contains several functional domains (Fig. 1). There are three nuclear localization signals, two “pat4” (HRRK; KKKK) and one “bipartite” (KKLEEAASRAAEEEKKR) (14, 15). Nuclear localization signals are required for proteins bigger than 70 kDa to translocate to the nucleus through the nuclear pore (14). SWAP-70 also has a putative nuclear export signal (NES) near its C terminus. Such signals are contained in proteins like HIV-1 Rev, MEKK, PKI, and in c-abl (16). Although mutation analysis is needed to address the activity of the putative NES in SWAP-70, its presence suggests that SWAP-70 shuttles to and from the nucleus. SWAP-70 also contains a PH domain between residues 211 and 306 (Fig. 1). The PH domain is found in many intracellular proteins that participate in signaling events and is thought to mediate membrane attachment (17). SWAP-70 also contains a number of putative phosphorylation sites and, indeed, it is tyrosine phosphorylated in the cytoplasm (R.J., unpublished data). The region encompassing amino acids 3–72 (Fig. 1) forms an EF hand, which is a binding site for calcium. These characteristics of SWAP-70 are all consistent with its putative role in signaling.
Figure 1.
Human SWAP-70 amino acid sequence in single-letter code, and comparison with mouse SWAP-70. The relevant domains are annotated: yellow boxes, nuclear localization signals; green, nuclear export signal; blue, pleckstrin homology; and brown, EF hand. Dashes, identity.
Tissue Expression.
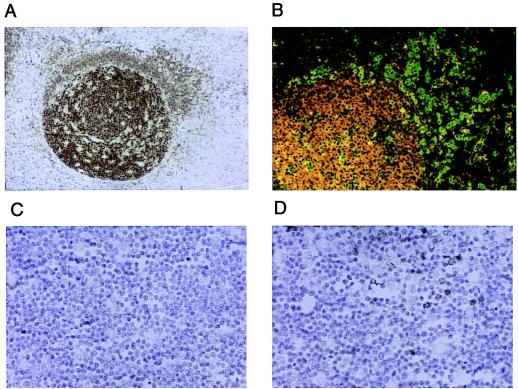
We have studied the distribution of SWAP-70 in various human tissues by staining tissue sections with a polyclonal anti-SWAP-70 antibody. In all tissue tested, SWAP-70 showed to be expressed only in mature B lymphocytes, including those associated with MALT (mucosa-associated lymphoid tissue) and BALT (bronchus-associated lymphoid tissue) structures. In Fig. 2A, B lymphocytes in a germinal center (GC) of a tonsil are strongly stained for SWAP-70, whereas B lymphocytes in the corona show a much weaker staining. When the polyclonal antibody is diluted 5-fold compared with the concentration used for this photograph, the staining of the corona disappears, whereas that of GC's B cell remains still strong (data not shown). Although the corona also contains plasma cells, they do not express SWAP-70, as shown in Fig. 2B, where the plasma cells are stained with an anti-VS38c antibody (green) and with the anti-SWAP-70 antibody (red). The expression of SWAP-70 does not superimpose with expression of VS38c antigen (the few yellow cells, with seemingly overlapping staining, are in fact high intensity green cells). SWAP-70 is also not expressed in pre-B lymphocytes, as staining of bone marrow was negative (not shown). In T lymphocytes, SWAP-70 is not expressed at any stage of development, neither in the spleen, nor in the thymus. Although the cortex of thymus is completely negative (Fig. 2C), some staining is observed in the medulla (Fig. 2D). These could be either B cells or dendritic cells; in any event, these are the only cells we found to stain outside of the B lymphocytic compartments; no other cell type has been found to express SWAP-70, including cells in brain, kidney, skin, prostate, and small intestine.
Figure 2.
Expression of human SWAP-70 in tonsil and thymus in paraffin-embedded tissue sections. (A and B) Germinal center of tonsil. (C and D) Cortex and medulla, respectively, of thymus. (A, C, and D) Stained with rabbit anti-SWAP-70 antibody. (B) Paraffin-embedded tissue section stained with anti-SWAP-70 antibody, followed by a Texas Red-coupled anti-rabbit Ig and FITC-coupled anti-VS38c.
Translocation on CD40 Activation.
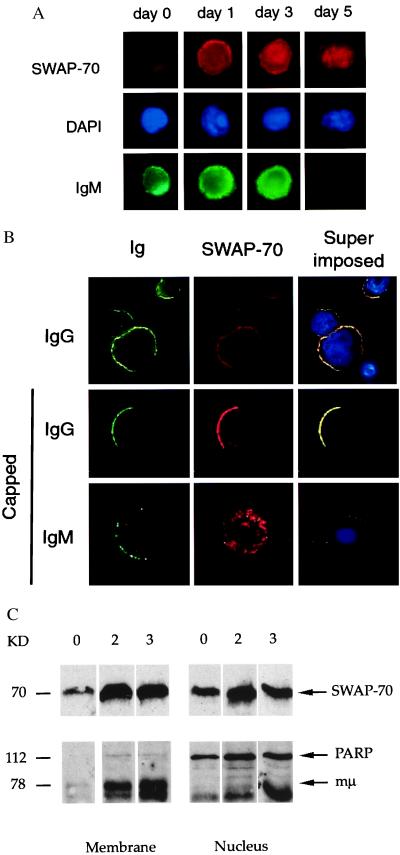
The presence of nuclear localization and export signals, the PH domain, and the specific expression of SWAP-70 on mature B lymphocytes suggest that its intracellular localization is regulated during B cell development. To study the localization of SWAP-70 in B cells, we set up two different cultures. One consisted of primary human peripheral blood B and T lymphocytes, cultured in the presence of T cell stimulatory factors (anti-CD2, IL-2) and IL-4, the other consisted of primary human peripheral blood purified B cells, cultured in the presence of anti-CD40 and IL-4. In both cultures, the presence of either T cells or anti-CD40, together with IL-4, promotes both B cell proliferation and Ig heavy chain class switching (8). Under these conditions, B cells start to proliferate at day 3, and secreted Ig of switched isotypes are detected starting at day 5. It should be remembered that, in these cultures, cells are not synchronized, and that, therefore, at any given time point, there are cells at different activation stage. In resting B cells, SWAP-70 remains at low level and it localizes diffusely over the cell (Fig. 3A, day 0). Immediately after activation, SWAP-70 expression strongly increases and, in addition to its cytoplasmic localization, it is also found associated with the plasma membrane (day 1). Following membrane association, SWAP-70 translocates to the nucleus (day 3) and, as cells lose IgM expression (i.e., in switched cells), it is detected almost exclusively in the nucleus (day 5). Later, at day 7 of activation, in switched cells, the expression of SWAP-70 in the nucleus greatly decreases, and it associates again with the plasma membrane. In these cells, SWAP-70 is found to colocalize with membrane IgG (Fig. 3B, Top). At this stage of activation, although to some degree SWAP-70 expression is associated with the plasma membrane also in IgM-positive cells, colocalization with the BCR was apparent only in IgG-positive cells, as shown in cocapping experiments (Fig. 3B, Middle and Bottom). We do not know whether SWAP-70 exits the nucleus to associate with membrane IgG, but the presence of a putative NES in the SWAP-70 sequence suggests this as a possibility.
Figure 3.
SWAP-70 translocation in in vitro-activated human peripheral blood B lymphocytes. (A) SWAP-70 translocation to the nucleus. Double immunofluorescence of B lymphocytes at days 0, 1, 3, and 5 of activation; red, SWAP-70; green, IgM; blue, 4′,6-diamidino-2-phenylindole (DAPI) nuclear staining. (B) SWAP-70 membrane localization and cocapping with membrane Ig at day 7 of activation; green, IgG or IgM; red, SWAP-70; blue, DAPI nuclear staining; and yellow, colocalization of SWAP-70 with membrane Ig. First row shows staining of noncapped cells, second and third rows show staining of cells with capped IgG or IgM. (C) SDS/PAGE of membrane (lanes 1–3) and nuclear (lanes 4–6) fractions of LPS-stimulated spleen cells. 0, 0 days; 2, 2 days; 3, 3 days. The gels were blotted with antibody to SWAP-70 (Upper) and antibody to poly(ADP-ribose) polymerase plus anti-Ig (Lower).
We have also attempted to follow the translocation of SWAP-70 by fractionation of cellular components and subsequent SDS gel electrophoresis. In Fig. 3C, the amount of SWAP-70 in membrane and nucleus fractions of LPS-activated B cells is assayed at days 0, 2, and 3. Although same cell numbers were applied, day 0 cannot be readily compared with days 2 and 3, as the resting B cells have much less surface, and, therefore, less membrane. To some degree, SWAP-70 is found both in the membrane and in the nucleus in unstimulated splenic B cells ex vivo (lanes 1 and 4); the amounts vary from spleen to spleen in that often there is no SWAP-70 in the nucleus. However, it is evident that on days 2 and 3, SWAP-70 is present both in the cell membrane (lanes 2 and 3) and in the nucleus (lanes 5 and 6). As internal markers for fraction identification, there is membrane μ-chain at 78 kDa, which is present in the membrane (lanes 2 and 3), but absent in the nucleus (lanes 5 and 6); and the enzyme poly(ADP-ribose) polymerase, at 112 kDa, which is absent in the membrane and present in the nucleus.
Binding to Ig.
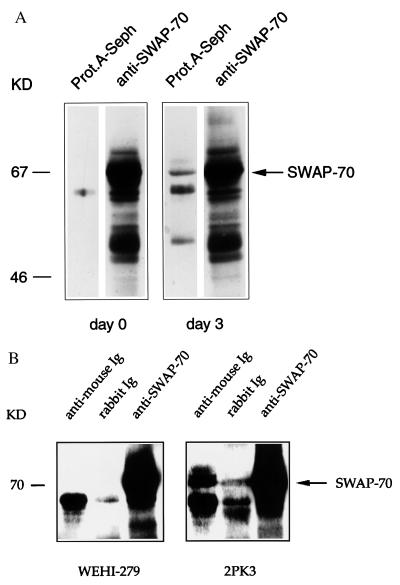
To confirm physical binding between SWAP-70 and membrane IgG, we performed coimmunoprecipitation experiments. In cell lysates, protein A-Sepharose, which binds to IgG but not to IgM, precipitated the SWAP-70-IgG complex in LPS-activated mouse splenocytes (Fig. 4A, lane 3), but not in resting splenocytes (lane 1). In a separate experiment, when using an antibody to total mouse Ig, we precipitated the SWAP-70-IgG complex only in LPS-activated, but not in resting, mouse splenocytes (data not shown). Similarly, we have immunoprecipitated SWAP-70 with anti-Ig antibody in the IgG-positive cell line 2PK3 (Fig. 4B, lane 4), but not with IgM in the IgM-positive cell line WEHI-279 (Fig. 4B, lane 1). These data indicate a physical link between the IgG isotype and SWAP-70, but not between IgM and SWAP-70. Our inability to find association with IgM does not exclude that SWAP-70, at some stage, is also linked to IgM, but we favor the interpretation that SWAP-70 prefers association with IgG. One of the differences between membrane μ- and γ-chains is their cytoplasmic tail (18, 19). Although we have not yet demonstrated that SWAP-70 indeed binds to the tail extension of γ-chain, it is clear from our experiments that the different isotypes do bind to at least one molecule—SWAP-70—differently.
Figure 4.
Coimmunoprecipitation of SWAP-70 and IgG. (A) Naive (lanes 1 and 2) and day-3 LPS-activated (lanes 3 and 4) spleen cells were immunoprecipitated with protein A-Sepharose (lanes 1 and 3) or anti-SWAP-70 antibody (lanes 2 and 4). Immunoprecipitates were run on an SDS/10% polyacrylamide gel and immunoblotted with anti-SWAP-70 antibody. (B) WEHI-279 (expressing μ-chain) (lanes 1–3) and 2PK3 (expressing γ-chain) (lanes 4–6) cell lysates were immunoprecipitated with rabbit anti-mouse Ig (lanes 1 and 4); with irrelevant rabbit antibody (lanes 2 and 5); or with rabbit anti-SWAP-70 antibody (lanes 3 and 6); the gels were immunoblotted with anti-SWAP-70 antibody.
Mutation Analyses.
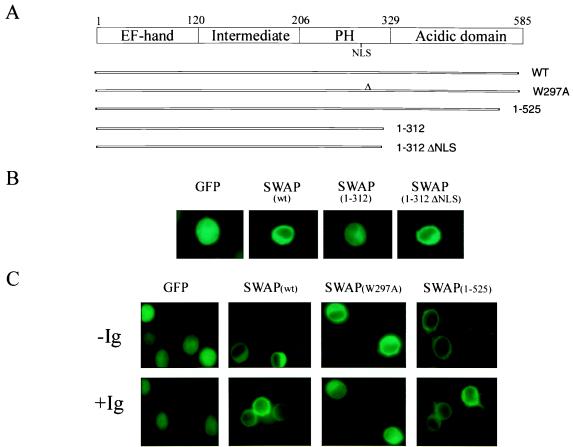
We attempted to identify the domains in SWAP-70 responsible for its differential subcellular localization. A series of SWAP-70 truncation mutants fused to GFP were generated (Fig. 5A). The constructs were stably expressed in the human IgM-positive line BJAB (12) and tested for their ability to localize the fused GFP to different compartments of the cell. Full-length wild-type SWAP-70 was predominantly localized in the cytoplasm, as compared with the uniform distribution of GFP alone throughout the cell (Fig. 5B). Truncating after the second NLS (mutant 1–312) caused uniform nuclear and cytoplasmic localization, whereas mutation of this NLS (mutant 1–312 ΔNLS) caused a notable cytoplasmic retention (Fig. 5B). These data suggest that the NLS within the C-terminal portion of the PH domain may localize SWAP-70 to the nuclear compartment in activated B cells. We next determined whether the intact functional PH domain is indeed responsible for the membrane localization of SWAP-70 in response to crosslinking of the BCR. As expected, a construct harboring a single point mutation in a conserved amino acid of the PH domain (tryptophan-297 to alanine) (20) could not associate with the plasma membrane (Fig. 5C, mutant W297A). Interestingly, the mutant lacking the C terminus from residues 525-end (mutant 1–525) was able to associate with the plasma membrane even in the absence of BCR crosslinking (Fig. 5C), suggesting that this portion of the protein might contain additional information involved in regulating SWAP-70 membrane association in response to B cell activation.
Figure 5.
Different domains of SWAP-70 control its subcellular localization. (A) SWAP-70 protein domains and truncation mutants. The mutants were fused in-frame with GFP and expressed in BJAB cells. (B) Fluorescent, inverted light microscopy of localization of wild-type SWAP-70 and several mutants in resting B cells. (C) Wild-type SWAP-70 and mutants 1–525 and W297A localization before (−Ig) and after (+Ig) crosslinking of the BCR. GFP, control vector expressing green fluorescent protein alone.
Conclusions
SWAP-70 was identified as a nuclear protein that may recruit a larger complex with biochemical properties of a putative Ig switch recombinase (6). In this report, we have shown that, in naive resting B lymphocytes, SWAP-70 is localized mainly in the cytoplasm and that, upon cell activation, either by means of the BCR or the CD40 pathways, it is recruited to the plasma membrane and it then translocates to the nucleus. After class switching has occurred, SWAP-70 is found associated to the membrane again, where it has so far been found linked only to IgG. In B cell activation, the antigen receptor initiates signal transduction in the cytoplasm, which promotes cell activation; but it can also serve as a high-affinity endocytic receptor for antigen, which leads to efficient antigen presentation. Because SWAP-70 ends up in the nucleus, we think that it is the cell activation that is mediated by SWAP-70. We suggest that the association of SWAP-70 with IgG, but not IgM, may allow previously activated (memory) B cells to signal faster and stronger, perhaps also giving rise to larger clone sizes. It may also reduce the frequency of further switch recombination.
Acknowledgments
We thank Dr. Giorgio Cattoretti for providing Fig. 2B, Nancy Abbey for excellent technical assistance, and D. G. Payan for continuous support. This work was supported by BioSTAR Grant 597-61 and National Institutes of Health Grant GM37699 (to M.W) and Center for AIDS Research Grant MH59037 (to B.H.). The Basel Institute was founded and is supported by Hoffmann–La Roche (Basel, Switzerland).
Abbreviations
- BCR
B cell receptor
- LPS
lipopolysaccharide
- GFP
green fluorescent protein
- NES
nuclear export signal
- PH
pleckstrin homology
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF210818).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040374497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040374497
References
- 1.DeFranco A L. Curr Opin Immunol. 1997;9:296–308. doi: 10.1016/s0952-7915(97)80074-x. [DOI] [PubMed] [Google Scholar]
- 2.Pleiman C M, D'Ambrosio D, Cambier J C. Immunol Today. 1994;15:393–399. doi: 10.1016/0167-5699(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 3.von Schwedler U, Jäck H M, Wabl M. New Biol. 1990;2:657–662. [PubMed] [Google Scholar]
- 4.Manis J P, Gu Y, Lansford R, Sonoda E, Ferrini R, Davidson L, Rajewsky K, Alt F W. J Exp Med. 1998;187:2081–2089. doi: 10.1084/jem.187.12.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casellas R, Nussenzweig A, Wuerffel R, Pelanda R, Reichlin A, Suh H, Qin X F, Besmer E, Kenter A, Rajewsky K S, Nussenzweig M C. EMBO J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borggrefe T, Wabl M, Akhmedov A T, Jessberger R. J Biol Chem. 1998;273:17025–17035. doi: 10.1074/jbc.273.27.17025. [DOI] [PubMed] [Google Scholar]
- 7.Borggrefe T, Masat L, Wabl M, Riwar B, Cattoretti G, Jessberger R. Eur J Immunol. 1999;29:1812–1822. doi: 10.1002/(SICI)1521-4141(199906)29:06<1812::AID-IMMU1812>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 8.Van der Pouw-Kraan C T, Rensink H J, Rappuoli R, Aarden L A. Clin Exp Immunol. 1995;99:473–478. doi: 10.1111/j.1365-2249.1995.tb05575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macias M J, Musacchio A, Ponstingl H, Nilges M, Saraste M, Oschkinat H. Nature (London) 1994;369:675–677. doi: 10.1038/369675a0. [DOI] [PubMed] [Google Scholar]
- 10.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier L L, Gorman D M, Nolan G P, Miyajima A, Kitamura T. Exp Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- 12.Marchini A, Longnecker R, Kieff E. Eur J Virol. 1992;66:4972–4981. doi: 10.1128/jvi.66.8.4972-4981.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotani H, Newton P B, III, Zhang S, Chiang Y L, Otto E, Weaver L, Blaese R M, Anderson W F, McGarrity G J. Hum Gene Ther. 1994;5:19–28. doi: 10.1089/hum.1994.5.1-19. [DOI] [PubMed] [Google Scholar]
- 14.Hicks G R, Raikhel N V. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- 15.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 16.Taagepera S, McDonald D, Loeb J E, Whitaker L L, McElroy A K, Wang J Y, Hope T J. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rebecchi M J, Scarlata S. Annu Rev Biophys Biomol Struct. 1998;27:503–528. doi: 10.1146/annurev.biophys.27.1.503. [DOI] [PubMed] [Google Scholar]
- 18.Reth M. Annu Rev Immunol. 1992;10:97–121. doi: 10.1146/annurev.iy.10.040192.000525. [DOI] [PubMed] [Google Scholar]
- 19.Achatz G, Nitschke L, Lamers M C. Science. 1997;276:409–411. doi: 10.1126/science.276.5311.409. [DOI] [PubMed] [Google Scholar]
- 20.Pitcher J A, Touhara K, Payne E S, Lefkowitz R J. J Biol Chem. 1995;270:11707–11710. doi: 10.1074/jbc.270.20.11707. [DOI] [PubMed] [Google Scholar]