Abstract
Background
Mg chelatase is a multi-subunit enzyme that catalyses the first committed step of chlorophyll biosynthesis. Studies in higher plants and algae indicate that the Mg chelatase reaction product, Mg-protoporphyrin IX plays an essential role in nuclear-plastid interactions. A number of Mg chelatase mutants have been isolated from higher plants, including semi-dominant alleles of ChlI, the gene encoding the I subunit of the enzyme. To investigate the function of higher plant CHLI, bacterial orthologues have been engineered to carry analogous amino acid substitutions to the higher plant mutations and the phenotypes examined through in vitro characterization of heterologously produced proteins. Here, we demonstrate the utility of a transient expression system in Nicotiana benthamiana for rapidly assaying mutant variants of the maize CHLI protein in vivo.
Results
Transient expression of mutant maize ChlI alleles in N. benthamiana resulted in the formation of chlorotic lesions within 4 d of inoculation. Immunoblot analyses confirmed the accumulation of maize CHLI protein suggesting that the chlorosis observed resulted from an interaction between maize CHLI and endogenous components of the N. benthamiana chlorophyll biosynthetic pathway. On the basis of this assay, PCR-based cloning techniques were used to rapidly recombine polymorphisms present in the alleles studied allowing confirmation of causative lesions. A PCR-based mutagenesis was conducted and clones assayed by transient expression. A number of novel allelic variants of maize ZmChlI were generated and analyzed using this assay, demonstrating the utility of this technique for fine mapping.
Conclusion
Transient expression provides a convenient, high-throughput, qualitative assay for functional variation in the CHLI protein. Furthermore, we suggest that the approach used here would be applicable to the analysis of other plastid-localized proteins where gain-of-function mutations will result in readily observable mutant phenotypes.
Background
Transient expression systems provide a rapid and flexible platform for the functional analysis of proteins in native, or near-native, environments [1]. In addition, expression can be localized to specific tissues at specific developmental stages. The use of Agrobacterium tumefaciens to transfer recombinant plant and viral sequences into plant leaves is now routine and has been applied to transgenic complementation, promoter analysis and protein production [2-7]. A liquid culture of the recombinant bacterial strain is infiltrated into the leaves of a host plant (typically Phaselous vulgaris or a member of the genus Nicotiana). Following infiltration, recombinant T-DNA molecules are transferred to the nucleus of host plant cells. Stable transformation requires integration of T-DNA sequences into the plant genome. However, transient expression may also result from non-integrated T-DNA copies that persist in the nuclei of transfected cells [4]. Transgene expression and protein accumulation is localized to the site of infiltration and usually peaks 40–72 h post-infiltration. A strikingly successful application of in planta transient expression has been the characterization of signaling events involved in the initiation of a type of programmed cell death termed the hypersensitive response that is associated with pathogen resistance (e.g. [8]). The easily scored necrotic phenotype associated with the hypersensitive response has facilitated the analysis of protein function among natural occurring allelic variants and both targeted and randomly generated mutants (e.g. [9,10]). In this study, we demonstrate the application of these techniques to the study of chlorophyll biosynthesis. We demonstrate the utility of agroinfiltration in the dissection of Mg chelatase I function and suggest that this technique will have broad application in dissecting the function of many plastid-localized proteins.
The first committed step of chlorophyll synthesis is catalysed by magnesium chelatase (Mg chelatase), a hetero-oligomeric enzyme that inserts a magnesium ion into the cyclic tetrapyrrole protoporphyrin IX [11,12]. The enzyme consists of three subunits that are designated ChlI, ChlD and ChlH in higher plants [13,14]. We have recently characterized a number of semi-dominant alleles of the maize Oil yellow1 (Oy1) mutant and linked the mutant phenotype to variation in the ZmChlI gene [15]. Homozygous Oy1 mutant seedlings are yellow in color and accumulate little or no chlorophyll, while heterozygous individuals exhibit a pale green phenotype intermediate to that of wild-type plants and homozygous mutants [16]. It has previously been suggested that the pale green phenotype of plants heterozygous for semi-dominant alleles of ChlI results from the formation of mixed oligomeric structures containing both mutant and wild-type CHLI subunits [15,17]. Thus, it was postulated that expression of mutant alleles in a wild-type background might induce chlorosis and provide an easily scored visual phenotype.
The use of transgenics to study gene function in maize is limited by existing transformation technology. The generation of stable transgenic lines is time consuming and costly and consequently is predominantly used to generate a small number of events, usually in the context of targeted agronomic manipulation. The use of a transient expression system is therefore particularly desirable. Although transient expression in leaf protoplasts [18] and microprojectile bombardment [19] have been successfully used in maize, they generate relatively small populations of transformed cells and are not necessarily applicable to the study of mature plant phenotypes. Therefore, we have used transient expression in N. benthamiana to study variation in maize ZmChlI. We show that expression of mutant forms of the maize CHLI protein will induce localized chlorosis in mature leaves of N. benthamiana and demonstrate the utility of this approach in precisely defining the genotypic variation underlying dominant mutant phenotypes. Additionally, we transiently express variants of the Synechocystis ChlI protein in N. benthamiana and show a degree of functional conservation between bacterial and plant Mg chelatase components. Finally, we demonstrate the utility of transient expression in the screening of a population of mutagenized clones and the identification of novel allelic variants.
Results and discussion
Transient expression of mutant variants of maize CHLI in Nicotiana benthamiana results in localized chlorosis
Previous characterisations of two semi-dominant alleles of the maize Oy1 locus established that the chlorotic phenotype associated with Oy1 mutants was due to variation in the ZmChlI gene [15]. In order to precisely define the polymorphisms within ZmChlI that were causative of the disruption in chlorophyll biosynthesis, the ZmChlI cDNAs were isolated from a wild-type maize line (W22 inbred) and two mutant lines (Oy1-N700 and Oy1-N1989) and expressed in N. benthamiana. ZmChlI sequences were cloned into a derivative of the pBIN61 binary vector that contains a single 35S CaMV promoter sequence and provides a C-terminal HA-eptitope tag to the recombinant fusion protein (Figure 1; [9]). Constructs were then transformed into Agrobacterium and introduced into mature leaves using infiltration (see Methods). Leaf tissues were inspected at 4 to 9 days post-inoculation for the development of localized chlorosis around the point of infiltration.
Figure 1.
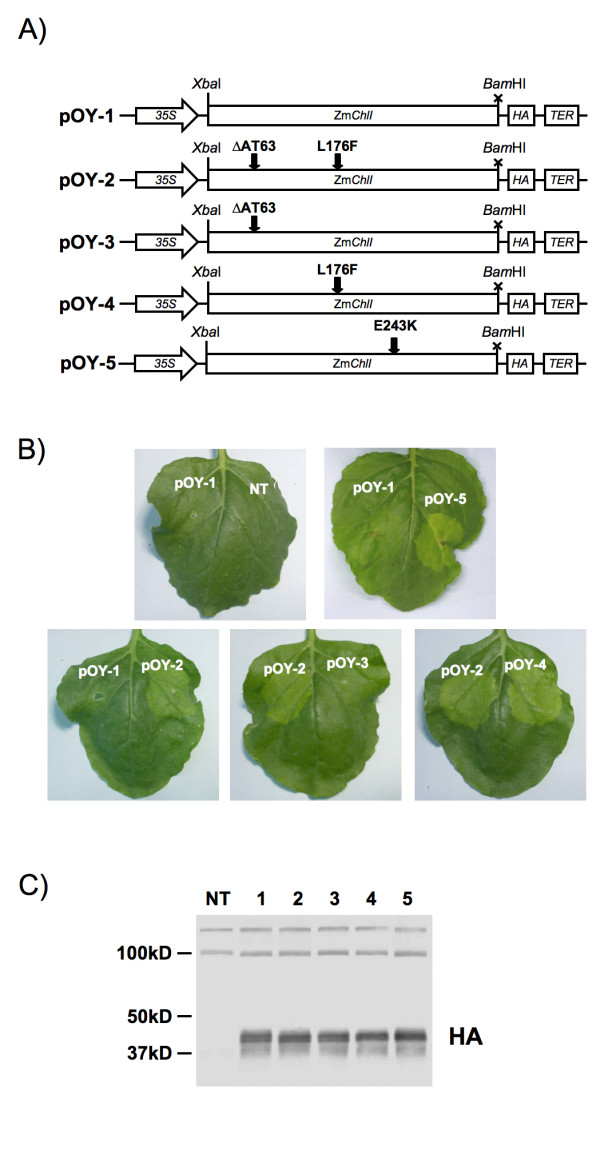
Transient expression of semi-dominant ZmChlI alleles in N. benthamina. A) Schematic of expression constructs, showing orientation of the 35S promoter (35S), maize chelatase I gene (ZmChlI), HA-epitope tag (HA) and 35S terminator (TER) sequences. Wild-type ZmChlI and a number of variants carrying polymorphisms present in semi-dominant mutant alleles were expressed in N. benthamiana. See text and Table 1 for details. B) Leaves of N. benthamiana 12 days post-infiltration. Each leaf was inoculated with two constructs. C) Immunoblot analysis of ZmCHLI accumulation following infiltration with pOY-1 to 5 (lanes 1–5, respectively) or from non-transformed (NT) tissues. Immunoblot was challenged with anti-HA antibodies. The predicted size of the mature CHLI-HA fusion is approximately 41 kDa. A cross-hybridizing protein recognized by the HA antibody at approximately 100 kDa was used as a loading control.
An initial concern was that expression of the wild-type ZmChlI might result in chlorosis, either via gene silencing of the endogenous N. benthamiana ChlI gene or through disruption of the proper stoichiometry of chlorophyll synthesis. However, no obvious phenotype was observed following expression of the wild-type maize sequence (Figure 1, pOY-1). Immunoblot analysis using an anti-HA antibody confirmed the accumulation of wild-type ZmCHLI following infiltration (Figure 1C). In contrast, expression of ZmChlI coding sequences of the semi-dominant alleles Oy1-N700 (pOY-5) and Oy1-N1989 (pOY-2) induced chlorosis (Figure 1). These observations are consistent with in vitro biochemical data obtained for Synechocystis ChlI variants that were engineered to contain substitutions homologous to those present in the maize mutant alleles [15]. The ability of mutant maize CHLI variants to induce chlorosis in N. benthamiana suggests that the mechanism of action is conserved and that maize CHLI can interact with components of the N. benthamiana chlorophyll biosynthetic machinery. This observation is consistent with the high degree of conservation of CHLI proteins between plant species (mature maize and tobacco CHLI proteins are ~85% identical). Additionally, these observations demonstrate that the kinetics of transgene product accumulation and chlorophyll turnover are such that this system is appropriate for the study of chlorophyll biosynthesis and metabolism. Furthermore, these data identify the Oy1 alleles as both visual markers and as effectors of a localized and synchronous block in chlorophyll production. That is, within four days after infiltrations, we observed a block in chlorophyll accumulation across a field of leaf cells that are at a similar developmental stage.
In planta expression allows rapid dissection of genetic variation
Transient expression of maize CHLI variants demonstrated that polymorphisms in the coding region of the maize ZmChlI gene between mutant and wild-type (W22 maize inbred line) alleles were sufficient to induce chlorosis in N. benthamiana. The allele ZmChlI-Oy1-N700 contains a single non-synonymous nucleotide change relative to wild-type sequence encoding the substitution of a glutamate residue at position 243 by lysine and therefore the results of transient expression are unambiguous. However, the allele ZmChlI-Oy1-N1989 contains two polymorphisms relative to the W22 wild-type, corresponding to the deletion of residues 63 and 64 and the substitution of a leucine residue at position 176 by phenylalanine. In this instance, the presence of two linked polymorphisms in the same allele make it difficult to determine the functionally important lesion. Although maize mutant collections represent a valuable genetic resource, information is not always available concerning the background from which mutants were derived and therefore there may be no well-defined reference wild-type sequence. While multiple single nucleotide polymorphisms may be separated by genetic recombination, the frequency of intragenic recombination events often makes such an approach impractical [20]. However, using a transient expression assay, it is trivial to independently test the phenotypic effects of linked polymorphisms. Transient expression of variants containing one of two polymorphisms present in ZmChlI-Oy1-N1989 (Figure 1, pOY-3 and pOY-4) demonstrated that the single L176F substitution was sufficient to induce chlorosis, while the two-residue deletion (ΔAT63) had no apparent effect. Interestingly, a dominant ChlI mutant in barley results from an analogous L to F substitution [17,21]. Furthermore, an equivalent L to F substitution in Synechocystis ChlI also results in a dominant mutant phenotype [15].
The two-residue deletion had not been previously tested using the Synechocystis system owing to poor conservation between maize and bacterial protein sequence in this region. In this instance, the utility of an in planta system to link genotypic and phenotypic variation is clear.
Transient expression of a mutant SschlI allele demonstrates inter-specific conservation of enzyme function
Biochemical characterization of Mg chelatase function has largely been restricted to the in vitro kinetic analysis of bacterial proteins. Previous studies have sought to confirm and characterize higher-plant mutations by introducing homologous changes into bacterial orthologues [15,17]. In the case of ChlI, these studies have been possible due to the high degree of sequence similarity between plant and bacterial proteins [15]. However, in the absence of biochemical or structural data for higher plant CHLI, it is difficult to estimate the degree of mechanistic conservation between bacterial and plant enzymes. To investigate this further, Synechocystis chlI variants were transiently expressed in N. benthamiana. The N-terminal region of ZmChlI predicted to encode the chloroplast tranist peptide (CTP) was fused to wild-type SsChlI (pOY-10) and to a SsChlI variant predicted to encode an amino acid substitution (SsChlI-L113F) homologous to the one observed in ZmChlI-Oy1-N1989 (pOY-11). SsChlI-L113F has previously been shown in an in vitro assay to be non-functional in magnesium chelation and to competitively inhibit wild-type SsChlI function [15]. Transient expression of pOY-11 in N. benthamiana was found to condition chlorosis in a manner qualitatively similar to the homologous ZmChlI-Oy1-N1989 variant (pOY-2) (Figure 2). This observation is consistent with mechanistic conservation between bacterial and higher-plant systems and suggests that both heterologous expression and homology modeling are valid approaches to the study of magnesium chelatase function.
Figure 2.
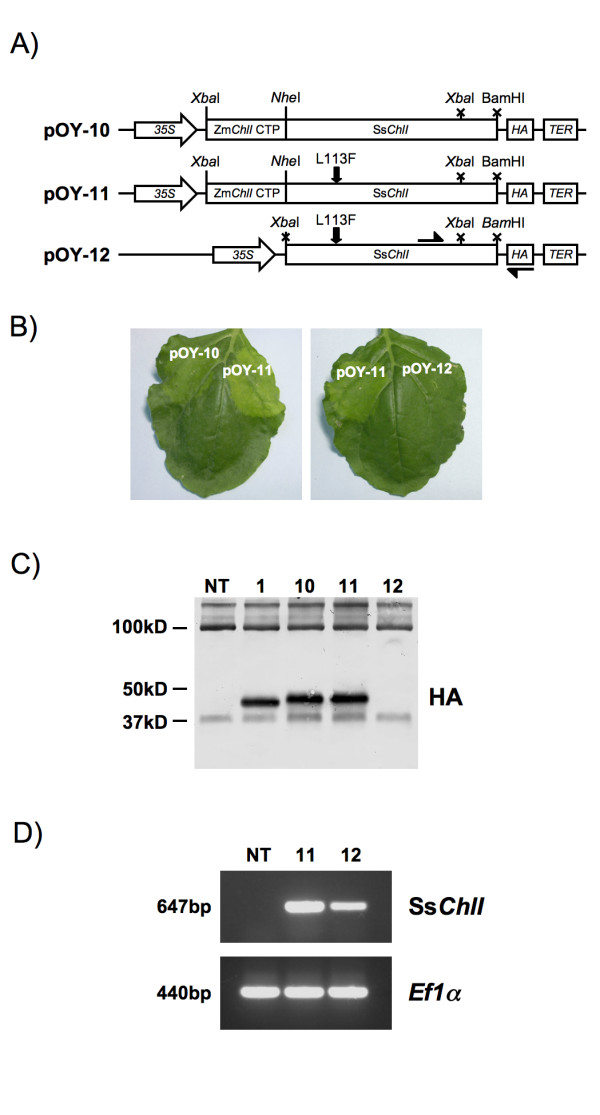
Transient expression of SsChlI in N. benthamiana. A) Schematic of expression constructs (abbreviations as in Figure 1). Wild-type and mutant variants of SsChlI were expressed in N. benthaminana with the addition of an N-terminal CTP sequence. See text and Table 1 for details. The binding-sites of SsChlI-specific primers used in RT-PCR analysis are shown as arrows on the schematic of pOY-12. B) Leaves of N. benthamina 12 days post-infiltration. No chlorosis was observed following infiltration with pOY-12. C) Immunoblot analysis of SsChlI protein accumulation following infiltration with pOY-1, pOY-10, pOY-11 and pOY-12 (lanes 1, 10, 11 and 12, respectively). A cross-hybridizing protein recognized by the HA antibody at approximately 100 kDa was used as a loading control. D) RT-PCR analysis of SsChlI transcripts in NT tissues or after infiltration with pOY-11 or pOY-12. Products amplified from an endogenous Ef1α transcript (440 bp) served as a control for cDNA synthesis.
Expression of SsChlI-L113F, lacking sequences encoding a CTP (pOY-12), did not result in chlorosis, suggesting that SsChlI-L113F disrupts chlorophyll synthesis by direct interaction with plastidic components, as is presumed to be the case for higher-plant dominant alleles [15,17]. However, immunoblot analysis revealed that levels of SsChlI protein accumulation were low in the absence of a CTP (Figure 2C). To confirm that DNA transfer had occurred and that the pOY-12 plasmid was functional, RT-PCR was used to assay the accumulation of SsChlI transcripts following infiltration with pOY-12 (Figure 2D). A product of 647 bp was amplified from cDNA prepared from pOY-12 infiltrated leaf tissue using a SsChlI-specific primer and a primer designed to the HA epitope sequence. A similar product was amplified from tissue infiltrated with pOY-11, but was not detected in non-transformed tissue. Amplification of Ef1α sequences served as a positive control for cDNA synthesis. These observations suggest that SsChlI transcripts are synthesized in pOY-12 transformed leaves but that SsChlI protein does not accumulate. It is interesting to note that, in this instance, the apparent stability and level of accumulation of a heterologous protein are dependent on translocation into the plastid and, potentially, on interaction with an endogenous protein complex in the stroma.
Somewhat surprisingly, expression of wild-type SsChlI with the N-terminal addition of the ZmCHLI CTP (pOY-10) resulted in mild chlorosis (Figure 2, pOY-10). Although, the effect was much stronger with the L113F mutation, it appears that the wild-type construct alone conditions a background level of chlorosis. In contrast, over-expression of maize wild-type CHLI does not result in chlorosis in this assay (pOY-1), despite its presumed incorporation into native complexes. Although inhibition of chelation has been observed following systemic, constitutive over-expression of wild-type ChlI [22], it appears unlikely that SsChlI would mimic this phenotype in a context where ZmChlI would not. Potentially, divergence between bacterial and plant proteins allows interaction of heterologous components but results in compromised function of the resulting complex. In addition, immunoblot analysis showed that the major species of SsChlI is slightly larger than ZmCHLI (Figure 2C).
Mature ZmCHLI-HA is predicted to have a molecular mass of approximately 41 kD and full-length SsChlI-HA approximately 39.5 kD. The higher than expected observed mass of SsChlI may be due to a failure in processing the heterologous maize CTP following import of SsChlI variants. Non-processed SsChlI fusion proteins would be predicted to have a mass of approximately 45 kD. Failure to remove additional N-terminal residues may result in a partially functional protein that acts as a weak dominant negative. The ZmCHLI CTP was defined by computer-based prediction and by alignment with Arabidopsis protein sequence and has not been experimentally determined [15]. Importantly, the sequence used may not contain residues C-terminal to the site of processing that are required for processing-site recognition. Thus, while we cannot eliminate the possibility that the SsChlI and tobacco ChlI proteins are not functionally divergent, it seems more likely that the subtle chlorosis observed with the wild-type ScChlI is due to the lack of N-terminal processing of the transit peptide. These observations highlight the potential for dominant ChlI variants as markers not just for plastid localization but also for assaying the correct processing functional integration into endogenous components of the plastid machinery.
Transient expression provides a system for mutational analysis of ChlI
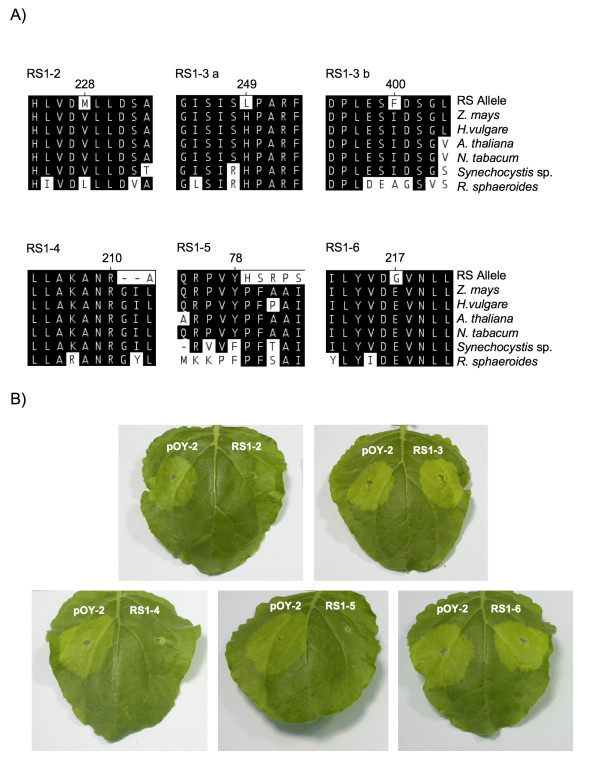
To identify novel allelic variation in maize ChlI, a PCR-based random mutagenesis procedure was used to introduce novel genetic variation (see Methods). Full-length ZmChlI cDNA sequence was amplified by PCR under conditions that introduced random base-pair changes. Resulting populations were cloned into the vector pGEM T-Easy and a small number of clones sequenced. In a pilot study, five clones were selected that contained non-synonymous nucleotide changes, subcloned into the pBin61 vector and transformed into Agrobacterium. The clones were then infiltrated into N. benthamiana and leaves scored for chlorosis. A schematic of the procedure is shown in Figure 3. The predicted protein sequence of the five mutagenized clones is shown in Figure 4A. Vectors RS1–2 and RS1–6 contain single non-synonymous nucleotide changes, RS1–3 contains two non-synonymous changes and vectors RS1–4 and RS1–5 contain frame shift mutations.
Figure 3.
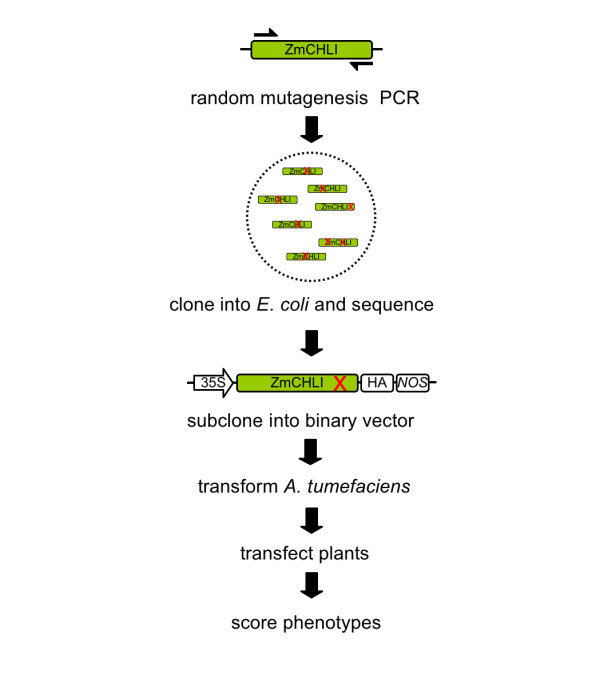
PCR-based mutagenesis of ZmChlI. Full-length ZmChlI was amplified using a mutagenic PCR procedure and fragments cloned into the pGEM T-Easy. Plasmids were electroporated into E. coli and DNA from several clones sequenced. Fragments were then subcloned into the pBIN61 binary vector and transformed into Agrobacterium. Clones were grown overnight and expressed in N. benthamiana using agroinfiltration. Following infiltration, plants were screened for the development of chlorosis indicating the generation of novel dominant mutant variants.
Figure 4.
Novel ZmChlI alleles obtained from PCR-based mutagenesis. A) Protein alignments showing lesions resulting from PCR-based mutagenesis. Recovered alleles compared to wild-type CHLI sequence from Zea mays (AAZ14052), Hordeum vulgare (DQ529207), Arabidopsis thaliana (NP_193583), Nicotiana tabacum (AAB97153), Synechocystis sp. (BAA17166), and Rhodobacter sphaeroides (ZP_00918218). Amino acid substitutions are denoted with the position of the change relative to the wild-type ZmCHLI sequence. B) Leaves of N. benthamiana 12 days post-infiltration.
As shown in Figure 4B, strong mutant phenotypes were observed in leaves of plants infiltrated with vectors RS1–3 and RS1–6. RS1–6 contains an E217G substitution within the highly conserved Walker B (DEAD box) domain [23] that likely disrupts the ATPase activity of the resulting protein. Significantly, the majority of previously characterized semi-dominant CHLI variants are deficient in ATPase activity [15]. RS1–3 contains both H249L and I400F substitutions. In this instance, it is not possible to unambiguously determine the causative lesion. However, PCR-based recombination, as described above for the ZmChlI-N1989, would provide a method to obtain these two polymorphisms independently. A very subtle mutant phenotype was observed in plants infiltrated with the RS1–2 construct. RS1–2 carries a V228M substitution. Again the proximity to the Walker B domain suggests an effect on ATP binding or hydrolysis. No phenotypic effects were observed with the truncated forms RS1–4 or RS1–5. In all cases, a greater understanding of these alleles would be gained by introduction of the homologous substitutions into a bacterial ChlI and subsequent in vitro analysis. This is especially relevant to the analysis of the apparently weak RS1–2 mutant allele.
This pilot study highlights the applicability of transient expression to the identification of novel protein variants but also demonstrates the limitations with regard to assessing quantitative differences. We envisage that the greatest utility of this system would be as an initial genetic screen for phenotypic variation. The system described here also allows for secondary mutagenesis of known dominant alleles. We have previously converted a dominant mutant variant CHLI into a recessive form by targeted amino acid substitution (Ruairidh Sawers, Jo Viney and Neil Hunter, University of Sheffield, UK, unpublished results). It is anticipated that other such events could be recovered by random mutagenesis.
General conclusions on the potential of this transient expression system for the study of chlorophyll biosynthesis
Although genetic studies have been instrumental in the identification chlorophyll biosynthetic genes, the analysis of chelatase function has largely been restricted to in vitro kinetic analysis of wild-type protein function. However, an extensive collection of photosynthetic mutants in barley and maize exist [15,17,21,24]. Of particular interest with regard to enzymatic function are a number of semi-dominant alleles of ChlI. The cloning of several of these alleles [15,17] and the subsequent introduction of observed variation into bacterial proteins for in vitro analysis has demonstrated the potential of mutant variants in extending understanding of these enzymes. The observation that transient expression of dominant Oy1 alleles is sufficient to induce chlorosis opens up the possibility for the identification of further alleles, not only of ChlI but also of other components of the pathway. As described here, this approach is most suitable for the identification of dominant and semi-dominant variants. Dominant variants are most likely to be isolated where there is some form of protein-protein interaction underlying the process. For this, the ChlD and ChlH subunits of Mg chelatase make attractive targets. Additionally, it has been suggested that GUN4 [25] and CHLM may also physically interact [26]. While such interactions may be transient and difficult to characterize in vitro using wild-type components, semi-dominant mutant forms may reveal, and even stabilize, complexed states.
Conclusion
Transient expression of dominant mutant alleles of ChlI in N. benthamiana demonstrated that such a system is applicable to the study chlorophyll synthesis and turnover. Causative lesions were confirmed for two maize alleles ZmChlI-Oy1N700 and ZmChlI-Oy1N1989. In addition, three potential novel alleles of ZmChlI were identified. Although in vitro kinetic analyses are required for quantitative characterization of enzymatic variants, the investment required makes such analyses most appropriate to the study of a small number of variants. In contrast, the transient expression system described here allows the rapid, qualitative analysis of a large number of variants. A combination of these two approaches offers the possibility for the large-scale isolation and characterization of allelic variation from plant and bacterial species.
Methods
Construction of pOY vectors
The pBIN61-pOY binary vectors contain a series of ChlI fusions and derivatives under the control of the CaMV 35S promoter. All constructs contain a C-terminal HA epitope fusion and 35S terminator sequence. Coding sequences of pOY vectors were assembled and sequenced in pGEM-T Easy (Promega, Madison, WI) prior to sub-cloning to the binary vector pBIN61-RxHA [27] to generate pBIN61-pOY vectors. pBIN61-RxHA was digested with XbaI and BamHI to release the Rx coding fragment. The pOY coding sequence was then introduced as an XbaI/BglII fragment.
Coding sequences for vectors pOY-1, pOY-2 and pOY-5 were PCR amplified directly from cDNA prepared from maize wild-type (W22 inbred), Oy1-N1989 and Oy1-N700 plants, respectively. Amplification reactions were performed using the primer CHLIFLF (5'-GGCACGACAGGTTTCCGACT-3') and the reverse primer CHLIFLR (5'-TGAAAGTAACAGAACAAGAGGGTTCC-3'). The full-length products were ligated into pGEM-T Easy (Promega), and subsequently used to transform E. coli. Plasmid DNA prepared from bacterial cultures containing the constructs was then used as template in PCR amplification using primers CHLIXBAFWD (5'-CTCCTCTCTAGAATGGCTTCCACCTTCTC-3') and the reverse primer CHLIBGLREV (5'-GAACAGATCTGCTAAAGACTTCATAAAACTTCTC-3') to introduce 5' XbaI and a 3' BglII sites (underlined).
The coding sequences for vectors pOY-3 and pOY-4 were generated using a two-step amplification strategy to engineer constructs with single lesions. To create pOY3, primers CHLIXBAFWD and CHLI55ATSEPMUT (5'-ATGGTGATCTTGGCGGTGGT-3') were used to amplify a 522 bp region from pOY2 incorporating the ΔAT63 lesion. In a separate reaction CHLIBGLREV and CHLI55FSEPMUT (5'-GTACCCATTCGCGGCCATC-3') were used to amplify a 1061 bp fragment from pOY1. To generate pOY-4, primers CHLIXBAFWD and CHLI55ATSEPMUT were used to amplify a 522 bp region from pOY1. In a separate reaction CHLIBGLREV and CHLI55FSEPMUT were used to amplify a 106 bp fragment from pOY2 to incorporate the L176F lesion. Products were then gel purified using a Gel Extraction kit (Qiagen, Valencia, CA) and resuspended in 50 μl of EB buffer. Approximately 20 ng of each purified product was used as template in a second amplification reaction together with primers CHLIXBAFWD and CHLIBGLREV to amplify a full-length synthetic gene. Products were sub-cloned to pBIN61-RxHA as described above.
Vectors pOY-10 and pOY-11 were generated by fusion of the ZmChlI CTP to Synechocystis sequence. The vectors were assembled in pGEM-T Easy prior to sub-cloning to pBIN61-RxHA as XbaI/Bglll fragments as described above. Initially, the ZmChlI CTP was amplified from wild-type template using the primer CHLIXBAFWD and the reverse primer CHLIXBABGL 5'-GTAGACAGATCTCAGATGGCTAGCGACATTGCAGACGGTGAATC-3' containing NheI and BglII restriction sites (underlined) and the fragment cloned to pGEM-T Easy. Wild-type (pOY-10) and mutant (pOY-11) sequence was amplified from previously described vectors [15] using the primers CHLINHE 5'-CATGTAGCTAGCATGACTGCCACCCTTGCCGCCCCCAGC-3' and CHLIBGLII 5'-CATGTAAGATCTAGCTTCATCGACAACGCCAAAAAC-3' to introduce 5' NheI and 3' BglII sites (underlined) used in sub-cloning to the vector containing the signal sequence. Prior to amplification, the template vectors were modified to remove an endogenous XbaI site present in the SsChlI gene in order to allow for the use of the common strategy of sub-cloning to pBIN61-RxHA as described above. The SsChlI coding sequences (wild-type and mutant) were excised from pET-9a [15] and cloned to the pGEM-T Easy (Promega) backbone as NheI/EcoRI fragments. A portion of the coding sequence was amplified from the 5' end to the endogenous XbaI site using the primers CHLIENDO 5'-AACCGGGAAATCCACTACCATTAGAGCTTT-3' and CHLISPE 5'-CATGTAACTAGTGGATCTTTCCGTAAACGATG-3' to introduce a SpeI site (underlined) at the position of the XbaI site. These products were then cloned as a BamHI/SpeI fragments back to the pGEM-T Easy (Promega) template digested with BamHI/XbaI. The compatible ligation of SpeI and XbaI ends resulted in a synonymous base pair change and the ablation of the endogenous site. The vector pOY-12 was built by digestion of pOY-11 with NheI and BglII and sub-cloning to XbaI/BamHI digested pBIN61-RxHA.
Transient expression
The pOY constructs, derived from pBIN61-RxHA, were transformed into Agrobacterium tumefaciens strain C58C1 carrying the virulence helper plasmid pCH32 [28]. Agrobacterium-mediated transformation was performed essentially as described by Bendahmane et al. [27]. N. benthamiana seedlings were sown in flats and grown under greenhouse conditions (24°C – 30°C) for 21 days. They were then transplanted to six inch pots and grown for an additional 7 to 10 days. Infiltrations were conducted in the laboratory under fluorescent light at 24°C – 28°C. After a 24 hr recovery in the laboratory, plants were returned to the greenhouse with supplemental lighting provided by 400 W metal halide lamps (average temperatures in the greenhouse varied between 26°C and 30°C). We found phenotypes were highly reproducible. In over 20 independent infiltrations using pOY-4 conducted on multiple leaves on multiple plants, we always observed chlorosis. Some subtle variation in phenotype was observed that was dependent on the developmental age of the leaf and temperature in the greenhouse. The most severe phenotypes were observed when leaf 6 or 7 was infiltrated when seedlings were 3 weeks old. At the time of infiltration leaves were approximately 2.5 to 4.0 cm in width.
RT-PCR
Total RNA was made from approximately 75 mg of tissue using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA). DNAse treatment was performed on a column during RNA extraction using the RNase-Free DNase Set (Qiagen) according to the manufacturer's protocol. Subsequent synthesis of cDNA was performed using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) with 2 μg total RNA in a volume of 10 μl. l μl of cDNA was used as template with the primers SsCHLIF (5'-CGAAGTTAACCTGTTGGACGATCAC-3') and HAR (5'-CGGCATAATCAGGTACATCATAAGGGTAG-3') to amplify a product of 647 bp. EF1α was amplified using the primers EF1aFor (5'-GGTGGTTTTGAAGCTGGTATCTCC-3') and EF1aRev (5'-CCAGTAGGGCCAAAGGTCACA-3'). The products were visualized on a 0.8% agarose gel run at 70 V for approximately 3 hours.
Immunoblot analysis
Protein was extracted by grinding 3 square centimeters of infiltrated leaf tissue in 500 μl of extraction buffer [100 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT, 0.1% TritonX 100] with Protease Inhibitor Cocktail (Sigma-Aldrich, St. Louis, MO). Samples were then centrifuged at 17,900 rcf at 4° for 10 min. The supernatant, containing the soluble protein fraction, was then mixed with 10× loading buffer Z [125 mM Tris pH 6.8, 12% SDS, 10% Glycerol, 0.001% Bromophenol Blue], boiled at 95° for 3–5 minutes and loaded onto a Criterion Tris-HCL 12.5% acrylamide gel (Bio-Rad, Hercules, CA). After separation, the samples were electroblotted to Protran BA83 0.2 μm nitrocellulose membrane (Whatman, Schleicher & Schuell, Keene NH). The blots were rinsed twice in 1× PBS pH 7.5 and gently rocked for at least 1 hour in blocking buffer [5% nonfat milk powder in PBST (0.05% Tween 20 in 1× PBS)]. A rabbit anti-HA antibody (Sigma-Aldrich) was diluted 1:10,000 in blocking buffer and used to challenge blots for at least 1 hour. A goat anti-rabbit secondary antibody conjugated to horseradish peroxidase (Bio-Rad) was then used to amplify the signal. Visualization was accomplished using the Opti-4CN Substrate kit (Bio-Rad).
PCR-based random mutagenesis
PCR-based mutagenesis was performed using the Diversify PCR Random Mutagenesis kit (Clontech, Mountain View, CA) according to the manufacturer's protocol for Buffer Condition 2. Wild-type ZmChlI plasmid was used as PCR template in conjunction with the primers CHLIXBAFWD and CHLIBGLREV (described above). These conditions were used to introduce mutations at a rate of 2.3 substitutions/1000 bp. The mutagenised products were inserted into pGEM-T Easy vector and a subset were sequenced, analyzed, and subcloned into the pBin-61 Rx HA expression vector. The resulting plasmids were used to transform Agrobacterium and transient expression was accomplished as described above.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
RS and PF constructed pOY vectors, conducted infiltration experiments and drafted the manuscript. PF conducted RT-PCR experiments and performed protein blot and sequence analysis. PM provided technical advice on the expression system and helped to draft the manuscript. TB coordinated the study and helped to draft the manuscript. All authors have read and approved the final manuscript.
Table 1.
pOy vectors
| Vector | Description | Residue alterations | Protein detected | Phenotype |
| pOY-1 | ZmChlI wild-type | - | yes | none |
| pOY-2 | ZmChlI-Oy1-N1989 | ΔAT63 L176F | yes | chlorosis |
| pOY-3 | ZmChlI-Oy1-N1989 derivative | ΔAT63 | yes | none |
| pOY-4 | ZmChlI-Oy1-N1989 derivative | L176F | yes | chlorosis |
| pOY-5 | ZmChlI-Oy1-N700 | E243K | yes | chlorosis |
| pOY-10 | SsChlI wild-type, N-terminal CTP fusion | ZmCHLI(1–54)-SsCHLI | yes | chlorosis |
| pOY-11 | SsChlI-L113F, N-terminal CTP fusion | ZmCHLI(1–54)-SsCHLI-L113F | yes | chlorosis |
| pOY-12 | SsChlI-L113F, no CTP fusion | L113F | no | none |
Acknowledgments
Acknowledgements
We would like to thank Dan Montiel for sequence analysis of mutagenized ZmChlI alleles, Dr. Rob Abramovitch for technical advice and Dr. Mats Hansson for critically reviewing the manuscript. We would also like to thank Ialon Irijimovich for growth and care of N. benthamiana. This work was supported by a National Science Foundation grant to T.P.B. (DBI-0211935).
Contributor Information
Ruairidh JH Sawers, Email: Ruairidh.Sawers@bioveg.unige.ch.
Phyllis R Farmer, Email: prf4@cornell.edu.
Peter Moffett, Email: pm99@cornell.edu.
Thomas P Brutnell, Email: tpb8@cornell.edu.
References
- Janssen BJ, Gardner RC. Localized transient expression of GUS in leaf discs following cocultivation with Agrobacterium. Plant Mol Biol. 1990;14:61–72. doi: 10.1007/BF00015655. [DOI] [PubMed] [Google Scholar]
- Wroblewski T, Tomczak A, Michelmore R. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotech J. 2005;3:259–273. doi: 10.1111/j.1467-7652.2005.00123.x. [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Querci M, Kanyuka K, Baulcombe DC. Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 2000;21:73–81. doi: 10.1046/j.1365-313x.2000.00654.x. [DOI] [PubMed] [Google Scholar]
- Kapila J, De Rycke R, Van Montagu M, Angenon G. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 1997;122:101–108. doi: 10.1016/S0168-9452(96)04541-4. [DOI] [Google Scholar]
- Vaquero C, Sack M, Chandler J, Drossard J, Schuster F, Monecke M, Schillberg S, Fischer R. Transient expression of a tumor-specific single-chain fragment and a chimeric antibody in tobacco leaves. Proc Natl Acad Sci U S A. 1999;96:11128–11133. doi: 10.1073/pnas.96.20.11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li R, Qi M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 2000;22:543–551. doi: 10.1046/j.1365-313x.2000.00760.x. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003;33:949–956. doi: 10.1046/j.1365-313X.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn RA, Laurent F, Roth R, De Wit PJ. Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol Plant Microbe Interact. 2000;13:439–446. doi: 10.1094/MPMI.2000.13.4.439. [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Farnham G, Moffett P, Baulcombe DC. Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 2002;32:195–204. doi: 10.1046/j.1365-313X.2002.01413.x. [DOI] [PubMed] [Google Scholar]
- Mestre P, Brigneti G, Durrant MC, Baulcombe DC. Potato virus Y NIa protease activity is not sufficient for elicitation of Ry-mediated disease resistance in potato. Plant J. 2003;36:755–761. doi: 10.1046/j.1365-313X.2003.01917.x. [DOI] [PubMed] [Google Scholar]
- Willows RD. Biosynthesis of chlorophylls from protoporphyrin IX. Nat Prod Rep. 2003;20:327–341. doi: 10.1039/b110549n. [DOI] [PubMed] [Google Scholar]
- Walker CJ, Willows RD. Mechanism and regulation of Mg-chelatase. Biochem J. 1997;327 ( Pt 2):321–333. doi: 10.1042/bj3270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Willows RD, Petersen BL, Vothknecht UC, Stummann BM, Kannangara CG, von Wettstein D, Henningsen KW. Structural genes for Mg-chelatase subunits in barley: Xantha-f, -g and -h. Mol Gen Genet. 1996;250:383–394. doi: 10.1007/BF02174026. [DOI] [PubMed] [Google Scholar]
- Gibson LC, Willows RD, Kannangara CG, von Wettstein D, Hunter CN. Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: reconstitution of activity by combining the products of the bchH, -I, and -D genes expressed in Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:1941–1944. doi: 10.1073/pnas.92.6.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers RJH, Viney J, Farmer PR, Bussey RR, Olsefski G, Anufrikova K, Hunter CN, Brutnell TP. The maize Oil Yellow1 (Oy1) gene encodes the I subunit of magnesium chelatase. Plant Mol Biol. 2006;60:95–106. doi: 10.1007/s11103-005-2880-0. [DOI] [PubMed] [Google Scholar]
- Mascia PN, Robertson DS. Genetic studies of the chlorophyll biosynthetic mutants of maize. J Hered. 1980;71:19–24. [Google Scholar]
- Hansson A, Willows RD, Roberts TH, Hansson M. Three semidominant barley mutants with single amino acid substitutions in the smallest magnesium chelatase subunit form defective AAA+ hexamers. Proc Natl Acad Sci U S A. 2002;99:13944–13949. doi: 10.1073/pnas.212504499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 2001;127:1466–1475. doi: 10.1104/pp.127.4.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein TM, Kornstein L, Sanford JC, Fromm ME. Genetic transformatin of maize cells by particle bombardment. Plant Physiol. 1989:440–444. doi: 10.1104/pp.91.1.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner HK. Extensive interallelic polymorphisms drive meiotic recombination into a crossover pathway. Plant Cell. 2002;14:1173–1183. doi: 10.1105/tpc.001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson A, Kannangara CG, von Wettstein D, Hansson M. Molecular basis for semidominance of missense mutations in the XANTHA-H (42-kDa) subunit of magnesium chelatase. Proc Natl Acad Sci U S A. 1999;96:1744–1749. doi: 10.1073/pnas.96.4.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J, Pfundel E, Mock HP, Grimm B. Decreased and increased expression of the subunit CHL I diminishes Mg chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants. Plant J. 2000;22:155–164. doi: 10.1046/j.1365-313x.2000.00724.x. [DOI] [PubMed] [Google Scholar]
- Fodje MN, Hansson A, Hansson M, Olsen JG, Gough S, Willows RD, Al-Karadaghi S. Interplay between an AAA module and an integrin I domain may regulate the function of magnesium chelatase. J Mol Biol. 2001;311:111–122. doi: 10.1006/jmbi.2001.4834. [DOI] [PubMed] [Google Scholar]
- Olsson U, Sirijovski N, Hansson M. Characterization of eight barley xantha-f mutants deficient in magnesium chelatase. Plant Physiol Biochem. 2004;42:557–564. doi: 10.1016/j.plaphy.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Davison PA, Schubert HL, Reid JD, Iorg CD, Heroux A, Hill CP, Hunter CN. Structural and biochemical characterization of Gun4 suggests a mechanism for its role in chlorophyll biosynthesis. Biochemistry. 2005;44:7603–7612. doi: 10.1021/bi050240x. [DOI] [PubMed] [Google Scholar]
- Shepherd M, McLean S, Hunter CN. Kinetic basis for linking the first two enzymes of chlorophyll biosynthesis. Febs J. 2005;272:4532–4539. doi: 10.1111/j.1742-4658.2005.04873.x. [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Kanyuka K, Baulcombe DC. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell. 1999;11:781–792. doi: 10.1105/tpc.11.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton CM, Frary A, Lewis C, Tanksley SD. Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc Natl Acad Sci U S A. 1996;93:9975–9979. doi: 10.1073/pnas.93.18.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]