Abstract
Background
Mycelium-to-yeast transition in the human host is essential for pathogenicity by the fungus Paracoccidioides brasiliensis and both cell types are therefore critical to the establishment of paracoccidioidomycosis (PCM), a systemic mycosis endemic to Latin America. The infected population is of about 10 million individuals, 2% of whom will eventually develop the disease. Previously, transcriptome analysis of mycelium and yeast cells resulted in the assembly of 6,022 sequence groups. Gene expression analysis, using both in silico EST subtraction and cDNA microarray, revealed genes that were differential to yeast or mycelium, and we discussed those involved in sugar metabolism. To advance our understanding of molecular mechanisms of dimorphic transition, we performed an extended analysis of gene expression profiles using the methods mentioned above.
Results
In this work, continuous data mining revealed 66 new differentially expressed sequences that were MIPS(Munich Information Center for Protein Sequences)-categorised according to the cellular process in which they are presumably involved. Two well represented classes were chosen for further analysis: (i) control of cell organisation – cell wall, membrane and cytoskeleton, whose representatives were hex (encoding for a hexagonal peroxisome protein), bgl (encoding for a 1,3-β-glucosidase) in mycelium cells; and ags (an α-1,3-glucan synthase), cda (a chitin deacetylase) and vrp (a verprolin) in yeast cells; (ii) ion metabolism and transport – two genes putatively implicated in ion transport were confirmed to be highly expressed in mycelium cells – isc and ktp, respectively an iron-sulphur cluster-like protein and a cation transporter; and a putative P-type cation pump (pct) in yeast. Also, several enzymes from the cysteine de novo biosynthesis pathway were shown to be up regulated in the yeast form, including ATP sulphurylase, APS kinase and also PAPS reductase.
Conclusion
Taken together, these data show that several genes involved in cell organisation and ion metabolism/transport are expressed differentially along dimorphic transition. Hyper expression in yeast of the enzymes of sulphur metabolism reinforced that this metabolic pathway could be important for this process. Understanding these changes by functional analysis of such genes may lead to a better understanding of the infective process, thus providing new targets and strategies to control PCM.
Background
The availability of great amounts of raw genomic and transcriptome data collected from several organisms has prompted the development of large-scale gene expression analysis which will ultimately help to unravel the function of many genes in diverse biological contexts. Different approaches such as cDNA microarrays [1-3], in silico ESTs subtraction [4,5] and serial analysis of gene expression – SAGE [6,7] are widely employed to assess differential gene expression patterns leading to the discovery of a great number of genes that are over or under expressed in each physiological context. The successful use of the cDNA microarray approach in fungal pathogens such as Candida albicans [8-13], Histoplasma capsulatum [14] and Cryptococcus neoformans [15] has resulted in the identification of genes involved in cell viability and opened new experimental perspectives to understand host-parasite interactions and thus develop new therapeutic approaches to systemic mycoses [8,11].
Paracoccidioidomycosis (PCM) is a human illness endemic to Latin America [16]; its area of incidence spreads non-uniformly from Mexico to Argentina [17], being higher in Brazil, Venezuela, Colombia and Argentina [18,19,16]. An estimation for Brazil points to an incidence rate between 1 and 3 and a mortality rate of 1.4 per million [20]. McEwen et al. [21] reported an overall infected population of 10 million individuals in Latin America, 2% of whom will eventually develop the disease. In nature, another important mammalian host is the armadillo Dasypus novemcinctus [22]. PCM affects the skin, lymph nodes and various internal organs, including the lungs – where it causes granulomatous processes – and the central nervous system [19,23]. Its clinical presentations range from a localised and benign disease to a progressive and potentially lethal systemic infection [24]. The disease is more frequent in adult males, who account for up to 90% of all cases. Healthy rural workers are the main targets, but PCM affects immunosuppressed individuals as well [25,26], including as much as 30% of AIDS patients [27]. All patients from whom the fungus is isolated must be treated and, in spite of new antifungal drugs, pulmonary fibrosis is still the most frequent sequel. The outcome of infection depends on several factors, including host responses and the virulence of the infecting isolate.
The causative agent of PCM, the thermo-regulated dimorphic fungus P. brasiliensis, is believed to be a free-living mycelium saprobe that undergoes transition to the yeast pathogenic form upon temperature change from the environmental 24–26°C to the mammalian body temperature of 37°C. This switch is necessary and sufficient to trigger morphotype interconversion in vitro, which makes this fungus an interesting model to study fungal cell differentiation at the molecular level. The biochemical events regulating dimorphic transition in P. brasiliensis are yet poorly defined, although relevant molecular-level information on this process has been partially described in the transcriptome analyses of two different P. brasiliensis isolates [28-30].
The exact ecological niche of this pathogen is still unknown [17], but P. brasiliensis can be retrieved from the soil. The fungus Penicillium marneffei is greatly similar in that it is a human opportunistic pathogen that also undergoes thermally-controlled dimorphic transition upon infection, can also infect a wild mammal (the bamboo rat) and has an yet unknown natural reservoir. Genomic data provided evidence that, in the case of P. marneffei, the fungus may have a sexual stage as a free-living organism [31].
Phylogenetic analysis of members of the order Onygenales demonstrated a close relationship of P. brasiliensis with the pathogenic fungi Blastomyces dermatitidis, Emmonsia parva and Histoplasma capsulatum [32]. P. brasiliensis can be fitted with B. dermatitidis and E. parva in the family Onygenacea [33]. Recently it was reported that P. brasiliensis is in fact a complex of at least three closely correlated phylogenetic species [34]. So far, the sexual phase of the ascomycete P. brasiliensis was not reported limiting our knowledge about the mechanisms that contribute to its dimorphism, pathogenicity, and virulence. P. brasiliensis isolates shows chromosomal polymorphism; it contains 4–5 chromosomal DNA molecules with molecular sizes ranging from 2–10 Mb [35,36]. The genome size was estimated to be around 30 Mb [37] and DNA sequencing of ~ 50 Kb revealed a density of one gene per 3.5–4.5 Kb, suggesting a total of 7,500–9,000 genes [38].
Recently, our group analysed the transcriptome of the Pb01 isolate, represented by a set of 6,022 clusters. The 16 genes that were then found to be differentially expressed by both methods used – in silico EST subtraction and cDNA microarray – were categorised by function. We chose to discuss in that work those that were involved in core metabolic pathways such as sugar metabolism [28]. Now, continued overlap analysis from raw data revealed 66 new genes that are differentially expressed in one or other morphotype. Upon categorisation by known databases we have selected two MIPS [39] classes, which were chosen to be confirmed by northern blotting. Here we present the result of this extended analysis, and discuss the putative roles the differential genes – related to cell organisation and ion metabolism and transport – play in the corresponding morphotype of this pathogen. One of the discussed pathways – de novo cysteine synthesis from inorganic sulphate, a branch of sulphur metabolism – was almost entirely up-regulated in the yeast form. The importance of sulphur metabolism to the life cycle of pathogenic fungi has been extensively reviewed elsewhere [40,41] and recently new data from microarray experiments have arisen from work in H. capsulatum that support a role of organic sulphate in the maintenance of the yeast phase [14]. In a previous report [42], the importance of organic sulphates to the growth and differentiation of P. brasiliensis was assessed. This phenomenon demanded further investigation and prompted us to assess up- and downregulation of sulphur metabolism genes in mycelium and yeast cells and also dimorphic transition in both directions without inorganic sulphate as a sulphur source. We have thus found that this compound is unnecessary for the process.
Results and discussion
Differentially expressed genes identified by in silico EST subtraction and cDNA microarray
Comparative gene expression profiling in dimorphic fungi can reveal key proteins involved in commitment to differentiation and gene regulation. From the 66 new PbAESTs (P. brasiliensis assembled expressed sequence tags) identified in this work, thirteen of which correspond to up-regulated genes in mycelium and fifty four which are differential for yeast cells (Tables 1 and 2). This set complements the one generated previously [28], which included 16 genes that were differential by the same overlap analysis and also 30 genes that were differential according to in silico EST subtraction alone. MIPS functional categories [43] were used to classify the 66 PbAESTs into 14 major groups (data not shown). Gene categorisation revealed some that are involved in energy production (11%) – this was expected considering the adaptation process that is required for the mycelium-to-yeast transition; control of cell wall organisation (10%); ion metabolism and transport (8%); transcription, translation and ribosome structure (8%); virulence and oxidative stress (4%). Manual annotation under stringent criteria of sequence alignment with other dimorphic fungi gene sets allowed us to ascribe a putative biological function to many of those genes. The genes that belonged in two categories – cell wall organisation and ion metabolism and transport – were selected for confirmation by northern blotting.
Table 1.
Mycelium up-regulated genes identified by in silico ESTs subtraction and cDNA microarray.
| PbAEST | Acession Numbers (GenBank) | Annotated function | Number of reads | P-valuea | Fold change | Accession Number/Best-hit organism/E-value | Functional categories | |
| M | Y | |||||||
| 202 | CA582032 | 1,3-beta-glucosidase* | 7 | 2 | 0.036942 | 12.3 | AAL09828.1/C. immitis/1.0E-132 | Control of cell organization: Cell wall and membrane |
| 2155 | CA582352 | Peroxisomal membrane protein PEX16 (peroxin-16) | 7 | 0 | 0.004174 | 1.4 | EAL88469.1/A. fumigatus/3.0E-64 | |
| 186 | CA583085 | HEX* | 13 | 8 | 0.049272 | 3.4 | EAL91716.1/A. fumigatus/3.0E-66 | |
| 2496 | CA583518 | Iron-sulphur cluster nifU-like protein* | 5 | 1 | 0.048854 | 1.7 | EAL90111.1/A. fumigatus/8.0E-58 | Ion transport |
| 4179 | CN245816 | Potassium transporter protein* | 0 | 1 | -b | 5.2 | CAA08814.1/N. crassa/4.0E-22 | |
| 1420 | CN247275 | U1 small nuclear ribonucleoprotein | 9 | 1 | 0.00526 | 1.6 | EAL91268.1/A. fumigatus/1.0E-60 | Transcription |
| 1029 | CA582332 | Methyltransferase | 32 | 1 | 0.000000 | 2.1 | EAL84975.1/A. fumigatus/1.0E-56 | Others |
| 2096 | CA581148 | Unkown | 20 | 1 | 0.000006 | 5.6 | - | |
| 514 | CA583322 | Unkown | 15 | 1 | 0.000138 | 23.4 | - | |
| 1045 | CA581951 | Unkown | 13 | 2 | 0.001769 | 24 | - | |
| 1178 | CN247241 | Unkown | 10 | 0 | 0.000535 | 8.5 | - | |
| 1664 | CN247289 | Unkown | 10 | 3 | 0.018648 | 2.5 | - | |
a FDR = 4,8% and Q-value < 5%.
b Not significant by Audic-Claverie's method.
* Up-regulated genes confirmed by northern blotting.
** Not assayed by cDNA microarray but confirmed as up-regulated in mycelium by northern blotting.
Table 2.
Yeast up-regulated genes identified by in silico ESTs subtraction and cDNA microarray.
| PbAEST | Acession Numbers (GenBank) | Annotated function | Number of reads | P-valuea | Fold change | Accession Number/Best-hit organism/E-value | Functional categories | |
| M | Y | |||||||
| 1422 | CA581980 | Alpha-1,2-mannosyltransferase (Alg11) | 4 | 11 | 0.019803 | 2.0 | EAL88400.1/A. fumigatus/1.0E-130 | Control of cellular organization: Cell wall and membrane |
| 4988 | CN253911 | Alpha 1,3-glucan synthase* | - | 1 | - | 5.7 | AAV52833.1/P brasiliensis/4.0E-93 | |
| 2162 | CN238153 | Putative WW domain protein (probable membrane protein) | 4 | 12 | 0.013092 | 3.6 | EAL85876.1/A. fumigatus/6.0E-17 | |
| 136 | CA582283 | Involved in cytoskeletal organization and cellular growth (verprolin)* | 4 | 10 | 0.029289 | 4.0 | NP_013441.1/S. cerevisiae/2.3 | |
| 667 | CA583397 | Adenylylsulphate kinase | 3 | 8 | 0.038949 | 2.1 | EAL90409.1/A. fumigatus/1.0E-88 | Ion transport and metabolism |
| 48 | CA582091 | ATP-sulphurylase | 10 | 18 | 0.023038 | 4.8 | EAL92915.1/A. fumigatus/0.0 | |
| 2031 | CA581274 | Outer mitochondrial membrane protein porin | 1 | 14 | 0.000207 | 1.3 | XP_323644.1/N. crassa/1.0E-108 | |
| 2724 | CA581633 | P-type Cu(2+) transporting ATPase* | 0 | 1 | -b | 3.8 | NP_009854.1/S. cerevisiae/1.7E-20 | |
| 635 | CN247312 | ATP citrate lyase | 1 | 7 | 0.014984 | 1.9 | EAL88915.1/A. fumigatus/0.0 | Energy |
| 2016 | CN242578 | ATPase inhibitor; Inh1 | 2 | 14 | 0.000835 | 2.7 | NP_010100.1/S. cerevisiae/4.0E-08 | |
| 563 | CA583982 | Cytochrome c oxidase subunit VII | 11 | 43 | 0.000002 | 2.1 | AAT77147.1/P. brasiliensis/3.0E-26 | |
| 2398 | CN240705 | Disulfide isomerase | 3 | 8 | 0.038949 | 2.1 | EAL91387.1/A.fumigatus/3.0E-61 | |
| 540 | CN240558 | Cytochrome C oxidase biogenesis protein | 0 | 5 | 0.015111 | 1.8 | XP_214182.2/R. norvegicus/1.0E-06 | |
| 578 | CA582837 | Pyruvate dehydrogenase e1 component beta subunit | 2 | 7 | 0.033994 | 1.6 | EAL86696.1/A. fumigatus/2.0E-99 | |
| 407 | CA583387 | Succinyl-CoA synthetase alpha subunit | 7 | 19 | 0.004468 | 2.6 | EAL91981.1/A. fumigatus/1.0E-155 | |
| 284 | CN239025 | Ubiquinol-cytochrome C reductase complex ubiquinonE-binding protein QP-C | 0 | 4 | 0.030475 | 1.5 | EAL90680.1/A. fumigatus/7.0E-29 | |
| 378 | CA580847 | Argininosuccinate synthase | 0 | 6 | 0.007492 | 1.7 | NP_229577.1/T. maritime/4.0E-77 | Amino acid metabolism and transport |
| 1618 | CA583639 | Aromatic-L-amino-acid decarboxylase | 1 | 33 | 0.00000 | 17.2 | EAL86509.1/A. fumigatus/0.0 | |
| 125 | CA583825 | Glycine cleavage system h protein | 4 | 9 | 0.042192 | 1.4 | EAL90537.1/A. fumigatus/6.0E-36 | |
| 1674 | CA583874 | Aldolase | 5 | 14 | 0.010368 | 37.9 | AAL34519.2/P. brasiliensis/0.0 | C-compound and carbohydrate metabolism |
| 42 | CA581699 | Phosphoglycerate kinase | 1 | 10 | 0.002512 | 2.6 | EAL90363.1/A. fumigatus/0.0 | |
| 9 | CA581893 | Beta-ketoacyl synthase (Cem 1) | 1 | 5 | 0.045709 | 2.2 | EAL87667.1/A. fumigatus/1.0E-88 | Lipid, fatty-acid and isoprenoid metabolism |
| 780 | CA581145 | GPR/FUN34 family protein | 0 | 11 | 0.000225 | 14.9 | EAL87502.1/A. fumigatus/6.0E-67 | |
| 1989 | CA581550 | Acetyl-coenzyme A synthetase (AcetatE – CoA ligase) (Acyl-activating enzyme) | 1 | 9 | 0.004605 | 2.0 | EAL89682.1/A. fumigatus/0.0 | |
| 1550 | CA582818 | NADH-cytochrome b5 reductase | 0 | 6 | 0.007492 | 5.4 | EAL88164.1/A. fumigatus/1.0E-86 | |
| 300 | CA581937 | Nucleoside diphosphate kinase | 6 | 58 | 0.00000 | 1.6 | AAP85295.1/A. fumigatus/2.0E-67 | Nucleotide metabolism |
| 547 | CA583473 | 6,7-dimethyl-8-ribityllumazine synthase | 0 | 6 | 0.007492 | 1.4 | AAD55372.1/A. fumigatus/9.0E-56 | Metabolism of vitamins, cofactors, and prosthetic groups |
| 924 | CN240624 | Coproporphyrinogen III oxidase | 2 | 7 | 0.033994 | 2.7 | EAL88456.1/A. fumigatus/0.0 | |
| 867 | CA580742 | NADH pyrophosphatase | 1 | 5 | 0.045709 | 5.7 | EAL85969.1/A. fumigatus/1.0E-159 | |
| 1490 | CA583063 | Pyridoxamine 5'-phosphate oxidase | 0 | 10 | 0.000453 | 3.5 | AAC28862.1/S. commune/2.0E-32 | |
| 447 | CA580589 | NADH:ubiquinone oxidoreductase B18 subunit | 1 | 10 | 0.002512 | 1.4 | EAL92195.1/A. fumigatus/9.0E-33 | |
| 488 | CA582788 | Exonuclease II | 1 | 5 | 0.045709 | 1.9 | EAL85993.1/A. fumigatus/1.0E-138 | Transcription, translation and ribosome structure |
| 165 | CN241393 | RNP domain protein | 3 | 13 | 0.003962 | 1.8 | EAL89070.1/A. fumigatus/5.0E-81 | |
| 2436 | CA580512 | Splicing factor u2af 35 kd subunit | 2 | 7 | 0.033994 | 2.5 | EAL86523.1/A. fumigatus/1.0E-103 | |
| 253 | CN240426 | Zinc finger, C3HC4 type | 0 | 5 | 0.015111 | 1.4 | NP_593329.1/S. cerevisiae/3.0E-10 | |
| 551 | CN239696 | Ribosomal protein L35** | 5 | 10 | 0.044755 | - | AAL08563.1/P. brasiliensis/1.0E-63 | |
| 979 | CA582579 | 60S ribosomal protein L7/L12 precursor | 1 | 8 | 0.008358 | 1.3 | EAL89813.1/A. fumigatus/4.0E-49 | |
| 175 | CA581863 | Complex I intermediatE-associated protein CIA30 precursor | 4 | 15 | 0.003399 | 5.6 | EAL92946.1/A. fumigatus/1.0E-114 | Protein fate and Secretion |
| 832 | CN242383 | Glutathione S transferase | 1 | 7 | 0.014984 | 2.0 | NP_588171.1/S. pombe/7.0E-42 | |
| 2387 | CA584103 | Non-classical export protein (Nce1) | 1 | 7 | 0.014984 | 55.6 | EAL87256.1/A. fumigatus/1.0E-29 | |
| 1823 | CA583903 | Profilin | 1 | 5 | 0.045709 | 1.3 | NP_014765.1/S. cerevisiae/8.0E-14 | |
| 4188 | CN245872 | Mating type protein (MAT1–2)* | 1 | 0 | - | 8.0 | EAL89707.1/A. fumigatus/2.0E-36 | Mating Type |
| 50 | CA581392 | Cu-Zn superoxide dismutasE-related* | 0 | 8 | 0.001842 | 2.1 | CAB97297.1/N. crassa/3.0E-30 | Virulence and oxidative stress |
| 2059 | CN241260 | Ribosome associated protein (Stm1) | 6 | 31 | 0.000007 | 1.7 | EAL92489.1/A. fumigatus/2.0E-32 | Others |
| 2005 | CA580764 | Signal peptide protein | 1 | 6 | 0.026442 | 2.3 | EAL93249.1/A. fumigatus/7.0E-68 | |
| 39 | CA581046 | Unknown | 0 | 6 | 0.007492 | 2.2 | - | |
| 33 | CA582496 | Unknown | 0 | 8 | 0.001842 | 3.1 | - | |
| 1442 | CA581846 | Unknown | 3 | 16 | 0.000836 | 4.5 | - | |
| 2399 | CA581839 | Unknown | 1 | 5 | 0.045709 | 2.5 | - | |
| 512 | CA583749 | Unknown | 0 | 6 | 0.007492 | 4.3 | - | |
| 639 | CA581506 | Unknown | 0 | 7 | 0.003715 | 1.7 | - | |
| 718 | CN247671 | Unknown | 0 | 6 | 0.007492 | 1.8 | - | |
| 765 | CA581478 | Unknown | 0 | 10 | 0.000453 | 3.9 | - | |
| 529 | CA580398 | Unknown | 1 | 5 | 0.045709 | 18.8 | - | |
a FDR = 4,8% and Q-value < 5%.
b Not significant by Audic-Claverie's method.
* Up-regulated genes confirmed by northern blotting.
** Not assayed by cDNA microarray but confirmed as up-regulated in yeast by northern blotting.
Mycelium and yeast up-regulated genes involved in cell organisation
The hex and bgl genes, which code for the hexagonal peroxisome protein and 1,3 β-glucosidase, respectively, were up-regulated in mycelium cells and are categorised as involved in cell wall, membrane and cytoskeleton organisation (Figure 1a). The hexagonal peroxisome protein has been identified in different filamentous ascomycetes such as the plant pathogen Magnaporthe grisea [44] and in Neurospora crassa [45], being the major protein of the Woronin body, a septal pore-associated organelle [46,47]. HEX1p has been shown to seal septal pores in response to cellular damage and is strongly implicated in cell integrity maintenance [45]. In M. grisea, hex1 mutants present delayed host penetration and subsequent disruption of invasive hyphal growth in plants. Inability of these mutants to survive under nitrogen starvation in vitro has also been observed [44]. Deletion of hex1 in N. crassa eliminates Woronin bodies from the cytoplasm and results in hyphae that exhibit a cytoplasmic-bleeding morphotype in response to cell lysis [45]. It was thus proposed that the Woronin body represents a new category of peroxisome acting in the maintenance of cellular integrity and virulence in filamentous fungi [45]. We hypothesise that these highly specialised vesicles are involved in the protection of P. brasiliensis against cellular damage as well as its survival during invasive growth and host colonisation in the process of infection. Future investigations are required to elucidate the role of Woronin bodies/HEX1 protein in P. brasiliensis.
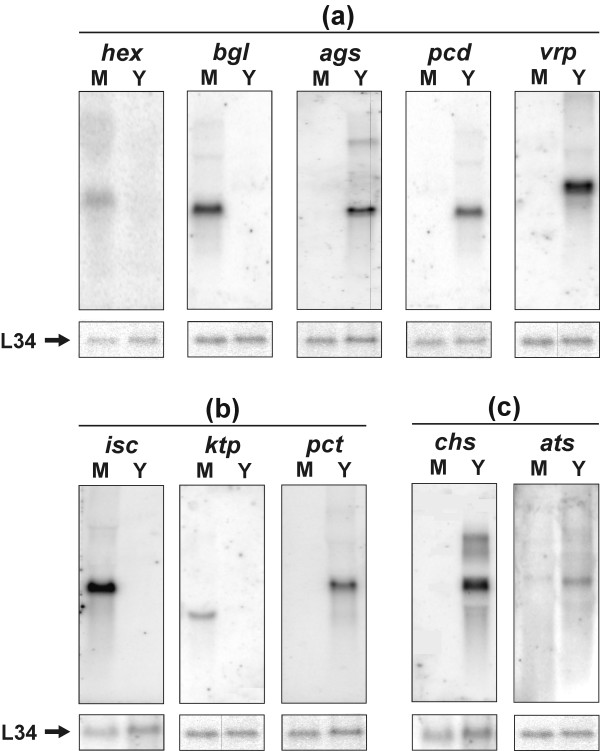
Figure 1.
Northern blot analysis of mycelium and yeastup-regulated genes of P. brasiliensis. Total RNA samples from both mycelium (M) and yeast (Y) were blotted onto nylon membranes and hybridised against gene-specific radiolabelled probes: (a) Control of cell organisation: hex – Hexagonal peroxisome protein, bgl –1,3 beta-glucosidase, ags – alpha 1,3-glucan synthase, cda – Chitin deacetylase, vrp – Verprolin; (b) Ion transporters: isc –Iron-sulphur cluster-like protein, ktp – Potassium transporter, pct – Putative P-type Cu(2+) transporting ATPase; (c) Sulphur metabolism: chs – Choline sulphatase, ats – ATP sulphurylase. The constitutive 60S ribosomal protein L34 was used as a loading control.
Another mycelium up-regulated gene codes for β-1,3-glucosidase, an enzyme that hydrolyses the O-glycosidic linkages of β-glucan. This polysaccharide is an important cell wall constituent in P. brasiliensis mycelium cells in contrast with α-glucans, which predominate in the yeast cell wall [48]. A hypothesis formulated by Kanetsuna et al. [49] and modified by San Blas and San Blas [50] explains the differentiation from mycelium to yeast and vice-versa based on a change on cell wall composition. At 37°C, there is an increased synthesis of chitin and α-glucan, and low levels of β-glucan, which results in the yeast form. In contrast, at 22°C, α-glucan synthesis occurs at low rates and long β-glucan fibrils are formed in the budding spots. In keeping with these morphological and biochemical events, 1,3-β-glucosidase increased levels are correlated to the shift to the mycelium phase.
Other three genes coding for proteins from the same category were confirmed to be up-regulated in yeast cells: ags (α-1,3-glucan synthase), cda (chitin deacetylase) and vrp-verprolin (Fig. 1a). The P. brasiliensis 1,3-α-glucan synthase gene was first described by Pereira et al. [51]. Recently, it was demonstrated that it is strongly up-regulated in yeast cells [28,52], which was confirmed in this work by northern blotting analysis. Rappleye et al. [53] silenced the 1,3-α-glucan synthase gene in H. capsulatum and demonstrated that α-(1,3)-glucan is an important virulence factor and affects the ability of H. capsulatum to kill macrophages and colonise murine lungs. In C. neoformans, mutants for 1,3-α-glucan synthase failed to assemble the capsule, which is an important virulence factor of this pathogen [54]. Morphogenetic transition is the essence of P. brasiliensis life cycle: for instance, low levels of α-1,3-glucan in the cell wall of the yeast form have been correlated with low virulence [55]. Virulent cultures of P. brasiliensis isolates grown i n vitro for long periods have thinner cell walls, low α-1,3-glucan levels and are consequently less virulent [56]. Our results suggest that α-glucan synthase is involved in the dimorphic transition of P. brasiliensis and possibly in its virulence. The cell wall is an essential and dynamic fungal structure that has been implicated in several pathogenic processes. Being absent in mammalian cells, it may be a relevant target to drug therapies. In this context, the gene that encodes α-1,3-glucan synthase was demonstrated to be a virulence factor using RNAi approaches in Cryptococcus neoformans [54] and H. capsulatum [53], and seems to be an ideal target for new antifungal drugs. In P. brasiliensis glucan polymers constitute 95% of yeast cell wall [49] and thus any interference in cell wall synthesis through glucan synthases is likely to affect virulence directly.
Chitin deacetylase enzyme (CDA) catalyses the conversion of chitin to chitosan by deacetylation of N-acetyl-D-glucosamine residues. Chitosan is a flexible, soluble polymer that integrates the cell wall of some fungi, such as S. cerevisiae [57] and C. neoformans [58]. In S. cerevisiae, chitosan is only found during sporulation [59]. The molecular characterisation of two sporulation-specific chitin deacetylase genes, CDA1 and CDA2, both of which contribute to spore wall rigidity, was described previously [59]. In S. cerevisiae, cda1 mutants present a more diffuse chitosan layer, while their surface layer remains intact. In cda2 mutant cells, by comparison, the chitosan layer is not detected at all. In the spore walls of cda1 and cda2 mutants both outer layers are missing due to defects on wall maturation. However, in C. neoformans, a study reported that chitin is present in the yeast cell wall and most of it is continually deacetylated to chitosan. Mutants for chitin deacetylase show suppression of growth due to the lack of chitosan and therefore have a reduced infection capability [58]. The same study hypothesized that this constant remodelling of the cell wall contributed to cellular integrity in this fungus. In P. brasiliensis, we identified a highly expressed cda gene in yeast cells that presents similarity to the C. neoformans. If the C. neoformans model is closer to what is found in P. brasiliensis, then chitin synthase and chitin deacetylase may be potential targets to antifungal therapy.
Verprolin is required for a fully polarised distribution of cortical actin patches and viability at high temperature. This is the first time that verprolin is described in P. brasiliensis, a pathogen that has as an intrinsic characteristic the ability to grow at the human body temperature, 37°C. The inability of vrp-1 mutants to grow at 37°C was reported by Naqvi et al. [60] in the non-pathogenic yeast S. cerevisiae. Likewise, we hypothesise that verprolin is involved in the ability of P. brasiliensis to grow at 37°C and in cell cytoskeleton organisation since this gene is over expressed in yeast cells. Considering that the actin cytoskeleton plays a crucial role on fundamental processes such as cell growth, differentiation and migration, localised membrane growth, endocytosis, and cell division [61], this protein is likely to play a key role in cell maintenance and viability of P. brasiliensis inside the host cell.
Mycelium and yeast up-regulated genes involved in ion metabolism and transport
Two genes putatively implicated in ion transport were confirmed to be highly expressed in mycelium cells: isc and ktp, an iron-sulphur cluster protein and a cation transporter, respectively. In contrast, a putative P-type cation pump (pct) was up-regulated in the yeast form (Figure 1b).
It has been reported that the ISC protein is responsible for mitochondrial uptake of iron and seems to monitor the cytoplasmic levels of this ion. In S. cerevisiae, the double knock-out of the homologues ISU1 and ISU2 is lethal. Defective mutants are distinguished by iron accumulation in the mitochondrial matrix and its respective decrease in the cytosol [62]. In C. neoformans, complementation, cloning and sequencing of such genes has recently been accomplished [63]. It has long been hypothesised that iron is a limiting factor for infectivity during cryptococcosis as well as in other systemic mycoses, in that the host normally provides only limited amounts of this compound. Arango and Restrepo [64]demonstrated iron availability to be essential for growth of mycelium and yeast of P. brasiliensis; but especially for mycelium, whose growth was totally prevented by the addition of the iron chelator phenanthroline to the medium, an effect observed only to a lesser extent in yeast. The effect of phenanthroline was reversed partially in mycelium and totally in yeast by addition of excess iron. This is in good agreement with the overexpression of the ISC protein in the mycelial phase. In P. brasiliensis it could be involved in monitoring the amount of iron in the environment and in providing a means of storage of this metal.
The ktp sequence from P. brasiliensis aligned best with potassium transporter proteins of the HAK family, which are mainly implicated in the resistance to potassium starvation. In N. crassa, the closest homolog of P. brasiliensis, KTP coexists with another potassium transporter of the TRK family [65]. It has been hypothesised that soil organisms are universally equipped with a powerful K+-concentrating apparatus, as these organisms are faced with a very diluted and variable environment, thus being forced to pump potassium in against a steep gradient [65]. This is likely to be the case of P. brasiliensis, whose ecological niche for the mycelium form is thought to be the soil.
Another yeast up-regulated gene is pct, a putative member of the E1-E2 (P-type) family of ATPases. These are ATP-dependent proteins which regulate transmembrane flow of all relevant cations, including Na+, H+, Mg2+, Ca2+, Cd2+, Cu2+ and K+ [66]. In C. albicans, the E1-E2 ATPase gene, CDR1, confers resistance to both copper and silver, the latter being used as an antimicrobial agent [67]. A similar function could be attributed to the P. brasiliensis pct gene, although alignment data are insufficient to identify which cation this protein transports.
Sulphur metabolism
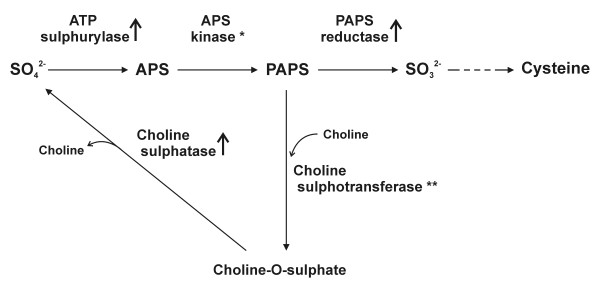
Several enzymes from the cysteine de novo biosynthesis pathway (Figure 2) were shown to be up-regulated in the yeast form of P. brasiliensis. Our previous analysis [28] had already confirmed over expression of paps reductase (the third in the pathway). In silico EST subtraction and cDNA microarray showed yeast up-regulation for atp sulphurylase and aps kinase; the former was confirmed by northern blotting (Figure 1c). Thus, we can strongly suggest that the yeast form synthesises cysteine actively from inorganic sulphate.
Figure 2.
Up-regulated genes encoding enzymes from the cysteine de novo biosynthesis pathway. Arrows indicate enzymes identified as up-regulated both by in silico subtraction, cDNA microarray and confirmed by northern blotting experiments. (*) enzyme identified as up-regulated by both in silico subtraction and cDNAs microarray but not assayed by northern blotting. (**) indicates an enzyme not found in the transcriptome of P. brasiliensis.
In order to reinforce these data, we have evaluated the importance of inorganic sulphate to growth and differentiation. Auxotrophy of P. brasiliensis yeast for several sources of organic sulphate – including cysteine itself and sulphydrylic compounds – has been reported before [42]. It was concluded then that organic sulphate deprivation suppressed growth in the yeast phase and prevented mycelium-to-yeast differentiation, whereas the mycelial phase is able to grow on either inorganic or organic sulphur [68]. Also, the saprophytic, mycelial form of H. capsulatum is prototrophic while the pathogenic yeast form requires cysteine [69]. It has been reported that exogenous cysteine is required for both yeast phase growth and morphological transition from mycelium-to-yeast of H. capsulatum [41,70]. In this work, both mycelium and yeast cells of P. brasiliensis were incubated in modified MVM medium without inorganic sulphate, apart from the negligible amounts present in the trace elements solution. Dimorphic transition was assessed in the mycelium to yeast direction and in the opposite way.Sustained growth was observed for both morphotypes (data not shown) and, upon the corresponding temperature shifts, differentiation was successfully triggered in both directions (Figure 3). Thus, inorganic sulphate seems to be unnecessary for the transition, quite contrarily to organic sulphate. In this context, it is interesting to consider a branch of the cysteine biosynthetic pathway (Fig. 2). In fungi and plants a fraction of PAPS, which is toxic to fungi if it reaches high cytosolic levels, is used by choline sulphotransferase to produce choline-O-sulphate [40], which serves as an osmoprotectant and cytosolic sulphur store in these organisms. We have not found a homologue of choline sulphotranferase in P. brasiliensis to date, but the enzyme choline sulphatase, which degrades its product to choline and sulphate, is also over expressed in the yeast morphotype, as confirmed here (Figure 1c) and previously reported [52]. The C. neoformans met3 mutant, which lacks ATP sulphurylase activity, had a substantial defect in melanin formation, significantly reducedgrowth rate, and greatly increased thermotolerance. In the murine inhalation infection model, the met3 mutant was avirulent and was deficient in its ability to survive in mice [71]. In this context, disrupting the genes encoding choline sulphatase or ATP sulphurylase in P. brasiliensis should reveal its role in the growth, maintenance of yeast cells and pathogenicity of this fungus. It is interesting that another intracellular pathogen of humans, the bacterium Mycobacterium tuberculosis, depends on sulphur compounds for expression of its full virulence, drug resistance and overall survival inside the macrophage. It has developed a very efficient sulphate activation pathway (SAC) that ensures constant synthesis of PAPS at high rates, from which sulphate may be distributed to other synthetic pathways [72]. The SAC includes the bacterial counterparts of ATP sulphurylase and APS kinase, the latter of which performs PAPS synthesis by coupling it with GTP hydrolysis by a GTPase that is also present in SAC. Whether similar mechanisms are present in pathogenic fungi such as P. brasiliensis remains to be investigated.
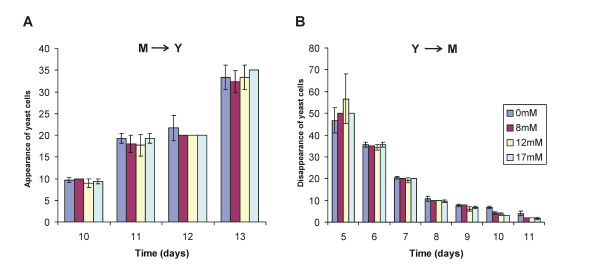
Figure 3.
Cell differentiation of P. brasiliensis in modified MVM medium without inorganic sulphate. The fungus was grown in four different concentrations of sulphate salts (0, 8, 12 and 17 mM; the latter is the original concentration of MVM medium). (A) The appearance of yeast cells was verified daily in the transition from mycelium to yeast after temperature shift to 37°C, (B) The disappearance of yeast cells was verified daily in the transition from yeast to mycelium after temperature shift to 22°C. Triple samples were counted for each time point. The coloured boxes indicate the average of the three samples and bars represent the standard deviation of the mean. As observed, the presence or absence of inorganic sulphate did not affect transition in either direction.
Conclusion
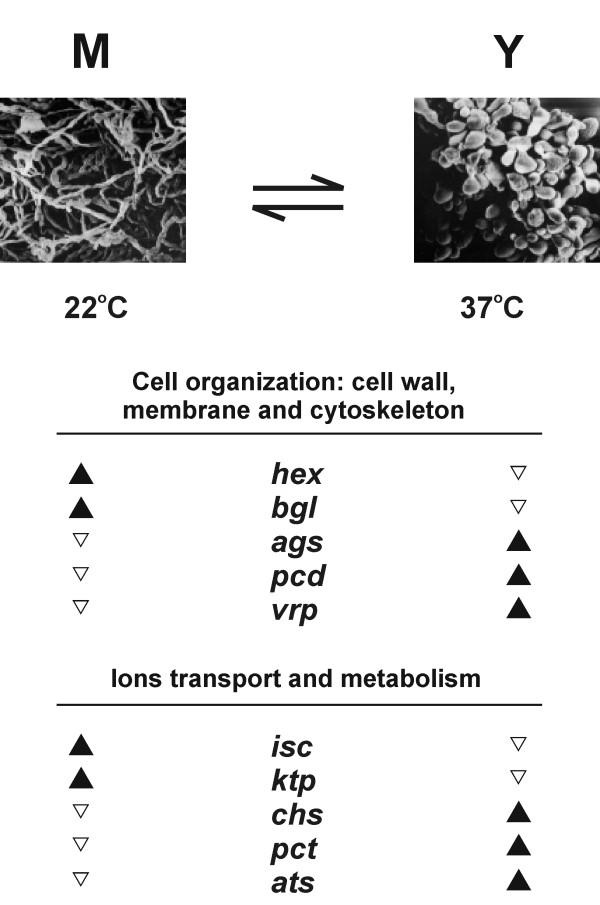
Taken together, these data show that several genes involved in cell organisation and ion metabolism/transport are differential in their expression along dimorphic transition, which is in accordance with the proposed model for this process in Figure 4. While α-glucan is synthesised during yeast phase due to high expression of 1,3 α-glucan synthase, β-glucan is degraded by the action of 1,3 β-glucosidase during hyphal growth. The cda gene is probably involved in the cell wall synthesis of yeast cells, since it is over expressed in this phase. In addition, genes related to septal sealing and cytoskeleton organisation (hex and vpr) are also probably implicated in the stabilisation and maintenance of mycelium and yeast cells in the environment and at 37°C in the human host. Also, the differential expression pattern of genes that control metabolism and transport of potassium, iron, copper and sulphur ions suggests that they may influence directly the pathogen adaptation to the host environment. Specifically, in spite of the undisturbed growth and differentiation on depletion of inorganic sulphate, the over expression of enzymes from de novo cysteine synthesis lends support to previous findings about the importance of this pathway to yeast metabolism. Understanding these changes by functional analysis of such genes may lead to a better understanding of the infective process, thus providing new targets and strategies to control PCM.
Figure 4.
Genes involved in cell organisation (cell wall, membrane and cytoskeleton), sulphur metabolism and ion transport. Genes that were identified as up-regulated in mycelium (22°C) or yeast (36°C) cells of P. brasiliensis are represented by black arrowheads. Electron microscopy was performed by Silva et al. [78].
Methods
Strains and cultures
P. brasiliensis clinical isolate Pb01 (ATCC-MYA-826) was used in this study. Cells from both mycelium and yeast forms were grown on semi-solid Fava Neto's medium [73] for 7 to 10 days at 22°C or 37°C, respectively.
Overlap analysis – in silico EST subtraction and cDNA microarrays
This work was based on the output of previous large-scale expression analysis experiments, as outlined in reference 28. Briefly, we have constructed a λZAP II® (Invitrogen) cDNA library from mycelium and yeast mRNA fractions and 5'-sequenced the mass-excised cloned fragments with the T7 vector primer. Raw sequence data were quality-assessed by PHRED and assembled by CAP3, thus generating a set of 6,022 PbAESTs (singlets and contigs). For functional annotation of sequences we used the nr (NCBI) database. In silico electronic subtraction was performed according to the Audic and Claverie [74] statistical approach, with a 95% confidence rate. For cDNA microarray 1,152 clones were selected and PCR-amplified for spotting onto nylon-membranes in triple experiments. Hybridisation against [α-33P] dCTP-labeled total RNA from mycelium or yeast and phosphor imager signal capture were performed as in [28]. After signal quantification and background subtraction [75], statistical analysis was carried out with the SAM (Significance Analysis of Microarrays) method [76]. Data from both experiments were overlapped to identify differential genes, thus generating the set of 66 sequences we used in this work.
Northern blot analysis
Total RNA (15μg) from mycelium and yeast cells of P. brasiliensis was separated on denaturing 1,5 % agarose gel and blotted onto a Hybond-N membrane (GE Healthcare). Probes were radiolabeled using [α-P32]dATP by random priming according to supplier's instructions (Invitrogen), purified and used in overnight hybridisation (50% formamide, 4X SSPE, 5X Denhardt's solution, 0,1% SDS, 100μg/ml herring sperm DNA) at 42°C. The membranes were then washed under stringency conditions of 2X SSPE-0.1% SDS at 65°C for 1h. Signal bands were visualised using the Typhoon 9210 Phosphor Imager (GE HealthCare).
Dimorphic transition without inorganic sulphate
We incubated both mycelium and yeast cells on modified versions of McVeigh and Morton's medium – MVM [77] where ammonium and magnesium sulphate salts were gradually replaced by their chloride counterparts, in the following chloride concentration set points: 0, 8, 12 and 17 mM, where the first corresponds to the original recipe and the last, to virtual absence of inorganic sulphate, apart from negligible amounts in the trace elements solution (~ 8 μM). Molar concentrations of both magnesium and ammonium were thus conserved. We have also evaluated whether dimorphic transition occurred normally in the medium without inorganic sulphur. To achieve this, five flasks containing 100 ml of modified MVM were inoculated with comparable amounts of mycelium (100 mg wet mass) and yeast (2.5 × 107 cells) previously grown on standard MVM. Samples were incubated in rotating shakers (120 rpm) at 36 and 22°C, respectively, thus triggering dimorphic transition. Fungal viability and progress of transition were assessed by serial 100 μl sampling every 24 hours (three independent samples). Each sample was coloured with Janus Green and the number of yeast cells was counted in a light microscope with the aid of a Neubauer counting chamber.
Accession numbers
The accession numbers of the EST sequences analysed in this work are shown in the Tables 1 and 2.
Abbreviations
ags alpha 1,3-glucan synthase
aps adenosine 5'-phosphosulphate
ats ATP sulphurylase
bgl 1,3 beta-glucosidase,
BLAST basic local alignment search tool
cda chitin deacetylase
cDNA complementary DNA
chs choline sulphatase
COG clusters of orthologous groups
e-value extreme value distributionESTs
ESTs expressed sequence tags
GO gene ontology
hex hexagonal peroxisome protein
isc iron-sulphur cluster-like protein
ktp potassium transporter
MIPS Munich information center for proteins sequences
PAPS phosphoadenylyl-sulfate reductase
PbAETs P. brasiliensis assembled EST sequences
PCM paracoccidioidomycosis
pct putative P-type Cu(2+) transporting ATPase
SAGE serial analysis of gene expression
SAM significance analysis of microarrays
vrp verprolin
Authors' contributions
RA and MF planned and designed the study, developed the experiments and the data analysis, wrote the main draft of the paper and support the preparation of the figures and tables. HP supports the discussion of the results and revised the manuscript. AN participated in the in silico ESTs subtraction analysis of the raw data generated by the transcriptome project. MC analysed the results of the microarray experiments, helped in the manuscript edition, and prepared the figures. AL executed the microarray experiments. MC, RC and MB participated in the normalization process of the microarray raw data and helped to make the statistical analyses. SS participated of the differentiation experiment involved of the inorganic sulphur, and of the preparation of the RNA of P. brasiliensis to make the microarray experiments. LF participated of the analysis of the cell wall organization. SP helped in the ESTs amplification and on the analysis of the sulphur metabolism. GP, ES, ED designed the microarray experiments. CS participated on the Pb ESTs annotation. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We are most indebted to Hugo Costa Paes for text draft revision, Maria Emilia M. T. Walter and Shana Santos for technical bioinformatics support. This work was supported by MCT/CNPq, CNPq, CAPES, FAP-DF, Fapesp, FUB, UFG.
Contributor Information
Rosângela V Andrade, Email: rvieira@unb.br.
Hugo C Paes, Email: sorumbatico@gmail.com.
André M Nicola, Email: a0058343@aluno.unb.br.
Maria José A de Carvalho, Email: mariajac@gmail.com.
Ana Lúcia Fachin, Email: analuf@rge.fmrp.usp.br.
Renato S Cardoso, Email: rscardoso@hotmail.com.
Simoneide S Silva, Email: simoneidesilva@yahoo.com.br.
Larissa Fernandes, Email: larissaf@unb.br.
Silvana P Silva, Email: petrofez@uol.com.br.
Eduardo A Donadi, Email: eadonadi@fmrp.usp.br.
Elza T Sakamoto-Hojo, Email: etshojo@usp.br.
Geraldo AS Passos, Email: passos@rge.fmrp.usp.br.
Célia MA Soares, Email: celia@icb.ufg.br.
Marcelo M Brígido, Email: brigido@unb.br.
Maria Sueli S Felipe, Email: msueli@unb.br.
References
- Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet. 2002;32:490–495. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- Lashkari DA, DeRisi JL, McCusker JH, Namath AF, Gentile C, Hwang SY, Brown PO, Davis RW. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc Natl Acad Sci USA. 1997;94:13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/14385. [DOI] [PubMed] [Google Scholar]
- Paillisson A, Dade S, Callebaut I, Bontoux M, Dalbies-Tran R, Vaiman D, Monget P. Identification, characterization and metagenome analysis of oocyte-specific genes organized in clusters in the mouse genome. BMC Genomics. 2005;6:76. doi: 10.1186/1471-2164-6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkovic A, Yan C, Klysik M, Matzuk MM. Discovery of germ cell-specific transcripts by expressed sequence tag database analysis. Fertil Steril. 2001;76:550–554. doi: 10.1016/S0015-0282(01)01966-5. [DOI] [PubMed] [Google Scholar]
- Steen BR, Lian T, Zuyderduyn S, MacDonald WK, Marra M, Jones SJ, Kronstad JW. Temperature-regulated transcription in the pathogenic fungus Cryptococcus neoformans. Genome Res. 2002;12:1386–1400. doi: 10.1101/gr.80202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen BR, Zuyderduyn S, Toffaletti DL, Marra M, Jones SJM, Perfect JR, Kronstad J. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot Cell. 2003;2:1336–1349. doi: 10.1128/EC.2.6.1336-1349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer MD, Ilyina T, Ma X, Vandoninck S, Luyten WHM, Bossche HV. Genomic profiling of the response of Candida albicans to Itraconazole treatment using a DNA microarray. Antimicrob Agents Chemother. 2001;45:1660–1670. doi: 10.1128/AAC.45.6.1660-1670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B, Nantel A, Whiteway M. Stress-induced gene expression in Candida albicans : Absence of a general stress response. Mol Biol Cell. 2003;14:1460–1467. doi: 10.1091/mbc.E02-08-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin C, Kretschmar M, Nichterlein T, Gaillardin C, d'Enfert C, Hube B. Stage-specific gene expression of Candida albicans in human blood. Mol Microbiol. 2003;47:1523–1543. doi: 10.1046/j.1365-2958.2003.03396.x. [DOI] [PubMed] [Google Scholar]
- Lane S, Birse C, Zhou S, Matson R, Liu H. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J Biol Chem. 2001;276:48988–48996. doi: 10.1074/jbc.M104484200. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel A, Dignard D, Bachevich C, Harcus D, Marcil A, Bouin AP, Sensen CW, Hogues H, Hoog MVH, Gordon P, Rigby T, Benoit F, Tessier DC, Thomas DY, Whiteway M. Transcription profiling, of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell. 2002;13:3452–3465. doi: 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Hocking-Murray D, Bahrami AK, Andersson M, Rine J, Sil A. Identifying phase-specific genes in the fungal pathogen Histoplasma capsulatum using a genomic shotgun microarray. Mol Biol Cell. 2003;14:2314–2326. doi: 10.1091/mbc.E03-01-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus PR, Boily MJ, Giles SS, Stajich JE, Allen A, Cox GM, Dietrich FS, Perfect JR, Heitman J. Identification of Cryptococcus neoformans Temperature-Regulated Genes with a Genomic-DNA. Microarray Eukaryot Cell. 2004;3:1249–1260. doi: 10.1128/EC.3.5.1249-1260.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns BJ, Baylis BW, Urbanski SJ, Gibb AP, Rabi HR. Paracoccidioidomycosis: case report and review. Clin Infect Dis. 1996;23:1026–1032. doi: 10.1093/clinids/23.5.1026. [DOI] [PubMed] [Google Scholar]
- Restrepo M. The ecology of Paracoccidioides brasiliensis : a puzzle still unsolved. Sabouradia. 1985;23:323–334. [PubMed] [Google Scholar]
- Blotta MH, Mamoni RL, Oliveira SJ, Nouer SA, Papaiordanou PM, Goveia A, Camargo ZP. Endemic regions of paracoccidioidomycosis in Brazil: a clinical and epidemiologic study of 584 cases in the southeast region. Am J Trop Med Hyg. 1999;61:390–394. doi: 10.4269/ajtmh.1999.61.390. [DOI] [PubMed] [Google Scholar]
- Brummer E, Castañeda E, Restrepo A. Paracoccidioidomycosis: an update. Clin Microbiol. 1993;6:89–117. doi: 10.1128/cmr.6.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo A, McEwen JG, Castañeda E. The habitat of Paracoccidioides brasiliensis : how far from solving the riddle? Med Mycol. 2001;39:233–241. doi: 10.1080/mmy.39.3.233.241. [DOI] [PubMed] [Google Scholar]
- McEwen JG, Garcia AM, Ortiz BL, Botero S, Restrepo A. In search of the natural habitat of Paracoccidioides brasiliensis. Arch Med Res. 1995;26:305–306. [PubMed] [Google Scholar]
- Bagagli E, Sano A, Coelho KI, Alquati S, Miyaji M, de Camargo ZP, Gomes GM, Franco M, Montenegro MR. Isolation of Paracoccidioides brasiliensis from armadillos (Dasypus noveminctus) captured in an endemic area of paracoccidioidomycosis. Am J Trop Med Hyg. 1998;58:505–512. doi: 10.4269/ajtmh.1998.58.505. [DOI] [PubMed] [Google Scholar]
- Tristano AG, Chollet ME, Willson M, Perez J, Troccoli M. Central nervous system paracoccidioidomycosis: case report and review. Invest Clin. 2004;45:277–288. [PubMed] [Google Scholar]
- De Camargo ZP, Franco MF. Current knowledge on pathogenesis and immunodiagnosis of paracoccidioidomycosis. Rev Iberoam Micol. 2000;17:41–48. [PubMed] [Google Scholar]
- Restrepo A, Salazar ME, Cano LE, Stover EP, Feldman D, Stevens DA. Estrogens inhibit mycelium-to-yeast transformation in the fungus Paracoccidioides brasiliensis : implications for resistance of females to paracoccidioidomycosis. Infect Immun. 1984;46:346–353. doi: 10.1128/iai.46.2.346-353.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo A, McEwen JG, Castaneda E. The habitat of Paracoccidioides brasiliensis : how far from solving the riddle? Med Mycol. 2001;39:233–241. doi: 10.1080/mmy.39.3.233.241. [DOI] [PubMed] [Google Scholar]
- Marques SA, Robles AM, Tortorano AM, Tuculet MA, Negroni R, Mendes RP. Mycoses associated with AIDS in the Third World. Med Mycol. 2000;38:269–279. [PubMed] [Google Scholar]
- Felipe MSS, Andrade RV, Arraes FBM, Nicola AM, Maranhão AQ, Torres FAG, Silva-Pereira I, Poças-Fonseca MJ, Campos EG, Moraes LMP, Andrade PA, Tavares AHFP, Silva SS, Kyaw CM, Souza DP, PbGenome Network, Pereira M, Jesuíno RSA, Andrade EV, Parente JA, Oliveira GS, Barbosa MS, Martins NF, Fachin AL, Cardoso RS, Passos GAS, Almeida NF, Walter MEMT, Soares CMA, Carvalho MJA, Brigido MM. Transcriptional profiles of the human pathogenic fungus Paracoccidioides brasiliensis in mycelium and yeast cells. J Biol Chem. 2005;280:24706–24714. doi: 10.1074/jbc.M500625200. [DOI] [PubMed] [Google Scholar]
- Felipe MSS, Andrade RV, Petrofeza SS, Maranhão AQ, Torres FAG, Albuquerque P, Arraes FB, Arruda M, Azevedo MO, Baptista AJ, Bataus LAM, Borges CL, Campos EG, Cruz MR, Daher BS, Dantas A, Ferreira MA, Ghil GV, Jesuíno RS, Kyaw CM, Leitão L, Martins CR, Moraes LM, Neves EO, Nicola AM, Alves ES, Parente JA, Pereira M, Poças-Fonseca MJ, Resende R, Ribeiro BM, Saldanha RR, Santos SC, Silva-Pereira I, Silva MA, Silveira E, Simões IC, Soares RB, Souza DP, De-Souza MT, Andrade EV, Xavier MA, Veiga HP, Venâncio EJ, Carvalho MJ, Oliveira AG, Inoue MK, Almeida NF, Walter ME, Soares CMA, Brigido MM. Transcriptome characterization of the dimorphic and pathogenic fungus Paracoccidioides brasiliensis by EST analysis. Yeast. 2003;20:263–271. doi: 10.1002/yea.964. [DOI] [PubMed] [Google Scholar]
- Goldman GH, dos Reis Marques E, Duarte Ribeiro DC, de Souza Bernardes LA, Quiapin AC, Vitorelli PM, Savoldi M, Semighini CP, de Oliveira RC, Nunes LR, Travassos LR, Puccia R, Batista WL, Ferreira LE, Moreira JC, Bogossian AP, Tekaia F, Nóbrega MP, Nóbrega FG, Goldman MH. Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot Cell. 2003;2:34–48. doi: 10.1128/EC.2.1.34-48.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K, Pascal G, Wong SSY, Glaser P, Woo PCY, Kunst F, Cai JJ, Cheung EYL, Médigue C, Danchin A. Exploring the Penicillium marneffei genome. Arch Microbiol. 2003;179:339–353. doi: 10.1007/s00203-003-0533-8. [DOI] [PubMed] [Google Scholar]
- Bailek R, Ibricevic A, Fothergill A, Begerow D. Small subunit ribosomal DNA sequence shows Paracoccidioides brasiliensis closely related to Blastomyces dermatitidis. J Clin Microbiol. 2000;38:3190–3193. doi: 10.1128/jcm.38.9.3190-3193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Blas G, Niño-Vega G, Iturriaga T. Paracoccidioides brasiliensis and paracoccidioidomycosis: molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med Mycol. 2002;40:225–242. doi: 10.1080/mmy.40.3.225.242. [DOI] [PubMed] [Google Scholar]
- Matute DR, McEwen JG, Puccia R, Montes BA, San-Blas G, Bagagli E, Rauscher JT, Restrepo A, Morais F, Niño-Vega G, Taylor JW. Cryptic Speciation and Recombination in the Fungus Paracoccidioides brasiliensis as Revealed by Gene Genealogies. Mol Biol Evol. 2006;23:65–73. doi: 10.1093/molbev/msj008. [DOI] [PubMed] [Google Scholar]
- Feitosa LS, Cisalpino PS, dos Santos MR, Mortara RA, Barros TF, Morais FV, Puccia R, da Silveira JF, de Camargo ZP. Chromosomal polymorphism, syntenic relationships and ploidy in the pathogenic fungus Paracoccidioides brasiliensis. Fungal Genet. 2003;39:60–69. doi: 10.1016/S1087-1845(03)00003-3. [DOI] [PubMed] [Google Scholar]
- Montoya AE, Moreno MN, Restrepo A, McEwen JG. Electrophoretic karyotype of clinical isolates of Paracoccidioides brasiliensis. Fungal Genet. 1997;21:223–227. doi: 10.1006/fgbi.1996.0955. [DOI] [PubMed] [Google Scholar]
- Cano MIN, Cisalpino PS, Galindo I, Ramírez JL, Mortara RA, da Silveira JF. Electrophoretic karyotypes and genome sizing of the pathogenic fungus Paracoccidioides brasiliensis. J Clin Microbiol. 1998;36:742–747. doi: 10.1128/jcm.36.3.742-747.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinoso C, Nino-vega G, San-Blas G, Dominguez A. Random sequencing of Paracoccidioides brasiliensis genes. Med Mycol. 2005;43:681–689. doi: 10.1080/13693780500129673. [DOI] [PubMed] [Google Scholar]
- Munich Information Center for Proteins Sequences http://mips.gsf.de/
- Marluf GA. Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu Rev Microbiol. 1997;51:73–96. doi: 10.1146/annurev.micro.51.1.73. [DOI] [PubMed] [Google Scholar]
- Boguslawski G, Akagi JM, Ward LG. Possible role for cysteine biosynthesis in conversion from mycelial to yeast form of Histoplasma capsulatum. Nature. 1976;261:336–338. doi: 10.1038/261336a0. [DOI] [PubMed] [Google Scholar]
- Medoff G, Painter A, Kobayashi GS. Mycelial-to yeast-phase transitions of the dimorphic fungi Blastomyces dermatitidis and Paracoccidioides brasiliensis. J Bacteriol. 1987;169:4055–4060. doi: 10.1128/jb.169.9.4055-4060.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munich Information Center for Protein Sequences http://mips.gsf.de/
- Soundararajan S, Jedd G, Li X, Ramos-Pamplona M, Chua NH, Naqvia NI. Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell. 2004;16:1564–1574. doi: 10.1105/tpc.020677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G, Chua NH. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat Cell Biol. 2000;2:226–231. doi: 10.1038/35008652. [DOI] [PubMed] [Google Scholar]
- Markham P, Collinge AJ. Woronin bodies in filamentous fungi. FEMS Microbiol Rev. 1987;46:1–11. doi: 10.1111/j.1574-6968.1987.tb02448.x. [DOI] [Google Scholar]
- Trinci AP, Collinge AJ. Occlusion of the septal pores of damaged hyphae of Neurospora crassa by hexagonal crystals. Protoplasma. 1974;80:57–67. doi: 10.1007/BF01666351. [DOI] [PubMed] [Google Scholar]
- Kanetsuna F, Carbonell LM. Cell wall Glucans of the yeast and mycelial forms of Paracoccidioides brasiliensis. J Bacteriol. 1970;101:675–680. doi: 10.1128/jb.101.3.675-680.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanetsuna F, Carbonell LM, Azuma I, Yamamura Y. Biochemical studies on the thermal dimorphism of Paracoccidioides brasiliensis. J Bacteriol. 1972;110:208–218. doi: 10.1128/jb.110.1.208-218.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Blas G. Paracoccidioides brasiliensis: cell wall glucans, pathogenicity, and dimorphism. Curr Top Med Mycol. 1985;1:235–257. doi: 10.1007/978-1-4613-9547-8_9. [DOI] [PubMed] [Google Scholar]
- Pereira M, Felipe MSS, Brígido MM, Soares CMA, Azevedo MO. Molecular cloning and characterization of a glucan synthase gene from the human pathogenic fungus Paracoccidioides brasiliensis. Yeast. 2000;16:451–462. doi: 10.1002/(SICI)1097-0061(20000330)16:5<451::AID-YEA540>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Marques ER, Ferreira MES, Drummond RD, Felix JM, Menossi M, Savoldi M, Travassos LR, Puccia R, Batista W, Carvalho KC, Goldman MHS, Goldman GH. Identification of genes preferentially expressed in the pathogenic yeast phase of Paracoccidioides brasiliensis, using suppression subtraction hybridization and differential macroarray analysis. Mol Genet Genomics. 2004;271:667–677. doi: 10.1007/s00438-004-1016-6. [DOI] [PubMed] [Google Scholar]
- Rappleye CA, Engle JT, Goldman WE. RNA interference in Histoplasma capsulatum demonstrates a role for α-(1,3)-glucan in virulence. Mol Microbiol. 2004;53:153–165. doi: 10.1111/j.1365-2958.2004.04131.x. [DOI] [PubMed] [Google Scholar]
- Reese AJ, Doering TL. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol Microbiol. 2003;50:1401–1409. doi: 10.1046/j.1365-2958.2003.03780.x. [DOI] [PubMed] [Google Scholar]
- Hallak J, San-Blas F, San-Blas B. Isolation and wall analysis of dimorphic mutants of Paracoccidioides brasiliensis. Sabouradia. 1982;20:51–62. doi: 10.1080/00362178285380081. [DOI] [PubMed] [Google Scholar]
- San Blas G, San Blas F. Paracoccidioides brasiliensis : cell wall structure and virulence. Mycopathologia. 1977;62:77–86. doi: 10.1007/BF01259396. [DOI] [PubMed] [Google Scholar]
- Zakrzewska A, Boorsma A, Brul S, Hellingwerf KJ, Klis FM. Transcriptional response of Saccharomyces cerevisiae to the plasma membrane-perturbing compound chitosan. Eukaryot Cell. 2005;4:703–715. doi: 10.1128/EC.4.4.703-715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks IR, Specht CA, Donlin MJ, Gerik KJ, Levitz SM, Lodge JK. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell. 2005;4:1902–1912. doi: 10.1128/EC.4.11.1902-1912.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulidou A, Bouriotis V, Thireos G. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J Biol Chem. 1996;271:31420–31425. doi: 10.1074/jbc.271.49.31420. [DOI] [PubMed] [Google Scholar]
- Naqvi SN, Feng Q, Boulton VJ, Zahn R, Munn AL. Vrp1p functions in both actomyosin ring-dependent and Hof1p-dependent pathways of cytokinesis. Traffic. 2001;2:189–201. doi: 10.1034/j.1600-0854.2001.020305.x. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/S0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Dutkiewicz R, Schilke B, Cheng S, Knieszner H, Craig EA, Marszalek J. Sequence-specific interaction between mitochondrial Fe-S scaffold protein Isu and Hsp70 Ssq1 is essential for their in vivo function. J Biol Chem. 2004;279:29167–29174. doi: 10.1074/jbc.M402947200. [DOI] [PubMed] [Google Scholar]
- Jacobson ES, Troy AJ, Nyhus KJ. Mitochondrial functioningof constitutive iron uptake mutations in Cryptococcus neoformans. Mycopathologia. 2005;159:1–6. doi: 10.1007/s11046-004-2223-8. [DOI] [PubMed] [Google Scholar]
- Arango R, Restrepo A. Growth and production of iron chelants by Paracoccidioides brasiliensis mycelial and yeast forms. J Med Vet Mycol. 1988;26:113–118. [PubMed] [Google Scholar]
- Haro R, Sainz L, Rubio F, Rodríguez-Navarro A. Cloning of two genes encoding potassium transporters in Neurospora crassa and expression of the corresponding cDNAs in Saccharomyces cerevisiae. Mol Microbiol. 1999;31:511–520. doi: 10.1046/j.1365-2958.1999.01192.x. [DOI] [PubMed] [Google Scholar]
- Smith DL, Tao T, Maguire ME. Membrane topology of a P-type ATPase. J Biol Chem. 1993;268:22469–22479. [PubMed] [Google Scholar]
- Riggle PJ, Kumamoto CA. Role of a Candida albicans P1-Type ATPase in resistance to copper and silver ion toxicity. J Bacteriol. 2000;182:4899–4905. doi: 10.1128/JB.182.17.4899-4905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris S, Durán-González S, Mariat F. Nutritional studies on Paracoccidioides brasiliensis : the role of organic sulfur in dimorphism. Sabouraudia. 1985;23:85–92. doi: 10.1080/00362178585380151. [DOI] [PubMed] [Google Scholar]
- Boguslawski G, Stetler DA. Aspects of physiology of Histoplasma capsulatum. (A review) Mycopathologia. 1979;67:17–24. doi: 10.1007/BF00436235. [DOI] [PubMed] [Google Scholar]
- Maresca B, Lambowitz AM, Kumar VB, Grant GA, Kobayashi GS, Medoff G. Role of cysteine in regulating morphogenesis and mitochondrial activity in the dimorphic fungus Histoplasma capsulatum. Proc Natl Acad Sci USA. 1981;78:4596–600. doi: 10.1073/pnas.78.7.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Pascon RC, Alspaugh A, Cox GM, McCusker JH. Molecular and genetic analysis of the Cryptococcus neoformans MET3 gene and a met3 mutant. Microbiology. 2002;148:2617–2625. doi: 10.1099/00221287-148-8-2617. [DOI] [PubMed] [Google Scholar]
- Sun M, Andreassi II JL, Liu S, Pinto R, Triccas JA, Leyh TS. The Trifunctional Sulfate-activating Complex (SAC) of Mycobacterium tuberculosis. J Biol Chem. 2005;280:7861–7866. doi: 10.1074/jbc.M409613200. [DOI] [PubMed] [Google Scholar]
- Fava-Neto C. Estudos quantitativos sobre a fixação do complemento na Blastomicose Sul-Americana com antígenos polissacarídicos. Arq Cirurg Clin Exp. 1955;18:97–254. [PubMed] [Google Scholar]
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tisbshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo A, Jiménez BE. Growth of Paracoccidioides brasiliensis yeast phase in a chemically defined culture medium. J Clin Microbiol. 1980;12:279–281. doi: 10.1128/jcm.12.2.279-281.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva SP, Borges-Walmsley MI, Pereira IS, Soares CM, Walmsley AR, Felipe MSS. Differential expression of an hsp70 gene during transition from the mycelial to the infective yeast form of the human pathogenic fungus Paracoccidioides brasiliensis. Mol Microbiol. 1999;31:1039–1050. doi: 10.1046/j.1365-2958.1999.01236.x. [DOI] [PubMed] [Google Scholar]