Abstract
The neuronal apoptosis inhibitory protein (NAIP) is a member of a novel family of inhibitor of apoptosis (IAP) proteins. The IAP genes are highly conserved from baculovirus to metazoans and suppress apoptosis induced by a variety of triggers both in vitro and in vivo. Here we describe the generation and characterization of mice with the targeted deletion of NAIP1. We demonstrate that the NAIP1-deleted mice develop normally. However, the survival of pyramidal neurons in the hippocampus after kainic acid-induced limbic seizures is greatly reduced in the NAIP1 knock-out animals. Thus, although NAIP1 is not necessary for normal development of murine central nervous system, the endogenous NAIP1 is required for neuronal survival in pathological conditions.
Apoptosis plays a critical role in the regulation of cell death in the peripheral and central nervous system (CNS) both in the physiological settings and in the neurological disease. Dysregulation of apoptosis occurs in chronic neurodegenerations such as Alzheimer's disease, amyotrophic lateral sclerosis, Parkinson's disease, as well as in epilepsy and ischemia (1). Spinal muscular atrophy (SMA) is a neurodegenerative disorder characterized by a progressive wasting of spinal cord motor neurons. Recently, we have proposed a link between inappropriate apoptosis and modulation of SMA severity. We have shown that the neuronal apoptosis inhibitory protein (NAIP) is deleted in a significant proportion of SMA patients (2). Full-length functional NAIP is absent in most individuals with type I SMA, and there exists a close correlation between the regional distribution of NAIP in the CNS and the neurodegenerative alterations in SMA (3). These observations are consistent with the neurodegenerative character of SMA. Although the causative gene involved in SMA is believed to be the SMN (survival motor neuron) gene (4), the role of NAIP in SMA is proposed to be a modulating one, exacerbating the severity of the disease (5).
The NAIP gene is a member of a recently identified family of intrinsic cellular regulators of apoptosis, the inhibitor of apoptosis (IAP) gene family (6). The IAP genes were initially discovered in baculoviruses, but their homologs have since been identified in other viruses, insects, birds, and mammals, suggesting a common evolutionary origin (7). Remarkably, IAP proteins suppress apoptosis induced by a diversity of triggers that is greater than that reported for any other antiapoptotic genes including the bcl-2 family (8). The mode of action of the IAP genes is believed to be by direct binding and inhibition of the key caspases and procaspases (primarily caspase 3 and 7) (9–12). Significantly, we have shown that transient forebrain ischemia selectively elevates levels of NAIP in neurons, which are resistant to ischemic injury. Moreover, the ectopic overexpression of NAIP in the CNS attenuated neuronal damage inflicted by transient forebrain ischemia, suggesting that NAIP plays a key role in neuroprotection (13).
The role of the IAP genes in the control and modulation of programmed cell death has been mainly inferred from the in vitro experimentation. The in vivo physiological roles of the endogenous IAP genes are less clear due to the lack of data from the knock-out models. In the present paper, we describe the generation and characterization of the NAIP1 knock-out mice. We show that the mice with the targeted deletion of Naip1 are morphologically normal and do not display any characteristics of SMA. However, the deletion of Naip1 results in increased sensitivity of the hippocampal pyramidal neurons to apoptotic death caused by kainic acid (KA)-induced epileptic seizures. Our data demonstrate that the endogenous, physiological levels of NAIP1 play a critical role in neuroprotection.
Materials and Methods
Generation of Naip1−/− Mice.
The 129/sv genomic clone (mg38; ref. 14) spanning the 5′ end of the Naip1 gene was used to construct a replacement-type targeting vector in which a IRES-lacZ and phosphoglycerate kinase (PGK)-neomycin (neo) cassette replaced exons 2 and 3 in the plasmid pKO (M.H. and R.G.K., unpublished data). The resultant targeting vector (pKO.naip1) was comprised of a 4.6-kb 5′ arm and a 6.5-kb 3′ arm bracketing the IRES-lacZ/PGK-neo insertion. RW4 embryonic stem cells were electroporated as described (15), and DNA from neomycin resistant clones was extracted and analyzed. Disruption of the Naip1 allele was confirmed by Southern blot analysis of EcoRI-digested genomic DNA after hybridization with a PCR-generated probe corresponding to exon 1 of the Naip1 gene. Chimeric mice were produced by morula aggregation (16) with targeted RW-4 cells. Chimeric male progeny were mated with 129/SvJ females, and heterozygous progenies were used as founders for two separate lines of mutant mice. Both the electroporation of embryonic stem cells and the generation of chimeric animals were performed at the Genome Systems facility (St. Louis). Mice were housed in a specific pathogen-free environment, and all experiments were performed in accordance with the guidelines of the Canadian Council on Animal Care and protocols approved by the University of Ottawa Animal Care Committee.
Induction of Limbic Seizures.
KA (Aldrich Chemical, Milwaukee, WI) was dissolved in 10 mM PBS and administered by i.p. injection. A dose of 45 mg/kg was established in a preliminary study as sufficient to elicit seizure activity in >95% of the mice with <20% mortality. The onset of seizure activity typically occurred within 10 min of injection and persisted for 2–3 hr.
Behavioral Assessment of Seizure Activity.
After KA injection, seizure activity was observed for a period of 2 hr and scored by a blinded observer according to the method of Morrison et al. (17). In brief, for each 5-min interval after the injection, an animal was given a score from 0 to 7, with the scores representing a progression of increasingly severe seizure-related behavior. Thus, a score of zero is indicative of normal behavior such as grooming, walking, and exploring. This behavior typically ceased after a few minutes and the animals became motionless (score = 1). The animals then adopted a rigid posture with forelimb and/or tail extension (score = 2). After this behavior, most animals became more active and displayed automatism such as circling, head bobbing, or repetitive scratching (score = 3). As the seizure activity became more severe, animals exhibited forelimb clonus as well as rearing and falling (score = 4). The animals were given a score of 5 when such behavior was repeated extensively. Some animals displayed more severe tonic-clonic seizures with barrel rolling and an inability to stand up (score = 6) and <20% of the animals died (score = 7). At the end of the 2-hr observation period, the 24 individual scores were totaled for the final seizure. Animals that did not exhibit any seizure activity were assigned a score of 0.
Assessment of Neurological Damage.
Seventy-two hours after KA injection, the mice were killed by i.p. injection of sodium pentobarbital, and the brains were rapidly removed and frozen by immersion in isopentane equilibrated on dry ice. Cryostat sections (12 μm) were collected from four different levels of the hippocampus, each level being separated by ≈400 μm. DNA fragmentation was demonstrated using the in situ end labeling (ISEL) method described by Fliss and Gattinger (18). In brief, frozen sections were thawed and fixed in 1% glutaraldehyde in PBS for 15 min at room temperature and washed three times for 5 min in PBS. The sections were permeabilized in 1:1 methanol–acetone for 10 min at room temperature and washed three times for 5 min in PBS. The sections were then treated with 20 μg/ml proteinase K in 25 mM Tris⋅HCl, pH 6.6, for 15 min at room temperature and washed two times for 15 min in deionized water. Following a 1-min rinse in PBS, the sections were incubated in a buffer solution containing 6.7 μl of terminal transferase, 12.7 μl of biotin-16-dUTP, 80 μl of CoCl2, 200 μl of 5× terminal deoxynucleotidyl transferase (TDT) buffer (all chemicals from Boehringer Mannheim), and 700.7 μl of dH20 per milliliter for 1 hr at 37°C in a humidified chamber. Sections were then washed three times for 1 min in PBS and labeled with fluorescein isothiocyanate by incubating in a solution containing 2.5 μg/ml avidin-fluorescein isothiocyanate in 4X saline-sodium citrate buffer with 0.1% Triton X-100 and 5% powdered milk for 45 min in the dark at room temperature. Sections were then washed three times in PBS for 1 min and coverslipped in a solution containing 1 mg/ml p-phenylenediamine and 90% glycerol in PBS. Positive control slides were prepared by incubating sections in 10 units/ml DNase I for 20 min at 37°C after treatment with proteinase K. Histofluorescence was monitored with a Zeiss Axioskop microscope. Both the ISEL assays and the assessment of the neurological damage were done blind.
Statistical Analysis.
It has been reported (19, 20) and also observed in our laboratory (data not shown) that there exists a strong correlation between the seizure intensity and the extent of neuronal damage; the low seizure scores (<50) produced no or minimal neuronal damage. Therefore, mice with a final seizure score of <50 were eliminated from the study (three wild-type and two Naip1-deleted mice). For the remaining animals, ISEL-positive CA3 pyramidal neuron nuclei were counted in each side of the brain in one section from each of the four different levels sampled and the eight numbers were added to give the total number of ISEL-positive nuclei. A one-way analysis of variance was performed on the cell count and the seizure score data using the sigmaplot statistical package (Jandel Scientific, San Rafael, CA).
Results and Discussion
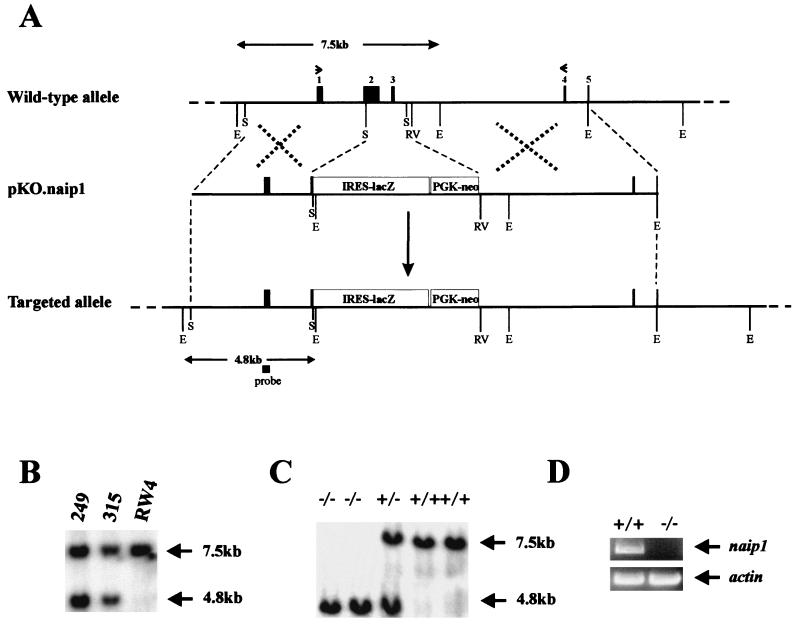
There exists at least three transcriptionally and translationally competent Naip loci in the mouse genome that have been proposed to be expressed in a tissue-specific manner (14). The Naip1 gene is expressed predominantly in the brain (21); consequently, a replacement-type targeting vector designed to inactivate Naip1 was constructed (Fig. 1A). Integration of the targeting vector results in the deletion of a 1.8-kb fragment containing exons 2 and 3 of the Naip1 gene. These exons encode a critical portion of the BIR (baculoviral inhibitor of apoptosis repeat) region, namely BIR1 and BIR2, a highly conserved region common to all IAPs that is required for the interaction with and inhibition of caspases (11). Of the 192 cells electroporated with this construct, 47 tested positive for the targeted Naip1 gene by Southern blot analysis of genomic DNA (Fig. 1B). Chimeric mice generated from two independent targeted embryonic stem cells transmitted the null allele to their offspring in a Mendelian fashion. Heterozygous littermates were mated to generate the Naip1−/− and Naip1+/+ mice described here. The mutant allele is characterized by an additional EcoRI site created by the insertion of the IRES-lacZ cassette. Southern blots of genomic DNA probed with an exon 1 clone revealed a 4.8-kb band in the case of Naip1−/− mice and a larger, 7.5-kb band in Naip1+/+ mice (Fig. 1C). The existence of multiple copies of the Naip gene in the mouse and the lack of an antibody that unambiguously recognizes endogenous murine NAIP renders Western blot analysis unsuitable for confirming the absence of Naip1 in null animals. We have therefore used reverse transcription-PCR analysis with primers specific to Naip1 (21) to confirm the absence of Naip1 mRNA in the brains of the Naip1−/− animals (Fig. 1D).
Figure 1.
Schematic diagram of the targeted disruption of the Naip1 gene in mice. (A) The structure of the 5′ end of the mouse Naip1 locus, targeting vector pKO.naip1, and the targeted Naip1 allele are shown. Solid boxes indicate exons; restriction sites are indicated by vertical black lines (E, EcoRI; RV, EcoRV; S, SstI). The position of the probe used for the genomic Southern blot analysis is indicated by the black box, and the positions of reverse transcription-PCR primers are indicated by arrowheads. (B) Southern blot analysis of EcoRI-digested genomic DNA from embryonic stem cells indicates the presence of the targeted (4.8 kb) and the wild-type (7.5 kb) alleles. (C) Southern blot analysis of EcoRI-digested genomic DNA from homozygous mutant (−/−), hetorozygous (+/−), and wild-type (+/+) Naip1 mice (line 249). (D) Reverse transcription-PCR analysis of brain mRNA for Naip1-null and wild-type mice. Naip1-specific PCR primers (21) were used to amplify Naip1 cDNA between exons 1 and 4. The 200-nt fragment of actin was amplified as a positive control.
Naip1−/− mice appeared normal and exhibited no consistent histological or morphological abnormalities. Appearance, sex ratios, litter size, and weight gain of Naip1−/− mice were similar to their Naip1+/+ littermates. In addition, brain sizes were indistinguishable between the two groups of animals, suggesting no gross neuronal loss (Table 1). Finally, extensive examination of behavior and spatial learning, including sensory and motor function tests, 24-hr activity, roto rod tests, the Morris water test, and nest building revealed no significant differences between the Naip−/− animals and their wild-type littermates (data not shown). These results indicate that the targeted disruption of Naip1 does not have any adverse effect on the development and proper function of the mouse CNS. The presence of multiple Naip homologs in mice makes it impossible to know whether the observed functional redundancy devolves from the other intact Naip loci, non-NAIP genes, or a combination thereof.
Table 1.
The effect of targeted deletion of Naip1 gene on selected parameters
| Naip1−/− | Naip1+/+ | |
|---|---|---|
| Body weight, g | 21.38 ± 2.66 | 21.21 ± 2.83 (n = 46) |
| Brain weight, g | 0.46 ± 0.06 | 0.47 ± 0.02 (n = 10) |
| Average litter size | 5.75 ± 1.56 | 5.82 ± 1.90 (n = 46) |
| Sex ratio (M/F) | 23/23 | 24/22 (n = 46) |
The average value for each characteristic ± SD for the distinct Naip genotypes is shown. No significant differences were observed (P > 0.1, one-way ANOVA).
We next investigated if Naip1 expression is essential for neuronal survival in pathological conditions. Peripheral administration of the l-glutamate analog KA to rodents leads to a characteristic seizure activity that is followed by specific excitotoxic neuronal damage within the limbic system. KA binds to members of both the α-amino-3-hydroxy-5-methyl-4-isoxazole-4-propionic acid and the kainate classes of glutamate receptors (22) and elicits prolonged depolarization in cells expressing these receptors. Although cells directly affected by KA may undergo extensive swelling and elevation of intracellular calcium ions, most of the damage elicited by KA is an indirect effect mediated by the limbic seizure activity characteristic of KA administration. The protracted depolarization is believed to result in an excitotoxic cell death (23). The widespread pattern of neuronal damage that follows KA administration in rats has been well characterized and includes areas in the olfactory cortex, amygdaloid complex, hippocampus, and related parts of the thalamus and neocortex (24). In contrast, damage of the murine CNS is chiefly restricted to the hippocampus with the CA3 and occasionally CA1 regions being affected (19). KA-induced seizures are used as a model system of human temporal lobe epilepsy due to the behavioral and pathological similarities (25). Although excitotoxic cell death differs from apoptosis, in many respects the two processes share several common features (26). Significantly, activation of caspase 3 following KA injection has been observed in the selectively vulnerable hippocampal regions (CA3 and CA1), suggesting a role for this member of the proapoptotic cysteine protease family (27, 28). Thus, an antiapoptotic protein that inhibits caspase 3 directly such as NAIP might be expected to reduce KA-induced excitotoxic cell death.
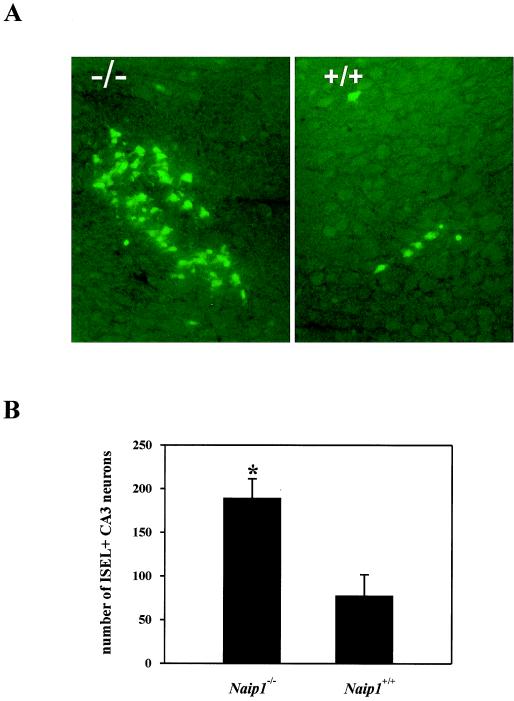
To test the hypothesis that Naip1 is required for neuronal apoptotic resistance, we subjected Naip1−/− and wild-type mice to a single i.p. injection of KA (45 mg/kg). KA produced limbic seizures with comparable intensity irrespective of their Naip1 status (Table 2), suggesting that the mechanism underlying seizure generation is independent of Naip1. ISEL-positive nuclei were observed only in pyramidal neurons of the CA3 subfield in the hippocampus in both groups of animals, and the remainder of the CNS appeared normal. In contrast to the similar seizure scores, there was a marked difference in the level of neuronal death as indicated by DNA fragmentation with a 2.5-fold increase in the number of ISEL-positive cells observed in Naip1−/− mice when compared with their wild-type littermates (Fig. 2).
Table 2.
The effect of Naip1 deletion on seizure severity after kainate administration
| Naip1−/− | Naip1+/+ | |
|---|---|---|
| Seizure score | 66.3 ± 21.1 (n = 11) | 66.5 ± 15.0 (n = 14) |
The behavioral response to KA injection was assessed as described in Material and Methods. The average seizure score ± SD for each group of animals is shown. No significant differences were observed among the two genotypes (P > 0.1, one-way ANOVA).
Figure 2.
Targeted disruption of Naip1 gene sensitizes CA3 neurons to KA-induced cell death. (A) Histopathologic changes in the CA3 region of dorsal hippocampus 72 hr after KA treatment (45 mg/kg; i.p.) of Naip1-deleted and wild-type mice. ISEL-positive CA3 neurons were detected as described in Material and Methods. Typical sections are shown. (B) Number of ISEL-positive CA3 neurons in the dorsal hippocampus differs between Naip1-deleted (n = 11) and wild-type (n = 14) mice. Cryostat sections from four different levels of the hippocampus in both hemispheres were ISEL-stained and the number of ISEL-positive CA3 pyramidal neurons nuclei was summed for each mouse [mean ± SD; *, P < 0.005 (one-way ANOVA)].
We have previously shown an up-regulation of NAIP in a number of CNS structures, including the hippocampus, following global ischemia in the rat. Moreover, expression of an adenoviral NAIP transgene in the rat hippocampus confers enhanced apoptotic resistance following global ischemia (13). Endogenous Naip1, including potentially its up-regulation, would therefore appear to form a central component of the neuronal survival of this vulnerable region based on our results showing a marked enhancement of cell death induced by KA in the absence of Naip1. Given the role caspase 3 is proposed to play in mediating neuronal death in kainate toxicity and the recently documented caspase 3–NAIP inhibitory interaction, it may be that the NAIP antagonism of the caspases is critical for neuronal survival in this vulnerable region of the hippocampus. This would suggest that pharmacological interventions that maintain or up-regulate NAIP expression may confer a clinically beneficial neuroprotection.
Acknowledgments
We thank Dr. Ian Whishaw (University of Lethbridge, Alberta, Canada) for assistance with the behavioral tests, and we thank the members of our laboratory for useful discussion. This work was supported by grants from the Medical Research Council of Canada (MRC), the Canadian Networks of Centers of Excellence (NCE), the Muscular Dystrophy Association (U.S.), and the Howard Hughes Medical Institute (HHMI). M.H. is a recipient of an MRC Postdoctoral Fellowship. A.E.M. is the recipient of an MRC Scientist Award and Burroughs Wellcome Clinical Translation Award. R.G.K. is a recipient of an MRC Senior Scientist Award, a Fellow of the Royal Society of Canada, and a HHMI International Research Scholar.
Abbreviations
- NAIP
neuronal apoptosis inhibitory protein
- ISEL
in situ end labeling
- KA
kainic acid
- BIR
baculoviral inhibitor of apoptosis repeat
- IAP
inhibitor of apoptosis
- CNS
central nervous system
- SMA
spinal muscular atrophy
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040469797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040469797
References
- 1.Savitz S I, Rosenbaum D M. Neurosurgery. 1998;42:555–572. doi: 10.1097/00006123-199803000-00026. [DOI] [PubMed] [Google Scholar]
- 2.Roy N, Mahadevan M S, McLean M, Shutler G, Yaraghi Z, Farahani R, Baird S, Besner-Johnston A, Lefebvre C, Kang X, et al. Cell. 1995;80:167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- 3.Xu D G, Korneluk R G, Tamai K, Wigle N, Hakim A, Mackenzie A, Robertson G S. J Comp Neurol. 1997;382:247–259. [PubMed] [Google Scholar]
- 4.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Nat Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 5.Gambardella A, Mazzei R, Toscano A, Annesi G, Pasqua A, Annesi F, Quattrone F, Oliveri R L, Valentino P, Bono F, et al. Ann Neurol. 1998;44:836–839. doi: 10.1002/ana.410440522. [DOI] [PubMed] [Google Scholar]
- 6.Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda J E, MacKenzie A, Korneluk R G. Nature (London) 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- 7.Deveraux Q L, Reed J C. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 8.LaCasse E C, Baird S, Korneluk R G, MacKenzie A E. Oncogene. 1998;17:3247–3259. doi: 10.1038/sj.onc.1202569. [DOI] [PubMed] [Google Scholar]
- 9.Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. Nature (London) 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 10.Roy N, Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi R, Deveraux Q, Tamm I, Welsh K, Assa-Munt N, Salvesen G S, Reed J C. J Biol Chem. 1998;273:7787–7790. doi: 10.1074/jbc.273.14.7787. [DOI] [PubMed] [Google Scholar]
- 12.Seshagiri S, Miller L K. Proc Natl Acad Sci USA. 1997;94:13606–13611. doi: 10.1073/pnas.94.25.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu D G, Crocker S J, Doucet J P, St-Jean M, Tamai K, Hakim A M, Ikeda J E, Liston P, Thompson C S, Korneluk R G, et al. Nat Med. 1997;3:997–1004. doi: 10.1038/nm0997-997. [DOI] [PubMed] [Google Scholar]
- 14.Yaraghi Z, Korneluk R G, MacKenzie A. Genomics. 1998;51:107–113. doi: 10.1006/geno.1998.5378. [DOI] [PubMed] [Google Scholar]
- 15.Wurst W, Joyner A L. In: Gene Targeting. Joyner A L, editor. Oxford: Oxford Univ. Press; 1993. pp. 33–62. [Google Scholar]
- 16.Wood S A, Allen N D, Rossant J, Auerbach A, Nagy A. Nature (London) 1993;365:87–89. doi: 10.1038/365087a0. [DOI] [PubMed] [Google Scholar]
- 17.Morrison R S, Wenzel H J, Kinoshita Y, Robbins C A, Donehower L A, Schwartzkroin P A. J Neurosci. 1996;16:1337–1345. doi: 10.1523/JNEUROSCI.16-04-01337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fliss H, Gattinger D. Circ Res. 1996;79:949–956. doi: 10.1161/01.res.79.5.949. [DOI] [PubMed] [Google Scholar]
- 19.Hu R Q, Koh S, Torgerson T, Cole A J. Brain Res. 1998;810:229–240. doi: 10.1016/s0006-8993(98)00863-4. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto K, Watanabe K, Nishimura T, Iyo M, Shirayama Y, Minabe Y. Brain Res. 1998;804:212–223. doi: 10.1016/s0006-8993(98)00708-2. [DOI] [PubMed] [Google Scholar]
- 21.Yaraghi Z, Diez E, Gros P, MacKenzie A. Mamm Genome. 1999;10:761–763. doi: 10.1007/s003359901087. [DOI] [PubMed] [Google Scholar]
- 22.Lerma J. Neuron. 1997;19:1155–1158. doi: 10.1016/s0896-6273(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Ari Y. Neuroscience. 1985;14:375–403. doi: 10.1016/0306-4522(85)90299-4. [DOI] [PubMed] [Google Scholar]
- 24.Schwob J E, Fuller T, Price J L, Olney J W. Neuroscience. 1980;5:991–1014. doi: 10.1016/0306-4522(80)90181-5. [DOI] [PubMed] [Google Scholar]
- 25.Coyle J T. J Neurochem. 1983;41:1–11. doi: 10.1111/j.1471-4159.1983.tb11808.x. [DOI] [PubMed] [Google Scholar]
- 26.Olney J W, Ishimaru M J. In: Cell Death and Diseases of the Nervous System. Koliatsos V E, Ratan R R, editors. Totowa, NJ: Humana; 1999. [Google Scholar]
- 27.Becker A J, Gillardon F, Blumcke I, Langendorfer D, Beck H, Wiestler O D. Brain Res Mol Brain Res. 1999;67:172–176. doi: 10.1016/s0169-328x(99)00060-1. [DOI] [PubMed] [Google Scholar]
- 28.Faherty C J, Xanthoudakis S, Smeyne R J. Brain Res Mol Brain Res. 1999;70:159–163. doi: 10.1016/s0169-328x(99)00143-6. [DOI] [PubMed] [Google Scholar]