Abstract
Many eukaryotic cells use RNA-directed silencing mechanisms to protect against viruses and transposons and to suppress endogenous gene expression at the posttranscriptional level. RNA silencing also is implicated in epigenetic mechanisms affecting chromosome structure and transcriptional gene silencing. Here, we describe enhanced silencing phenotype (esp) mutants in Arabidopsis thaliana that reveal how proteins associated with RNA processing and 3′ end formation can influence RNA silencing. These proteins were a putative DEAH RNA helicase homologue of the yeast PRP2 RNA splicing cofactor and homologues of mRNA 3′ end formation proteins CstF64, symplekin/PTA1, and CPSF100. The last two proteins physically associated with the flowering time regulator FY in the 3′ end formation complex AtCPSF. The phenotypes of the 3′ end formation esp mutants include impaired termination of the transgene transcripts, early flowering, and enhanced silencing of the FCA-β mRNA. Based on these findings, we propose that the ESP-containing 3′ end formation complexes prevent transgene and endogenous mRNAs from entering RNA-silencing pathways. According to this proposal, in the absence of these ESP proteins, these RNAs have aberrant 3′ termini. The aberrant RNAs would enter the RNA silencing pathways because they are converted into dsRNA by RNA-dependent RNA polymerases.
Keywords: aberrant RNA, polyadenylation, siRNAs, epigenetics, RNA polymerase
RNA silencing provides defense against viruses and transposons and regulates genetic functions at transcriptional and posttranscriptional levels (1). A defining feature of RNA silencing is the processing of long double-stranded (ds)RNA into 20- to 24-bp short interfering (siRNA) or microRNA (miRNA) duplexes by an RNase III known as Dicer or Dicer-like (DCL). One of the two strands of the short RNA duplex is incorporated as a guide RNA into an RNA-silencing effector complex that includes an Argonaute (AGO). The targets of these complexes are RNA or possibly DNA molecules with partial or complete sequence complementarity to the miRNA or siRNA. If the AGO complex cleaves mRNA or represses productive translation, it is known as RISC (RNA-induced silencing complex) (2); if the complex suppresses expression at the DNA level, it is known as the RITS (RNA-induced transcriptional silencing) complex (3).
Other components of RNA-silencing pathways in plants include RNA-dependent RNA polymerases (RDRs) (4–6), a putative DNA-dependent RNA polymerase IV (PolIV) (7), dsRNA-binding proteins (8–11), and proteins of unknown function including SGS3 (4) and the helicase-like SDE3 (12). These additional proteins operate together with different DCL proteins in multiple RNA-silencing pathways. The DCL1 pathway in Arabidopsis, for example, results in production of miRNAs from a precursor-miRNA in which a double-stranded region forms via folding of a partially mismatched inverted repeat (13, 14). The dsRNA in a DCL2 pathway is formed by annealing of complementary regions in converging transcripts of adjacent genes and the resulting (natural antisense) nat-siRNAs target mRNAs in a salt stress response (15). The DCL3 pathway involves RDR2 (6) and PolIV (7). It is thought that RDR2 uses a PolIV transcript as a template for dsRNA production and that the siRNAs, including some from transposons and repetitive sequences, may mediate transcriptional silencing as a result of DNA methylation and heterochromatin formation. A fourth pathway uses DCL4 and RDR6 (16–18). Like the miRNAs and nat-siRNAs, the DCL4-generated siRNAs target mRNAs in trans and can affect growth and development of the plant.
In Arabidopsis, there is additional complexity because these RNA-silencing pathways are interdependent: the DCL4 pathway, for example, is initiated by miRNA-mediated cleavage of a ssRNA (19), and the DCL2 pathway overlaps with several other pathways (15). It requires PolIV from the DCL3 pathway, RDR6 from the DCL4 pathway and DCL1. There is ample scope for further complexity in these pathways because the Arabidopsis genome encodes 10 different AGO proteins, and there are many thousands of different 20- to 24-nt short RNAs (20).
Genetic analysis of RNA silencing helped to identify many features in these silencing pathways through the characterization of the loss of silencing mutants. An alternative genetic approach, involving a screen for mutants with enhanced silencing, also has the potential to be informative regarding RNA-silencing pathways and has been used to identify siRNA-degrading nucleases (21). Enhanced silencing screens also could identify endogenous silencing suppressors or proteins, like RRF-3 or SMG-2 in C. elegans that diverts other proteins or RNA molecules away from or between RNA-silencing pathways. RRF-3 is thought to be required for an endogenous RNA-silencing pathway so that a wild-type worm has limited availability of proteins for silencing with exogenously added dsRNA (22). In an rrf-3 mutant, the endogenous-silencing pathways are not active and their proteins are available to mediate silencing by exogenous RNA. SMG-2 is a component of the nonsense-mediated decay pathway that antagonizes RNA silencing so that smg-2 mutants also exhibit enhanced responses to exogenous dsRNA (23). Mutants also may have an enhanced silencing phenotype if negative feedback of RNA silencing is inactivated. Such negative feedback occurs because silencing protein mRNAs may be miRNA targets (24, 25) or because the pathway involves RNA species that are both a template of siRNAs and a target of the silencing mechanism (1).
Here we describe four mutants in a screen for enhanced silencing phenotype (esp) mutants in Arabidopsis. They are in genes that encode RNA-processing components including putative members of the cleavage polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CstF) complexes. The mutants link RNA silencing with the control of flowering because they have early flowering phenotypes and exhibit modified processing of an RNA that encodes the FCA enhancer of flowering. There is a further link with flowering control because two of these RNA processing components (ESP4, a symplekin/PTA1 homologue, and ESP5, the CPSF100 of Arabidopsis) are present in a complex with the FY protein that also regulates FCA processing. Finally, the reduction in the level of one FCA RNA species depends on RNA-silencing components. Based on these data, we speculate that defects to RNA processing can lead to aberrant RNA substrates that play a role in the initiation or propagation of silencing.
Results
Use of a PVX Amplicon to Identify Enhanced Silencing Mutants.
Potato Virus X (PVX) transgene amplicons comprise a cDNA copy of the replicating viral RNA coupled to a transgene promoter. Viral amplicon transgenes initiate siRNA production in transgenic plants because the viral RNA is a substrate for the RNA-silencing machinery, and they can be used to target silencing in trans by insertion of nonviral sequence. When a GFP cDNA insert was inserted into a PVX amplicon (AGFP) (26), there was efficient silencing of a functional GFP transgene. However, when amplicons with inserts from the endogenous phytoene desaturase (PDS) gene were tested, the photobleaching silencing phenotype was restricted to small spots on leaves of the transgenic plants (Apds2; Fig. 1A). The difference between this inefficient silencing of PDS and strong GFP silencing is most likely related to the production of secondary siRNAs. The GFP transgene target RNAs support amplification of the silencing mechanism through an RDR6-dependent mechanism of secondary siRNA production, whereas PDS and other endogenous RNA species do not (27).
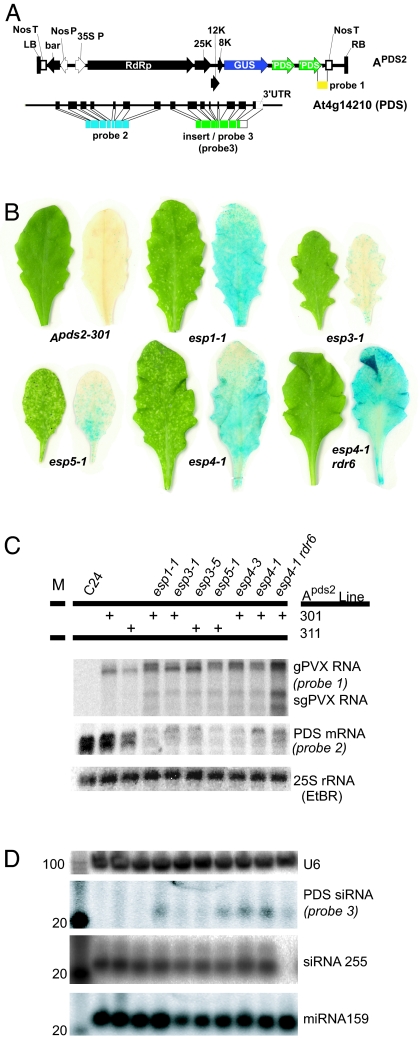
Fig. 1.
Screen for esp mutants. (A) Initiator (Apds2) and target (At4g14210; PDS) of silencing. LB, left border; Nos T, nopaline synthase transcriptional terminator; bar, basta herbicide resistance; Nos P, nopaline synthase promoter; 35S P, Cauliflower mosaic virus 35S promoter; RdRp, PVX replicase; 25K, PVX 25-kDa protein; 12K, PVX 12-kDa protein; 8K, PVX 8-kDa protein; GUS, β-glucuronidase (only expressed during virus replication); ′PDS, 3′ sense fragment of PDS cDNA; RB, Right border. (B) Silencing and virus replication phenotypes of esp mutants. C24, nontransgenic parental line; Apds2-301 and Apds-311, the two independent parental lines; rdr6, RNA-dependent RNA polymerase 6. (C) Northern blot analysis of transgene and endogenous PDS mRNA. (D) Northern blot analysis of enriched low molecular weight RNA. Numbers to left of figure correspond to length of RNA in nucleotides.
To identify factors limiting the silencing phenotype, we mutagenized two independent Apds2 lines with ethyl-methanesulfonate (Fig. 1B; Apds2-301 and Apds-311) and identified esp mutant plants by an increase in spots of photobleaching (Fig. 1B). One class of mutants represented by esp3 had reduced stature, early flowering (see below), and altered leaf morphology (Fig. 6, which is published as supporting information on the PNAS web site). A second class represented by esp1, esp4, and esp5 also were early flowering (see below) but had normal stature and leaf morphology. In both classes, relative to wild-type plants, there was 2- to 3-fold fewer of the endogenous PDS mRNA than in the wild-type plants (Fig. 1C) and 10- to 50-fold higher levels of PDS siRNA (Fig. 1D).
The PVX amplicons included a GUS reporter gene of virus replication in addition to a tandem repeat PDS insert. Thus, by Northern blotting (Fig. 1C) and GUS staining (Fig. 1B), we established that the enhanced silencing in the esp mutants was correlated with increased replication and accumulation of PVX::PDS genomic and subgenomic RNAs. We also showed that the enhanced silencing involved a previously characterized silencing pathway because it depended on the RDR6 RNA-dependent RNA polymerase that also was required for transgene silencing of GFP (5, 7, 12). In esp4-1 rdr6, photobleaching was reduced relative to esp4-1 (Fig. 1B) and the level of PVX::PDS RNA increased (Fig. 1C), whereas the level of PDS siRNA decreased (Fig. 1D).
Our initial conclusion from these analyses was that in esp4-1 and most probably the other esp mutants, there was enhancement both of virus replication and RDR6-dependent RNA silencing. In the single esp mutants, the enhanced replication would have been partially counteracted because the viral RNA is a target of silencing. However, in the double esp4-1 rdr6 mutant when the RDR6-silencing pathway would not have been active, the enhancement of virus replication would have been unrestrained.
To determine whether the enhanced silencing phenotype was specific to the tandem sense PDS insert found in Apds2, we crossed the esp1-1, 3-1, 4-1, and 5-1 alleles into plants with a PVX amplicon containing a single PDS insert in the antisense orientation (Apds1as) or with a sense fragment from Albino3 (Aalb3) (Fig. 7A, which is published as supporting information on the PNAS web site). Albino3 is a chloroplast membrane protein that produces a photobleached phenotype when defective (28). In the WT background, both amplicons produced higher virus levels than Apds2, but as with Apds2 lines, visible silencing was minimal (Fig. 7B). However, in the esp backgrounds, the silencing was enhanced. With Apds1as mutants and Aalb3 esp3-1, the enhanced silencing phenotype was on true leaves, as with Apds2. However, with Aalb3 esp1-1, 4-1, and 5-1, the enhanced photobleaching was manifested only in the cotyledons (Fig. 7B). Because both Apds1as and Aalb3 exhibited enhanced silencing phenotypes in the mutants, we considered that further characterization of esp1, esp3, esp4, and esp5 would be informative regarding the mechanisms limiting silencing.
ESP Loci Encode RNA-Processing Proteins.
ESP3 mapped to a gene previously identified as essential for embryonic development in Arabidopsis (At1g32490/EMB2733; Fig. 6 A and B; ref. 29). ESP3 is a homologue of PRP2 (Fig. 2A), one of four related DEAH RNA helicases identified as essential cofactors for RNA splicing in yeast (PRP2, PRP16, PRP22, and PRP43; refs. 30 and 31). ESP3 is likely to be the principle PRP2 homologue in Arabidopsis because it has all six of the required helicase motifs (Fig. 6C) and is expressed in a wider set of tissues than the two other helicases of the PRP2 class (At2g35340 and At4g16680; Fig. 2B). All seven esp3 mutations introduce changes to the protein primary structure because they result in amino acid changes in the encoded protein or disrupt RNA splice junction sites (Fig. 6).
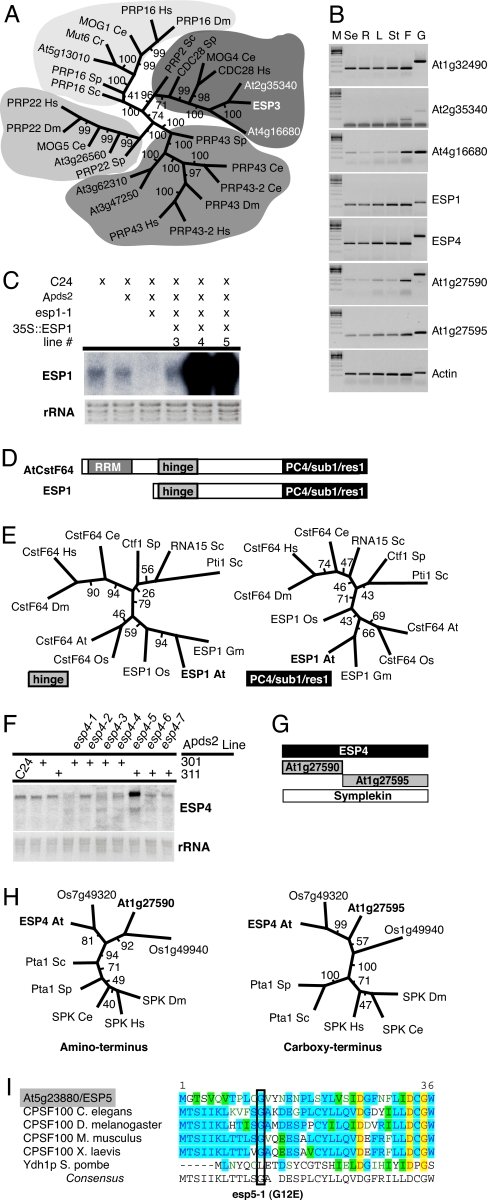
Fig. 2.
ESP genes encode RNA processing components. Bootstrap values for each internodal length corresponds to the frequency the two groups are separated in a population of 100 independent trees. Hs, Homo sapiens; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; At, Arabidopsis thaliana; Cr, Chlamydomonas reinhardtii; Os, Oryza sativa; Gm, Glycine max. (A) Phylogenetic tree of ESP3 homologues by using the helicase domain and the neighbor-joining method. (B) Expression of ESP genes and closest Arabidopsis homologues (se, seedling; R, roots; L, leaves; St, stems; F, flowers; G, genomic DNA). (C) Northern blot analysis of ESP1 mRNA. (D) ESP1 is a CstF64 homologue. RRM, RNA recognition motif; Hinge, interacts with Symplekin and CstF77 in mammals; PC4/Sub1/Res1, interacts with polymerase processivity factors in mammals and yeast. (E) Phylogenetic trees of CstF64-related proteins based on the hinge and PC4/sub1/res1 domains (maximum parsimony). (F) ESP4 RNA levels. (G) There are three Arabidopsis Symplekin homologues. At1g27590 and At1g27595 have homology to the 5′ and 3′ halves of Symplekin and ESP4. RT-PCR detects expression from all 3 genes (see Fig. 2B). (H) Phylogenetic trees of ESP4 family members generated by a neighbor-joining method. (H Left) Amino terminus, amino acids 115–215 of ESP4. (H Right) Carboxyl terminus, amino acids 755–830. (I) Alignment of amino terminus of CPSF100 and esp5-1 substitution: red letters highlighted in yellow, absolute conservation; blue letters in aqua, conserved residues; black letters in green, similar residues; green letters (no highlighting), weakly similar residues.
ESP1 mapped to At1g73840 (Fig. 8A, which is published as supporting information on the PNAS web site). The only esp1allele from our screen (esp1-1) carried a premature nonsense codon in exon 4. We confirmed the identity of ESP1 by fully complementing the enhanced silencing and virus replication phenotypes with a genomic fragment of ESP1 transcribed from the 35S promoter (Fig. 2C and 8B). ESP1 was expressed in all developmental stages (Fig. 2B). Consistent with a role in RNA metabolism, ESP1 resembled the CstF64 family of RNA processing factors that are conserved between yeast and mammals (Fig. 8C). In mammals, CstF64 is a component of the CstF complex. CstF is required for mRNA 3′ end formation in mammals (32) along with CPSF, PolyA polymerase, cleavage factors I and II, and the carboxyl-terminal domain of RNA polymerase II (reviewed in ref. 33). CPSF and CstF physically interact and bind cooperatively to the cleavage sites in mRNA precursors.
ESP1 differs from a canonical Arabidopsis homologue of CstF64 (At1g71800; hereafter denoted AtCstF64) in that it lacks an RNA recognition motif (RRM) (Figs. 2D and 8C). However, ESP1 does contain two key domains found in other CstF64 homologues (Figs. 2D and 8C). The first domain, the hinge, interacts with CstF77 and with Symplekin/Pta1 (34, 35). The second domain is recognized by PC4 (mammals), Sub1(budding yeast) (36), or the ankyrin repeat protein, res1 (fission yeast) (37). The PC4/Sub1/res1 domain controls transcription elongation in these organisms. PC4/Sub1 homologues exist in Arabidopsis (At5g09250/KIWI and At4g10920/KELP) suggesting that this function is retained (38) in plants. Phylogenetic analysis by using the hinge and PC4/Sub1/res1 domains indicated that AtCstF64 and ESP1 diverged after the establishment of the plant lineage but before the separation of monocots and dicots (Fig. 2E). The conservation of these two domains suggests that, despite the absence of the RRM, ESP1 has the potential to interact with the 3′ end formation apparatus.
The identity of ESP4 also is consistent with the idea that ESP1 may be an RNA processing component. All seven esp4 alleles mapped to a Symplekin/Pta1 homologue (At5g01400; Fig. 9 A and B, which is published as supporting information on the PNAS web site) that, as described above, would have the potential to interact with the hinge domain of ESP1 or AtCstF64. The esp4 mutants would all have affected the encoded protein either by modification of the coding sequence or by disrupting RNA splice sites (Fig. 9). The splice site mutations affected the size of the esp4 mRNA and, with esp4-5, a misspliced mRNA accumulated at a much higher level than its wild-type equivalent (Fig. 2F).
The yeast ESP4 homologue, PTA1, is essential for growth (39) and is one of several components of the cleavage polyadenylation factor (CPF), which is analogous to mammalian CPSF (40). However the esp4 plants are fully viable, suggesting that esp4 may be functionally redundant with its closest homologues in the Arabidopsis genome (At1g27590/At1g27595). These adjacent genes are similar to the amino and carboxyl termini of ESP4 (Figs. 2G and 9D) and are both expressed throughout development (Fig. 2B). We were unable to detect a transcript spanning both genes either experimentally (data not shown) or in cDNA databases (www.arabidopsis.org), even though there is a single predicted structural homologue (Os1g49940) of these adjacent genes in rice (Fig. 2H).
The ESP5 locus also encodes a protein that is likely to be part of the mRNA3′ end formation apparatus because it encodes the orthologue of mammalian CPSF100 (AtCPSF100/At5g23880/EMB1265) (Fig. 2I; Fig. 10 A–C, which is published as supporting information on the PNAS web site). This protein would be a key component of the CPSF complex that acts together with CstF in 3′ end formation. AtCPSF100 interacts with PolyA polymerase (41) and is essential for embryonic development (29). The esp5 allele (G12E) obtained from our screen did not have a strong effect on plant development or morphology, and it is likely to encode a protein with partial loss of function. We confirmed the identity of ESP5 by transgenic complementation of the mutant phenotype, in three independent lines, with a genomic ESP5 construct transcribed by the ESP5 promoter (Fig. 10B).
ESP Mutations Affect RNA Processing.
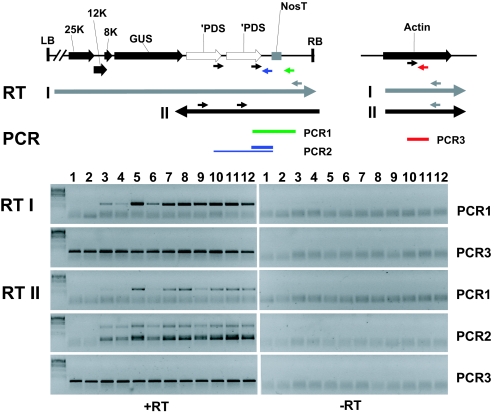
Based on the identity of the ESP loci, we considered that both the virus replication and enhanced silencing esp1, esp4, and esp5 phenotypes could be due to modified 3′ end formation of the amplicon transgene transcripts. To test this possibility, we used RT-PCR to monitor transcriptional read-through of the nopaline synthase terminator (NosT) at the end of the Apds2 transgene. Using primers to detect sense RNA transcripts downstream of NosT (RTI and PCR1; Fig. 3), we detected a low level of transcription through NosT in both Apds2 parental lines (Fig. 3, lanes 3 and 4) that increased markedly in esp1-1 (lane 5), esp4-1 (lanes 7 and 10), esp4-3 (lane 8), and esp5-1 (lane 9). The abundance of the read-through product in esp4-1 was not affected by RNA silencing mutations rdr6 (lane 11) or sgs3 (lane 12).
Fig. 3.
ESP mutants elevate transcriptional read-through of the transgene initiator of silencing. Strand-specific RT-PCR analysis (Upper) Schematic of 3′ end of Apds2 transgene: LB, left border; 25K, 12K, 8K all encode viral proteins; GUS, β-glucuronidase; ′PDS, 3′ half of PDS cDNA; RB, Right border; arrows, locations of primers used in RT I, RT II, and PCR 1–3. (Lower) RT I: RT reaction used gray primers to prime cDNA synthesis from (+) sense transgene transcripts and actin mRNA. RT II: RT reaction used black primers for cDNA synthesis from (−) sense transgene transcripts and the gray actin primer for actin cDNA; The two PCR products in PCR2 with RTII are due to the presence of duplicate primer binding sites. Samples: 1, C24; 2, Apds1as (see Fig. 7); 3, Apds2-301; 4, Apds2-311; 5, esp1-1; 6, esp3-1; 7, esp4-1; 8, esp4-3; 9, esp5-1; 10, esp4-1; 11, esp4-1 rdr6; 12, esp4-1 sgs3.
NosT antisense RNA from the read-through region (detected by RTII and PCR1: Fig. 3) was present in wild-type plants and at elevated levels in esp1-1, esp4-1, esp4-3, and esp5-1 (lanes 5 and 7–9). This antisense RNA is not likely to be a product of RDR6 because the levels in esp4-1 were not affected by rdr6 or sgs3 mutations (compare lanes 7 and 10 with lanes 11 and 12). Apds2 antisense RNA corresponding to the double pds insert (RT2 and PCR2; Fig. 3) also was more abundant in the esp mutants than in the wild-type plants.
We can rule out that the increase in abundance of the read-through transcripts is a consequence of the enhanced silencing because they were rare in esp3-1. Instead, it is likely that the enhanced silencing phenotypes of esp1, esp4, and esp5 are a consequence of these read-through transcripts. A plausible scenario is that the read-through transcripts arise because transgene transcript 3′ end formation is impaired in these esp mutants. If there are read-through transcripts from both DNA strands, the sense and antisense RNAs then could anneal to form a dsRNA DCL substrate, as for natural antisense RNA involved in salt stress response (15). Alternatively, the read-although transcripts may be converted to dsRNA by an RDR other than RDR6.
To explain the enhanced replication of the PVX amplicon RNA phenotype in the esp mutants, we propose that there are cryptic 3′ end formation sites within the viral genome so that many of the transgene transcripts are incomplete viral RNAs in the wild-type plants. In the esp1, esp4, and esp5 mutants, the reduced frequency of 3′ end formation would mean that that there is more full-length or greater-than-full-length viral RNA and, consequently, more replicating viral RNA.
The ESP3 protein differs from ESP1, ESP4, and ESP5 in that it is implicated in RNA splicing rather than 3′ end formation. It therefore is not surprising that there was no enhancement of read-through transcription in esp3-1 (Fig. 3, lane 6). To account for the esp3 phenotypes, we propose that there may be cryptic splice sites in the PVX amplicon transcripts. An ESP3-based mechanism would splice out regions of the transcript to generate RNA that would be a poor initiator of silencing and replicate inefficiently. According to this idea, RNA silencing and PVX amplicon replication would be enhanced in the esp3 plants because these spliced RNAs would be less abundant than in the wild-type plants.
ESP5 and ESP4 Are Components of CPSF.
The molecular characteristics of ESP1, ESP4, and ESP5/AtCPSF100 suggest that they are part of a complex that processes amplicon transcripts and influences their ability to initiate RNA silencing. Given that ESP5 is expected to be a core component of AtCPSF, we wanted to determine whether ESP1 and ESP4 were also core components of this complex. We purified AtCPSF by using a FLAG-CPSF100 fusion protein expressed from the endogenous AtCPSF100 promoter. This transgene construct complemented esp5-1, and the plants expressed a protein of the predicted size that could be detected with FLAG antibody (Fig. 4A).
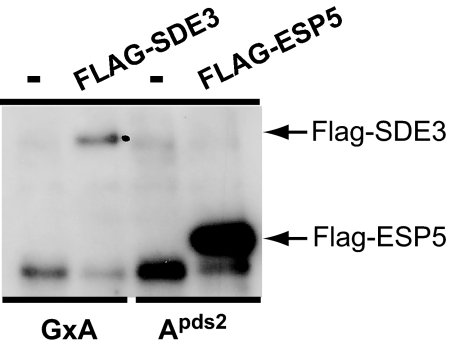
Fig. 4.
ESP5 and ESP4 are components of AtCPSF. FLAG IP of FLAG-ESP5. IPs were carried out from extracts from a line bearing a FLAG-ESP5 transgene that complements esp5-1 and three control lines: a FLAG-SDE3 line that complements sde3-1 and two lines not transformed with a FLAG-expressing transgene.
We immunoprecipitated FLAG-CPSF100 from 3-week-old seedling extracts, eluted protein bound to the beads with FLAG peptide, digested the samples with trypsin and identified eluted proteins by mass spectrometry. As negative controls, we performed the same procedure by using plants expressing an unrelated FLAG-fusion protein (FLAG-SDE3) and plants that did not express a FLAG fusion protein (GxA) (Fig. 4A). The analysis, done in duplicate for each sample, revealed numerous FLAG-CPSF100-specific peptides corresponding to ESP5/CPSF100, CPSF160, CPSF73, ESP4, and FY. FY is a homologue of yeast Pfs2p, which is part of the CPF complex in Saccharomyces cerevisiae (Fig. 4B; see Fig. 11, which is published as supporting information on the PNAS web site), the counterpart of mammalian CPSF (42).
This collection of interacting proteins confirms that ESP5 is an integral component of AtCPSF. The copurification of ESP4 with ESP5 and the absence of peptides from the ESP4 homologues (At1g27590/95) suggest that ESP4 is the principal Symplekin-like component of AtCPSF in vegetative organs. We did not identify peptides from homologues of CPSF30 (At1g30460) and Fip1 (At5g58040; 133 kDa), which both are found in mammalian CPSF. AtCPSF30 may be too small to obtain sufficient peptides for detection under our purification conditions, whereas Fip1 may not be an integral component of CPSF in plants. In vitro, AtFip1 interacted with Arabidopsis polyA-polymerase and members of AtCPSF, AtCstF, and AtCF1 (43), suggesting that AtFip1 may associate preferentially with a larger complex of 3′ end formation factors similar to that identified in yeast (34, 35). This larger complex is not stable under our purification conditions because we did not detect any AtCstF-like components, including ESP1 (CstF64 homologue), in our preparations.
Modified RNA Silencing Influences the Control of Flowering.
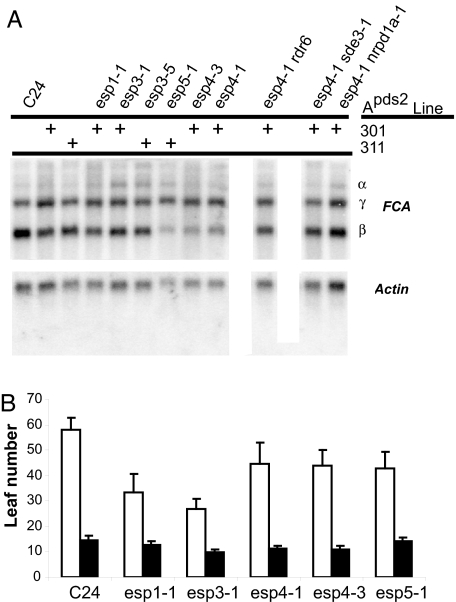
The AtCPSF complex described here is likely required for processing of most, if not all, mRNAs in vegetative tissues. However, because 3′ end formation is essential and the esp4 and esp5 mutant phenotypes are relatively mild, much of the cellular mRNA must be properly processed and expressed. Nevertheless, there may be endogenous mRNAs where misprocessing leads to silencing in the esp mutants. One well characterized example of RNA misprocessing occurs when FY is defective. FY normally binds the RNA-binding protein FCA and mediates an autoregulatory mechanism involving alternate 3′ end formation. In this mechanism, FCA guides 3′ end formation within intron 3 of the FCA pre-mRNA so that there is accumulation of a truncated RNA referred to as FCA-β mRNA, which is reduced in fy. As ESP4 and ESP5 bind to FY in AtCPSF, we expected that levels of FCA-β mRNA also would be lower in esp4 and esp5 mutants. Our results confirmed that expectation (Fig. 5A). However, other aspects of the esp4 and esp5 phenotype differed from fy. For example, the full-length FCA-γ mRNA was equally abundant in the esp4, esp5, and wild-type plants (Fig. 5A), whereas in fy, it was more abundant (44). There was also a difference in flowering time: esp4 and esp5 plants were early flowering (Table 1; see Fig. 12, which is published as supporting information on the PNAS web site), whereas fy genotype plants flowered late. The difference between the fy and esp phenotypes could be because mutants were not in the same genotypes. However, it also remains possible that ESP4/ESP5 do not act in exactly the same way as FY.
Fig. 5.
ESP mutants influence alternate RNA processing of the flowering time regulator FCA. (A) Altered FCA RNA processing in esp mutants. (B) Flowering time measured in number of true leaves before the first inflorescence. Open bars, 8 h of light in a 24-h cycle; filled bars, 16 h of light in a 24-h cycle.
Table 1.
Summary of mass-spectral sequencing analysis of FLAG-ESP IP-specific peptides
| Protein | Predicted Mr, Da | Total peptides | Unique peptides | P | Sf | XCorr |
|---|---|---|---|---|---|---|
| FLAG-ESP5 (CPSF100/At5g23880) | 85,124 | 153 | 21 | 2.22e−15 | 16.42 | 200.31 |
| CPSF160/At5g51660 | 158,064 | 37 | 12 | 1.18e−11 | 11.03 | 116.27 |
| CPSF73/At1g61010 | 77,395 | 25 | 10 | 3.25e−12 | 8.35 | 90.33 |
| ESP4/Symplekin/At5g01400 | 159,469 | 21 | 8 | 4.15e−9 | 2.71 | 30.26 |
| FY/Pts2/At5g13480 | −79,195 | 12 | 4 | 2.90e−7 | 2.64 | 30.17 |
Because ESP4 and ESP5 have an effect on transgene RNA silencing, we explored the possibility that their effect on FCA might involve enhanced silencing of an endogenous RNA. We therefore recombined esp4-1 separately with the RNA silencing mutants rdr6, sde3-1, and nrpd1a-1 and monitored the profile of FCA mRNAs. The results, shown in Fig. 5A, illustrate that these silencing mutants reversed the hypoaccumulation of FCA-β mRNAs in esp4. It therefore is likely that the reduced accumulation of FCA-β mRNA in esp4 is due, directly or indirectly, to RNA silencing.
Discussion
Aberrant RNA.
We have described esp mutations in core RNA processing components that enhance the ability of a viral transgene to initiate both viral replication and RNA silencing (Fig. 1). To explain these phenotypes, we propose that the esp mutations result in an increase in the pool of transgene RNA to initiate viral replication or silencing. The enhanced silencing phenotype is likely to involve transcription of these transgene transcripts by RDR6 (Fig. 1 B and C). We further propose that, because the silencing in the 3′ end formation mutants is correlated with an increase in transcriptional read-through, the mechanisms of enhanced silencing involve RNAs that lack a polyA-tail and are perceived as aberrant (Fig. 3). In keeping with the view that misprocessing may create aberrant RNA for silencing, we also have shown that FCA-β mRNA levels in esp4 are reduced in a manner that depends on silencing components (Fig. 5).
The concept of “aberrant” RNA has been a long standing abstraction in the field of RNA silencing (45). The aberrant RNA species would have regions that either form double-stranded structures or would be converted to double-stranded RNA by RDR proteins. “Nonaberrant” or “normal” RNAs would be those that do not enter silencing pathways. At present, there is no molecular definition of aberrant RNA, although decapped mRNA has the characteristics of aberrant RNA in that it activates RNA silencing through an RDR6-dependent pathway (46). Now, from the identification of mutations in genes affecting RNA splicing or 3′ end formation as described here, we infer that misspliced or misterminated RNAs also may be aberrant. It seems likely therefore that there are several routes to RNA aberrancy and that the defining characteristic is the absence rather than the presence of any particular feature. A plausible scenario is that cap-, poly(A)- and other RNA-binding proteins normally prevent RDR and RNA-silencing proteins from interacting with mRNAs. In misprocessed RNAs with an aberrant characteristic these RNA-binding proteins might be bound inefficiently so that siRNAs can be produced by the RNA-silencing cofactors.
Transgene RNAs would be particularly prone to aberrancy according to this definition, especially if they have non-plant-derived elements, because they may not have the precise structures necessary for efficient interactions with the full complement of mRNA-binding proteins associated with most cellular mRNAs. In addition, if transcription terminates prematurely or late, they would produce truly aberrant RNAs. Premature or late termination of transgene transcription may be affected by structural features of the transgene DNA or RNA or, as suggested many years ago, by DNA methylation within the transcribed region (45).
Viral or transposon RNAs also may be aberrant because they are transcribed from parasitic genetic elements that that have not adapted to the host. This nonadapted parasitic RNA may not interact efficiently with the host-encoded mRNA-binding proteins and, consequently, as suggested above for transgene RNAs, the RDR and other RNA silencing cofactors would mediate silencing of the parasitic RNAs. Transposon RNAs also may be silenced if they are integrated into the genome as incomplete copies that would not produce RNAs with a normally processed 3′ end. In these scenarios, the RNA silencing would provide a defense against the viral or transposon parasites. There also may be situations in which parasites recruit RNA silencing as part of an “invasion by stealth” strategy. These stealthy parasites might have conserved “aberrancy” so that their level is reduced by silencing to be below a level that is detected by other defense mechanisms, including innate immunity.
Recent analyses in Arabidopsis indicate that there are >500 protein coding genes corresponding to clusters of siRNAs (20) (F. Schwach, R. Mosher, and D.C.B., unpublished data). It is likely that many of these siRNAs will be generated by RDR-dependent mechanisms like those involved in the esp-dependent transgene silencing described here and in RNA silencing of viruses and transposons. These siRNA loci could be viewed as producing naturally aberrant RNAs. According to this idea, the normal function of the ESP proteins would be to suppress formation of these naturally aberrant RNAs. Silencing would occur if expression of the ESP proteins is suppressed or if transcription of an siRNA locus is perturbed, perhaps by stress or development processes, so that the transcripts do not enter the ESP pathway.
A candidate locus with the potential to form naturally aberrant transcripts is FCA because the FCA-β mRNA is silenced in an RDR6-dependent manner in the esp4 and esp5 mutants (Fig. 5). To explain this observation, we propose that aberrantly terminated FCA-β RNA in these mutant plants is a substrate for production of FCA-β-specific siRNAs by RDR6 and a DCL protein. These siRNAs then would target RNA silencing to the remaining FCA-β polyadenylated transcripts. An alternative model is that esp-dependent naturally aberrant RNAs from other loci would produce siRNA suppressors of FCA-β mRNA. These siRNAs could be targeted directly at FCA-β mRNA or at mRNAs for proteins required for FCA-β mRNA formation. If these esp4-1- and esp5-1-dependent siRNAs also silence genes that suppress flowering, we can explain the early flowering phenotype of the esp mutants. It should now be possible, through the use of high-throughput sequencing technologies (20, 47), to identify esp-specific siRNAs by comparison of wild-type and esp mutant plants and determine whether the effects on FCA-β are direct or indirect. This approach also should identify other loci that produce esp-dependent siRNAs.
3′ End Formation Complexes.
The processing pathway of mRNAs is clearly similar in plants and other organisms. However, there are also some differences and variations on the basic mechanism indicated by the analysis of the ESP proteins and their homologues in plants. For example, it is likely that there are at least two complexes that contain CstF64-like homologues. They could both be CstF-like complexes that bind downstream of the cleavage site. One of these complexes would be the standard CstF, which uses the RNA recognition motif of AtCstF64 to bind downstream of the mRNA3′ end formation sites. The other would use ESP1 and a separate RNA-binding protein to recognize alternate 3′ end formation sites. Alternatively, it could be that plants, like S. cerevisiae, have two complexes containing CstF-64 homologues that operate at different steps of 3′ end formation (33). In yeast, Pti1p is a member of the CPF complex, analogous to the plant AtCPSF complex described here. Another homologue, RNA15p, operates in the CFIA complex, which is analogous to the CstF complex of mammals (33). We do not have evidence for an interaction of ESP1 or AtCstF64 with AtCPSF; however, such an interaction might be revealed under different purification conditions.
There also may be variants of the CPSF complex in Arabidopsis. One of these complexes was detected through our analysis of proteins bound to CPSF100 (ESP5) and resembles the yeast version of CPSF (CPF) because it is associated with a homologue of yeast Pfs2p (FY) (ref. 42; Fig. 4). A second type of CPSF may be more similar to the mammalian complex in that it lacks Pfs2p. In yeast, Pfs2p is thought to mediate interactions between CPF and the downstream 3′ end formation complex CFIA (42). In mammals, the interaction between upstream and downstream 3′ end formation complexes requires CstF50. Pfs2p and CstF50 proteins both have a WD domain and interact with orthologues of CstF-77 but they are otherwise distinct (42). The existence of CstF50 (At5g60940) in Arabidopsis suggests that a CPSF complex lacking Pfs2p likely would interact with the downstream 3′ end formation machinery in a similar manner as mammalian CPSF.
Heterogeneity in CPSF complexes also is implied by absence of a growth impairment phenotype in loss of function mutants at esp4. The yeast ESP4 homologue, PTA1, is essential for growth (39). If ESP4 was functionally identical to PTA1, the esp4-null mutants (e.g., esp4-3) would have a growth impairment phenotype. Because they did not, it is likely that there is a second CPSF in Arabidopsis that involves the ESP4 homologues (At1g27590/At1g27595) (Fig. 2G) instead of ESP4. It could be that these proteins are functionally redundant or that ESP4 processes a subset of RNAs with nonessential functions.
Our experiments do not address the reason for heterogeneity in 3′ end formation complexes. In many instances, it is likely that these different complexes facilitate the production of more than one RNA species from a given locus, expanding the coding potential of the genome. In some cases, these additional RNAs may have a regulatory role to play in that they have evolved to regulate RNA-silencing pathways.
Materials and Methods
Amplicon Constructs and Plant Lines.
Apds2 is a derivative of PVX/GUS (48) containing two copies of PDS in the sense orientation. This construct was transformed into the Arabidopsis thaliana C24 ecotype to create the transgenic lines Apds2-301 and Apds2-311. Segregation analysis showed Apds2-301 and Apds2-311 contained single insertions of the transgene. T3 Apds2-301 and Apds2-311 seeds were mutagenized with ethyl-methanesulfonate, and M2 seedlings were screened for mutants with an esp phenotype. Apds1as contains a single antisense PDS insert and was transformed into C24 to generate line Apds1as-1; Aalb3, containing a sense fragment from ALB3, was used to create Aalb3-TD36 (C24). Both Apds1as-1 and Aalb3-TD-36 lines were crossed into esp mutant lines in which the Apds2 previously had been crossed out and mutant individuals were identified in the F2 generation. Leaves were stained for GUS expression as described in ref. 49.
The rdr6 allele used in these experiments corresponds to sde1-1 (5). The sgs3 allele corresponds to sde2-1 (5), a 3.6-kb deletion of the entire SGS3 coding sequence (A.J.H., unpublished data). The sde3-1 (12) and nrpd1a-1 (7) alleles have been described. Additional information on amplicon construction, mapping, genetic crosses, complementation of esp1 and esp5, and PCR genotyping assays is found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Phylogenetic Analysis.
Full-length cDNA sequences for ESP1, ESP3, and ESP5/CPSF100 were available from the Arabidopsis sequence database (www.arabidopsis.org) and confirmed by our own sequencing of cDNAs. Only a partial 3′ cDNA sequence was available for ESP4, so we determined the full-length sequence from RT-PCR and 5′ RACE products and then deduced the complete amino acid sequence for ESP4. Details of phylogenetic analysis can be found in Supporting Materials and Methods.
Northern Blot Analysis.
RNA was prepared from 2.5-week-old seedlings (7, 50). For analysis of high molecular weight total RNA, 10 μg of RNA was separated on 1% formaldehyde gels and blotted to Hybond-NX (Bio-Rad, Hercules, CA). For analysis of PVX and PDS RNA, the membrane first was probed with random prime-labeled probe 2. The membrane then was stripped and reprobed with sense-specific probe 1. The small RNA blot was prepared by using enriched small RNA fractions purified from 200 μg (1 μg/1 μl) total RNA as described in ref. 7. The blot first was probed for miRNA159 and siRNA 255 (7) and then PDS siRNA by using a PDS sense-specific riboprobe. ESP1 and ESP4 samples were probed by using random prime ESP1 or ESP4 cDNA probes. For FCA analysis, polyA-RNA was purified from 300 μg of total RNA by using the MicroPolyA-RNA purist mRNA purification kit (Ambion, Austin, TX). The resulting membrane first was probed for FCA (51), then stripped and reprobed for actin. Details of probes can be found in Supporting Materials and Methods.
RT-PCR.
Expression of ESP genes and their homologues was assessed by amplifying cDNA made from polyA RNA purified from seedlings, roots, rosette leaves, stems, and flowers. Gene-specific primers were selected that amplified across exon-exon junctions, except in the case of At4g16680, which has a single small intron. Genomic DNA in each case was included as an amplification and size control.
Strand-specific RT-PCR analysis of PVX transcripts was performed on total RNA that was pretreated with Turbo DNA-free (Ambion). Reverse transcription for RT1 was primed with a mixture of two primers: Actin 2exR (Table 2, which is published as supporting information on the PNAS web site) and M13R, which is present downstream of the Nos terminator in Apds2. Reverse transcription for RT2 was primed with Actin 2exR and PVX1 (Table 2), which is a sense primer upstream of the double PDS-insert in Apds2. PCR1 was performed with PDS5502F and 0179fp2006 (Table 2). PCR2 used PDS5502F and PVX2 (Table 2). PCR3 used ACTmaiFW and ACTmaiRV (7). PCR1 and 2 were carried out for 30 cycles, whereas PCR3 went for 25 cycles.
Purification of ESP5-Specific Peptides.
The starting material for the mass spectral analysis of FLAG-ESP5 interacting proteins was a line in which the esp5-1 mutation was complemented by an ESP5 promoter-driven FLAG-ESP5 transgene (73ESP5-2). Controls were GxA (ecotype C24) and sde3-1 35S-FLAG-SDE3 GxA, which complements the sde3-1 mutation (A.J.H., unpublished data; ecotype C24). Homozygous T4 seedlings were grown under long days (70% humidity; cycles of 16 h light/8 h dark) for 2.5 weeks. Final purification conditions were worked out by using smaller volumes of tissue and Western analysis as depicted in Fig. 7a. For the final purifications, FLAG immunoprecipations were performed on extracts from 100 g of vegetative tissue as described in Supporting Materials and Methods. Eluted proteins were precipitated by using methanol/chloroform and the pellet resuspended in 50 mM ammonium bicarbonate for tryptic digestion (sequencing grade modified trypsin, Promega, Madison, WI).
Mass Spectrometry.
Nano flow LC-MS/MS analysis was performed by using a LTQ mass spectrometer (Thermo Electron Corp.) employing automated data-dependent acquisition as described in Supplementary. Raw data were processed by using BioWorks 3.2 and TurboSEQUEST (Thermo Electron, San Jose, CA) and searched against the Arabidopsis genome supplemented with common contaminants (trypsin and keratins; sequences collated by Thermo Electron) with oxidized M as a variable modification. Peptide hits were filtered by Xcorr and charge state [xc (+1, 2, 3) 2.0, 2.5, 3.5] and protein hits by probability (1e−3).
Flowering Time Assays.
Apds2-free plants for flowering time assays were obtained from the progeny of lines that were homozygous for the esp mutant and hemizygous for Apds2 (described above). To exclude that the enhanced flowering phenotype was due to a background mutation, plants were assayed from multiple independent lines for each esp mutant. Flowering time was determined by counting the number of true leaves (rosette and cauline leaves) until emergence of the first florescence.
Supplementary Material
Acknowledgments
We thank David Brice for assistance in the latter stages of mapping esp1. D.C.B., A.J.H., A.M., and A.J. were supported by the Gatsby Charitable trust. A.J.H. was also supported by a Hitchin's-Elion Burrough's Wellcome Fund Fellowship, and A.M. received a European Molecular Biology Organization long-term fellowship.
Abbreviations
- CPF
cleavage polyadenylation factor
- CPSF
cleavage polyadenylation specificity complex
- CstF
cleavage stimulation factor
- esp
enhanced silencing phenotype
- rdr
RNA-dependent RNA polymerase.
Footnotes
The authors declare no conflict of interest.
References
- 1.Baulcombe D. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 2.Hammond SM, Bernstein E, Beach D, Hannon G. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 3.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel J-B, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 5.Dalmay T, Hamilton AJ, Rudd S, Angell S, Baulcombe DC. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 6.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. PLoS Biol. 2004 doi: 10.1371/journal.pbio.00201040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herr AJ, Jensen MB, Dalmay T, Baulcombe D. Science. 2005 doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 8.Vazquez F, Gasciolli V, Crete P, Vaucheret H. Curr Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 9.Kurihara Y, Takashi Y, Watanabe Y. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han M-H, Goud S, Song L, Fedoroff N. Proc Natl Acad Sci USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T. Plant Mol Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- 12.Dalmay TD, Horsefield R, Braunstein TH, Baulcombe DC. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinhart BJ, Weinstein EG, Rhoades M, Bartel B, Bartel DP. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park W, Li J, Song R, Messing J, Chen X. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu J-K. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Xie Z, Allen E, Wilken A, Carrington JC. Proc Natl Acad Sci USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshikawa M, Peragine A, Park MY, Poethig RS. Genes Dev. 2005 doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen E, Xie Z, Gustafson AM, Carrington JC. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- 21.Kennedy S, Wang D, Ruvkun G. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 22.Lee RC, Hammell CM, Ambros V. RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domeier ME, Morse DP, Knight SW, Portereiko M, Bass BL, Mango SE. Science. 2000;289:1928–1930. doi: 10.1126/science.289.5486.1928. [DOI] [PubMed] [Google Scholar]
- 24.Vaucheret H, Mallory AC, Bartel DP. Mol Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Z, Kasschau KD, Carrington JC. Curr Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 26.Dalmay T, Hamilton AJ, Mueller E, Baulcombe DC. Plant Cell. 2000;12:369–379. doi: 10.1105/tpc.12.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaistij FE, Jones L, Baulcombe DC. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sundberg E, Slagter JG, Fridborg I, Cleary SP, Robinson C, Coupland G. Plant Cell. 1997;9:717–730. doi: 10.1105/tpc.9.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, Meinke D. Plant Physiol. 2004;135:1206–1220. doi: 10.1104/pp.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potashkin J, Li R, Frendewey D. EMBO J. 1989;8:551–559. doi: 10.1002/j.1460-2075.1989.tb03409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vijayraghavan U, Company M, Abelson J. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 32.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 33.Proudfoot N. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 35.Dichtl B, Blank D, Ohnacker M, Friedlein A, Roeder D, Langen H, Keller W. Mol Cell. 2002;10:1139–1150. doi: 10.1016/s1097-2765(02)00707-4. [DOI] [PubMed] [Google Scholar]
- 36.Calvo O, Manley JL. Cell. 2001;7:1013–1023. doi: 10.1016/s1097-2765(01)00236-2. [DOI] [PubMed] [Google Scholar]
- 37.Aranda A, Proudfoot N. Mol Cell. 2001;7:1003–1011. doi: 10.1016/s1097-2765(01)00235-0. [DOI] [PubMed] [Google Scholar]
- 38.Cormack RS, Hahlbrock K, Somssich I. Plant J. 1998;14:685–692. doi: 10.1046/j.1365-313x.1998.00169.x. [DOI] [PubMed] [Google Scholar]
- 39.O' Connor JP, Peebles CL. Mol Cell Biol. 1992;12:3843–3856. doi: 10.1128/mcb.12.9.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preker PJ, Ohnacker M, MinvielleSebastia L, Keller W. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elliott B, Dattaroy T, Meeks-Midkiff LR, Forbes KP, Hunt AG. Plant Mol Biol. 2003;51:373–384. doi: 10.1023/a:1022035219500. [DOI] [PubMed] [Google Scholar]
- 42.Ohnacker M, Barabino SML, Preker PJ, Keller W. EMBO J. 2000;19:37–47. doi: 10.1093/emboj/19.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forbes KP, Addepalli B, Hunt AG. J Biol Chem. 2006;281:176–186. doi: 10.1074/jbc.M510964200. [DOI] [PubMed] [Google Scholar]
- 44.Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. Cell. 2003;113:777–787. doi: 10.1016/s0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- 45.English JJ, Mueller E, Baulcombe DC. Plant Cell. 1996;8:179–188. doi: 10.1105/tpc.8.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 47.Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobson SE. Nat Genet. 2006 doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 48.Angell SM, Baulcombe DC. EMBO J. 1997;16:3675–3684. doi: 10.1093/emboj/16.12.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Angell S, Baulcombe D. Plant J. 1995;7:135–140. doi: 10.1046/j.1365-313x.1995.07010135.x. [DOI] [PubMed] [Google Scholar]
- 50.White JL, Kaper JM. J Virol Methods. 1989;23:83–94. doi: 10.1016/0166-0934(89)90122-5. [DOI] [PubMed] [Google Scholar]
- 51.Quesada V, Macknight R, Dean C, Simpson GG. EMBO J. 2003;22:3142–3152. doi: 10.1093/emboj/cdg305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.