Abstract
The origin recognition complex (ORC) was originally identified in the yeast Saccharomyces cerevisiae as a protein that specifically binds to origins of DNA replication. Although ORC appears to play an essential role in the initiation of DNA replication in the cells of all eukaryotes, its interactions with DNA have not been defined in species other than budding yeast. We have characterized a Schizosaccharomyces pombe homologue of the ORC subunit, Orc4p. The homologue (Orp4p) consists of two distinct functional domains. The C-terminal domain shows strong sequence similarity to human, frog, and yeast Orc4 proteins, including conserved ATP-binding motifs. The N-terminal domain contains nine copies of the AT-hook motif found in a number of DNA-binding proteins, including the members of the HMG-I(Y) family of chromatin proteins. AT-hook motifs are known from biochemical and structural studies to mediate binding to the minor groove of AT-tracts in DNA. Orp4p is essential for viability of Sc. pombe and is expressed throughout the cell cycle. The Orp4 protein (and its isolated N-terminal domain) binds to the Sc. pombe replication origin, ars1. The DNA binding properties of Orp4p provide a plausible explanation for the characteristic features of Sc. pombe origins of replication, which differ significantly from those of Sa. cerevisiae.
The initiation of DNA replication in bacteria, bacteriophages, and eukaryotic viruses occurs at well-defined origins of DNA replication that are recognized by specific initiator proteins (1). Recent work indicates that initiation of chromosomal DNA replication in the budding yeast Saccharomyces cerevisiae proceeds by a similar mechanism (2–4). Sa. cerevisiae origins of DNA replication [autonomously replicating sequence (ARS)] are short (≈100 bp) segments consisting of two essential regions: the A domain, which contains a conserved 11-bp ARS consensus sequence (ACS) that is essential for origin function, and the B domain, which contains several stimulatory sequence elements (5). The ACS is required for binding of the origin recognition complex (ORC), the Sa. cerevisiae initiator protein (6, 7). ORC consists of six subunits (Orc1p–Orc6p), each of which is essential for viability of Sa. cerevisiae (8–11). It is not yet clear which subunits of ORC determine its specificity of binding, but the strongest candidates are Orc1p, Orc2p, and Orc4p, because protein-DNA cross-linking studies indicate that these subunits interact with the major groove of the DNA and bind within 10 Å of the ACS (12). Genetic and biochemical data strongly suggest that ORC is required for efficient initiation of Sa. cerevisiae DNA replication in vivo, in part by recruiting essential initiation factors to replication origins before S phase (11, 13, 14). The complex has also been implicated in other cellular processes, such as the establishment of gene silencing and progression through M phase (11, 15).
Homologues of ORC subunits have now been identified in a variety of other eukaryotic species, including mammals (16). In addition, protein complexes containing ORC-related subunits have been purified from extracts of Xenopus laevis eggs and Drosophila melanogaster embryos (17, 18). In several cases (Schizosaccharomyces pombe, Xenopus, and Drosophila), there is genetic or biochemical data indicating that specific ORC subunits are required for initiation of DNA replication (19–21). Thus, ORC appears to be highly conserved in eukaryotes, suggesting the existence of common mechanisms for initiating eukaryotic DNA replication. However, studies aimed at identifying origins of replication in eukaryotes other than Sa. cerevisiae have revealed unexpected complexity and raised some questions about the generality of the budding yeast paradigm (22). In metazoans, for example, there is evidence that the sequence requirements for initiation of chromosomal DNA replication may be more relaxed than those in Sa. cerevisiae. Although a number of studies have localized efficient origins of bidirectional replication to discrete chromosomal sites as in budding yeast, other studies indicate that initiation occurs with some efficiency at many sites within so-called “initiation zones.” In special cases, such as Xenopus eggs, ORC-dependent initiation of DNA replication on virtually any DNA molecule can occur with high efficiency. Collectively, these studies suggest that the interaction between ORC and DNA in metazoan cells may differ in some ways from that observed in Sa. cerevisiae.
The fission yeast Sc. pombe represents an excellent model system for analysis of the regulation of DNA replication during the cell cycle. It is evolutionarily distant from S. cerevisiae, and some aspects of its chromosomal organization more closely resemble those of metazoan cells (23). Sc. pombe chromosomal sequences capable of supporting the autonomous replication of plasmids (ARSs) have been identified, and a number of them have been shown to function as origins of DNA replication at their normal chromosomal loci (24–29). Fission yeast ARS elements are rich in AT base pairs and have a minimal size of 500-1000 bp, which is considerably larger than budding yeast origins. In previous studies, we carried out a detailed genetic analysis of Sc. pombe ars1 (27). This study, together with similar analyses of two additional fission yeast ARS elements, ars3001 and ars3002, revealed several interesting features of fission yeast replication origins not shared by those of Sa. cerevisiae (29, 30). Each Sc. pombe ARS element contains one or more sequence blocks of 20 to 50 bp that are important for ARS function as determined by deletion analysis. These blocks exhibit some similarity to one another: they are all extremely AT-rich, and they display a tendency toward clustering of A or T residues. However, they do not share a common consensus sequence comparable with the ACS of Sa. cerevisiae replication origins. Sc. pombe ARS elements are also characterized by a high degree of functional redundancy. In many cases sequence blocks that are important for function appear to be composed of smaller AT-rich sequence elements that can be deleted individually without significantly affecting origin activity. Consistent with these clear differences in sequence organization, Sc. pombe replication origins do not function in Sa. cerevisiae and vice versa (27).
We report here the identification and characterization of a Sc. pombe homologue of the Orc4 protein (Orp4p) that consists of two similarly sized domains. The C-terminal domain shows high sequence similarity to human, Xenopus and Sa. cerevisiae Orc4 (31–33). The N-terminal domain contains nine repeats of the so-called “AT-hook” motif, which includes the core sequence, RGRP (34–37). The AT-hook motif is characteristic of the HMG-I(Y) family of mammalian nonhistone chromosomal proteins, and is found in a variety of other proteins in organisms from yeast to humans (37). We have demonstrated that full-length Orp4p or its isolated N-terminal domain binds to sequences in the Sc. pombe replication origin, ars1. We suggest that the Orp4 protein targets Sc. pombe ORC to replication origins and that the characteristic features of ars1 and other Sc. pombe origins of DNA replication can be understood in terms of the unique DNA binding properties of the protein. We further speculate that HMG-I(Y)-like proteins may play a role in targeting ORC to replication origins in other species.
MATERIALS AND METHODS
Cloning of Sc. pombe orp4+.
The design of degenerate PCR primers was based upon two highly conserved amino acid sequences (GPRG/QSG/YKT and EKRVKSRF found in human and Sa. cerevisiae Orc4 proteins; refs. 31–33). A 500-bp fragment of orp4+ was amplified by PCR from Sc. pombe genomic DNA and cloned into the plasmid pKS (Stratagene). The fragment was radiolabeled with 32P and used as a probe to screen Sc. pombe genomic and cDNA libraries (a gift of G. Hannon, Cold Spring Harbor Laboratory, Plainview, NY). One genomic clone (4.8 kb) and several cDNA clones (up to 3.2 kb) were isolated.
Sc. pombe Strains and Plasmids.
Strain YRC1 expressing Orp4p with a triple hemagglutinin (HA) epitope tag at the C terminus was constructed as follows. A DNA fragment containing the 3′-terminal 1.1 kb of the orp4+ gene was synthesized by PCR and inserted into the NotI site of the integration vector pRCI272 in frame with the HA epitope tag. [pRCI272 was derived from the expression vector pSLF272 (38) by removing the nmt1+ promoter and the ars1 origin of DNA replication.] The resulting plasmid was linearized at a unique NcoI site in the orp4+ sequence and introduced into strain TK3 (h− leu1–32 ura4-D18). The YRC1 strain, harboring a single integration of the plasmid at the orp4+ locus (h− leu1–32 ura4-D18 orp4∷3HA[ura4+]) was obtained by selection for ura+ transformants. The structure of YRC1 was verified by PCR and Southern blot analysis of restriction enzyme digests of purified genomic DNA.
Strain YRC3 (h− ura4-D18 pRCE3X-orp4+), containing a plasmid expressing the full-length orp4+ gene under the control of the nmt1+ promoter, was constructed as follows: The plasmid pRCE3X was derived from the plasmid pREP81X (39) by replacing the 2.2-kb nmt1+ promoter/terminator DNA fragment with the nmt1+ promoter/terminator DNA fragment of pSLF172 (38). The orp4+ gene was synthesized by PCR and inserted into the NotI site of pRCE3X. Strain YRC4 (h− ura4-D18 pRCE3X-orp4N), expressing the N-terminal half of Orp4 protein (amino acids 1–486), and strain YRC5 (h− ura4-D18 pRCE3X-orp4C), expressing the C-terminal terminal half of Orp4 protein (amino acids 436–972), were constructed in a similar way to pRCE3X-orp4+.
Cell Cycle Arrest.
Strains carrying the HA epitope-tagged orp4 gene and cdc mutant alleles were constructed by standard genetic crosses. Strain YRC15 contained the cdc10-129 allele (h− cdc10–129 leu1–32 ura4-D18 ade6-M210 orp4∷3HA[ura4+]), and strain YRC16 contained the cdc25–33 allele (h− cdc25–22 leu1–32 ura4-D18 ade6-M210 orp4∷3HA[ura4+]). The cells were arrested in G1 phase (YRC15) or G2 phase (YRC16) by incubation for 4 hr at the nonpermissive temperature of 36°C. Strain YRC1 was arrested in S phase or M phase, by growth in media containing 25 mM hydroxyurea for 4 hr or 25 μg/ml benomyl for 3 hr, respectively.
Disruption of the orp4+ Gene.
The entire ORF of the orp4+ gene was replaced by the ura4+ gene by using standard gene replacement methods (40). For this purpose a DNA fragment containing ura4+ flanked by a 1-kb genomic fragment (from the 5′ side of the orp4+ ORF) and a 1.5-kb genomic DNA fragment (from the 3′ side of the orp4+ ORF) was introduced into a diploid strain (h+/h− leu1–32/leu1–32 ura4-D18/ura4-D18 ade6-M210/ade-M216). Heterozygous orp4+/orp4∷ura4+ diploids were identified among ura+ transformants by colony PCR. Southern hybridization analysis of several restriction enzyme digests of genomic DNA confirmed that the orp4+ gene had been replaced by ura4+.
Preparation of Extracts.
Yeast extracts were prepared as described (41) with slight modifications. Approximately 9 ml of logarithmically growing culture (OD600 ≈ 0.5–0.7) was fixed by adding 1 ml of 100% of trichloroacetic acid. Cells were harvested by centrifugation and resuspended in 100 μl of 20% trichloroacetic acid. After addition of equal volume of glass beads, the cells were disrupted by vortexing for 5 min at room temperature.
Southwestern Blot Analysis.
Cellular proteins were resolved by 10% SDS/PAGE and transferred to a nitrocellulose membrane. The membrane was incubated in denaturation buffer (10 mM Tris, pH 7.5/50 mM NaCl/2 mM EDTA/0.5 mM DTT/4 M urea) at room temperature for 2 hr. Binding buffer (10 mM Tris⋅HCl, pH 7.5/1 mM EDTA/1 mM 2-mercaptoethanol/50 mM NaCl) was added to reduce the urea concentration in 2-fold steps (42). When the concentration of urea was reduced to 0.125 M, the membrane was incubated in binding buffer plus 5% nonfat milk at room temperature for 2 hr. Gel purified radiolabeled DNA, prepared by PCR methods (43), was added (1 × 106 cpm/ml), and after incubation for an additional 2 hr, the membrane was washed and autoradiographed. For competition assays, a 100-fold excess of gel purified nonradioactive DNA was mixed with radiolabeled DNA before the incubation. The ars1 DNA fragment contained nucleotides 800-1129, which are essential for origin activity (27). Equivalent data were obtained with a fragment containing the complete ars1 element. The nonspecific competitor DNA fragment contained nucleotides 1773–2109 of the orp4+ coding region.
RESULTS
Identification of an Sc. pombe Homologue of ORC4.
To identify a fission yeast homologue of Orc4p, we carried out PCR on Sc. pombe genomic DNA with degenerate primers corresponding to sequence blocks conserved in Sa. cerevisiae and human Orc4p (31, 33). The initial PCR product was used as a probe to screen cDNA and genomic libraries. A cDNA clone with a single open reading frame capable of coding for a protein of 972 amino acids was obtained (Fig. 1). A genomic clone containing a partial copy of the same protein coding sequence (residues 241–972) was also obtained. We refer to the encoded protein as Orp4p and to the corresponding gene as orp4+.
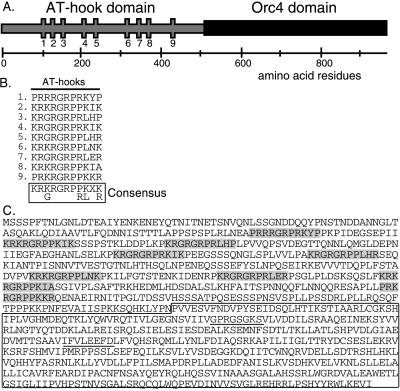
Figure 1.
Identification of Orp4. (A) Diagram of the sequence organization of the Orp4 protein. The locations of the nine AT-hooks in the N-terminal domain are indicated by vertical bars. (B) Alignment of the sequences of the AT-hook motifs in Orp4p. (C) Predicted amino acid sequence of the Orp4 open reading frame. The nine AT-hooks in the N-terminal domain are shaded and the putative ATP/GTP binding motifs in the C-terminal domain are underlined. The region of the protein with homology to human and yeast Orc4 proteins is boxed.
Inspection of the sequence of Orp4p revealed that the C-terminal half (≈460 amino acids) exhibited strong sequence similarity to human, Xenopus, and Sa. cerevisiae Orc4 proteins, which range in size from 432 to 529 amino acids (Fig. 2). The amino acid sequence of the fission yeast protein was ≈35% identical and 63% similar to the human and Xenopus Orc4 proteins over their common lengths. Similar, but slightly lower, values (29% identity/58% similarity) were obtained when the fission yeast sequence was compared with that of Sa. cerevisiae. All four Orc4 proteins contain a consensus purine nucleotide binding motif as well as several additional highly conserved sequence blocks.
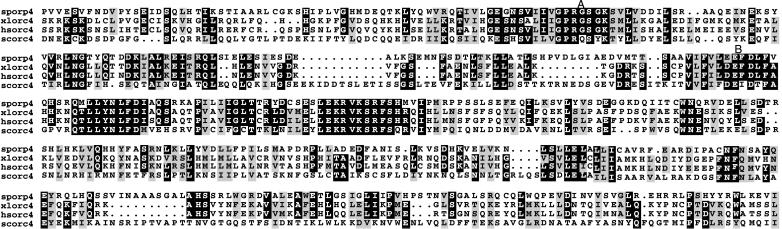
Figure 2.
Sequence alignment of Orc4 proteins. The amino acid sequence of the C-terminal domain of Orp4 protein (sporp4) is aligned with the sequences of the Orc4 proteins of human (hsorc4), frog (xlorc4), and budding yeast (scorc4). Identical amino acids shared by the majority of sequences are shaded black and similar amino acids are shaded gray. The Walker A and B sites are indicated.
The Sc. pombe Orp4 protein contains an N-terminal extension of ≈500 amino acids that is absent from Orc4 proteins of the other species. This N-terminal domain has an unusual amino acid composition that is weighted toward hydrophilic amino acids and contains greater than average amounts of proline, serine, aspartic acid, glutamic acid, and arginine. Distributed along the length of the domain are nine AT-hook motifs, consisting of a perfectly conserved central core sequence, RXRGRP, with flanking regions containing additional basic amino acids (Fig. 1). The AT-hook motif is known to bind to the minor groove of AT tracts 4–8 nucleotides in length (36, 37). The best-studied examples of proteins containing AT-hooks are the HMG-I(Y) proteins that are known to facilitate the assembly of protein complexes at transcriptional enhancers (37). The HMG-I(Y) proteins contain three AT-hook motifs, and it has been demonstrated that a single protein molecule can simultaneously bind with high affinity to three appropriately spaced AT tracts (44). On the basis of these properties, it would be expected that the N-terminal domain of Orp4p would target the molecule to genomic segments that are rich in AT-residues, such as origins of replication (see below).
Expression of Sc. pombe Orp4 Protein.
An HA epitope tag was introduced at the C terminus of Orp4p by means of chromosomal gene targeting methods. Whole cell extracts prepared from the resulting strain contained a protein, detectable with anti-HA antibody, that migrated in SDS gels with an apparent molecular mass of ≈135 kDa (Fig. 3). The protein was not detectable in control strains expressing untagged Orp4 protein. The predicted molecular mass of the tagged Orp4 protein is 112 kDa, so the protein appears to migrate slightly more slowly than expected, possibly as a result of the unusual amino acid composition of the N-terminal domain.
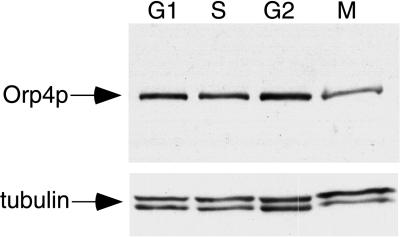
Figure 3.
Orp4 protein is expressed throughout the cell cycle. Fission yeast cells expressing epitope-tagged Orp4 protein were blocked in the G1, S, G2, or M phases of the cell cycle as described in Materials and Methods. Extracts prepared from blocked cells were fractionated by SDS/PAGE and Orp4 protein was detected by Western blot analysis. The same Western blots were probed for tubulin protein as a control for equal loading of the gels.
To determine whether the expression of Orp4p varied during the cell cycle, extracts were prepared from cells blocked in the G1, S, G2, and M phases of the cell cycle and assayed for Orp4p by Western blot analysis. To confirm that approximately equal amounts of protein were loaded in each lane, the same Western blot was probed with anti-tubulin antibodies. As shown in Fig. 3, expression of Orp4p was nearly the same at all phases of the cell cycle, with the possible exception of a small reduction in expression in M phase. This finding is consistent with observations in most other species, indicating that ORC subunits are generally expressed throughout the cell cycle (16).
Phenotypic Effects of Deletion or Overexpression of Orp4 Protein.
By using standard genetic methods, one copy of the orp4+ gene in a diploid strain of Sc. pombe was deleted and replaced with the ura4+ gene. The orp4 deletion strain was induced to sporulate and tetrads were analyzed. Two spores in each tetrad failed to form colonies. Microscopic examination revealed that these spores gave rise to 1–10 vegetative cells of variable size, but failed to progress further. The two remaining spores in each tetrad produced colonies of normal size that were invariably ura− in phenotype. We conclude that the orp4+ gene is essential for viability in Sc. pombe. The ability of cells lacking the orp4+ gene to undergo two or three cell divisions before arresting suggests that some active Orp4 protein is sequestered in the spores derived from the heterozygous diploid parent. To further examine the terminal phenotype of cells lacking Orp4 protein, a population of spores derived from the orp4 deletion strain was germinated in medium lacking uracil. Under these conditions only the ura+ spores carrying the orp4 deletion (orp4Δ) are capable of germinating. Flow cytometric analysis confirmed that orp4Δ cells synthesized DNA during at least the first cell cycle after germination. Microscopic examination of the cells at later times revealed the accumulation of elongated cells with abnormal nuclear morphology (n) as well as some “cut” cells that had undergone abnormal mitosis (c). These terminal phenotypes are similar to the phenotypes previously observed with cells lacking Sc. pombe Orp1 protein (45) (Fig. 4 A and B).
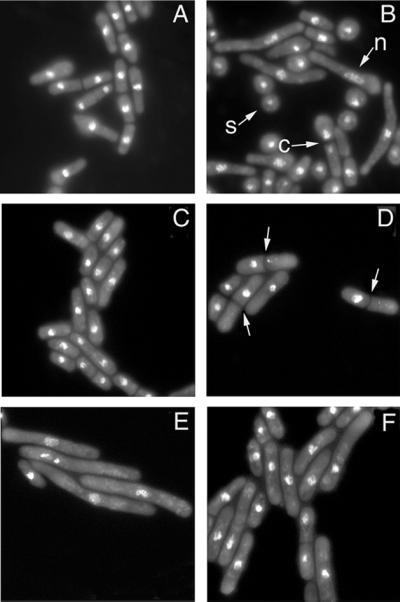
Figure 4.
Phenotypic effects of deletion or over-expression of Orp4 protein. (A and B) Deletion of Orp4. Spores from a control diploid strain (ura4+/ura4-D18) or a diploid strain in which one copy of the orp4+ gene replaced by the ura4+ gene (ura4-D18/ura4-D18 orp4+/orp4∷ura4+) were incubated in medium lacking uracil for 15 hr and stained with DAPI. The control spores yielded a population of growing vegetative cells with mostly normal morphology (A). Spores lacking the orp4+ gene yielded a variety of terminal phenotypes, including elongated cells with abnormal nuclei (n) and “cut” cells (c) with the septum separating unequal nuclear masses (B). An ungerminated spore is marked by (s). (C–F) Over-expression of Orp4 protein or its domains. Wild-type Sc. pombe cells were transformed with vector alone (C) or with plasmids expressing the N-terminal domain of Orp4p (D), the C-terminal domain of Orp4p (E) or the complete Orp4p (F) under the control of the inducible nmt1+ promoter. Twenty hours after induction the cells were fixed and stained with DAPI. Approximately 30% of cells over-expressing the N-terminal domain of Orp4p contained a septum with a single nucleus (D). Arrows indicate septa. The majority of cells over-expressing the C-terminal domain of Orp4p were elongated and many exhibited abnormal nuclear morphology (E). Cells over-expressing the complete Orp4p showed both phenotypes (F).
To determine the phenotypic consequences of over-expressing Orp4 protein or either of its two constituent domains, cDNAs encoding each protein were inserted into Sc. pombe expression plasmids in frame with a C-terminal HA epitope tag, and expression of the fusion proteins was driven by the strong inducible nmt1+ promoter. Over-expression of Orp4 protein or either of its domains markedly slowed cell growth. Microscopic examination revealed that the two domains of Orp4p had dramatically different phenotypic effects. Overexpression of the N-terminal AT-hook domain resulted in the accumulation of septated cells that contained only a single nucleus, suggesting a failure of chromosome disjunction before septation or a derangement of the regulatory mechanism that normally couples septation to the completion of mitotic events (Fig. 4D). A similar phenotype has been observed in the case of the cut9 mutant that is defective in anaphase (46). Flow cytometry showed that roughly half of the cells had a normal 2C DNA content, and the remainder had a wide range of DNA contents from <1C to >2C, consistent with a segregation defect (data not shown). Over-expression of the C-terminal Orc4 homology domain resulted in the accumulation of highly elongated cells, indicative of cell cycle arrest or delay (the cdc phenotype). The nuclei of such cells were grossly abnormal and contained DAPI-staining material that was distributed in fragments over a significantly larger volume than that of normal interphase nuclei (Fig. 4E). Approximately half of the cells had a 2C DNA content by flow cytometry. Most of the remainder had DNA contents >2C, suggesting that DNA replication was partially uncoupled from cell division. A small fraction of the cells (5%) had a 1C DNA content indicative of failure to enter S phase. Cells over-expressing the full-length Orp4 protein exhibited phenotypes similar to those observed with over-expression of either of its domains, but the extent of cellular elongation was less marked and the frequency of abnormal nuclear division was lower (Fig. 4F). The majority of the cells had a normal 2C DNA content, but a fraction (≈30%) had DNA contents >2C. Western blot analysis demonstrated that the level of accumulation of the full-length protein was somewhat less than that of either isolated domain, perhaps accounting for this less extreme phenotype.
Binding of Orp4 Protein to the ars1 Replication Origin.
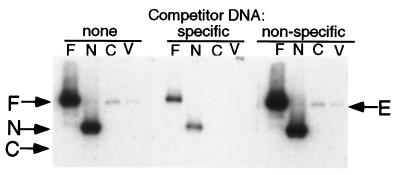
To assess the ability of Orp4p to bind to DNA we carried out southwestern blot analysis. Orp4 protein or its N- and C-terminal domain was expressed from Sc. pombe plasmids under the control of the nmt1+ promoter. Whole cell extracts were prepared after induction of expression and subjected to SDS/PAGE. After transfer to nitrocellulose membranes and incubation under renaturing conditions, the cellular proteins were incubated with a 32P-labeled DNA fragment derived from a region of the ars1 origin of replication known to be required for origin function (27). Strongly radioactive bands at the positions of the full-length Orp4 protein and the N-terminal domain were observed (Fig. 5, lanes F and N). Extracts from cells expressing the C-terminal Orc4 homology domain or control extracts from cells harboring the vector alone did not exhibit similarly intense radioactive bands (Fig. 5, lanes C and V). However, the latter extracts (as well as extracts expressing the N-terminal domain) contained low levels of an ars1-binding protein that migrated slightly faster than the epitope-tagged Orp4p (Fig. 5, lanes N, C, and V). It is likely that this protein represents endogenous untagged Orp4p. The binding of radioactive ars1 DNA to Orp4p or the N-terminal domain was almost completely eliminated by an excess of non-radioactive ars1 DNA, but not by a similar amount of an unrelated DNA fragment of the same size. We conclude from these data that Orp4 protein specifically binds to ars1 DNA and that the N-terminal AT-hook domain is necessary and sufficient for such binding.
Figure 5.
Binding of Orp4 protein to the ars1 replication origin. Cell extracts were prepared from haploid cells expressing full length Orp4p (F), the N-terminal domain of Orp4p (N), or the C-terminal domain of Orp4p (C). Extracts from cells containing vector alone (V) served as a control. After SDS/PAGE the cellular proteins were transferred to a nitrocellulose membrane and subjected to a renaturation protocol. The membranes were incubated with a radiolabeled fragment of the ars1 replication origin in the presence or absence of a 100-fold excess of nonradioactive competitor DNA. The competitor DNAs consisted of the identical ars1 fragment (specific) or an unrelated fragment of similar size (nonspecific). The positions of full-length Orp4p, the N-terminal domain and the C-terminal domain are indicated as F, N, and C, respectively. (The expression of all three proteins was verified by Western blot analysis.) The position of endogenous Orp4p is indicated by E.
DISCUSSION
Orc4 protein is an essential subunit of the Sa. cerevisiae origin recognition complex and has been identified in other eukaryotes, including mouse, frog, and human (31–33). The Sc. pombe Orc4 protein shares all of the sequence blocks conserved in other species, suggesting that it is a true homologue. We have demonstrated that the orp4+ gene encoding Orp4p is essential for viability of fission yeast. Cells lacking Orp4p arrest as elongated cells with deranged nuclear morphology, similar to fission yeast cells lacking other ORC subunits. The protein is expressed throughout the cell cycle, consistent with observations in Sa. cerevisiae and other species that ORC subunits are relatively stable proteins whose levels do not fluctuate significantly during the cell cycle (16).
Among the sequence blocks conserved in Orc4 proteins is a bipartite purine nucleotide binding motif (47). The Sc. pombe nucleotide binding motif is more closely related to that of the vertebrate species than to that of Sa. cerevisiae. The Orc4 proteins of Sc. pombe, human, Xenopus, and mouse show a very strong match to the consensus Walker A motif, whereas Sa. cerevisiae Orc4 proteins (ScOrc4) has only a weak match with other amino acids replacing two highly conserved glycine residues (Fig. 2). Although the ScOrc4 subunit can be cross-linked to ATP in the context of the complete origin recognition complex, there is evidence that the interaction with ATP may be indirect (12). Mutations in the Walker A or Walker B motifs of ScOrc4 do not reduce the extent of ATP cross-linking and have no discernable phenotypic effects in vivo. Thus, the nucleotide binding motif of ScOrc4 is probably inactive, whereas the motifs of Sc. pombe and higher eukaryotes may well be functional.
Orp4p has a 500-amino acid N-terminal extension that contains nine AT-hook motifs (34–37). We suggest that the function of this domain is to target Sc. pombe Orc to origins of replication. In support of this hypothesis we have demonstrated that the N-terminal domain of Orp4p mediates in vitro binding to ars1 origin DNA, which like other Sc. pombe ars elements is extremely AT-rich. The AT-hook motif consists of a core sequence of RGRP flanked by basic residues and has previously been observed in proteins with a variety of functions, including transcriptional regulation, chromatin remodeling, and mitotic control (37). Two known ORC subunits, Sc. pombe Orp1p and Sa. cerevisae Orc2p, contain single AT-hook motifs (31, 45). Abp-2, a previously described Sc. pombe protein with one AT-hook motif, binds to ars DNA, but is not essential for viability (48).
The best studied examples of proteins with AT-hooks are the mammalian HMG-I(Y) proteins, which contain three motifs separated by flexible spacer sequences (37). Structural and biochemical studies of HMG-I(Y) protein-DNA complexes indicate that the conserved RGR motif adopts an extended configuration and lies deep within the minor groove of AT-tracts that are a minimum of four residues in length (34–37). The flanking basic residues mediate electrostatic and hydrophobic interactions with the DNA backbone (36). The DNA bound to HMG-I(Y) protein is essentially B-type with a minor groove that is only slightly wider than normal (36). It has been clearly demonstrated that the three AT-hooks in HMG-I(Y) proteins bind in a multivalent fashion to sequential AT-tracts along the DNA (44). The affinity of HMG-I(Y) for AT-rich DNA has been measured in the nanomolar range (34). Thus, Orp4p could have a very high binding affinity (and a long lifetime on the DNA) if all nine motifs simultaneously engage AT-tracts in the DNA. A high binding affinity would be consistent with the possibility that Orp4p (and other ORC subunits) remain bound to DNA throughout the cell cycle as has been observed in Sa. cerevisiae. On the other hand, the organization of the DNA binding domain of Orp4p into a set of independent modules, each with relatively low affinity on its own, could facilitate the progressive displacement of the protein from the DNA when necessary, e.g., for the passage of a replication fork.
Our competition binding data indicate that Orp4p has a significantly higher affinity for ars1 DNA than for a randomly chosen DNA fragment of normal base composition. This result is perhaps not surprising given the fact that ars1 contains many lengthy AT-tracts. However, we suspect that Orp4p may recognize some AT-rich sequences in preference to others and that this may be an important factor in determining the specificity of initiation of DNA replication. Recent studies have demonstrated that the spacing between successive AT-tracts is an important factor in determining the affinity of HMG-I(Y) binding (44). The binding affinity decreases rapidly if the AT-tracts are too close or too distant. These considerations suggest that the affinity of Orp4p for a particular DNA segment may be strongly dependent upon the detailed organization and spatial arrangement of the AT-tracts within the segment. Thus, it is possible that Orp4p recognizes origins of replication with high specificity and that the relative affinity of Orp4p for different origins could be one factor that determines their relative efficiency of utilization. Further work will be required to assess these possibilities.
The DNA binding properties of Orp4p provide a satisfying explanation for the characteristics of Sc. pombe replication origins deduced from genetic studies (27, 29, 30). All fission yeast origins studied carefully to date are strikingly AT-rich, but lack a short consensus sequence that is absolutely essential for origin function. These properties are consistent with the finding that the DNA binding domain of Orp4p is comprised of repeated DNA binding motifs each recognizing a sequence whose only distinguishing feature is a core of AT base pairs. Thus, as noted above, the specificity of Orp4p binding is likely determined by the spatial arrangement of sequential AT-tracts, rather than by a short consensus sequence such as the budding yeast ACS. The fact that Orp4p consists of repeated DNA binding modules can also explain the functional redundancy that is such a striking characteristic of Sc. pombe origins of replication. High affinity binding may depend on the total number of AT-tracts that can be effectively bound by the nine AT-hooks in the protein and not on any particular AT-tract. Thus, the deletion of an AT-tract or a small set of AT-tracts is likely compensated for by the presence of other AT-tracts that can bind to the protein with similar affinity. The fact that the novel structure of Orp4p can explain the characteristic features of Sc. pombe origins provides strong additional support for our hypothesis that the N-terminal domain of the protein targets ORC to origins of replication.
As noted above, the sequence requirements for initiation of chromosomal DNA replication in higher eukaryotes have not yet been clearly defined, but there is some suggestive evidence that they may not be as stringent as in Sa. cerevisiae (22). Our findings raise the question of whether HMG-like proteins might be involved in initiation of DNA replication in organisms other than Sc. pombe. Although Orp4p is so far unique among ORC subunits in containing a recognizable DNA binding domain, it is possible that targeting of ORC to origins in higher eukaryotes involves specific protein–protein interactions with chromatin factors that have similar properties to the N-terminal domain of Orp4p. It is known that HMG-I(Y) proteins play a key role in the cooperative assembly of specific protein complexes at enhancers (37). We speculate that HMG or other chromatin proteins may play a similar role in assembly of the replication initiation complex. In this regard, it is interesting that the protein in the sequence data bases that is most similar in organization to the N-terminal domain of Orc4p is the Drosophila D1 protein that contains seven consensus AT-hooks (49–51). Immunofluorescence studies have shown that the D1 protein binds sites throughout the chromosomes, but is especially abundant in regions of heterochromatin that contain AT-rich satellite DNA (49). Comparison of the distribution of D1 protein with the recently described distribution of Drosophila Orc2 protein (DmOrc2p) reveals a striking similarity. Like the D1 protein, the Drosophila Orc2 protein (DmOrc2p) exhibits a preferential, though not exclusive, association with heterochromatin in interphase and mitotic chromosomes (52).
Several lines of evidence suggest that ORC may have functions other than its role in initiation of DNA replication. In Sa. cerevisiae ORC is involved in the establishment of gene silencing at the silent mating type loci (9, 11, 53). In addition, genetic studies have demonstrated that certain orc5 mutants arrest in early M phase, suggesting a mitotic role for ORC (15). Finally, recent studies have shown that mutations in the Drosophila Orc2 gene can suppress position effect variegation, consistent with some role in the formation or maintenance of heterochromatin (52). Although there is no direct evidence for non-replicative functions of ORC in Sc. pombe, it is intriguing that over-expression of the N-terminal domain Orp4p can lead to derangement of the normal process of nuclear division and septation. Thus, it is possible that the binding of Orp4p to replication origins or other AT-rich DNA sequences could have some more general role in chromosome dynamics.
Acknowledgments
This work was supported by grants from the National Cancer Institute and the National Institute of General Medical Sciences.
Footnotes
The sequence reported in this paper has been deposited in the GenBank database (accession no. AF125185).
References
- 1.Kornberg A, Baker T A. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 2.Diffley J F. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- 3.Stillman B. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 4.Bell S P. Curr Opin Genet Dev. 1995;5:162–167. doi: 10.1016/0959-437x(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 5.Campbell J L. Annu Rev Biochem. 1986;55:733–771. doi: 10.1146/annurev.bi.55.070186.003505. [DOI] [PubMed] [Google Scholar]
- 6.Rao H, Marahrens Y, Stillman B. Mol Cell Biol. 1994;14:7643–7651. doi: 10.1128/mcb.14.11.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowley A, Cocker J H, Harwood J, Diffley J F. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell S P, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 9.Bell S P, Kobayashi R, Stillman B. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- 10.Li J J, Herskowitz I. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 11.Fox C A, Loo S, Dillin A, Rine J. Genes Dev. 1995;9:911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- 12.Lee D G, Bell S P. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang C, Weinreich M, Stillman B. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 14.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 15.Dillin A, Rine J. Science. 1998;279:1733–1737. doi: 10.1126/science.279.5357.1733. [DOI] [PubMed] [Google Scholar]
- 16.Dutta A, Bell S P. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- 17.Gossen M, Pak D T, Hansen S K, Acharya J K, Botchan M R. Science. 1995;270:1674–1677. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- 18.Rowles A, Chong J P, Brown L, Howell M, Evan G I, Blow J J. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- 19.Carpenter P B, Mueller P R, Dunphy W G. Nature (London) 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- 20.Grallert B, Nurse P. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- 21.Landis G, Kelley R, Spradling A C, Tower J. Proc Natl Acad Sci USA. 1997;94:3888–3892. doi: 10.1073/pnas.94.8.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePamphilis M L. In: DNA Replication in Eukaryotic Cells. Depamphilis M L, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 45–86. [Google Scholar]
- 23.Zhao Y, Lieberman H B. DNA Cell Biol. 1995;14:359–371. doi: 10.1089/dna.1995.14.359. [DOI] [PubMed] [Google Scholar]
- 24.Maundrell K, Hutchison A, Shall S. EMBO J. 1988;7:2203–2209. doi: 10.1002/j.1460-2075.1988.tb03059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wohlgemuth J G, Bulboaca G H, Moghadam M, Caddle M S, Calos M P. Mol Biol Cell. 1994;5:839–849. doi: 10.1091/mbc.5.8.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubey D D, Zhu J, Carlson D L, Sharma K, Huberman J A. EMBO J. 1994;13:3638–3647. doi: 10.1002/j.1460-2075.1994.tb06671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clyne R K, Kelly T J. EMBO J. 1995;14:6348–6357. doi: 10.1002/j.1460-2075.1995.tb00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez J A, Kim S M, Huberman J A. Exp Cell Res. 1998;238:220–230. doi: 10.1006/excr.1997.3835. [DOI] [PubMed] [Google Scholar]
- 29.Kim S M, Huberman J A. Mol Cell Biol. 1998;18:7294–7303. doi: 10.1128/mcb.18.12.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubey D D, Kim S M, Todorov I T, Huberman J A. Curr Biol. 1996;6:467–473. doi: 10.1016/s0960-9822(02)00514-6. [DOI] [PubMed] [Google Scholar]
- 31.Bell S P, Mitchell J, Leber J, Kobayashi R, Stillman B. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 32.Tugal T, Zou-Yang X H, Gavin K, Pappin D, Canas B, Kobayashi R, Hunt T, Stillman B. J Biol Chem. 1998;273:32421–32429. doi: 10.1074/jbc.273.49.32421. [DOI] [PubMed] [Google Scholar]
- 33.Quintana D G, Hou Z, Thome K C, Hendricks M, Saha P, Dutta A. J Biol Chem. 1997;272:28247–28251. doi: 10.1074/jbc.272.45.28247. [DOI] [PubMed] [Google Scholar]
- 34.Reeves R, Nissen M S. J Biol Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 35.Solomon M J, Strauss F, Varshavsky A. Proc Natl Acad Sci USA. 1986;83:1276–1280. doi: 10.1073/pnas.83.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huth J R, Bewley C A, Nissen M S, Evans J N, Reeves R, Gronenborn A M, Clore G M. Nat Struct Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 37.Bustin M, Reeves R. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 38.Forsburg S L, Sherman D A. Gene. 1997;191:191–195. doi: 10.1016/s0378-1119(97)00058-9. [DOI] [PubMed] [Google Scholar]
- 39.Forsburg S L. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno S, Klar A, Nurse P. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 41.Muzi-Falconi M M, Piseri A, Ferrari M, Lucchini G, Plevani P, Foiani M. Proc Natl Acad Sci USA. 1993;90:10519–10523. doi: 10.1073/pnas.90.22.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, Lemire B D, Cass C E, Weiner J H, Michalak M, Singh D, Fliegel L. Arch Biochem Biophys. 1997;346:193–202. doi: 10.1006/abbi.1997.0293. [DOI] [PubMed] [Google Scholar]
- 43.Schowalter D B, Sommer S S. Anal Biochem. 1989;177:90–94. doi: 10.1016/0003-2697(89)90019-5. [DOI] [PubMed] [Google Scholar]
- 44.Maher J F, Nathans D. Proc Natl Acad Sci USA. 1996;93:6716–6720. doi: 10.1073/pnas.93.13.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muzi-Falconi M, Kelly T J. Proc Natl Acad Sci USA. 1995;92:12475–12479. doi: 10.1073/pnas.92.26.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samejima I, Yanagida M. J Cell Biol. 1994;127:1655–1670. doi: 10.1083/jcb.127.6.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker J E, Saraste M, Runswick M J, Gay N J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez J P, Murakami Y, Huberman J A, Hurwitz J. Mol Cell Biol. 1998;18:1670–1681. doi: 10.1128/mcb.18.3.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez Alfageme C, Rudkin G T, Cohen L H. Chromosoma. 1980;78:1–31. doi: 10.1007/BF00291907. [DOI] [PubMed] [Google Scholar]
- 50.Levinger L, Varshavsky A. Proc Natl Acad Sci USA. 1982;79:7152–7156. doi: 10.1073/pnas.79.23.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashley C T, Pendleton C G, Jennings W W, Saxena A, Glover C V. J Biol Chem. 1989;264:8394–8401. [PubMed] [Google Scholar]
- 52.Pak D T, Pflumm M, Chesnokov I, Huang D W, Kellum R, Marr J, Romanowski P, Botchan M R. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 53.Foss M, McNally F J, Laurenson P, Rine J. Science. 1993;262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]