Abstract
Certain E2F transcription factor species play a pivotal role in regulating cell-cycle progression. The activity of E2F1, a protein with neoplastic transforming activity when unregulated, is tightly controlled at the transcriptional level during G0 exit. In addition, during this interval, the stability of endogenous E2F1 protein increased markedly. E2F1 stability also was dynamically regulated during myogenic differentiation and in response to gamma irradiation. One or more retinoblastoma family proteins likely participate in the stability process, because simian virus 40 T antigen disrupted E2F1 stability regulation during G1 exit in a manner dependent on its ability to bind to pocket proteins. Thus, endogenous E2F1 function is regulated by both transcriptional and posttranscriptional control mechanisms.
E2F1 was the first identified member of a family of transcription factors generically referred as E2F (1–3). Six different E2F species (E2F1–E2F6) and two DP members (DP1 and DP2) now have been identified and characterized. E2Fs and DPs form heterodimers, and the complexes bind to and regulate the transcription of several genes involved in the control of the cell cycle regulation and of DNA replication (for review see refs. 4–6). One of them (E2F-5) also plays a unique role in the behavioral control of a postmitotic cell, the cerebrospinal fluid secretory cell of the choroid plexus (7).
Overproduction of E2F1 can elicit a state of neoplastic transformation in certain lines of immortalized rodent fibroblasts (8–10). Induction of ectopic E2F1 overproduction in serum-deprived, quiescent cells promotes S phase entry followed by apoptosis (11–14), and primary murine fibroblasts (MEFs) from embryos nullizygous for E2F1 (E2F1−/−) exit from G0 more slowly than normal MEFs, indicating that E2F1 plays a major role in the timing of the G0 exit process (15). Studies of the same mice reveal that E2F1 has a tumor suppressor function and an apoptosis-inducing activity, two functions that appear to be linked to one another (16–19).
The retinoblastoma (RB) protein, p107, and p130, referred to collectively as pocket proteins, constitute a nuclear protein family that share a common structural unit (the pocket) dedicated to binding certain proteins, such as certain members of the E2F family (for review, see refs. 6, 20, and 21). Generally, when an E2F species interacts with a pocket protein, its ability to activate certain genes is suppressed, and the pocket protein/E2F complex acquires transrepression function (22–24). All known pocket proteins bind certain viral oncoproteins, i.e., papovavirus T antigen, adenovirus E1A, and Human Papilloma virus E7 (20, 21).
E2F1 transcription activity is closely regulated during the cell cycle. In G1, it is under retinoblastoma protein (pRb) control, and pRb/E2F1-DP complexes can induce a state of G1 arrest (6). In late G1, cyclin-dependent kinases phosphorylate pRb, leading to the dissociation of E2F1/DP heterodimers from pRb with the reappearance of their transctivation function. Indeed, overall E2F transactivation activity peaks at G1/S and early S and decreases in late S (25, 26). One explanation for this decrease is that, in late S, cyclin A/cdk2 binds to E2F1 (as well as to E2F2 and E2F3) and phosphorylates the relevant DP partner, thereby suppressing E2F DNA binding activity (27–31).
Another level of regulation of E2F1 function is reflected by its cell cycle-dependent synthesis. E2F1 gene expression is barely detectable in G0. By contrast, it rises as cells, exiting from G0, approach late G1. One or more E2F species activate(s) the E2F1 promoter during the G0 exit process, resulting an increase in E2F1 RNA and protein synthesis (32–35). Conversely, transcription of the E2F1 gene decreases in late S, possibly because of negative regulation of the DNA binding activity of E2F1–3 by cyclin A/cdk2 (27–30).
In addition to transcriptional control of the E2F1 gene, E2F1 abundance may be affected by yet another process. The protein is unstable, and its degradation is mediated by the ubiquitin proteasome pathway (36–38). Ectopic overproduction of underphosphorylated pRb can bind and stabilize E2F1, protecting it from efficient ubiquitination. These findings suggest a hypothesis in which timely, pRb-regulated changes in E2F1 stability contribute to the periodic rise and fall of E2F1 function during Go/G1 exit and S phase progression. Here, we present evidence that supports this hypothesis.
MATERIALS AND METHODS
Cell Cultivation, Cell Cycle Analysis, and Transfection.
All cells were grown in a 10% CO2-containing atmosphere. IMR90 primary human fibroblasts (I90 PO4, I90 PO5, or I90 PO6, Coriell Cell Repositories, Camden, NJ) were grown in DMEM containing 10% bovine serum. NIH 3T3 (American Type Culture Collection), RB−/− 3T3 (39), BOSC23 (40), and 293T (41) cells were grown in DMEM containing 10% FCS. C2/C12 (American Type Culture Collection) growth and differentiation conditions have been described (42). To synchronize fibroblasts, exponentially growing cells were treated with 10 mM lovastatin, 0.5 mM hydroxyurea, or 5 mg/ml of aphidicolin for 18 hr and then released, as needed, into drug-free medium.
Gamma irradiation was delivered by using a Gammacell 40 apparatus (Atomic Energy of Canada Limited, Commercial Products, Ottawa, Canada) at a dose rate of 114 Rad/min. The conditions of cell cycle analysis and N-acetyl-l-leucinyl-l-leucinyl-N-norleucinal (LLnL) treatment were described by Hofmann et al. (36).
Cells were transfected by the calcium phosphate precipitation method (43). pRbG4-His6-myc-Ub (44) was a gift of R. Kopito (Stanford University, Stanford, CA), and pCDNA3 mE2F1 (45), an expression vector encoding mouse E2F1, was a gift of Peggy Farnham (University of Wisconsin Medical School, Madison).
Retroviral Infection.
The pBABE-puro retroviral vector (46) was a kind gift of J. P. Morgenstern and H. Land (Imperial Cancer Research Fund, London). pBABE-puro T and pBABE-puro K1 retroviruses were the generous gift of J. A. DeCaprio (Dana Farber Cancer Institute and Harvard Medical School, Boston, MA). BOSC23 cells were cultivated and transfected as described by Pear et al. (40). The medium containing the emerging retrovirus was harvested 36–48 hr after transfection. To assay for infectious virus, NIH 3T3 cells were infected as described by Pear et al. (40) and selected in puromycin-containing medium (2 mg/ml).
Western Blotting and Normalization Method for Protein Accumulation Analysis.
These procedures were performed as described in Hofmann et al. (36). Antibodies to E2F1 (C20, Santa Cruz Biotechnology), pRb (G3–245, PharMingen), simian virus 40 T antigen (Pab419, ref. 47), and the c-myc epitope tag (9E10, Santa Cruz Biotechnology) were used to detect the proteins of interest. The specificity of the antibodies used was determined by analyzing cell lysates that did not contain the relevant antigens.
Immunoprecipitation and Pulse–Chase Analysis.
Immunoprecipitation and pulse–chase experiments were performed as described by Hofmann et al. (36) with the following modifications for pulse–chase experiments. Two 100-mm dishes of cells were analyzed at each time point. Lovastatin or hydroxyurea treatment was performed before and during the pulse–chase protocol, as indicated. To obtain a better signal-to-noise ratio, each sample was immunoprecipitated twice, sequentially (immunoprecipitation/reimmunoprecipitation), using the same antibody (C20, Santa Cruz Biotechnology) each time, as described by Krek et al. (28). The intensity of E2F1-specific bands was determined by quantification in a Storm 860 PhosphorImager (Molecular Dynamics) after 5 days of exposure.
RESULTS
Endogenous E2F1 Is Degraded by the Ubiquitin-Proteasome Pathway.
To determine whether endogenous E2F1 is degraded by the ubiquitin proteasome pathway, we compared the abundance of endogenous E2F1 protein in cells that had and had not been exposed to a proteasome-specific inhibitor, LLnL (48). 293T (human) cells were treated for 12 hr with either 50 mM LLnL or with the solvent DMSO (0.25%). To rule out transcriptional effects on E2F1 abundance, the quantities of protein analyzed were normalized for the corresponding level of E2F1 mRNA present in each cell extract, as determined by Northern blotting (see Materials and Methods). 293T cells synthesize both E1A and simian virus 40 large T antigen (T). This cell line was chosen, because its cell cycle profile was not greatly affected by LLnL (data not shown). Moreover, the presence of T and E1A, both of which disrupt pocket protein/E2F complexes (6, 20, 21), insured that any effect observed on E2F1 stability was likely not caused by pocket protein binding (see below).
As shown in Fig. 1A, LLnL treatment led to a marked enhancement in E2F1 abundance. Similar results were obtained in U2OS cells, a line devoid of any known viral oncoprotein (data not shown). These results are consistent with the hypothesis, derived from experiments performed on cells that ectopically overproduce human E2F1 (36–38), that endogenous E2F1 is degraded by the ubiquitin-proteasome pathway. To test this notion further, we measured the E2F1 half-life in pulse–chase experiments. 293T cells were treated either with LLnL or with DMSO, before and during the pulse and chase periods. As shown in Fig. 1B, the endogenous E2F1 half-life was 1–2 hr in DMSO-treated cells and more than 5 hr in LLnL-treated cells, confirming that the LLnL effect on endogenous E2F1 abundance is linked to its protein-stabilizing effect.
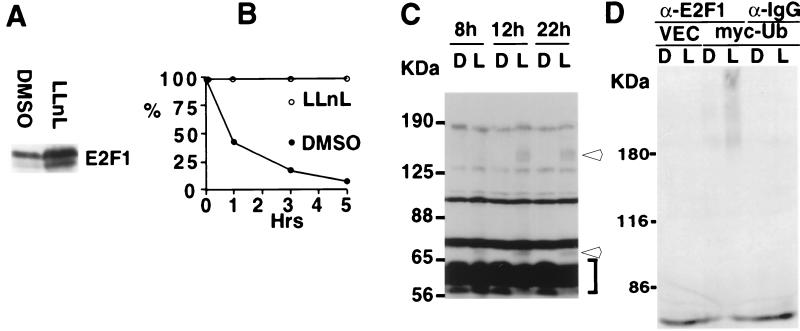
Figure 1.
Endogenous E2F1 is degraded by the ubiquitin-proteasome pathway. (A) 293T cells were incubated for 12 hr either with DMSO or 50 mM LLnL. Cell extracts were prepared, and protein accumulation was analyzed by Western blotting using an antibody to E2F1 (C20, Santa Cruz Biotechnology). The quantities of protein used in the Western blot were normalized for the amount of E2F1 mRNA present in each cell extract, as described in Materials and Methods. (B) Cells, treated as above, were pulse-labeled for 30 min and chased for 0, 1, 3, or 5 hr, followed by immunoprecipitation/reimmunoprecipitation with an antibody to E2F1, as described in Materials and Methods. Data obtained from the quantification of the E2F1-specific bands were plotted as a function of time. (C) U2OS cells were treated either with DMSO or 50 mM LLnL for the indicated periods of time. E2F1-specific bands were analyzed by Western blotting using an antibody to E2F1 (C20, Santa Cruz Biotechnology). The arrows refer to abnormally migrating E2F1 species. The bracket indicates migration positions of the ≈60-kDa E2F1 bands. (D) Cell extracts were obtained from 293T cells transfected with backbone vector (VEC) or with pRbG4-His6-myc-Ub (myc-Ub) and treated with either DMSO (D) or LLnL (L) for 15 hr. The extracts were matched for E2F1 content and immunoprecipitated with either a polyclonal antibody to E2F1 (C20, Santa Cruz Biotechnology) or a polyclonal antibody to mouse IgG (Cappel), followed by Western blotting analysis using an anti-myc tag mAb (9E10, Santa Cruz Biotechnology).
A key step in the ubiquitin-proteasome proteolytic pathway is represented by the covalent attachment of multiple ubiquitin polypeptide chains to a substrate. This modification may target a substrate for rapid degradation by the proteasome. Often, these ubiquitinated substrate conjugates are unstable and can be detected only after exposure of cells to proteasome inhibitors (for review see ref. 49). Western blotting of extracts of LLnL-treated U2OS cells were incubated with an E2F1-specific antibody. After long film exposure higher molecular weight E2F1 species were detected (Fig. 1C, arrows), along with the 60-kDa doublet typical of human E2F1 (refs. 1–3; Fig. 1C, bracket). These high molecular weight forms were absent from DMSO-treated cell extracts.
To determine whether these high molecular weight forms were ubiquitinated E2F1 species, we transfected 293T cells with an expression plasmid encoding a ubiquitin species fused to a myc epitope and an oligohistidine tag (myc-Ub) (44). After treatment of these cells with either LLnL or DMSO, extracts were prepared, and aliquots were immunoprecipitated with a polyclonal, affinity-purified antibody to E2F1 followed by Western blotting analysis using monoclonal anti-myc epitope antibody as probe. Myc-immunoreactive bands were readily observed in the E2F1 immunoprecipitate derived from LLnL-treated cells transfected with myc-tagged ubiquitin (Fig. 1D). In the E2F1 immunoprecipitate derived from similarly transfected, DMSO-treated cells, there was a very weak signal, which was detected only after long exposure. By contrast, no anti-myc reactive bands were detected in E2F1 immunoprecipitates from cells not transfected with myc-Ub or in control immunoprecipitates from myc-Ub transfected cells (Fig. 1D). From these results, it was concluded that endogenous E2F1 is ubiquitinated and degraded by the ubiquitin-proteasome pathway.
It is worth noting that the above-noted myc-immunoreactive species observed in myc-Ub-transfected 293T cells migrated differently than the higher molecular weight forms of E2F1 observed in LLnL-treated U2OS cells (compare C and D of Fig. 1). One possible explanation for this discrepancy is the presence of two tags (the myc epitope and a stretch of six histidines) on each of the multiple ubiquitin molecules bound to E2F1 in 293T-transfected cells. Moreover, the presence of the tags might have altered the quality of E2F1 ubiquitination by comparison with modification by endogenous ubiquitin.
Regulation of E2F1 Stability During the Cell Cycle.
In G1, pRb, binds, in its hypophosphorylated form, to E2F1. Not long before the G1/S transition, cyclin-dependent kinases phosphorylate pRb, leading to the dissociation of pRb from E2F1 (4–6, 20, 21). In experiments focusing on ectopically overexpressed E2F1 and RB protein, it was shown that pRb binding can stabilize E2F1 (36–38). These observations prompted us to investigate whether there is a change in the stability of E2F1 as cells pass from G1 into S. For these experiments, primary human diploid fibroblasts (IMR90) were used. In these cells, the RB pathway of G1 exit control is likely to be intact, whereas, in nearly all cancer cell lines, it is compromised (20).
Exponentially growing IMR90 were exposed to either lovastatin, an agent that induces G1 arrest, or to hydroxyurea, which induces S phase arrest (data not shown). As expected, in lovastatin-treated cells, pRb was largely hypophosphorylated and, in hydroxyurea-treated cells, much of it was hyperphosphorylated (Fig. 2A, Lower). In Western blots normalized for E2F1 mRNA content, it appeared that E2F1 accumulated to a much greater extent in lovastatin-treated than in hydroxyurea-treated cells (Fig. 2A, Upper).
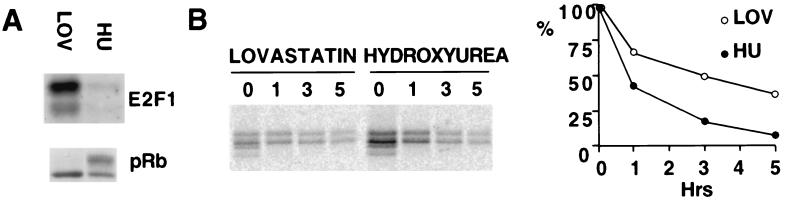
Figure 2.
E2F1 stability in G1 and in S phase primary cells. IMR90 fibroblasts were incubated for 18 hr with either 10 mM lovastatin (LOV) or 0.5 mM hydroxyurea (HU). (A) E2F1 Western blot normalized for E2F1 mRNA content (Upper) and pRb Western blot from matched quantities of the two relevant cell extracts (lovastatin-treated and hydroxyurea-treated; Lower). (B) IMR90 cells, treated either with lovastatin or hydroxyurea before the pulse–chase periods, as well as throughout them, were pulse-labeled for 30 min and chased for 0, 1, 3, or 5 hr, followed by immunoprecipitation/reimmunoprecipitation with an antibody to E2F1, as described in Materials and Methods. (Left) Autoradiography. (Right) Data obtained by the quantification of E2F1-specific bands were plotted as a function of time.
To determine whether this difference in protein accumulation was caused by a major difference in protein stability, pulse–chase experiments were performed. IMR90 were treated either with lovastatin or hydroxyurea before the pulse–chase periods, as well as throughout them. The results showed that the half-life of E2F1 in lovastatin-treated cells was in the 3- to 4-hr range, whereas, in hydroxyurea-treated cells, it was approximately 1 hr (Fig. 2B).
Effect of Pocket Proteins on E2F1 Stability.
To address the role of pocket proteins in the regulation of E2F1 stability, E2F1 abundance, normalized for that of E2F1 mRNA, was measured in cells that do and do not synthesize T. Cells were infected with retroviral vectors encoding either wild-type T or K1, a T missense mutant that is unable to bind pocket proteins and is transformation defective. K1 T retains all other known T functions (50, 51).
NIH 3T3 cells were infected with backbone virus, wild-type T virus, or K1 virus and then exposed to either lovastatin or aphidicolin. These drugs induced similar increases in G1 or S phase cells in all three cell populations, as determined by fluorescence-activated cell sorter analysis (not shown). However, the abundance of E2F1 protein, normalized for that of E2F1 mRNA, was much higher in G1 than in S phase for K1 or for backbone virus- infected cells, By contrast, in wild-type T-expressing cells, the E2F1 level was similar in these two states (Fig. 3A). This finding suggests that T can disrupt E2F1 stability regulation and that the ability of T to bind endogenous pocket proteins is necessary for this function.
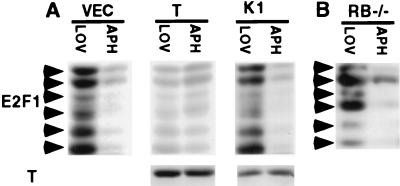
Figure 3.
Effect of pocket proteins on E2F1 stability. The arrows refer to different phosphorylation forms of mouse E2F1. (A) NIH 3T3 cells were infected with identical quantities of pBABE-puro (VEC), pBABE-puro T, or pBABE-puro K1 retrovirus, and synchronized in G1 by lovastatin (LOV) or in S by aphidicolin (APH) treatment followed by E2F1 Western blotting, normalized for E2F1 mRNA content. The same extracts were analyzed for T by Western blotting (Lower). (B) RB−/− 3T3 cells were synchronized in G1 by lovastatin or in S phase by aphidicolin treatment, followed by E2F1 Western blotting normalized for E2F1 mRNA content.
Given that E2F1 binds preferentially to pRb, as opposed to either of the other known pocket proteins (20, 21), we asked whether the regulation of E2F1 stability was pRb dependent. In 3T3 cells derived from mice nullizygous for RB (RB−/−) (39), the G1 to S phase change in E2F1 abundance corrected for its mRNA level again was observed (Fig. 3B). Because wild-type T binds all three pocket proteins and K1 T is defective for binding to these polypeptides (50, 51), one explanation is that, in the absence of pRb, at least one pocket protein(s) other than pRb itself can perform this putative pRb function.
E2F1 Is Stabilized After Irradiation of S Phase Cells.
The experiments described above imply a significant role for pocket proteins in the regulation of E2F1 stability in cells exiting G0/G1. We also asked whether E2F1 stability is affected during a cell cycle interval when pRb/E2F1 complexes are not known to exist.
During S phase, pRb is phosphorylated and E2F1 exists free of bound pocket protein (4–6, 20, 21) and is relatively unstable (see above). Because gamma irradiation in S phase is known to transiently inhibit S phase progression (52), we asked whether it affected E2F1 stability. Hence, S phase IMR90 cells were gamma-irradiated, and the abundance of E2F1 and phosphorylated pRb was measured. Six hours after irradiation of S phase, primary human fibroblasts (IMR90), S phase progression had slowed (data not shown) and was accompanied by significant pRb dephosphorylation (ref. 53; Fig. 4; F.M., F. Hofmann, and D.M.L., unpublished work). No such changes were observed in unirradiated cells. Whereas the E2F1 protein level fell in the unirradiated culture, a much lower decrease was observed in the irradiated culture. More importantly, when E2F1 protein levels were normalized for E2F1 mRNA level much more of this protein was detected in the 6-hr postirradiated culture than in the unirradiated control. This finding suggests that E2F1 is stabilized after gamma irradiation of S phase cells, in parallel with the above-noted change in pRb phosphorylation status. These data imply that E2F1 stabilization is related to the accumulation of un(der)phosphorylated pRb, a known E2F1 binding partner, capable of promoting its stabilization.
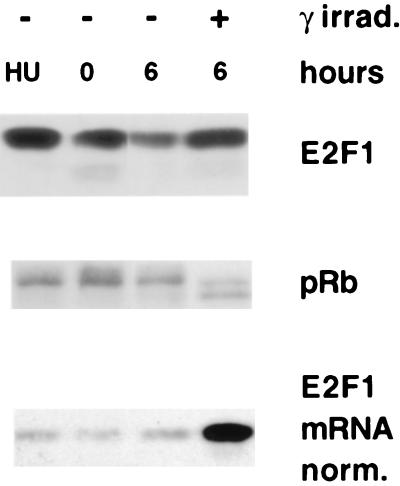
Figure 4.
pRb dephosphorylation and E2F1 stabilization after gamma irradiation of S phase cells. IMR90 cells were synchronized at the G1/S transition with hydroxyurea (HU), released, and, 2 hr later (0), irradiated with 2,000 Rad or left untreated. Six hours after irradiation, aliquots of irradiated and unirradiated cells were removed from their respective cultures. Lysates of these cells were analyzed for E2F1 abundance (Top) and pRb phosphorylation pattern (Middle) by Western blotting of lysate volumes containing equivalent amounts of total protein. For an indication of E2F1 stability (Bottom), E2F1 Western blotting was performed on volumes of these lysates that contained identical quantities of E2F1 mRNA.
E2F1 Stability During Myogenic Differentiation.
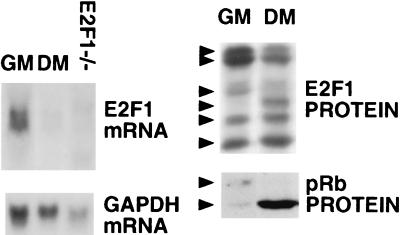
In C2/C12 myoblasts stimulated to differentiate into myotubes, pRb becomes permanently hypophosphorylated, and its expression is highly up-regulated (Fig. 5, refs. 42 and 54). These findings suggested that the myogenic differentiation system might be a relevant model in which to study the regulation of E2F1 stability by RB.
Figure 5.
E2F1 stability during myogenic differentiation. C2/C12 cells were grown as undifferentiated myoblasts in growth medium (GM) or, for 48 hr, in differentiation medium (DM). (Left) E2F1 Northern blot using equivalent amounts of total RNA derived from C2/C12 myoblasts and myotubes. RNA from E2F1−/− murine embryonic fibroblasts served as a negative control. (Right) E2F1 and pRb Western blots of identical quantities of cell extract protein derived from C2/C12 myoblasts (GM) and myotubes (DM), respectively. The arrows indicate differentially phosphorylated E2F species.
We observed that E2F1 is most likely stabilized on myogenic differentiation. In an exponentially growing population of asynchronous C2/C12 myoblasts, E2F1 mRNA was readily detectable. Conversely, after these cells formed myotubes, E2F1 RNA was nearly undetectable. By contrast, analysis of E2F1 protein levels in extracts of myoblasts and myotubes revealed no significant change (Fig. 5). These data, too, are consistent with a model in which E2F1 stability is elevated when there is a significant accumulation of hypophosphorylated pRb. A change in the E2F1 band pattern was noted during differentiation, likely reflecting a change in the phosphorylation state of the protein. This finding raises speculation regarding a possible link between E2F1 phosphorylation state and the state of differentiation of at least one cell type.
DISCUSSION
In this report, we have shown that endogenous E2F1 is an unstable protein, targeted for proteolysis by the ubiquitin proteasome system. In this regard, incubation of cells with a specific inhibitor of proteasome action led to E2F1 accumulation. Furthermore, in cells treated with a proteasome inhibitor, newly detectable E2F1-polyubiquitin conjugates appeared. Additional evidence strongly suggests that E2F1 stability is dynamically regulated during cell cycle progression, myogenic differentiation, and in response to S phase gamma irradiation.
E2F1 was more stable in G0/G1 than in S phase in unirradiated cells. Enhanced stability in G0/G1 was particularly striking in the case of differentiating myoblasts, where, on withdrawal from the cell cycle, no change in E2F1 protein level accompanied a dramatic reduction in E2F1 mRNA. Clearly there is not enough information available to understand the detailed molecular mechanisms regulating E2F1 stability. However, multiple lines of evidence imply a significant role for pocket proteins in this process.
First, we observed a positive correlation between pRb phosphorylation status and E2F1 stability. In all of the relevant experiments, when pRb existed in a largely hyphophosphorylated state, and was, thus, capable of binding E2F1, E2F1 stability increased. By contrast, in S phase, where pRb is hyperphosphorylated and cannot bind to E2F-1, E2F-1 stability decreased relative to that observed in G0/G1. Similar relationships were detected in myoblasts undergoing differentiation and in gamma-irradiated S phase fibroblasts where E2F1 stability increased as hypophosphorylated pRb accumulated.
Moreover, in cells synthesizing T, which disrupts the binding of E2F to pocket proteins and of E2F1 to pRb (4–6, 20, 21), E2F1 was equally unstable in G1 and S. By contrast, in cells synthesizing K1 T, a point mutant that is unable to bind pocket proteins but retains most other T functions (50, 51), E2F1 stability fell as cells emerged from G1 into S, much as in cells that lack a viral oncoprotein. Again, the simplest interpretation of these findings is that pocket protein/E2F1 binding enhances E2F1 stability.
Among the pocket proteins, pRb is believed to be the main in vivo partner of E2F1 (6). Hence, it was surprising that E2F1 stability was not deregulated in RB−/− cells as they exited G1. Given that one pocket protein can substitute for another as an E2F binding partner in G0/G1 (55), one might argue that at least one other pocket protein can substitute for pRb in promoting G1 stability of E2F1 in RB−/− cells.
All of these results fit well with earlier evidence indicating that overproduction of un(der)phsphorylated pRb enhances the stability of ectopically overproduced E2F1 (36–38). Taken together, they suggest a function of at least a subset of the pocket protein family, i.e., stabilizing E2F1 (and possibly other E2F species) in a timely, cell cycle-dependent manner. One implication of the proposed functional overlap among certain pocket protein species is that this activity is a biologically important one.
Little is known of the significance of E2F1 stability control, although two hypotheses can be proposed: E2F1 is both a potent oncoprotein and cell-killing element (8–14, 18, 19, 56). Therefore, one might imagine that efficient E2F1 degradation contributes to normal cellular homeostasis, at least in part, through a premature death and transformation avoidance mechanism. In this regard, E2F1 stabilization might contribute to induction of apoptosis after DNA damage. Indeed, mice lacking both E2F1 alleles, are defective in the induction of apoptosis (16, 18, 19). Moreover loss of E2F1 function decreased the frequency of pituitary and thyroid tumor development in mice lacking only one RB allele, suggesting that E2F1 acts as an oncogene in this setting (56).
In G1 cells, pRb binds to E2F1, and the ensuing transcription repression function of this complex appears to be translated, in part, into pRb-mediated growth suppression (22–24). Indeed, mice lacking both E2F1 alleles develop late-onset tumors (17). In this regard, one might imagine that a critical amount of pRb-E2F1/DP complex must be present for pRb to perform its G1 exit control function. In particular, pRb might stabilize E2F1 during a phase of the cell cycle in which E2F1 mRNA is scarce, allowing the protein to reach a threshold level where the abundance of pRb/E2F complexes is sufficient to exert effective proliferation control, a process that possibly contributes to the late-onset tumor suppression function of E2F-1 (17). At the same time, one might imagine that any E2F1 molecules that remain uncomplexed with pRb or another pocket protein are, likely, degraded, protecting cells from any unscheduled E2F1 transcription activity that could translate into apoptosis or the evolution of neoplastic behavior.
In RB−/−, but not wild type, cells blocked in G2/M by either p21 overexpression or gamma irradiation, Niculescu et al. (57) demonstrated the development of DNA endoreduplication. This finding suggests that functional RB is necessary to prevent the onset of a new S phase in the absence of an intervening mitosis. Although little is known of the nature of E2F target gene expression after S phase gamma irradiation, it is intriguing to speculate that newly formed and increasingly abundant E2F1/pRb complexes might down-regulate the transcription of a critical set of genes, contributing to the avoidance of a new round of DNA replication.
Acknowledgments
We thank Francesco Hoffman, Jim DeCaprio, Stefan Gaubatz, and our other divisional colleagues for their helpful suggestions and critical analysis. We are grateful to Drs. Peggy Farnham, R. R. Kopito, and J. A. DeCaprio for the gift of plasmids and to Dr. Y. S. Chao for providing us with lovastatin. This work was supported by funds provided to D.M.L. by the National Cancer Institute and to F.M. by the Leukemia Society of America.
ABBREVIATIONS
- LLnL
N-acetyl-l-leucinyl-l-leucinyl-N-norleucinal
- RB
retinoblastoma
- pRb
retinoblastoma protein
- T
simian virus 40 large T antigen
References
- 1.Kaelin W, Jr, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A, et al. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 2.Helin K, Lees J A, Vidal M, Dyson N, Harlow E, Fattaey A. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- 3.Shan B, Zhu X, Chen P L, Durfee T, Yang Y, Sharp D, Lee W H. Mol Cell Biol. 1992;12:5620–5631. doi: 10.1128/mcb.12.12.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 5.Farnham P J. Transcriptional Control of Cell Growth: The E2F Gene Family. Heidelberg: Springer; 1996. [Google Scholar]
- 6.Nevins J R. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 7.Lindeman G J, Dagnino L, Gaubatz S, Xu Y, Bronson R T, Warren H B, Livingston D M. Genes Dev. 1998;12:1092–1098. doi: 10.1101/gad.12.8.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson D G, Cress W D, Jakoi L, Nevins J R. Proc Natl Acad Sci USA. 1994;91:12823–12827. doi: 10.1073/pnas.91.26.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh P, Wong S H, Hong W. EMBO J. 1994;13:3329–3338. doi: 10.1002/j.1460-2075.1994.tb06635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu G, Livingston D M, Krek W. Proc Natl Acad Sci USA. 1995;92:1357–1361. doi: 10.1073/pnas.92.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson D G, Schwarz J K, Douglas Crees W, Nevins J R. Nature (London) 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 12.Qin X Q, Livingston D M, Kaelin W G, Jr, Adams P D. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan B, Lee W H. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X, Levine A J. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z M, Yang H, Livingston D M. Proc Natl Acad Sci USA. 1998;95:15583–15586. doi: 10.1073/pnas.95.26.15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Field S J, Tsai F Y, Kuo F, Zubiaga A M, Kaelin W, Jr, Livingston D M, Orkin S H, Greenberg M E. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 17.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson N J. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 18.Tsai K Y, Hu Y, Macleod K F, Crowley D, Yamasaki L, Jacks T. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- 19.Pan H, Yin C, Dyson N J, Harlow E, Yamasaki L, Dyke T V. Mol Cell. 1998;2:283–292. doi: 10.1016/s1097-2765(00)80273-7. [DOI] [PubMed] [Google Scholar]
- 20.Sellers W R, Kaelin W J. J Clin Oncol. 1997;15:3301–3312. doi: 10.1200/JCO.1997.15.11.3301. [DOI] [PubMed] [Google Scholar]
- 21.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 22.Weintraub S J, Prater C A, Dean D C. Nature (London) 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 23.Weintraub S J, Chow K N, Luo R X, Zhang S H, He S, Dean D C. Nature (London) 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 24.Sellers W R, Rodgers J W, Kaelin W., Jr Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Means A L, Slansky J E, McMahon S L, Knuth M W, Farnham P J. Mol Cell Biol. 1992;12:1054–1063. doi: 10.1128/mcb.12.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wade M, Kowalik T F, Mudryj M, Huang E S, Azizkhan J C. Mol Cell Biol. 1992;12:4364–4374. doi: 10.1128/mcb.12.10.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dynlacht B D, Flores O, Lees J A, Harlow E. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 28.Krek W, Ewen M E, Shirodkar S, Arany Z, Kaelin W G, Livingston D M. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 29.Krek W, Xu G, Livingston D M. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 30.Kitagawa M, Higashi H, Suzuki-Takahashi I, Segawa K, Hanks S K, Taya Y, Nishimura S, Okuyama A. Oncogene. 1995;10:229–236. [PubMed] [Google Scholar]
- 31.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slansky J E, Li Y, Kaelin W G, Farnham P J. Mol Cell Biol. 1993;13:1610–1618. doi: 10.1128/mcb.13.3.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuman E, Flemington E K, Sellers W R, Kaelin W J. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsiao K M, McMahon S L, Farnham P J. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 35.Johnson D G, Ohtani K, Nevins J R. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 36.Hofmann F, Martelli F, Livingston D M, Wang Z. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 37.Hateboer G, Kerkhoven R M, Shvarts A, Bernards R, Beijersbergen R L. Genes Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- 38.Campanero M R, Flemington E K. Proc Natl Acad Sci USA. 1997;94:2221–2226. doi: 10.1073/pnas.94.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zalvide J, DeCaprio J A. Mol Cell Biol. 1995;15:5800–5810. doi: 10.1128/mcb.15.10.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DuBridge R B, Tang P, Hsia H C, Leong P M, Miller J H, Calos M P. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martelli F, Cenciarelli C, Santarelli G, Polikar B, Felsani A, Caruso M. Oncogene. 1994;9:3579–3590. [PubMed] [Google Scholar]
- 43.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward C L, Omura S, Kopito R R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Slansky J E, Myers D J, Drinkwater N R, Kaelin W G, Farnham P J. Mol Cell Biol. 1994;14:1861–1869. doi: 10.1128/mcb.14.3.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harlow E, Crawford L V, Pim D C, Williamson N M. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 49.Hochstrasser M. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 50.Kalderon D, Smith A E. Virology. 1984;139:109–137. doi: 10.1016/0042-6822(84)90334-9. [DOI] [PubMed] [Google Scholar]
- 51.DeCaprio J A, Ludlow J W, Figge J, Shew J Y, Huang C M, Lee W H, Marsilio E, Paucha E, Livingston D M. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 52.Larner J M, Lee H, Hamlin J L. Cancer Surv. 1997;29:25–45. [PubMed] [Google Scholar]
- 53.Linke S P, Harris M P, Neugebauer S E, Clarkin K C, Shepard H M, Maneval D C, Wahl G M. Oncogene. 1997;15:337–345. doi: 10.1038/sj.onc.1201200. [DOI] [PubMed] [Google Scholar]
- 54.Gu W, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 55.Hurford R J, Cobrinik D, Lee M H, Dyson N. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 56.Yamasaki L, Bronson R, Williams B O, Dyson N J, Harlow E, Jacks T. Nat Genet. 1998;18:360–364. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- 57.Niculescu A R, Chen X, Smeets M, Hengst L, Prives C, Reed S I. Mol Cell Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]