Abstract
A 65-bp “core” sequence is dispersed in hundreds of thousands copies in the human genome. This sequence was found to constitute the central segment of a group of short interspersed elements (SINEs), referred to as mammalian-wide interspersed repeats, that proliferated before the radiation of placental mammals. Here, we propose that the core identifies an ancient tRNA-like SINE element, which survived in different lineages such as mammals, reptiles, birds, and fish, as well as mollusks, presumably for >550 million years. This element gave rise to a number of sequence families (CORE-SINEs), including mammalian-wide interspersed repeats, whose distinct 3′ ends are shared with different families of long interspersed elements (LINEs). The evolutionary success of the generic CORE-SINE element can be related to the recruitment of the internal promoter from highly transcribed host RNA as well as to its capacity to adapt to changing retropositional opportunities by sequence exchange with actively amplifying LINEs. It reinforces the notion that the very existence of SINEs depends on the cohabitation with both LINEs and the host genome.
Almost 30% of the human genome consists of copies of interspersed repeats that are amplified by retroposition (1), a process widely spread among eukaryotic taxa (2). Retroposition involves reverse transcription of the transcribed copies and reintegration of the resulting cDNAs into the host genome. In this way, numerous copies of the propagating elements accumulated in host genomes over extended evolutionary periods. These copies eventually decayed by mutations and, because these were random (3), we can reconstruct the once-active elements from the alignment of their genomic fossil record elements as the average or consensus sequence. In other words, ancestral sequences that do not exist in an active form any more are inferred from bits and pieces of their mutated copies still found in the genome. This so-called paleogenomic approach has been used to reconstruct the complex evolution of Alu subfamilies (4, 5). Although the consensus is often only an approximation, it is the best available representation of the ancestrally active elements.
Retroposition requires specific activities in addition to the enzymatic machinery commonly found in the host cells. The reverse transcriptase (6, 7) as well as the endonuclease (8–10) involved in cDNA synthesis and the integration, respectively, are coded for by the actively retroposing long elements such as long interspersed elements (LINEs). In contrast, short interspersed elements (SINEs) do not encode any protein facilitating their proliferation. However, considering thousands of SINE copies dispersed in vertebrate’s genomes, these elements must have evolved other adaptations to secure their efficient amplification, using both host-specific and retroposition-specific activities provided in trans.
What are the structural attributes of efficiently amplifying SINEs? Recruitment of the internal promoter of RNA polymerase III (Pol III) from abundantly transcribed cellular RNAs such as 7SL or transfer RNAs seems to be one such adaptation. The use of Pol III promoter assures easy access to the transcription machinery of the host. The fact that it is carried within the transcript is convenient and advantageous for a mobile element that might need to be expressed from changing genomic locations (11). In 7SL RNA-derived retroposons, such as primate Alu and rodent B1 elements, their proliferative efficiency correlates well with Pol III promoter and with the presence of the conserved RNA secondary structure, still resembling that of their 7SL RNA from which these elements originated (12, 13). It is the folding of these SINEs RNA and the resulting interactions that are believed to facilitate their access to retropositional machinery (12, 14, 15). Such retroposition promoting mechanism, though still hypothetical, cannot be generalized. In the majority of SINEs spread in extant eukaryotic taxa, including plants and invertebrates, transfer RNAs were at the origin of their 5′ Pol III promoter region (16–18). However, the similarity to tRNA in these elements remains often limited to boxes A and B of the split Pol III promoter only, because the original base pair interactions stabilizing tRNA cloverleaf structure were lost (16). Earlier works suggested that SINEs evolved as “satellites” of LINEs by using their retropositional machinery (14, 15). Okada and colleagues proposed (19) that LINEs were at the origin of tRNA-like SINEs through a mechanism assembling tRNA acting as a primer with the LINE template (typically its 3′ end segment involved in the initiation of cDNA synthesis). This hypothesis suggested that different tRNA-like SINEs originated independently from diverse LINEs. Here, we show that rather than by numerous de novo origins, a variety of eukaryotic SINEs can be ancestrally related to an ancient tRNA-like element. We find that this generic element, whose conserved moiety included the promoter and the central region, had the ability to diversify by recruiting its 3′ end from LINE elements, capturing by the same token sequence segments able to facilitate its retroposition.
MATERIALS AND METHODS
Sequence Data.
Consensus sequences of CORE-SINEs from different mammalian species (see text) were obtained from the alignment of genomic elements sequenced in our laboratory (n = 107) and were extracted from the GenBank release 105.0 (n = 43). Details of this study are available elsewhere (N.G. and D.L., unpublished work). CORE-SINEs sequences of nonmammalian species were obtained through GenBank searches and from published literature (see figures for the list of these sequences). Sequence were aligned by using multalin 4.0 (20), and the alignments were refined manually. Sequence identity was calculated from the alignment as follows:
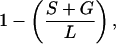 |
where S denotes the number of nucleotide substitutions, G (gaps) denotes the number of insertions/deletions (counted once irrespective the size), and L denotes the length of the compared Ther-1 reference sequence segment (gaps introduced into the reference sequence are not added to L to keep the estimation conservative). Dot matrix comparison was done by using mac dnasis pro v3.5 (Hitachi, Tokyo), with 9 matches required in a window of 15 nucleotide positions (except for AvaIII:HpaI comparison, where 10 matches were required).
GenBank Searches.
Nuclear DNA sequence databases were created based on the GenBank release 105.0 for reptile, birds, and fish by using the lookup program from Wisconsin Package version 9.0 (Genetic Computer Group, Madison, WI). By using fasta from the Genetic Computer Group, with default parameters and the conserved or the variable regions of CORE-SINE consensus sequences (see Fig. 1), these databases were examined for the presence of the corresponding sequences.
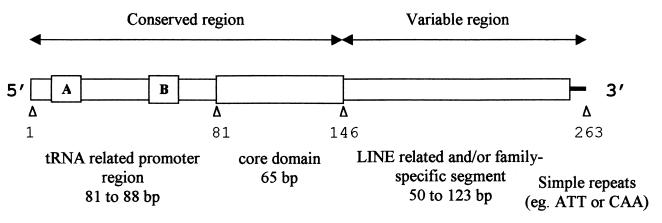
Figure 1.
Schematic structure of MIR (CORE-SINE) elements based on Ther-1 family consensus. The tRNA-related region and the central core are common to all sequence families, distinguished by their distinct 3′ terminal segments. Boxes A and B represent elements of the split Pol III RNA promoter.
RESULTS AND DISCUSSION
We analyzed a class of tRNA-like retroposons, which, after Alu repeats, constitute the most prominent SINEs of the human genome. However, in contrast to Alu, these elements, which were called mammalian-wide interspersed repeats (MIRs) [see Repbase 1992 (http://www.girinst.org)] can be found in all mammalian orders (21, 22). Their tRNA-like Pol III promoter region is followed by a central sequence domain of 65 bp, named “core” (23), and a variable 3′ sequence segment (Fig. 1). Recently, we sequenced MIR elements from monotremes and marsupials, representing nonplacental mammals. By including these elements (n = 107) in the analysis, we reconstructed consensus sequences defining five MIR families (N.G. and D.L., unpublished work). Each family refers to a group of genomic copies sharing the same 3′ segment. These were named Ther-1 and Ther-2 to indicate their initial identification in therians (i.e., marsupials and placentals), Mon-1 in monotremes, Mar-1 in marsupials, and Opo-1 in the opossum genome. Ther-1 represents a modified consensus of the MIR sequences previously described in the human genome (22). In general, the within family divergence—i.e., between the consensus and the contributing repeats of up to >30% (see also refs. 21 and 22)—was greater than that between consensus sequences of the families, excluding their 3′ end nonhomologous segments.
The 3′ segments in four of the five MIR families were found homologous (based on > 80% sequence identity) to different vertebrate LINEs (Table 1). As shown in Fig. 2a, Ther-1 and Mon-1 shared ≈50 bp with the 3′ end of the L2 family of ancient mammalian, and presumably pan-vertebrate, LINE repeats (http://www.girinst.org); Mon-1 elements differed from Ther-1 in the proximal part of their 3′ terminal portion (Fig. 2a). The identity between MIR (Ther-1) and L2 was already recognized earlier (1, 22). Ther-2 sequences (Fig. 2b), in turn, shared their 3′ segments with the turtle PsCR1 elements (19), representing a trans-specific LINE family originally described in chicken (as “CR1”) and reptiles (24–26). Finally, we found a high degree of sequence identity (Fig. 2c) between the 3′ segments of Mar-1 elements and the 5′ and 3′ portion of Bov-B sequences (also known as Art2 or BDDF), again trans-specific, apparently pan-vertebrate LINEs described so far in artiodactyls and snakes (27–29). Our analysis shows that the portion of MIR sequence shared with LINEs was derived from these latter elements, adding to similar examples of LINEs/SINEs sequence sharing reported earlier (19, 26) (Table 1). The retropositional efficiency of tRNA-like SINEs could be explained by their structural identity with 3′ end segments of actively retroposing LINE elements. This molecular mimicry of SINEs can be considered as an adaptive response to changing retropositional opportunities over time and in different lineages and can also reflect the need of using the reverse transcriptase activity in trans.
Table 1.
Identity among sequence segments between LINEs and SINEs
| SINE name | SINE ref. | LINE name | LINE ref(s). | Shared segment, bp | Identity, % | SINE/LINE relationship ref(s). |
|---|---|---|---|---|---|---|
| Bov-tA and Bov-A2 | 40 | Bov-B or BDDF | 28, 40 | 111 | 75–89 | 1, 41 |
| Pol III/SINE | 42 | CR1-like | 43–45 | 100 | 65–70 | 19, 26 |
| HpaI | 30 | RSg-1 | 46 | 57 | 77 | 19 |
| Tobacco SINE | 47 | Solanaceous LINE | 19 | 105 | 77 | 19 |
| SmaI | 48 | Eel LINE-like | 19 | 40 | 94 | 19 |
| MIR general consensus | 22 | L2 | http://www.girinst.org | 52 | 90 | 1 |
| Ther-1 | This study | L2 | http://www.girinst.org | 52 | 86.5 | This study |
| Mon-1 | L2 | 56 | 82 | |||
| Ther-2 | PsCr1 | 45 | 53 | 83 | ||
| Mar-1 | Bov-B or BDDF | 28 | 95 | 80 |
Figure 2.
Sequences shared between MIRs (CORE-SINEs) and LINEs. (a) 3′ ends of human Ther-1 and Mon-1 families compared with the 3′ end of L2 LINE (http://www.girinst.org). (b) 3′ terminal portion of wallaby Ther-2 compared with the 3′ end of PsCR1 LINE (19). (c) Mar-1 3′ end compared with the 5′/3′ end sequences of Bov-B LINE (28), where both 5′ (left) and 3′ ends (right) of the LINE contributed to Mar-1 3′ end. Sequence segments distinguishing Ther-1 and Mon-1 families are in italic. Simple sequence repeats at the 3′ extremities are underlined. Symbols M and R denotes A or C and C or G, respectively. Identical nucleotides to the sequence shown on the top are represented by dots, whereas gaps are represented by dashes.
The constant feature of MIR sequence is the 5′ tRNA-like promoter region followed by its particular central core segment. The core, first considered a hallmark of MIRs (21), turns out to identify a more widely spread class of short retroposons found beyond mammalian genomes as well. fasta searches and inter-MIR PCR amplification experiments (21) documented the presence of the truncated Ther-1 elements in birds and reptiles, suggesting that these elements proliferated in nonmammalian vertebrates as well (Table 2). Taking into account that ≈0.33% of the bird genome is represented in GenBank release 105.0, we obtained the genomic copy number of these SINEs of the order of 5,000.
Table 2.
CORE-SINEs in nonmammalian genomes
| Genomes | Sequence (ref.) | Length | Similarity with Ther-1* (segments included) | Identity,† % | |
|---|---|---|---|---|---|
| Invertebrate | Octopodidae | OR2 central segment (35) | 65 | 81–145 (core) | 63.1 |
| OR1 central segment (35) | 44 | 81–124 (core) | 50.0 | ||
| Fish | Salmonidae | HpaI central segment (30) | 65 | 81–145 (core) | 64.6 |
| AvaIII central segment (33) | 44 | 82–124 (core) | 68.2 | ||
| Cichlidae | AFC central segment (31) | 42 | 81–122 (core) | 73.8 | |
| Fundulus heteroclitus | U59855 | 200 | 8–207 (all) | 61.5 | |
| Birds | Gallus gallus | J02839 | 71 | 118–188 (core + variable region) | 77.5 |
| D82080 | 104 | 77–179 (core + variable region) | 66.3 | ||
| U46503 | 89 | 65–153 (tRNA region + core) | 67.4 | ||
| L10232 | 80 | 73–150 (tRNA region + core) | 68.7 | ||
| X83246 | 86 | 148–231 (variable region) | 60.4 | ||
| D10737 | 67 | 97–163 (core + variable region) | 61.2 | ||
| X69491 | 132 | 21–148 (tRNA region + core) | 56.1 | ||
| U25125 | 92 | 11–98 (tRNA region + core) | 63.0 | ||
| L13208 | 125 | 96–209 (core + variable region) | 64.8 | ||
| L39766 | 72 | 178–244 (variable region) | 72.2 | ||
| Y14342 | 126 | 44–165 (all) | 62.7 | ||
| M88072 | 61 | 158–216 (variable region) | 68.9 | ||
| U49693 | 66 | 99–158 (core + variable region) | 71.2 | ||
| Eopsaltria australis | U40495 | 120 | 24–140 (tRNA region + core) | 61.7 | |
| Anas platyrhynchos | X68810 | 164 | 94–253 (core + variable region) | 59.8 | |
| Columba livia | M36969 | 149 | 44–190 (all) | 60.4 | |
| Reptile | Anolis carolinensis | L31503 | 115 | 8–124 (tRNA region + core) | 72.3 |
Numbering used is that of Ther-1 consensus (Fig. 1).
Reported sequence identities were calculated as described in Materials and Methods.
Additional searches of databanks, using the conserved sequence region of Ther-1 family (positions 1–145; Fig. 1), identified a number of related sequences. In Fig. 3a, similarity between these sequences and the core can be appreciated in the alignment with the corresponding segment of Ther-1 element. In addition, pairwise comparisons using the dot matrix approach, which are shown in Fig. 3b, illustrate that the degree of sequence identity observed between the central core segments of different elements is not inferior to that between their tRNA-like portions upstream or their 3′ variable segments downstream if the latter is the case [e.g., Ther-1:AFC 3′end identity reported by Terai et al. (32)]. This suggests that selection constraints concerning the core segment over the evolution are not weaker than those maintaining the integrity of the RNA Pol III promoter and that both of these segments are similarly vital for the element survival. We found HpaI SINEs from Salmonidae (30) and AFC SINEs from Cichlidae (31) to be related to MIR elements (Table 2 and Fig. 3). As in MIRs, the core segment in HpaI is preceded by a tRNA-like promoter region and is followed by a LINE-related 3′ portion (19). The same is true for AFC element (32). The AvaIII repeat from Salmonidae (33) also should be related to MIR based on its sequence similarity to the HpaI family (Table 2 and Fig. 3). By fasta searches, we also found a putative related element of so far uncharacterized SINE family from a teleost fish, Fundulus heteroclitus. This element, located in the 5′ flanking region of Ldh-B gene (34), was identified by the DNA segment with similarity to core and by the presence of two intact boxes of the RNA Pol III promoter in tRNA-like portion located upstream (Fig. 3b and Fig. 4). Moreover, its 3′ end exhibits some sequence identity with Ther-1-specific segment (immediately downstream of the core) defined in Fig. 2. More importantly, this element is not more divergent from the Ther-1 consensus than Ther-1 repeats from placental genomes. Finally, the analysis of published sequences of tRNA-like SINEs from invertebrates revealed substantial identity between core and the central portion of OR2 SINE (35) from DNA of octopus (Cephalopoda) as shown in Fig. 3 (Table 2). A related OR1 SINE from the same species (35) was included in the comparison principally because of its similarity with OR2 element (Fig. 3 and Table 2). Cephalopods that belong to Mollusca and vertebrate lineages diverged ≈550 million years ago whereas fish lines diverged 400 million years ago, reptile and bird lines diverged 300 million years ago, and mammal lines diverged 250 million years ago (36).
Figure 3.
(a) Sequence identity between the core domain of Ther-1 family and non-tRNA segments of HpaI and AvaIII SINEs from Salmonidae, AFC SINE from Cichlidae, and OR2 SINE from Octopodidae (30, 31, 33, 35). OR1 SINE from Octopodidae was included in the alignment because of its identity with OR2. Nucleotide numbering corresponds to that of Ther-1 consensus (see Fig. 1). See also legend to Fig. 2. (b) Dot matrix comparison. SINE names or U59855, representing the genomic element from F. heteroclitus we identified here, are indicated on the top or on the left of the squares. Thin lines indicate sequence segments corresponding to core. In the OR1:OR2 comparison, the last 120 nucleotides of OR1 consensus were discarded.
Figure 4.
Sequence identity between Ther-1 consensus and part of the 5′ untranslated region of F. heteroclitus Ldh-B gene (GenBank accession no. U59855). Putative A and B boxes of the RNA Pol III promoter are boxed, and the general consensus is indicated on the top. The thicker box signal core segment and the specific Ther-1 segment is underlined. See also legend to Fig. 2.
Taken together, all of these data point to the existence of a generic SINE element that we propose to call CORE-SINE. This element equipped with Pol III promoter region followed by the core segment is capable of exchanging its 3′ end. Remarkably, the central core segment was maintained over long evolutionary time periods in SINEs proliferating in extant eukaryotic genomes. In the five MIR families discussed here, the core conservation was in average almost twice greater than the conservation of their corresponding 5′ regions (N.G. and D.L., unpublished work). Lowest conservation of the tRNA related segment could be explained by the selection acting primarily on the A and B boxes of the split RNA Pol III promoter or by different origin of tRNAs contributing this region to the evolving SINEs. The integrity of t-RNA cloverleaf secondary structure seems unnecessary for the transcription and for the retroposition. In contrast, the core sequence appears to have been maintained because of a more stringent selection that would be related to its possible importance for the integrity of the element (either by contributing directly to its proliferating activity or by promoting its long-term survival). In general, serving as an “assembly unit” of potentially functional segments, the core could help to reconstruct promiscuous SINEs from the pool of genomic copies that are continuously modified by mutations. Facilitating the exchange of 3′ ends with actively retroposing LINEs would be of particular importance because mimicking the active LINEs appears vital for tRNA-like SINEs surviving through amplification (37). LINEs evolve in time and in different lineages; with successive LINE families observed in eukaryotic lineages, one would predict the presence of SINEs with homologous 3′ ends in the corresponding genomes, and vice versa. Indeed, the presence of Mar-1 family with 3′ end similarity to Bov-B LINE implies the presence of the latter in the marsupial genome, which we confirmed experimentally (N.G. and D.L., unpublished data). In light of the data on a variety of tRNA-like SINEs (ref. 38 and this paper), this mechanism seems to generally apply to this class of elements. Here again, the notorious human Alu as well as rodent B1 elements, which represent a novel 7SL RNA rather than the tRNA connection, should be considered as exception. These SINEs do not share its 3′ end with L1 and presumably any other LINE in the primate genome. Similarity of poly-A tails shared between Alu, rodent B1, and L1 elements is nonspecific given the presence of poly-A ends in the majority of host mRNAs. Thus, the specific liaison between LINEs and Alu might be of different nature and could be related to particular conservation of the Alu RNA folding as mentioned above (14, 39).
In this study, we have documented the structural and presumably evolutionary continuity among SINEs in different vertebrate genomes, and we have found examples showing that this continuity extends beyond the vertebrate phyla. tRNA-like SINEs are found in all eukaryotic kingdoms and can be almost as old as the actively retroposing LINEs. CORE-SINEs are structural mosaics assembling distinct functional domains necessary for retroposition. The evolutionary persistence of CORE-SINEs can be related to the conservation of the central core segment. CORE-SINEs appear to have survived for more than hundreds of millions of years in different eukaryote lineages, given the fact that their recent amplification is documented in fish and in nonplacental mammals.
Acknowledgments
We thank Arian Smit for sharing the L2 sequences, Pierre Chartrand, Jean-Marc Deragon, François-Joseph Lapointe, Daniel Sinnett, and Ewa Zietkiewicz for their comments and discussions, Jennifer M. A. Graves, Chris Collet, and Clément Lanthier for DNA samples, and Raffaela Ballarano for excellent secretarial assistance. N.G. had a studentship from the Fondation de l’Hôpital Sainte-Justine. This investigation was supported by grant from the Medical Research Council of Canada.
ABBREVIATIONS
- LINE
long interspersed element
- SINE
short interspersed element
- MIR
mammalian-wide interspersed repeats
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Smit A F. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 2.Xiong Y, Eickbush T H. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Labuda D, Striker G. Nucleic Acids Res. 1989;17:2477–2491. doi: 10.1093/nar/17.7.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jurka J. In: Origin and Evolution of Alu Repetitive Elements. Maraia R J, editor. Austin, TX: R. G. Landes; 1995. pp. 25–41. [Google Scholar]
- 5.Zietkiewicz E, Richer C, Sinnett D, Labuda D. J Mol Evol. 1998;47:172–182. doi: 10.1007/pl00006374. [DOI] [PubMed] [Google Scholar]
- 6.Deragon J M, Sinnett D, Labuda D. EMBO J. 1990;9:3363–3368. doi: 10.1002/j.1460-2075.1990.tb07537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathias S L, Scott A F, Kazazian H H J, Boeke J D, Gabriel A. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 8.Feng Q, Moran J V, Kazazian H H J, Boeke J D. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 9.Jurka J. Proc Natl Acad Sci USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tatout C, Lavie L, Deragon J M. J Mol Evol. 1998;47:463–470. doi: 10.1007/pl00006403. [DOI] [PubMed] [Google Scholar]
- 11.Van Arsdell S W, Denison R A, Bernstein L B, Weiner A M, Manser T, Gesteland R F. Cell. 1981;26:11–17. doi: 10.1016/0092-8674(81)90028-3. [DOI] [PubMed] [Google Scholar]
- 12.Labuda D, Sinnett D, Richer C, Deragon J M, Striker G. J Mol Evol. 1991;32:405–414. doi: 10.1007/BF02101280. [DOI] [PubMed] [Google Scholar]
- 13.Labuda D, Zietkiewicz E. J Mol Evol. 1994;39:506–518. doi: 10.1007/BF00173420. [DOI] [PubMed] [Google Scholar]
- 14.Sinnett D, Richer C, Deragon J M, Labuda D. J Mol Biol. 1992;226:689–706. doi: 10.1016/0022-2836(92)90626-u. [DOI] [PubMed] [Google Scholar]
- 15.Boeke J D. Nat Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- 16.Okada N, Ohshima K. In: Evolution of tRNA-Derived SINEs. Maraia R J, editor. Vol. 4. Austin, TX: R. G. Landes; 1995. pp. 61–79. [Google Scholar]
- 17.Deragon J M, Landry B S, Pelissier T, Tutois S, Tourmente S, Picard G. J Mol Evol. 1994;39:378–386. doi: 10.1007/BF00160270. [DOI] [PubMed] [Google Scholar]
- 18.Bradfield J Y, Locke J, Wyatt G R. DNA. 1985;4:357–363. doi: 10.1089/dna.1985.4.357. [DOI] [PubMed] [Google Scholar]
- 19.Ohshima K, Hamada M, Terai Y, Okada N. Mol Cell Biol. 1996;16:3756–3764. doi: 10.1128/mcb.16.7.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurka J, Zietkiewicz E, Labuda D. Nucleic Acids Res. 1995;23:170–175. doi: 10.1093/nar/23.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smit A F, Riggs A D. Nucleic Acids Res. 1995;23:98–102. doi: 10.1093/nar/23.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donehower L A, Slagle B L, Wilde M, Darlington G, Butel J S. Nucleic Acids Res. 1989;17:699–710. doi: 10.1093/nar/17.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stumph W E, Kristo P, Tsai M J, O’Malley B W. Nucleic Acids Res. 1981;9:5383–5397. doi: 10.1093/nar/9.20.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva R, Burch J B. Mol Cell Biol. 1989;9:3563–3566. doi: 10.1128/mcb.9.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandergon T L, Reitman M. Mol Biol Evol. 1994;11:886–898. doi: 10.1093/oxfordjournals.molbev.a040171. [DOI] [PubMed] [Google Scholar]
- 27.Duncan C H. Nucleic Acids Res. 1987;15:1340. doi: 10.1093/nar/15.3.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szemraj J, Plucienniczak G, Jaworski J, Plucienniczak A. Gene. 1995;152:261–264. doi: 10.1016/0378-1119(94)00709-2. [DOI] [PubMed] [Google Scholar]
- 29.Kordis D, Gubensek F. Eur J Biochem. 1997;246:772–779. doi: 10.1111/j.1432-1033.1997.00772.x. [DOI] [PubMed] [Google Scholar]
- 30.Kido Y, Aono M, Yamaki T, Matsumoto K, Murata S, Saneyoshi M, Okada N. Proc Natl Acad Sci USA. 1991;88:2326–2330. doi: 10.1073/pnas.88.6.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K, Terai Y, Nishida M, Okada N. Mol Biol Evol. 1998;15:391–407. doi: 10.1093/oxfordjournals.molbev.a025936. [DOI] [PubMed] [Google Scholar]
- 32.Terai Y, Takahashi K, Okada N. Mol Biol Evol. 1998;15:1460–1471. doi: 10.1093/oxfordjournals.molbev.a025873. [DOI] [PubMed] [Google Scholar]
- 33.Kido Y, Himberg M, Takasaki N, Okada N. J Mol Biol. 1994;241:633–644. doi: 10.1006/jmbi.1994.1540. [DOI] [PubMed] [Google Scholar]
- 34.Schulte P M, Gomez-Chiarri M, Powers D A. Genetics. 1997;145:759–769. doi: 10.1093/genetics/145.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohshima K, Okada N. J Mol Biol. 1994;243:25–37. doi: 10.1006/jmbi.1994.1627. [DOI] [PubMed] [Google Scholar]
- 36.Futuyma D J. Evolutionary Biology. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 37.Zuckerkandl E, Latter G, Jurka J. J Mol Evol. 1989;29:504–512. doi: 10.1007/BF02602922. [DOI] [PubMed] [Google Scholar]
- 38.Okada N, Hamada M, Ogiwara I, Ohshima K. Gene. 1997;205:229–243. doi: 10.1016/s0378-1119(97)00409-5. [DOI] [PubMed] [Google Scholar]
- 39.Sinnett D, Richer C, Deragon J M, Labuda D. J Biol Chem. 1991;266:8675–8678. [PubMed] [Google Scholar]
- 40.Lenstra J A, van Boxtel J A F, Zwaagstra K A, Schwerin M. Anim Genet. 1993;24:33–39. doi: 10.1111/j.1365-2052.1993.tb00916.x. [DOI] [PubMed] [Google Scholar]
- 41.Okada, N. & Hamada, M. (1997) J. Mol. Evol.44, Suppl. 1, S52–S56. [DOI] [PubMed]
- 42.Endoh H, Nagahashi S, Okada N. Eur J Biochem. 1990;189:25–31. doi: 10.1111/j.1432-1033.1990.tb15455.x. [DOI] [PubMed] [Google Scholar]
- 43.Burch J B, Davis D L, Haas N B. Proc Natl Acad Sci USA. 1993;90:8199–8203. doi: 10.1073/pnas.90.17.8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haas N B, Grabowski J M, Sivitz A B, Burch J B. Gene. 1997;197:305–309. doi: 10.1016/s0378-1119(97)00276-x. [DOI] [PubMed] [Google Scholar]
- 45.Kajikawa M, Ohshima K, Okada N. Mol Biol Evol. 1997;14:1206–1217. doi: 10.1093/oxfordjournals.molbev.a025730. [DOI] [PubMed] [Google Scholar]
- 46.Winkfein R J, Moir R D, Krawetz S A, Blanco J, States J C, Dixon G H. Eur J Biochem. 1988;176:255–264. doi: 10.1111/j.1432-1033.1988.tb14276.x. [DOI] [PubMed] [Google Scholar]
- 47.Yoshioka Y, Matsumoto S, Kojima S, Ohshima K, Okada N, Machida Y. Proc Natl Acad Sci USA. 1993;90:6562–6566. doi: 10.1073/pnas.90.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsumoto K, Murakami K, Okada N. Proc Natl Acad Sci USA. 1986;83:3156–3160. doi: 10.1073/pnas.83.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]