Abstract
Site-specific proteases play critical roles in regulating many cellular processes. To identify novel site-specific proteases, their regulators, and substrates, we have designed a general reporter system in Saccharomyces cerevisiae in which a transcription factor is linked to the intracellular domain of a transmembrane protein by protease cleavage sites. Here, we explore the efficacy of this approach by using caspases, a family of aspartate-specific cysteine proteases, as a model. Introduction of an active caspase into cells that express a caspase-cleavable reporter results in the release of the transcription factor from the membrane and subsequent activation of a nuclear reporter. We show that known caspases activate the reporter, that an activator of caspase activity stimulates reporter activation in the presence of an otherwise inactive caspase, and that caspase inhibitors suppress caspase-dependent reporter activity. We also find that, although low or moderate levels of active caspase expression do not compromise yeast cell growth, higher level expression leads to lethality. We have exploited this observation to isolate clones from a Drosophila embryo cDNA library that block DCP-1 caspase-dependent yeast cell death. Among these clones, we identified the known cell death inhibitor DIAP1. We showed, by using bacterially synthesized proteins, that glutathione S-transferase–DIAP1 directly inhibits DCP-1 caspase activity but that it had minimal effect on the activity of a predomainless version of a second Drosophila caspase, drICE.
Site-specific proteolysis plays a critical role in regulating a number of cellular processes. An important class of site-specific proteases are a group of cysteine proteases known as caspases (1). Extensive genetic and biochemical evidence indicates that caspases play roles as cell death signaling and effector molecules in a number of different contexts, thus making them attractive potential therapeutic targets. Caspases identified to date have been found primarily based on homology to the Caenorhabditis elegans caspase CED-3 and mammalian caspase 1 and through biochemical purification (reviewed in refs. 2–5). Viral and cellular activators and inhibitors of caspase function also have been identified in genetic and biochemical screens for regulators of apoptosis (reviewed in refs. 6–8). These approaches to isolating caspases and their regulators are limited by the fact that some proteases that cleave a caspase target site and their regulators may not share primary sequence homology with the proteins identified to date or they may be expressed only in specific tissues with limited availability for biochemical purification. Furthermore, it is clear that caspases regulate processes other than cell death, including cytokine secretion in mammals (9–13) and cell proliferation and oogenesis in Drosophila (14, 15). It seems likely, given the early stage of the field, that more roles exist. Caspases and caspase regulators involved in these processes may be missed in screens that focus strictly on cell death-related phenotypes. Thus, molecules that possess caspase or caspase regulatory activity may not have been identified yet. As an alternative approach to identifying novel caspases or caspase regulators, it would be useful to have assays for caspase function that are based strictly on protease activity.
Because of the importance of site-specific proteolysis, we sought to develop a versatile system that would allow the identification of novel site-specific proteases, regulators of the activity of known site-specific proteases, or their substrates. Because caspase cleavage sites have been well defined, and activators and inhibitors of caspases have been identified, we set out to establish a prototype system that would allow positive selection for caspase-like proteases, their activators, and their inhibitors. Our approach to identifying these molecules uses reporters for caspase activity that function in living cells. Yeast, though eukaryotic, lacks many of the specialized proteolytic systems found in cells of higher eukaryotes. Thus, it constitutes an ideal background in which to carry out function-based screens for these proteases, their regulators, and their targets. Reporters for the activity of specific proteases in bacteria and eukaryotes have been developed by using several strategies that involve cleavage-dependent alterations in the activity of specific proteins (16–21). To visualize caspase activity, we created a fusion protein in which a transcription factor is linked to the intracellular domain of a transmembrane protein by caspase cleavage sites. Expression of this protein in yeast, in the presence of an active caspase, should result in release of the transcription factor from the membrane, followed by transcriptional activation of a reporter. As described below, using such a reporter system, we can visualize caspase activity in yeast and can identify proteins that act as caspase activators and inhibitors. Caspase inhibitors also can be identified by virtue of their ability to suppress caspase overexpression-dependent yeast cell death.
MATERIALS AND METHODS
Constructs
Yeast Strains.
The W303α strain (MATα, can1–100, leu2–3,-112, his3–11,-15, trp1–1, ura3–1, ade2–1) was used to monitor caspase activity by using the lacZ reporter system. EGY48 (MATa, ura3, trp1, his3, LexAop6-LEU2) (Invitrogen) was used to monitor caspase-dependent cell killing.
Construction of Caspase Target Site Fusion Proteins.
The reporter, CLBDG6, was generated by using PCR and standard techniques (details provided on request). This protein consists of, from N to C termini, amino acids 1–401 of a type 1 transmembrane protein, human CD4 (22), a linker consisting of six tetrapeptide caspase target sites that bracket the specificity of known caspases and granzyme B (23)—DEVDG-WEHDG-IEHDG-IETDG-DEHDG-DQMDG—, each of which is followed by a glycine residue, which acts as a stabilizing residue in the N-end rule degradation pathway in yeast (reviewed in refs. 24), and finally, a transcription factor containing the LexA DNA binding domain (25). A second construct, designated CLBGG6, was generated that encodes a protein identical to CLBDG6 except that the essential P1 aspartates of the six caspase cleavage sites are replaced with glycines, rendering them nonfunctional.
Construction of Yeast Expression Plasmids.
Plasmids for expression of genes in yeast were derived from the pRS series (26). To express genes in yeast under galactose-inducible control, we used two GAL1 promoter fragments: a long version, extending from base 1–815, called GALL, and a shorter and somewhat weaker version, extending from base 406–815, called GALS (27). The yeast actin terminator, bases 2,107–2,490 (GenBank accession no. L00026), was used for all constructs. GALL promoter and actin terminator fragments were inserted into pRS313 (HIS3), pRS314 (TRP1), and pRS315 (LEU2), generating pGALL-(HIS3), pGALL-(TRP1), and pGALL-(LEU2). CLBDG6 and CLBGG6 coding regions were cloned into pGALL-(TRP1). GALS promoter and actin terminator fragments were inserted into pRS315, generating pGALS-(LEU2). To express genes under the control of the copper-inducible CUP1 promoter, we used a promoter fragment extending from bases 1,079–1,533 of the CUP1 locus (GenBank accession no. K02204). Site-directed mutagenesis was used to mutate the CUP1 promoter to prevent activation in response to glucose starvation (28). The mutated CUP1 promoter and actin terminator fragments were inserted into pRS315 (LEU2), generating pCUP1-(LEU2).
The coding region for the C. elegans caspase CED-3 (29) was introduced into pCUP1-(LEU2). Site-directed mutagenesis was used to generate an inactive version of CED-3 in which the active site cysteine was changed to serine (CED-3CS). Full length human caspase 7 (caspase 7FL) (30), caspase 7 lacking the N-terminal 53-aa prodomain (caspase 753), and the caspase 8 isoform corresponding to MACHα2 (31) or Mch5 (32) (caspase 8FL) were introduced into pCUP1-(LEU2). Full length DCP1 (14) was introduced into pGALS-(LEU2) and pGALL-(LEU2). Site-directed mutagenesis was used to change the active site cysteine to serine (DCP-1CS). The resulting full length coding region was introduced into pGALL-(LEU2). Full length drICE (33) was inserted into pGALL-(LEU2). Full length human caspase 9 (34, 35) was introduced into pGALL-(LEU2). The region encoding amino acids 1–530 of Apaf-1 (36) (Apaf-1530) was introduced into pGALL-(HIS3). Full length DIAP1 (FLDIAP1) and a version of DIAP1 C-terminally truncated following residue 381 (DIAP1BIR) (37) were introduced into pGALL-(HIS3). The mouse IAP MIHA (38) and baculovirus p35 (39) were introduced into pGALL-(HIS3).
Yeast Transformation and Characterization.
Plasmids were introduced into yeast by lithium acetate transformation. For caspase activity assays in which lacZ expression was monitored, yeast were transformed with pSH18–34 (URA3) (Invitrogen), which carries a LexA-responsive lacZ gene. These cells then were transformed with either pGALL-CLBDG6-(TRP1) or pGALL-CLBGG6-(TRP1). Caspase expression plasmids or an empty expression vector were introduced into these backgrounds and were characterized as described below. For caspase activation and inhibition assays, a fourth plasmid also was introduced, either expressing Apaf-1530, p35, or MIHA or with no insert. To carry out X-gal filter assays for β-galactosidase (β-gal) activity, transformants were plated on selective plates with glucose (2%) as the sugar source. After 3 days, duplicate colonies were picked and resuspended in 1 ml of sterile Tris⋅EDTA (pH 8.0). One microliter of each sample was streaked on a minimal medium glucose plate. After 2 days, a nylon membrane was used to lift the streaked yeast (yeast side upwards) onto a complete medium plate containing 2% galactose and 1% raffinose (gal/raf media) and 3 μM copper sulfate. After various periods of induction, the filters were processed for X-gal staining (40). To quantitate β-gal activity, three tubes of liquid selective gal/raf medium were inoculated with single colonies from each transformation plate, were grown for 24 hr, then were diluted 1:10 into fresh gal/raf selective medium containing the indicated concentration of copper sulfate and were grown for a further 10 hr. o-nitrophenyl-β-d-galactoside assays were performed as described by Miller (41). To assay caspase-dependent cell death and protection by inhibitors, colonies carrying the relevant plasmids were streaked from 2% glucose selective media plates onto gal/raf selective media plates. The plates were photographed after 3 days.
Expression and Purification of Recombinant Drosophila IAPs and Caspases.
The DIAP1 coding region was amplified by PCR using primers that generated an N-terminal myc epitope (EQKLISEEDL) and was introduced into the glutathione S-transferase (GST) expression vector pGEX4T-1 (Amersham Pharmacia). The GST-myc-DIAP1 fusion protein was expressed in Escherichia coli strain BL21(DE3)pLysS (Novagen) and was affinity-purified on glutathione-Sepharose. The eluted protein was dialyzed against buffer A (25 mM Tris, pH 8.0/50 mM NaCl/10 mM DTT). After dialysis, the protein was frozen in aliquots after addition of glycerol to 10%.
DCP-1, initiating at codon 31 (DCP-131), was introduced into pET23a(+) (Novagen), generating a DCP-131-His6 fusion. A similar procedure was used to generate a drICE-His6 fusion protein in which DrICE initiates at codon 81 (drICE81-His6). DCP-131-His6 and drICE81-His6 were expressed in the E. coli strain BL21(DE3)pLysS. Protein expression and affinity purification from the soluble fraction were carried out using ProBond resin (Invitrogen). Eluted protein was dialyzed against buffer A and subsequently was snap frozen in buffer A containing 10% glycerol.
Drosophila cDNA Library Construction and DCP-1 Inhibitor Screening.
Drosophila embryonic polyA+ mRNA (CLONTECH) was converted into cDNA by using a Superscript cDNA synthesis kit (GIBCO). cDNAs larger than 600 bp were ligated into a modified version of pGALL-(HIS3) in which the polylinker was expanded to contain XhoI and NotI sites. ElectroMax DH10B cells (GIBCO) were transformed with the ligation mix and were used to amplify the library, which contained 5 × 106 primary transformants.
W303α yeast carrying pGALL-DCP-1-(LEU2) were transformed with 26 μg of library plasmid DNA, were grown in yeast extract/peptone/dextrose for 3 hr, were washed twice to remove glucose, and were plated on gal/raf selective plates. A total of 140,000 transformants were screened. Colonies were picked after 4 days of growth at 30°C. PCR was carried out on DNA isolated from individual colonies by using DIAP1-specific primers.
In Vitro Protease Assays.
DCP-131-His6 and DrICE81-His6 caspase activity were measured fluorometrically by following the release of 7-amino-4-trifluoromethyl-coumarin (AFC) from Ac-DEVD-AFC (Enzyme Systems Products, Livermore, CA) using the fmax fluorescence microplate reader (Molecular Devices) with an excitation wavelength of 405 nm and an emission wavelength of 510 nm. The ability of GST-DIAP1 to inhibit caspase activity was determined from caspase activity assay progress curves in which substrate hydrolysis (100 μM) by DCP-131-His6 (0.2 nM) or drICE81-His6 (0.62 nM) was measured in the presence of GST (0.48 μM) or GST-mycDIAP1 (0.16 μM), in caspase activity buffer (50 mM Hepes, pH 7.5/100 mM NaCl/1 mM EDTA/0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate/10% sucrose/5 mM DTT).
RESULTS AND DISCUSSION
Our approach to monitoring caspase activity in vivo was to create cells in which caspase activity stimulates transcriptional activation of a reporter. We created a fusion protein substrate for caspase cleavage in which the transcription factor LexA-B42 (LB) is linked to the truncated cytoplasmic domain of a membrane protein, CD4 (C), by a short linker (DG6) consisting of six different caspase cleavage sites that bracket the specificities of known caspases and the serine protease granzyme B, which cleaves caspases and other targets at sites of similar sequence (Material and Methods for details). When this molecule, referred to as CLBDG6, is expressed in a reporter strain in which a LexA-dependent promoter drives lacZ expression (LexA/β-gal reporter), levels of β-gal activity should depend on the presence of an active caspase able to cleave one or more of the introduced target sites, thereby releasing LexA-B42 from membrane association. (Fig. 1 A and B).
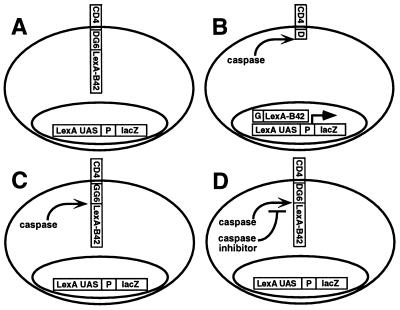
Figure 1.
A genetic system for monitoring caspase activity in yeast using a transcriptional reporter. Yeast were created that express a chimeric type-1 transmembrane protein (CLBDG6) in which the N-terminal signal sequence and transmembrane domain (CD4) is followed by a linker consisting of six tetrapeptide caspase target sites (indicated in bold) that bracket the specificity of known caspases and granzyme B (23)—DEVDG-WEHDG-IEHDG-IETDG-DEHDG-DQMDG—each of which is followed by a glycine residue, which acts as a stabilizing residue in the N-end rule degradation pathway in yeast (reviewed in ref. 24). C-terminal to the caspase target site linker is a transcription factor domain, LexA-B42. The LexA-dependent transcriptional reporter consists of LexA binding sites (LexA UAS) and a promoter (P) upstream of the bacterial lacZ gene (lacZ) (A). The cells in A act as caspase activity reporters because expression of an active caspase results in CLBDG6 cleavage at the caspase target sites, releasing LexA-B42, which enters the nucleus and activates lacZ transcription (B). A version of CLBDG6 in which the P1 aspartates are changed to glycines (CLBGG6) cannot be cleaved by caspases. Cells expressing CLBGG6 act as false positive reporters for molecules that activate lacZ expression independent of cleavage at caspase target site (C). If cells in B express a caspase inhibitor as well as an active caspase, caspase activity, and thus caspase-dependent release of LexA-B42, is inhibited, and β-gal levels are decreased compared with cells that express the caspase alone (D).
A Reporter for Caspase Activity in Yeast.
We introduced CLBDG6 into the LexA/β-gal reporter strain in a plasmid, pGALL-CLBDG6, in which expression is induced in response to galactose. We introduced into this background a copper-inducible expression plasmid, pCUP1, containing either no insert or different versions of the caspase CED-3. Transformants initially were streaked on glucose medium. Colonies from these streaks then were replica plated onto gal/raf medium containing 3 μM copper to induce expression of CLBDG6 and from the pCUP1 plasmid. After 12 hr of induction, levels of β-gal activity were determined by using an X-gal assay in which cells that do not express β-gal remain white whereas those that do turn shades of blue. Reporter cells that expressed CLBDG6 alone remained white in this assay (Fig. 2A), indicating that yeast contains negligible amounts of proteases capable of cleaving caspase target sites under standard growth conditions. However, when expression of the C. elegans caspase CED-3 (pCUP1-CED-3) was induced, a high level of β-gal activity was observed (Fig. 2A), which increased in a copper concentration-dependent manner (Fig. 2B). Of importance, caspase activity was required for reporter activation because expression of an inactive CED-3 mutant in which the active site cysteine had been changed to serine (CED-3CS) did not result in β-gal expression (Fig. 2A). Finally, expression of wild-type CED-3 in a reporter strain in which the essential P1 aspartates of the caspase target sites in CLBDG6 had been mutated to glycines (CLBGG6) (Fig. 1C) did not result in β-gal activity (Fig. 2A), arguing that the CED-3-dependent induction of β-gal activity was a direct result of cleavage of CLBDG6 at the caspase target sites.
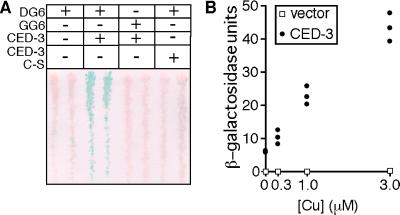
Figure 2.
Yeast expressing CLBDG6 act as reporters for CED-3 caspase activity. W303α yeast were transformed with pSH18–34, which carries a LexA-responsive lacZ transcriptional cassette (the LexA/β-gal reporter strain). These cells were transformed with pGALL expression plasmids carrying CLBDG6 (DG6) or CLBGG6 (GG6). These cells also carry a copper-inducible pCUP1 plasmid, which contains either wild-type CED-3 (CED-3), an inactive C to S mutant version of CED-3 (CED-3 C-S), or nothing. Duplicate colonies from each transformation were streaked onto gal/raf medium to induce GAL1-dependent expression of the caspase substrates and then were lifted onto complete media plates with 3 μM copper sulfate to induce caspase expression. After a 12-hr induction, an X-gal assay was performed on the filter. Only cells expressing CLBDG6 and wild-type CED-3 have significant β-gal activity (A). Cultures from three transformants carrying pSH18–34, pGAL-CLBDG6, and either the empty pCUP1 vector or pCUP1-CED-3 were grown to stationary phase, then were diluted into medium containing the indicated levels of copper sulfate and grown for a further 10 hr. o-nitrophenyl-β-d-galactoside assays were performed, and β-gal activity was determined. β-gal activity in the CED-3-expressing cells increased as a function of copper concentration (filled circles, CED-3). No β-gal activity was found in the cultures carrying only the empty pCUP1 vector (open boxes, vector) (B).
These results establish that yeast can be used as a cell-based reporter system for caspase activity. In order for a caspase to be identified in this assay, the caspase must be active in yeast. Physiological activation of caspases occurs through multiple mechanisms, including recruitment and oligomerization at the plasma membrane, cleavage by caspases or other proteases able to recognize a caspase target site, interactions with members of the CED-4/Apaf-1 family of proteins, and autoactivation. In some cases, overexpression alone is sufficient to induce autoactivation whereas, in other cases, significant activation requires interactions with other proteins (reviewed in refs. 2–5). Thus, it is likely that only proteases in which the primary translation product is active, or in which the protease is able to autoactivate, will be identified in the simplest reporter-based caspase screen. However, more complex screens for caspases that can activate after forced oligomerization or association with potential caspase activators (42–46) can be envisioned. We have tested several other caspases in this reporter system. Expression of mammalian caspase 753 (below) and full length caspase 8 (data not shown) resulted in reporter-dependent lacZ expression. Expression of human caspase 3, caspase 9, or Drosophila drICE failed to activate reporter expression (data not shown), even though active forms of these caspases are known to efficiently cleave peptides with the same sequence as the target sites introduced into CLBDG6 (23, 33). Moreover, although overexpression of wild-type but not an inactive mutant of CED-3 induced yeast cell death (below), similar overexpression of caspase 3, caspase 9, or drICE had no effect on cell growth. Based on these observations, it is likely that, in yeast, the procaspase forms of these caspases do not autoprocess to generate active caspase heterodimers. This result is expected: Caspase 9 is thought to function as an upstream caspase, in which a major mechanism of activation requires association with Apaf-1 (42, 43), whereas caspase-3 is thought to act as a downstream caspase, in which a principal mechanism of activation is cleavage by other caspases (reviewed in refs. 2–5). drICE activation may be regulated by either of these mechanisms.
Activators of Caspase-Dependent Reporter Activation.
The fact that certain caspases do not activate in yeast suggests that it should be possible to screen for their activators as molecules that induce reporter expression in the presence of an otherwise inactive caspase. To demonstrate this, we carried out an experiment in which caspase activity was monitored in yeast that expressed full length caspase 9, alone or in combination with a fragment of Apaf-1 that is constitutively active with respect to caspase 9 processing activity in vitro (43). Transformants of the pGALL-CLBDG6 LexA/β-gal reporter strain were generated that carried either two empty vectors, an empty vector and pGALL-Apaf-1530, an empty vector and pGALL-caspase 9, or pGALL-caspase 9 and pGALL-Apaf-1530. Transformants initially were streaked on glucose medium. Colonies from these streaks then were replica plated onto gal/raf medium to induce GALL-dependent expression. After 16 hr of induction, levels of β-gal activity were determined by using an X-gal assay. Reporter cells that expressed either nothing, caspase 9, or Apaf-1530 alone remained white in this assay, indicating that caspase activity was not induced (Fig. 3). However, colonies that expressed both caspase 9 and Apaf-1530 showed robust β-gal activity, suggesting the occurrence of Apaf-1530-mediated activation of the otherwise inactive procaspase 9 (Fig. 3).
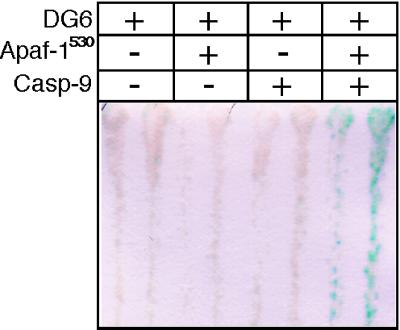
Figure 3.
Expression of Apaf-1530 induces caspase 9-dependent reporter activation. W303α containing pSH18–34 and pGALL-CLBDG6 were transformed with pGALL plasmids to carry either two empty pGALL vectors, Apaf-1530 and an empty pGALL vector, caspase 9 and an empty pGALL vector, or Apaf-1530 and caspase 9. Two colonies from each transformation were streaked onto selective glucose medium plates, were grown for several days, and then were replica plated onto complete gal/raf medium. After 16 hr, an X-gal assay was performed. Only cells expressing both Apaf-1530 and caspase 9 show significant β-gal activity.
We used β-gal activity, as assayed after replica plating of colonies, as the basis for our caspase reporter assay because caspases are expressed conditionally after replica plating. Thus, their identification is feasible even if their expression is toxic to cells. However, caspase activity screens also can be adapted to positive, survival-based screening assays by requiring LexA-dependent expression of a yeast auxotrophic marker such as HIS3 or URA3. False positives in these caspase reporter assays could arise because introduced proteins bind to the LexA binding sites and activate transcription directly or because they encode proteases that cleave CLBDG6, but not at the caspase target sites. Both classes of false positives can be identified by the fact that they should still activate lacZ expression when introduced into a LexA/β-gal reporter strain that expresses CLBGG6, the false positive reporter strain (Fig. 1C).
Suppression of Caspase-Dependent Reporter Activation by Caspase Inhibitors.
Once a reporter for the activity of a specific caspase has been established in yeast, it should be possible to screen for inhibitors of that activity by identifying cells that express the protease but in which reporter activity is repressed (Fig. 1D). Here, we demonstrate the feasibility of this approach by showing that two different families of caspase inhibitors, exemplified by baculovirus p35 and the murine IAP MIHA, suppress caspase-dependent reporter activation. p35 is a broad specificity caspase inhibitor in which inhibition is associated with cleavage (reviewed in ref. 6). The IAPs comprise a second class of caspase inhibitors distinct from p35 in which cleavage of the inhibitor does not play a role. IAPs originally were identified in the baculovirus system by virtue of their ability to substitute for baculovirus p35 as suppressers of viral infection-induced cell death (47). IAP homologs have now been found in other viruses, Drosophila, C. elegans, mammals and yeast and are characterized by one or more N-terminal repeats known as baculovirus IAP repeats (BIRs). Many also have a C-terminal RING finger motif (reviewed in refs. 6–8 and 48). Several mammalian IAPs—XIAP, cIAP1, and cIAP2—bind to and directly inhibit caspases-3 and -7 in vitro (49, 50). We introduced pGALL-CLBDG6 and either an empty pCUP1 plasmid or a pCUP plasmid containing CED-3 or caspase 753 into the LexA/β-gal reporter strain along with either an empty pGALL expression vector, a pGALL expression vector carrying p35, or a pGALL expression vector carrying MIHA (murine XIAP). Transformants were grown in galactose-containing medium to induce expression of CLBDG6 and the caspase inhibitor and then were transferred to medium containing galactose and 3 μM copper to induce expression of the caspase. β-gal activity was determined after a 10-hr copper induction. As shown in Fig. 4 A and B, expression of p35 inhibited both CED-3 and caspase 753 activity ≈5-fold. A similar though somewhat weaker (≈2-fold) inhibition of caspase 753 activity was seen in the presence of MIHA (Fig. 4B), indicating that caspase-IAP interactions can be detected in this assay.
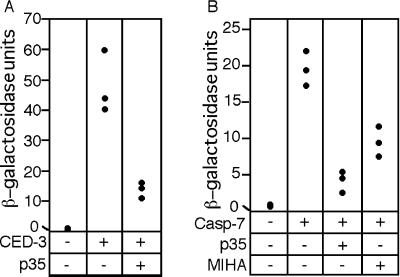
Figure 4.
Expression of caspase inhibitors suppresses caspase-dependent reporter activation in yeast. The LexA/β-gal reporter strain carrying pGALL-CLBDG6 was transformed with either an empty pCUP1 plasmid or pCUP1-CED-3 and either an empty pGALL vector or pGALL-p35. Three colonies from each transformation were grown for 24 hr in selective gal/raf medium. Cultures were diluted 1:10 into fresh gal/raf medium containing 3 μM copper sulfate and were grown for a further 10 hr, after which o-nitrophenyl-β-d-galactoside assays for β-gal activity were performed. Cultures from caspase transformants showed significant β-gal activity, which was suppressed by GALL-dependent expression of baculovirus p35 (A). In an experiment similar to that described in A, expression of caspase 753 was induced in cells that express baculovirus p35 or the mouse IAP MIHA. Expression of caspase 753 resulted in a significant increase in cellular β-gal activity, which was suppressed by GALL-dependent expression of p35 or MIHA (B).
Caspase Overexpression-Dependent Yeast Cell Death.
In the above assay, the presence of a caspase inhibitor is indicated by a decrease in β-gal activity. However, in many situations, it would be useful if caspase inhibition was coupled to a positive reporter output. A direct approach to identifying caspase inhibitors rests on the observation that high level expression of active caspase causes yeast cell death. Low level expression of the Drosophila caspase DCP-1 from the induced CUP-1 promoter did not significantly compromise yeast cell growth. Higher level expression from the GAL promoter, however, did result in cell lethality. Cells were able to grow on galactose-containing media if they carried an empty pGALL expression vector but not if they carried a pGALL-DCP-1 expression construct (Fig. 5A). This effect of DCP-1 expression was attributable to cell death because a >250-fold decrease in the number of colony forming units was seen when cells carrying the pGALL-DCP-1 expression plasmid were grown for 12 hr in liquid gal/raf medium, thus inducing high level DCP-1 expression, and then were plated on glucose-containing medium (data not shown). Of importance, GALL-DCP-1-expression-dependent cell killing depended on caspase activity because expression of an inactive mutant form of DCP-1 (DCP-1 C285S) did not cause cell death (Fig. 5A). Caspase-mediated cell death in yeast may be a general phenomenon because other caspases that are active in yeast, including full length CED-3, full length caspase 8, and caspase 753, block colony formation when expressed under control of the strong GALL promoter (data not shown).
Figure 5.
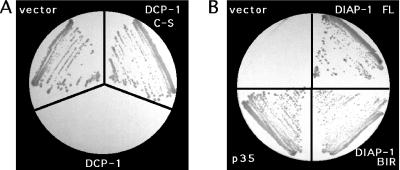
High level expression of the Drosophila caspase DCP-1 kills yeast and is prevented by coexpression of baculovirus p35 or DIAP1. EGY48 yeast were transformed with pGALL plasmids containing either full length DCP-1 (FLDCP-1), an active site C-S mutant of DCP-1 (DCP-1 C-S), or no insert (vector). Transformants were streaked from selective glucose-containing medium onto selective gal/raf-inducing medium. Cells expressing either an empty pGALS vector or the DCP-1 C-S active site mutant grow on galactose-containing medium whereas cells expressing full length DCP-1 do not (A). EGY48 yeast carrying pGALS-FLDCP-1 were transformed with pGALL vectors carrying full length DIAP1 (DIAP1), the DIAP1 BIR repeats (DIAP1 BIR), baculovirus p35 (p35), or nothing (vector). GALL-dependent expression of p35, full length DIAP1, and, to a somewhat lesser extent, the DIAP1 BIR repeats block DCP-1-dependent cell death (B).
Caspase Inhibitors Suppress Caspase Overexpression-Dependent Cell Death.
To demonstrate that caspase inhibitors can be identified as proteins that restore cell viability to yeast expressing an active caspase, we carried out an experiment in which the broad specificity caspase inhibitor p35 was coexpressed with full length DCP-1. We introduced pGALL expression plasmids that either had no insert, or that carried p35, into cells carrying a pGALS-DCP-1 plasmid. Cells from colonies carrying these plasmids were grown on glucose-containing media and then were streaked onto gal/raf media to induce expression of the caspase and the potential inhibitor. As shown in Fig. 5B, pGALS-dependent expression of DCP-1 in the presence of an empty pGALL expression vector resulted in no cell growth. In contrast, coexpression of baculovirus p35 with DCP-1 resulted in a dramatic rescue of cell growth.
To determine whether a yeast survival-based screen can be used to identify novel caspase inhibitors, we transformed yeast carrying pGALS-DCP-1 with a Drosophila pGALL embryonic cDNA expression library and plated these cells on gal/raf medium. In a screen of ≈1.4 × 105 transformants, ≈50 positives were obtained. These were tested by PCR, and all were found to correspond to DIAP1, which was originally identified as an inhibitor of reaper or hid overexpression-induced, caspase-dependent cell death in the fly eye (37). In the fly eye, and in cell culture (51–54), it also was found that cell death could be suppressed by expression of an N-terminal DIAP1 fragment containing the two BIR repeats but lacking the C-terminal RING finger domain. To determine whether the same fragment of DIAP1 was sufficient to block DCP-1-dependent cell killing in yeast, we carried out an experiment in which full length DIAP1, an N-terminal fragment of DIAP1, or an empty vector was expressed under GALL control in the presence of GALS-driven DCP-1. As shown in Fig. 5B, inhibition of DCP-1-dependent cell death was seen when DIAP1 or the N-terminal fragment of DIAP1 containing only the DIAP1 BIR repeats was coexpressed with DCP-1. To determine whether the observed interaction between DIAP1 and DCP-1 was direct, we generated bacterially synthesized GST-DIAP1 as well as His6-tagged versions of prodomainless DCP-1 (DCP-131-His6) and drICE (drICE81-His6). As shown in Fig. 6, GST-DIAP1 inhibited DCP-131-His6 caspase activity but had little if any effect on that of drICE81-His6. Thus, these results demonstrate that caspase inhibitors can be identified as molecules that block GAL-driven, caspase-dependent cell death.
Figure 6.
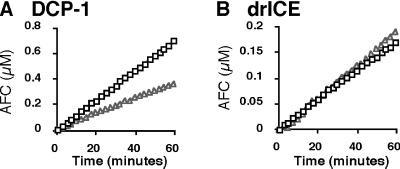
GST-DIAP1 inhibits the caspase activity of bacterially synthesized DCP-131-His6, but not drICE81-HIS6. Purified GST-DIAP1 (0.16 μM) (open triangles) or GST (0.48 μM) (open squares) was incubated with a fixed amount of DCP-131His6 (0.2nM) in caspase activity assay buffer containing 100 μM of the Ac-DEVD-AFC substrate. Release of AFC was monitored fluorometrically over time. GST-DIAP1 inhibits DCP-1-dependent caspase activity (A). In similar experiments in which 0.62nM drICE81His6 was incubated in caspase activity buffer with GST (0.48 μM) or GST-DIAP1, (0.16 μM), no inhibition of drICE activity by GST-DIAP1was seen (B).
Concluding Remarks.
We have developed a yeast cell-based assay for the activity of one group of proteases, the caspases, in which caspase activity is monitored either by the cleavage-dependent release of a transcription factor from its transmembrane anchor and subsequent activation of a reporter or by induction of cell killing. Both reporter activation and cell killing are suppressed by known caspase inhibitors. We have exploited this fact to directly isolate caspase inhibitors from a Drosophila embryo cDNA library.
Yeast carrying the transcription-based caspase reporter should be useful as a background in which to carry out screens for proteins that cleave a caspase target site and for their regulators. Because yeast can be transformed with high efficiency, it also constitutes an ideal system in which to carry out large scale mutagenesis studies of particular proteases or their regulators. It may also be possible to screen for cellular targets of specific caspases by using artificial substrate libraries in which cDNA fragments substitute for the caspase target site linker in the caspase substrate fusion protein CLBDG6. A transcription-based reporter strategy similar to that described here also may provide a way to monitor caspase activity in cells of higher eukaryotes. Finally, substituting caspase cleavage sites for those of other site-specific proteases in the CLBDG6 reporter should enable the identification and study of these proteins and their regulators.
Acknowledgments
We thank the following for providing plasmids and strains: J. Adams (CD4), J. Yuan (pβactced38Z), L. K. Miller (pBS p35), H. Steller (DCP-1), D. Vaux (MIHA and caspases 3 and 9), X. Wang (Apaf-1), and R. Deshaies (yeast strains and plasmids). R. Deshaies, P. Sternberg, M. Guo, and W. Y. Shou provided helpful discussions and comments. C.J.H. is supported by a fellowship from the Human Frontiers Science Program. B.A.H. is a Searle Scholar. This work was supported by National Institutes of Health Grant GM057422-01 and a Burroughs Wellcome Fund New Investigator Award in the Pharmacological Sciences to B.A.H.
ABBREVIATIONS
- β-gal
β-galactosidase
- GST
glutathione S-transferase
- AFC
7-amino-4-trifluoromethyl-coumarin
- BIR
baculovirus IAP repeats
- gal/raf medium
medium containing 2% galactose and 1% raffinose
References
- 1.Alnemri E S, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson D W, Thornberry N. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 3.Salvesen G S, Dixit V M. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 4.Thornberry N A, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 5.Cryns V, Yuan J. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 6.Teodoro J G, Branton P E. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villa P, Kaufmann S H, Earnshaw W C. Trends Biochem Sci. 1997;22:388–393. doi: 10.1016/s0968-0004(97)01107-9. [DOI] [PubMed] [Google Scholar]
- 8.Clem R J, Duckett C S. Trends Biochem Sci. 1997;7:337–339. doi: 10.1016/S0962-8924(97)01088-X. [DOI] [PubMed] [Google Scholar]
- 9.Cerretti D P, Kozlosky C J, Mosley B, Nelson N, Ness K V, Greenstreet T A, March C J, Kronheim S R, Druck T, Cannizzaro L A, et al. Science. 1992;256:97–100. doi: 10.1126/science.1373520. [DOI] [PubMed] [Google Scholar]
- 10.Thornberry N A, Bull H G, Calaycay J R, Chapman K T, Howard A D, Kostura M J, Miller D K, Molineaux S M, Weidner J R, Aunins J. Nature (London) 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 11.Ghayur T, Banerjee S, Hugunin M, Butler D, Herzog L, Carter A, Quintal L, Sekut L, Talanian R, Paskind M, et al. Nature (London) 1997;386:619–623. doi: 10.1038/386619a0. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Center D M, Wu D M H, Cruikshank W W, Yuan J, Andrews D W, Kornfeld H. J Biol Chem. 1998;273:1144–1149. doi: 10.1074/jbc.273.2.1144. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Miura M, Jung Y-K, Zhu H, Li E, Yuan J. Cell. 1998;92:501–509. doi: 10.1016/s0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 14.Song Z W, McCall K, Steller H. Science. 1997;275:536–540. doi: 10.1126/science.275.5299.536. [DOI] [PubMed] [Google Scholar]
- 15.McCall K, Steller H. Science. 1998;279:230–234. doi: 10.1126/science.279.5348.230. [DOI] [PubMed] [Google Scholar]
- 16.McCall J O, Kadam S, Katz L. Bio/Technology. 1994;12:1012–1016. doi: 10.1038/nbt1094-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Gerard A L V, Huang B C B, Anderson D C, Payan D G, Luo Y. Nucleic Acids Res. 1998;26:2034–2035. doi: 10.1093/nar/26.8.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sices J J, Kristie T M. Proc Natl Acad Sci USA. 1998;95:2828–2833. doi: 10.1073/pnas.95.6.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Struhl G, Adachi A. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 20.Lecourtis M, Schweisguth F. Curr Biol. 1998;8:771–774. doi: 10.1016/s0960-9822(98)70300-8. [DOI] [PubMed] [Google Scholar]
- 21.Stagljar I, Korostensky C, Johnsson N, te Heesen S. Proc Natl Acad Sci USA. 1998;95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madden P J, Littman D R, Godfrey M, Maddon D E, Chess L, Axel R. Cell. 1985;42:93–104. doi: 10.1016/s0092-8674(85)80105-7. [DOI] [PubMed] [Google Scholar]
- 23.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt J P, et al. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 24.Varshavsky A. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horii T, Ogawa T, Ogawa H. Cell. 1981;23:689–697. doi: 10.1016/0092-8674(81)90432-3. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston M, Davis R W. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamai K T, Liu X L, Silar P, Sosinowski T, Thiele D J. Mol Cell Biol. 1994;14:8155–8165. doi: 10.1128/mcb.14.12.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miura M, Zhu H, Rotello R, Hartweig E A, Yuan J. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 30.Duan H J, Chinnaiyan A M, Hudson P L, Wing J P, He W W, Dixit V M. J Biol Chem. 1996;271:1621–1625. doi: 10.1074/jbc.271.3.1621. [DOI] [PubMed] [Google Scholar]
- 31.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 32.Fernandes-Alnemri T, Armstrong R C, Krebs J, Srinivasula S M, Wang L, Bullrich F, Fritz L C, Trapani J A, Tomaselli K J, Litwack G, et al. Proc Natl Acad Sci USA. 1996;93:7464–7469. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraser A G, Evan G I. EMBO J. 1997;16:2805–2813. doi: 10.1093/emboj/16.10.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duan H, Orth K, Chinnaiyan A M, Poirier G G, Froelich C J, He W W, Dixit V M. J Biol Chem. 1996;271:16720–16724. doi: 10.1074/jbc.271.28.16720. [DOI] [PubMed] [Google Scholar]
- 35.Srinivasula S M, Fernandes-Alnemri T, Zangrilli J, Robertson N, Armstrong R C, Wang L, Trapani J A, Tomaselli K J, Litwack G, Alnemri E S. J Biol Chem. 1996;271:27099–27106. doi: 10.1074/jbc.271.43.27099. [DOI] [PubMed] [Google Scholar]
- 36.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 37.Hay B A, Wassarman D A, Rubin G M. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 38.Uren A G, Pakusch M, Hawkins C J, Puls K L, Vaux D L. Proc Natl Acad Sci USA. 1996;93:4974–4978. doi: 10.1073/pnas.93.10.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clem R, Fechheimer M, Miller L K. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- 40.Breeden L, Nasmyth K. Cold Spring Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 41.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 42.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 43.Srinivasula S, Ahmed M, Fernandes-Alnemri T, Alnemri E S. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 44.Yang X, Chang H Y, Baltimore D. Science. 1998;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- 45.Yang X, Chang H Y, Baltimore D. Mol Cell. 1998;1:319–325. doi: 10.1016/s1097-2765(00)80032-5. [DOI] [PubMed] [Google Scholar]
- 46.Muzio M, Stockwell B R, Stennicke H R, Salvesen G S, Dixit V M. J Biol Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 47.Crook N E, Clem R J, Miller L K. J Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uren A G, Coulson E J, Vaux D L. Trends Biochem Sci. 1998;23:159–162. doi: 10.1016/s0968-0004(98)01198-0. [DOI] [PubMed] [Google Scholar]
- 49.Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. Nature (London) 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 50.Roy N, Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harvey A J, Soliman H, Kaiser W J, Miller L K. Cell Death Differ. 1997;4:733–744. doi: 10.1038/sj.cdd.4400294. [DOI] [PubMed] [Google Scholar]
- 52.Vucic D, Kaiser W J, Harvey A J, Miller L K. Proc Natl Acad Sci USA. 1997;94:10183–10188. doi: 10.1073/pnas.94.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vucic D, Kaiser W J, Miller L K. Mol Cell Biol. 1998;17:667–676. doi: 10.1128/mcb.17.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hawkins C J, Ekert P G, Uren A G, Holmgreen S P, Vaux D. Cell Death Differ. 1998;5:569–576. doi: 10.1038/sj.cdd.4400389. [DOI] [PubMed] [Google Scholar]