Abstract
We have constructed a library of DNA fragments heavily methylated in human adenocarcinomas of the lung to permit the comprehensive isolation of methylated CpG islands in cancer. Heavily methylated genomic DNA fragments from tumors of nine male patients were enriched using a methylated DNA binding column and used for construction of the library. From this library, DNA fragments having properties of CpG islands were isolated on the basis of their reduced rate of strand dissociation during denaturing gradient gel electrophoresis. Approximately 1,000 clones, corresponding to 0.3% of the library were analyzed, and nine DNA fragments were identified as being associated with CpG islands that were methylated in tumor DNA. One CpG island was methylated specifically in tumor DNA, whereas the remaining eight CpG islands were methylated both in normal and tumor DNA derived from the same patients. Our results suggest that the number of CpG islands methylated specifically in tumors is not large. The library, which contains DNA fragments from methylated CpG islands comprehensively, is expected to be valuable when elucidating epigenetic processes involved in carcinogenesis.
Keywords: segregation of partly melted molecules, epigenetics, gene silencing
Epigenetic gene silencing is known to be associated with many biological phenomena. In physiological settings, such as X inactivation and genomic imprinting, one of the two alleles is transcriptionally inactive without any apparent alteration of nucleotide sequences. Although the molecular mechanism of epigenetic gene silencing has yet to be elucidated, accumulating evidence strongly suggests that DNA methylation is closely associated with long-term repression of transcription. It has been proposed that binding of the repressor protein(s) to methylated CpG pairs (mCpGs) induces gene silencing, possibly by altering chromatin structure and subsequently eliminating basal transcriptional machinery and RNA polymerase activities (for a recent review, see ref. 1).
Cancer is caused by the accumulation of both genetic and epigenetic changes. Although genetic changes, that is, alteration of DNA sequences, have been extensively investigated, epigenetic changes remain less well examined. In cancer, methylation of CpG islands is considered to be one of the major epigenetic aberrations that causes gene inactivation (for recent reviews, see refs. 2 and 3). However, only limited information is currently available on the nature of those CpG islands. Comprehensive identification of methylated CpG islands in tumors may lead to the understanding of the epigenetic process that underlies tumorigenesis.
Methylated DNA binding column is an affinity matrix that contains a polypeptide derived from the methyl-CpG binding domain (MBD) of the rat chromosomal protein MeCP2, which preferentially binds DNA at symmetrical mCpG sites (4). A polypeptide, termed HMBD, consists of 85 aa of the MBD domain and a histidine tag at the N-terminal end, which can then be attached to a solid support (5). Because the stoichiometry of binding is reported to be 1 HMBD to 1 mCpG (4), heavily methylated DNA fragments have increased affinity with the column when compared with poorly or moderately methylated fragments. This property provides an opportunity for the efficient enrichment of heavily methylated DNA fragments.
Segregation of partly melted molecules (SPM) is a method for the preferential isolation of DNA fragments associated with CpG islands from cloned DNA fragments (6). SPM takes advantage of the reduced rate of strand dissociation in a denaturing gradient gel of DNA fragments derived from CpG islands (6). We have shown that many DNA fragments containing CpG islands can be detected efficiently from anonymous regions of cloned DNA by employing SPM methodology (6–8). The overall procedure is relatively simple and can be performed without prior knowledge of nucleotide sequences.
In this study, we have attempted a comprehensive isolation of CpG islands that are methylated in human adenocarcinomas of the lung by the combined use of MBD column chromatography and SPM.
MATERIALS AND METHODS
DNA Fragments and High Molecular Weight DNA.
DNA fragments with known nucleotide sequences subjected to in vitro methylation are summarized in Table 1. Methylation of DNA fragments using HpaII methylase or SssI methylase (New England Biolabs) were performed following the manufacturer’s recommendation. Methylation status of modified fragments was examined by incubation with appropriate restriction endonucleases. Isolation of high molecular weight DNA from surgical specimens of human lung cancer and those from subjects obtained at autopsy has been reported previously (9). To digest 1 μg of genomic DNA completely, 2.5 units of Tsp509 I (New England Biolabs) were used under the condition recommended by the supplier.
Table 1.
DNA fragments with known nucleotide sequences
| Name | Enzyme | Size, kb | No. of methyl-CpGs before and after in vitro methylation | Accession no. | CpG island |
|---|---|---|---|---|---|
| R15-4 | AatII* | 3.5 | 0, 205† | X65308, D31748 | − |
| pUC19 | NdeI* | 2.7 | 0, 173† | L09137 | − |
| pEG | MluI/XhoI | 1.8 | 0, 32‡, 192† | M58602 | + |
| R16-2 | SalI§ | 1.0 | 0, 71† | D31750 | + |
| pS5¶ | KpnI/SacI‖ | 0.9 | 0, 35† | D45254 | − |
Linearized with the endonuclease.
Methylated using SssI methylase.
Methylated using HpaII methylase.
A short stretch of multiple cloning site of pGEM-5Zf(+) (accession no. X65308) is attached.
pS5 contains a region of the nucleotide sequence of the cDNA for rat cellular nucleic acid binding protein CNBP (accession no. D45254) (31).
A short stretch of multiple cloning site of pBluescript II SK(+) (accession no. X52328) is attached.
Preparation of MBD Column.
Polypeptide HMBD was produced in bacteria transformed with pET6HMBD (5). The MBD column was prepared following a published procedure (10). A stepwise gradient of salt (0.4–1.0 M NaCl by 40 mM per step, in 20 mM Hepes (pH 7.9), 10% glycerol, 0.1% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride, and 10 mM 2-mercaptoethanol) was applied to elute bound DNA fragments as described (10), with minor modifications. The relative elution profile is reproducible, but absolute salt concentration for elution of a specific DNA fragment requires calibration with known DNA fragments.
PCR Experiments.
The PCR primers used to detect DNA fragments containing a CpG island of the human CDH1 gene were 5′-AGAGGGTCAACGCGTCTATG-3′ and 5′-TCACAGGTGCTTTGCAGTTC-3′. The product size was 200 bp. The PCR primers used to detect DNA fragments R16–2 and pS5 were 5′-CCTTCCCCAAAAGTCCTCTC-3′ and 5′-GGAAAGGAGATAGATGGGGG-3′ (product size, 150 bp) and 5′-TATCGCTGTGGTGAGTCTGG-3′ and 5′-GCATTGCTCTCGCTCTCTCT-3′ (product size, 129 bp), respectively. Thirty cycles of amplification (30 sec at 94°C; 30 sec at 55°C; and 1 min at 72°C) using AmpliTaq DNA Polymerase Stoffel fragment (PerkinElmer) was performed. The PCR primers used to analyze DNA fragments associated with CpG islands isolated in this study are summarized in Table 2. Reactions were performed at conditions similar to those above except with an annealing temperature of 66°C and 35 cycles of amplification. In some cases, PCR experiments were repeated at least twice to confirm reproducibility.
Table 2.
Isolated fragments derived from CpG islands by MBD column chromatography and SPM analysis
| Clone name | Length, bp | Accession no. | Tumors with methylated allele* | Identical cDNA or EST sequence (accession no.) | PCR primer | Product size, bp |
|---|---|---|---|---|---|---|
| pSPM39 | 441 | AB017141 | NTSM† | yn55c04.r1 (H20090) | 5′-TCAGTGACCTGTGCTGCTCT-3′ | |
| yn55c04.s1 (H20019) | 5′-GGAAGGAACTGAGACCACGA-3′ | 78 | ||||
| pSPM125 | 482 | AB017143 | NTSM | — | 5′-ACTGGAGCCCCTGCTTACTC-3′ | |
| 5′-TCCGATCTCAAACTAACCCG-3′ | 26 | |||||
| pSPM210 | 497 | AB017142 | 56, 59‡ | ENDOG (X79444) | 5′-TTATTCACCTTCCTCCTGCG-3′ | |
| 5′-CAGCCTTCTTCTGCTGTGTG-3′ | 90 | |||||
| pSPM639 | 480 | AB017144 | NTSM | — | 5′-GAACTGCTGGAGGAACTTGG-3′ | |
| 5′-CTTATCTCCGGACTTCGCTG-3′ | 86 | |||||
| pSPM769 | 380 | AB017145 | NTSM | zd82e12.r1 (W80610) | 5′-TTGGAGCGGATAGAAAGGAA-3′ | |
| 5′-AGACACACTCAGCTGCCCTT-3′ | 10 | |||||
| pSPM789 | 486 | AB017146 | NTSM | — | 5′-GGCACTGTCAAATGGTTCAA-3′ | |
| 5′-GACATCTTCCTTGGCGTCAT-3′ | 72 | |||||
| pSPM814 | 644 | AB017147 | NTSM | zw85f07.s1 | 5′-GGACTTGTTTTCTTCCGGGT-3′ | |
| (AA446967) | 5′-TTCGCTTCCCGTAGCTCTAA-3′ | 122 | ||||
| pSPM845B | 707 | AB017148 | NTSM | — | 5′-CATGGAATCTGCCCAAGACT-3′ | |
| 5′-CCTGGGTATGAGAACGCATC-3′ | 105 | |||||
| pSPM997 | 467 | AB017149 | NTSM | cDNA for KIAA0445 | 5′-GGGTTAGGTGACACCCACTG-3′ | |
| protein (AB007914) | 5′-TTCCTGAAATGGGTCTGGTT-3′ | 87 |
Samples used to construct the DNA library of heavily methylated fragments are tumors from patients 44, 56, 59, 108, 113, 116, 120, 202, and 209.
NTSM, non-tumor-specific methylation, meaning that methylated and nonmethylated alleles were present in both normal and tumor DNA in all patients.
Presence of both methylated and nonmethylated alleles in normal DNA.
Cloning of the Fragments and SPM Analysis.
Highly methylated genomic DNA fragments enriched by MBD column chromatography were cloned using a lambda Zap II vector (Stratagene). A 1-μl aliquot of bacteriophage suspension recovered from a single plaque was subjected to PCR to select clones possessing long inserts. The PCR primers used were 5′-TAACCCTCACTAAAGGGAAC-3′ and 5′-GTAATACGACTCACTATAGG-3′ on the basis of the nucleotide sequence of the vector. Reactions were performed as described above (annealing temperature at 55°C and 30 cycles of amplification). In vivo excision of phagemid DNA from the lambda Zap II vector was performed following the manufacturer’s recommendation. SPM analysis and nucleotide sequence analysis of the retained fragments were carried out as described (6). The algorithm of Gardiner-Garden and Frommer (11) as implemented for computation (http://www.no.embnet.org/cpg/cpg.html) was helpful in recognition of potential CpG islands.
Southern Analyses.
High molecular weight DNA from six human lung adenocarcinoma cell lines, A549, RERF-LC-OK, RERF-LC-MS, ABC-1, LC-2/ad, and VMRC-LCD were subjected to Southern analysis as described (12). Northern analysis was carried out following published procedures (13).
RESULTS
Separation of Differentially Methylated DNA Fragments with an MBD Column.
Digestion of genomic DNA with restriction endonuclease Tsp509 I is expected to maintain the integrity of CpG islands because the occurrence of Tsp509I sites (AATT) within CpG islands is rare. Preliminary investigation of 22 sequenced human genes with CpG islands revealed that the sizes of Tsp509I fragments containing a CpG island were 0.8–2.8 kb, and the number of CpG pairs contained in these fragments was 60–180. CpG sequences are considered to be densely methylated in reported CpG islands, although some recent reports have indicated the existence of regions of DNA having few mCpGs in CpG islands that are methylated as a whole (14–17).
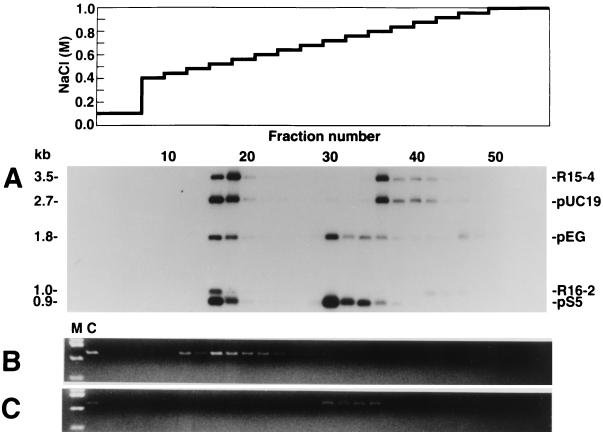
We first examined whether DNA fragments having 60–180 mCpGs could be separated from those having 30 mCpGs by MBD column chromatography. Eleven cloned DNA fragments with or without in vitro methylation described in Table 1 were pooled and loaded on an MBD column. Aliquots of eluates from appropriate fractions were then subjected to Southern analysis (Fig. 1A).
Figure 1.
The elution profiles of cloned DNA fragments with various levels of methylation. Four milliliters of elution buffer was applied at each step, and 1.3-ml aliquots were collected for each fraction. (A) Twenty nanograms each of DNA corresponding to cloned fragments in Table 1 with and without in vitro methylation were applied to the column. One hundred-microliter aliquots were subjected to Southern analysis. Fifty micrograms of Tsp509 I digests of DNA from human adult male tissue (B) and 50 μg of Tsp509 I-digests of DNA from MKN1 cell line (C) were applied to the column. Three hundred microliters of aliquot was subjected to PCR to detect DNA fragments containing a CpG island of the human CDH1 gene. M, HaeIII digests of pUC19 DNA; C, PCR products from each Tsp509 I-digested genomic DNA. Elution at the same salt concentration in separate experiments does not necessarily guarantee similar methylation status, even when the same column was used. After extensive use, an alteration of the retention capacity of the column was observed. The retention capacity was monitored by elution profile of some fragments shown in Table 1 (data not shown). Thus, methylation status of DNA fragment in fraction 30 in A may not be identical to that of fraction 30 in B or C.
pEG fragments of 1.8 kb from fraction 16 were cleaved with HpaII or HhaI, those from fraction 30 were cleaved with HhaI but not with HpaII, and those from fraction 46 were resistant to HpaII or HhaI. These results showed that the DNA fragments from fraction 16 were nonmethylated, those from fraction 30 were methylated at HpaII sites (32 mCpGs), and those from fraction 46 were extensively methylated (192 mCpGs) (data not shown). This result indicates that DNA fragments with the same nucleotide sequence but different methylation status are separated by MBD column chromatography.
All nonmethylated fragments were eluted in fractions 16–18 (0.52–0.56 M NaCl). Fragments R15–4 (3.5 kb) and pUC19 (2.7 kb) differed in size and nucleotide sequence, but had a similar number of mCpGs after treatment with Sss I methylase (207 and 173, respectively). Both of the methylated fragments were eluted predominantly in fraction 36 (0.80 M NaCl). Fragments pEG, treated with HpaII methylase (1.8 kb, 32 mCpGs), and pS5, treated with SssI methylase (0.9 kb, 35 mCpGs), were eluted predominantly in fraction 30 (0.72 M NaCl). These results demonstrate that DNA fragments with a similar number of mCpGs are eluted at a similar salt concentration independent of their nucleotide sequences and size. DNA fragments denser in mCpGs (pEG fragment with 192 mCpGs and R16–2 fragment with 71 mCpGs) were eluted at higher salt concentrations than compatible with monotonic increase based on the absolute number of mCpGs.
Separation of Genomic DNA Fragments with an MBD Column.
We then examined whether MBD column chromatography enables the separation of methylated CpG islands from corresponding nonmethylated CpG islands in genomic DNA. The CpG island of the human CDH1 gene has been reported to be heavily methylated in a gastric cancer cell line, MKN1 (18). The Tsp509I fragment containing this CpG island is 1.7 kb in size and has 126 CpG dinucleotides. Tsp509I digests of high molecular weight DNA derived from normal human tissue and from MKN1 cells were analyzed by MBD column chromatography.
Tsp509I fragment containing the CpG island derived from normal somatic tissue was eluted in fraction 16 (0.52 M NaCl) (Fig. 1B) and surrounding fractions. However, in the case of MKN1 DNA, the corresponding fragment was eluted in fractions 30–36 (0.72–0.80 M NaCl) (Fig. 1C). These results indicate that genomic fragments from methylated and corresponding nonmethylated CpG islands can be separated by MBD column chromatography.
Similar differences in elution profiles were observed when we analyzed male and female DNAs. DNA fragments containing CpG island of the human HPRT gene on X chromosome from male DNA were detected only in lower salt fractions, whereas the corresponding fragments of female DNA were detected both in lower and higher salt fractions (data not shown).
Enrichment and Cloning of Highly Methylated DNA Fragments from Tumors.
Fifty-two micrograms of high molecular weight DNA derived from nine male patients with adenocarcinomas of the lung (≈6 μg from each patient) was digested with Tsp509I and subjected to MBD column chromatography. Differentially methylated DNA fragments (R16-2 with 72 mCpGs and pS5 with 35 mCpGs, Table 1) were added to the digest as internal standards. The MBD column chromatography was repeated, and two pools of methylated DNA fragments from a total of 104 μg of tumor DNA were combined. The DNA fragments were subjected to two more rounds of MBD column chromatography to eliminate contaminating moderately methylated DNA fragments. The recovered fragments were ligated to the arms of vector lambda Zap II, and 3 × 105 phages were obtained.
Isolation of Methylated CpG Islands.
Phage clones with long inserts were detected by using PCR. Of 1,067 clones examined, only 60 had Tsp509I fragment inserts longer than 0.8 kb. The majority of clones, some of which were CpG-rich, had shorter inserts (data not shown).
Some CpG-rich multicopy sequences, such as subtelomeric repeat and ribosomal DNA (rDNA) nontranscribed spacer (NTS) region are known to be methylated in somatic cell DNA (19–21). The presence of these sequences was investigated before SPM analysis, and the results revealed that 9 of the 60 clones were derived from such regions (data not shown).
The remaining 51 clones were digested with Tsp509 I/MseI/BfaI/NlaIII and subjected to SPM analysis. A total of 25 retained fragments were obtained from 23 clones. A representative SPM result is shown in Fig. 2 as bands retained on a denaturing gradient gel after prolonged field exposure. Nucleotide sequence analysis of retained fragments revealed that 9 of the 25 retained fragments were derived from CpG islands. The features of these retained DNA fragments are summarized in Table 2. Because our previous study (6, 8) showed that most CpG islands expected in the analyzed fragments could be recovered by using SPM, only few DNA fragments from CpG islands, if any, would be contained in nonretained ones.
Figure 2.
SPM analysis of plasmid clones. Approximately 5 μg of plasmid DNA was serially digested with Tsp509I (AATT), MseI (TTAA), BfaI (CTAG), and NlaIII (CATG) and loaded on an 8% polyacrylamide gel containing a linear gradient of chemical denaturant in a bath maintained at 59.5°C. The denaturant concentration was 9% at the top and 90% at the bottom; 100% denaturant is 7 M urea/40% formamide in 1× TAE buffer (40 mM Tris/20 mM NaOAc/1 mM EDTA, pH 7.4). An electric field of 10 V/cm was applied for 11 h. Lanes 1 and 25, HindIII digests of λ DNA; lanes 2–24, Tsp509I/MseI/BfaI/NlaIII digests of independent plasmid clones.
Clones pSPM210 and pSPM997 had nucleotide sequences identical to the human ENDOG cDNA and the human cDNA for KIAA0445 protein, respectively. Clones pSPM39, 769, and 814 had nucleotide sequences identical to those of human expressed sequence tags (ESTs). Clone pSPM814 was closely similar to the mouse Wnt-5b cDNA. Clone pSPM125 had a sequence identical to that of a reported CpG island clone 31a1 (accession nos. Z58195 and Z58196). Clone pSPM789 had a region similar to an EST sequence vi68e08.r1 (accession number AA497761), which is similar to the nucleotide sequence of human CCAAT-binding transcription factor I subunit A. The remaining two clones, pSPM639 and 845B, did not show any similarity to reported gene-associated sequences, but their nucleotide sequences were identical to part of a human chromosome 5 P1 clone 1307e8 (accession no. AC005355) and a P1 artificial chromosome clone pDJ778a2 sequence derived from human chromosomal region 15q11-q13 (accession no. AC004583), respectively.
Two of the 16 fragments carrying non-island sequences were derived from rDNA regions that were not analyzed and therefore not eliminated before SPM analysis. The majority of the retained non-island fragments were derived from repetitive elements, such as Alu medium reiteration frequency (MER) families and long terminal repeat (LTR) sequences (data not shown). These results suggest that the sequences with repetitive elements are highly methylated.
Investigation of Methylation Status in Genomic DNA.
Tsp509I digests of tumor DNA used in constructing the methylated-DNA library and DNA from the noncancerous portion of the lung of the same patients were subjected to MBD column chromatography to determine their methylation status. Calibration of the MBD column using appropriate DNA fragments was performed before and after separation of genomic fragments.
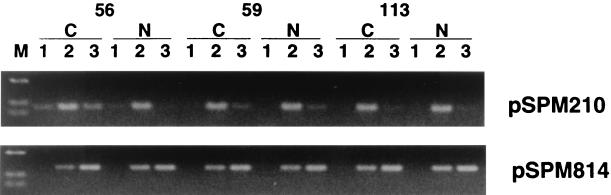
DNA from the flow-through fraction, DNA from lower salt fractions, and DNA from higher salt fractions were subjected to PCR. The representative results from clones pSPM210 and pSPM814 are shown in Fig. 3. In the case of the CpG island corresponding to clone pSPM210, PCR fragments were observed from lower salt fractions but not from the higher salt fractions of normal DNA of patient 56 (Fig. 3, pSPM210, lanes 2 and 3 of 56N). However, in tumor DNA of the same patient, a PCR product was detected from higher salt fractions in addition to the lower salt fractions (Fig. 3, pSPM210, lane 3 of 56C). These results indicate that the CpG island corresponding to pSPM210 is methylated specifically in some tumor cells of this patient. In contrast, no tumor-specific methylation of the CpG island was observed from DNAs of patients 59 and 113.
Figure 3.
Methylation status of CpG islands corresponding to clones pSPM210 and pSPM814. Genomic DNA derived from a cancerous and noncancerous portion of the same patients was digested, fractionated on an MBD column, amplified by PCR, and applied to agarose gel. Numbers indicate a specific patient. C, fractionated template DNA from cancerous portion; N, from apparent noncancerous portion of the lung; M, HaeIII digest of pUC19; 1, flow-through fraction; 2, lower salt fractions (0.40–0.56 M NaCl); 3, higher salt fractions (0.60–1.0 M NaCl).
As observed in CpG islands corresponding to clone pSPM814 (Fig. 3, pSPM814), the remaining eight CpG islands were highly methylated in both normal and tumor DNAs of all nine patients.
Investigation of Methylation Status in DNA from Cell Lines.
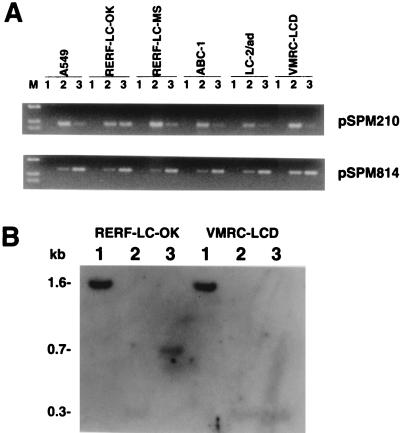
The methylation status of CpG islands corresponding to the observed clones in the genome of several cell lines of human adenocarcinomas of the lung also was analyzed. In the majority of cell lines, with the CpG island corresponding to clone pSPM210 (Fig. 4A), which showed tumor-specific methylation, the level of PCR product from higher salt fractions was very low when compared with that from the lower salt fraction. However, in RERF-LC-OK, similar amounts of PCR products were observed from both lower and higher salt fractions. This result suggests that in RERF-LC-OK cells, one allele of this gene is methylated, whereas the other is not.
Figure 4.
Methylation status of CpG islands corresponding to fragments pSPM210 and pSPM814 in cultured cancer cells. (A) M, HaeIII digest of pUC19; 1, flow-through fraction; 2, lower salt fractions (0.40–0.48 M NaCl); 3, higher salt fractions (0.52–1.0 M NaCl) (B) Southern hybridization experiment using the 0.4-kb ApaI/BstXI insert of clone pSPM210 as a probe. 1, Tsp509I digest; 2, Tsp509I/MspI digest; 3, Tsp509I/HpaII digest.
Sensitivity to methylation-sensitive restriction endonucleases confirms the presence of the methylated CpG island in RERF-LC-OK cells (Fig. 4B). In both RERF-LC-OK and VMRC-LCD DNAs, 1.6-kb Tsp509I fragments were observed in lane 1. Further digestion of VMRC-LCD DNA with MspI or HpaII gave a single 0.3-kb band hybridization pattern. However, in contrast, HpaII digests of RERF-LC-OK DNA gave a 0.7-kb band that was not observed in HpaII digest of VMRC-LCD DNA. These results further indicate the presence of methylated alleles in RERF-LC-OK cells.
With respect to CpG islands corresponding to clone pSPM814 (Fig. 4A), approximately the same level of PCR products was detected from lower and higher salt fractions of cell line VMRC-LCD, as was observed from the DNA derived from tissues (Fig. 3, pSPM814), suggesting methylation of one of the two alleles. However, in the other five cell lines, PCR products were observed at greater levels from higher salt fractions, indicating that the methylated allele of this gene is predominant in five of the cell lines. We confirmed that the absence of signal in the lower salt fraction did not reflect a gene dosage effect associated with sex chromosomes (data not shown). In the case of CpG island corresponding to clone pSPM997, a reduced amount of PCR product from higher salt fractions of cell line ABC-1 was detected, suggesting demethylation, whereas approximately the same level of PCR products was detected from lower and higher salt fractions of the other five cell lines (data not shown).
In the case of CpG islands corresponding to clones pSPM39 and 769, PCR products were observed from both lower and higher salt fractions in all six cell lines (data not shown). The methylation status of the CpG islands corresponding to other clones was not analyzed.
Northern analysis was carried out to correlate methylation to gene silencing, but mRNA corresponding to clones pSPM210 or pSPM814 were not observed in the six cell lines regardless of methylation status (data not shown).
DISCUSSION
We have constructed a library of DNA fragments that are highly methylated in human adenocarcinomas of the lung. DNA fragments associated with CpG islands were then isolated from this library using SPM methodology. Separation of DNA fragments by MBD column chromatography depends primarily on the number of mCpGs within the fragments, as previously reported (5). However, some fragments denser in mCpGs were eluted at greater salt concentrations than might be expected based on the absolute numbers of mCpGs. Hypothetically this could be attributed to protein–protein interactions, which can strengthen binding.
The methylation status of genomic DNA can be conventionally analyzed by digestion with restriction endonucleases that are sensitive to methylation. One of the benefits of MBD column chromatography and SPM is that the separation of DNA fragments is independent of restriction sites within target sequences. Accumulating evidence suggests that the presence of mCpGs among CpG residues within CpG islands in cancer is not necessarily uniformly distributed (15–17). Therefore, an analysis based on the presence of restriction sites can underestimate the extent of methylation. PCR-based methods for isolation of methylated CpG islands or methylated DNA fragments after digestion of genomic DNA with methylation-sensitive restriction endonucleases (22–26) have been reported. However, the usefulness of these approaches remains dependent on the methylation status of restriction sites within the region of DNA. In a method based on arbitrarily primed PCR using a single primer, nucleotide sequences of the primer become critical for detection. Our method is clearly advantageous for an unbiased, comprehensive isolation of methylated CpG islands in cancer cell-derived DNA. Methylation-specific PCR (27) is a robust method to discriminate methylated and nonmethylated alleles. However, when the distribution of mCpGs is not uniform, design of PCR primers becomes difficult, and in many cases, different PCR conditions are required to amplify DNA fragments derived from the methylated and nonmethylated alleles.
Our results demonstrate that there are two types of methylated CpG islands in tumors. One type of CpG island is methylated specifically in tumor DNA and the other type is methylated in normal and tumor DNAs. The former may be associated directly with tumorigenesis. The presence of a methylated allele together with a nonmethylated allele in the same male individual suggests that the corresponding genes are imprinted. Therefore, one possibility to explain the presence of bands from both lower and higher salt fractions is that CpG islands corresponding to clones pSPM39, 125, 639, 769, 789, 814, 845B, and 997 could be associated with imprinted genes. However, allele-specific methylation remains to be elucidated because of the difficulty in distinguishing between the two alleles.
CpG islands of the human ER gene in normal colon epithelial cells has been reported to undergo methylation associated with aging (28). In some cases, for both normal and tumor DNAs, we found that a minor population of DNA fragments containing CpG islands of the human ENDOG gene was eluted in higher salt fractions, for example, in patient 59 (Fig. 3, pSPM210, lane 3 of 59N). The age of this patient was 71, allowing the possibility that this increase in methylation may be associated with aging.
From 1,067 clones, we isolated 3 CpG-rich fragments derived from the NTS region of the rRNA genes (positions 18,644–19,590, accession no. U13369) and 5 from different segments of the same region (positions 41,637–42,486). CpG pairs of the NTS region have been reported to be heavily methylated in lymphocyte and sperm genomic DNA (19). We have found that the NTS regions are extensively methylated both in lung cancer DNA and in normal DNA, and the methylation status of the rDNA genes in the lung cancer DNAs used in the present study does not undergo gross alteration (29). In this study we have analyzed ≈0.3% of the total number of clones in the library (3 × 105), and on the average, four clones derived from the specific segment of the NTS region were detected. From these results, 1,200 clones from the specific NTS region would be expected to be contained in the library. The number of rDNA clones predicted to be in the library corresponds to a three-genome equivalency, because copy numbers of the human rRNA genes are reported to be ≈400 per haploid genome (30). Therefore, the enriched library should contain, on average, three molecules of each mCpG clone of the genome if their methylation status is constant in most tumor DNA samples. Assuming no aberration of methylation signals of imprinted genes in the majority of cancer, we should expect that the CpG islands of many imprinted genes are represented in our library.
In summary, we generated a library of 3 × 105 phage clones from a pool of nine human adenocarcinomas of the lung. From this library, we found 5.6% (60 of 1,067 clones) had long inserts. In further analysis of the 60 clones, 9 had subtelomeric repeat or rRNA NTS region sequences. SPM analysis of the remaining 51 clones resulted in retention of 25 fragments, 9 of which were found to be derived from CpG islands. Of these 9 CpG islands, 1 was methylated in the tumor but not in normal tissue in 1 of the 9 tumor specimens. The other 8 CpG islands were highly methylated, possibly monoallelically, both in normal and tumor tissue. Our results suggest that the number of CpG islands methylated specifically in tumor DNA is much lower than the total number of imprinted genes. However, in our analysis, one CpG island methylated specifically in tumor DNA was identified from a total of 1,067 clones. Therefore, one would expect that hundreds of clones containing CpG islands methylated specifically in tumor DNA would be represented in the complete library. Further characterization of such CpG islands will facilitate understanding of epigenetic aberration in cancer.
Acknowledgments
We thank S. Cross and A. Bird for providing the plasmid clone pET6HMBD and for helpful advice. We also thank J. Graff for providing the unpublished nucleotide sequence data of the human CDH1 gene. Cell line MKN1 was kindly provided by Y. Kanai and S. Hirohashi. Other cell lines used in this study were obtained from the Health Science Research Resources Bank (Osaka) and Riken Cell Bank (Tsukuba). This work was supported in part by a Grant-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan and a Grant-in-Aid for Cancer Research from the Ministry of Health and Welfare of Japan (to M.S.) and by a Grant-in-Aid from the Ministry of Health and Welfare of Japan for the Second-Term Comprehensive 10-Year Strategy for Cancer Control and a Research Grant on Human Genome and Gene Therapy from the Ministry of Health and Welfare, Japan (to T.S.); Y.H.C. was a Research Fellow aided by the Massachusetts Institute of Technology Japan Program.
ABBREVIATIONS
- EST
expressed sequence tag
- MBD
methyl-CpG binding domain
- mCpG
methylated CpG pair
- NTS
nontranscribed spacer
- SPM
segregation of partly melted molecules
Footnotes
References
- 1.Kass S U, Pruss D, Wolffe A P. Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 2.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J-P. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 3.Schmutte C, Jones P A. Biol Chem. 1998;379:377–388. doi: 10.1515/bchm.1998.379.4-5.377. [DOI] [PubMed] [Google Scholar]
- 4.Nan X, Meehan R R, Bird A P. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross S H, Charlton J A, Nan X, Bird A P. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- 6.Shiraishi M, Lerman L S, Sekiya T. Proc Natl Acad Sci USA. 1995;92:4229–4233. doi: 10.1073/pnas.92.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiraishi M, Sekiya T. Proc Japan Acad B. 1996;72:101–103. [Google Scholar]
- 8.Shiraishi M, Oates A J, Xu L, Hosoda F, Ohki M, Alitalo T, Lerman L S, Sekiya T. Nucleic Acids Res. 1998;26:5544–5550. doi: 10.1093/nar/26.24.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiraishi M, Noguchi M, Shimosato Y, Sekiya T. Cancer Res. 1989;49:6474–6479. [PubMed] [Google Scholar]
- 10.John R M, Cross S H. In: Genome Analysis: A Laboratory Manual. Birren B, Hieter P, Myers R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. pp. 217–285. [Google Scholar]
- 11.Gardiner-Garden M, Frommer M. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 12.Shiraishi M, Alitalo T, Sekiya T. DNA Res. 1996;3:425–429. doi: 10.1093/dnares/3.6.425. [DOI] [PubMed] [Google Scholar]
- 13.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 14.Hornstra I K, Yang T P. Mol Cell Biol. 1994;14:1419–1430. doi: 10.1128/mcb.14.2.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stirzaker C, Millar D S, Paul C L, Warnecke P M, Harrison J, Vincent P C, Frommer M, Clark S J. Cancer Res. 1997;57:2229–2237. [PubMed] [Google Scholar]
- 16.Qian X C, Brent T P. Cancer Res. 1997;57:3672–3677. [PubMed] [Google Scholar]
- 17.Dodge J E, List A F, Futscher B W. Int J Cancer. 1998;78:561–567. doi: 10.1002/(sici)1097-0215(19981123)78:5<561::aid-ijc6>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 18.Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S. Proc Natl Acad Sci USA. 1995;92:7416–7419. doi: 10.1073/pnas.92.16.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brock G J, Bird A P. Hum Mol Genet. 1997;6:451–456. doi: 10.1093/hmg/6.3.451. [DOI] [PubMed] [Google Scholar]
- 20.Brown W R A, MacKinnon P J, Villasanté A, Spurr N, Buckle V J, Dobson M J. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- 21.de Lange T, Shiue L, Myers R M, Cox D R, Naylor S L, Killery A M, Vermus H E. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ushijima T, Morimura K, Hosoya Y, Okonogi H, Tatematsu M, Sugimura T, Nagao M. Proc Natl Acad Sci USA. 1997;94:2284–2289. doi: 10.1073/pnas.94.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalgo M L, Liang G, Spruck C H, III, Zingg J-M, Rideout W M, III, Jones P A. Cancer Res. 1997;57:594–599. [PubMed] [Google Scholar]
- 24.Huang T H-M, Laux D E, Hamlin B C, Tran P, Tran H, Lubahn D B. Cancer Res. 1997;57:1030–1034. [PubMed] [Google Scholar]
- 25.Kohno T, Kawanishi M, Inazawa J, Yokota J. Hum Genet. 1998;102:258–264. doi: 10.1007/s004390050689. [DOI] [PubMed] [Google Scholar]
- 26.Liang G, Salem C E, Yu M C, Nguyen H D, Gonzales F A, Nguyen T T, Nicholas P W, Jones P A. Genomics. 1998;53:260–268. doi: 10.1006/geno.1998.5502. [DOI] [PubMed] [Google Scholar]
- 27.Herman J G, Graff J R, Myöhänen S, Nelkin B D, Baylin S B. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Issa J-P, Ottaviano Y L, Celano P, Hamilton S R, Davidson N E, Baylin S B. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 29.Shiraishi M, Sekiguchi A, Chuu Y H, Sekiya T. Biol Chem. 1999;380:81–84. doi: 10.1515/BC.1999.010. [DOI] [PubMed] [Google Scholar]
- 30.Schmickel R D. Pediatr Res. 1973;7:5–12. doi: 10.1203/00006450-197301000-00002. [DOI] [PubMed] [Google Scholar]
- 31.Yasuda J, Mashiyama S, Makino R, Ohyama S, Sekiya T, Hayashi K. DNA Res. 1995;2:45–49. doi: 10.1093/dnares/2.1.45. [DOI] [PubMed] [Google Scholar]