Abstract
Heterozygosity for germ-line mutations in the DNA mismatch repair gene MSH2 predisposes humans to cancer. Here we use a highly sensitive reporter to describe a spontaneous mutator phenotype in diploid yeast cells containing a deletion of only one MSH2 allele. We also identify five MSH2 missense mutations that have dominant mutator effects in heterozygous cells when expressed at normal levels from the natural MSH2 promoter. For example, a 230-fold mutator effect is observed in an MSH2/msh2 diploid strain in which Gly693, which is invariant in MutS homologs and involved in ATP hydrolysis, is changed to alanine. DNA binding data suggest that mismatch repair is suppressed by binding of a mutant Msh2–Msh6 heterodimer to a mismatch with subsequent inability to dissociate from the mismatch in the presence of ATP. A dominant mutator effect also is observed in yeast when Gly693 is changed to serine. An early onset colorectal tumor is heterozygous for the analogous Gly → Ser mutation in hMSH2, and a second hMSH2 mutation was not found, suggesting that this missense mutation may predispose to cancer via a dominant mutator effect. The mutator effects of the deletion mutant and the Gly → Ala missense mutant in yeast MSH2 are enhanced by heterozygosity for a missense mutation in DNA polymerase δ that reduces its proofreading activity but is not a mutator in the heterozygous state. The synergistic effects of heterozygosity for mutations in two different genes that act in series to correct replication errors may be relevant to cancer predisposition.
Keywords: mismatch repair, microsatellite instability, frameshift mutation
DNA mismatch repair (MMR) contributes to genome stability in a dividing cell by correcting replication errors. In Escherichia coli, the first step in MMR is binding of MutS protein to mismatches (for review, see ref. 1). Three eukaryotic MutS homologs (2) also participate in mismatch binding, as either a MutSα heterodimer of Msh2 and Msh6 (3–8) or a MutSβ heterodimer of Msh2 and Msh3 (3, 8–10). Germ-line mutations in one of the two MSH2 alleles predispose humans to hereditary nonpolyposis colon cancer (for review, see ref. 11), with the tumors often containing inactivating mutations in the second allele. An important but unresolved question is whether a mutation in the first allele predisposes heterozygous cells to further mutation. This question has not yet been examined in human or yeast cells. In murine cells, no difference was observed in mutation rates in MSH2 (+/+) and (+/−) cells (12–14). However, mutagenesis was examined at only three loci, whereas eukaryotic genomes contain many genes that can be at greatly different risk of mutation because of loss of MMR (15–16). Even a small increase in mutation rate may be significant, given the many rounds of replication that occur in the lifetime of an organism (for reviews, see refs. 17 and 18). Moreover, the MMR capacity of a cell may not be constant; it may be saturated during periods of rapid proliferation (19–20) or be limited during quiescence (21–22). For these reasons, here we have examined the effect of MSH2 heterozygosity on mutation rates.
Consistent with Knudson’s two-hit hypothesis for inactivation of tumor-suppressor genes (23), tumors arising in hMSH2-mutant hereditary nonpolyposis colon cancer patients often contain nonsense, frameshift, or deletion mutations in the second hMSH2 allele, resulting in complete loss of function. However, missense mutations whose functional consequences are uncertain have also been found, both as germ-line mutations and as somatic mutations in tumors (ref. 11; see also http://www.nfdht.nl/). As one example, the tumor of a patient with early onset colorectal cancer contains a glycine → serine change in hMSH2 (24). A second hMSH2 mutation was not found, leading to the suggestion (24) that wild-type hMSH2p might still be present but its activity suppressed by the mutant protein. The glycine (italicized) is in the consensus sequence of the Walker A motif (GxxxxGKS/T), found in many proteins known to hydrolyze ATP (for review, see refs. 25 and 26). In some ATPases, these residues are known to coordinate the phosphates of ATP (e.g., ref. 27), hence this motif is called the P loop. Mutation of P loop residues in E. coli MutSp and Mshp can inactivate function and are dominant mutators when the genes are overexpressed in wild-type cells (7, 28–30). However, no dominant negative mutators have yet been reported when mutant MutS or Msh proteins are expressed at normal levels, and no MSH gene mutations reported in tumors have been demonstrated to confer a dominant mutator phenotype.
Inactivation of MMR yields a mutator phenotype whose magnitude depends on the reporter system used. In one of the most sensitive mutation detection systems, the rate of loss of a single base pair from an (A–T)14 run in the yeast LYS2 gene is 10,000-fold higher in an msh2 yeast strain than in wild-type yeast cells (32). The high sensitivity of this system allows the detection of subtle losses in MMR activity. By using this system, we provide evidence for a spontaneous mutator phenotype in diploid cells in which one allele is disrupted or contains dominant missense mutations. We further show that the mutator phenotypes are enhanced by a missense mutation in the proofreading exonuclease of DNA polymerase δ.
MATERIALS AND METHODS
Strains and Plasmids.
For all comparisons made in this study, all yeast strains were isogenic. Yeast strains E134 (wild type) and DAG60 (msh2) containing the A14 homonucleotide run in the LYS2 gene have been described (32, 34). Yeast strains containing the msh3, msh6,or msh3 msh6 disruption are isogenic to E134, except for the disruption of the respective mismatch repair gene. The entire ORF of MSH6 was deleted by using a PCR disruption technique with the kanMX module (33) and primers described below. The lower case indicates nucleotide sequences that belong to the kanMX cassette; DNA sequences belonging to the genes are written in upper case. MSH6–kanMX–5′: 5′-CTACCCCTAAAACTTCTAAGACTGCACACTTCGAAAATGGatcgatgaattcgagctcg-3′ and MSH6–kanMX–3′: 5′-GTCCATCTCCGTACGCAATTCGAACGAAATCACTTTGTAA cgtacgctgcaggtcgac-3′. Disruption was verified by PCR using the following primers: MSH6–test–5′: 5′-CAGCTACCCCTAAAACTTC-3′ and MSH6–test–3′: 5′-TTCCAATCATAGTTCAAGACCCC-3′. msh3Δ-hisG was introduced by transformation with EcoRI fragment (msh3Δ∷hisG-URA3-hisG) and subsequent pop-out of the URA3 marker on 5-fluoroorotic acid media. The msh3 disruption was verified by PCR using primers: msh3-1 (5′-ATT AGA GTA GGC TAC AAG TAC-3′) and msh3-2 (5′-AAC ATA CGT ACC ATC CGC ATC-3′). Amplification was for 35 cycles (30 sec at 96°C, 1 min at 55°C, and 6 min at 68°C) by using Taq polymerase (PerkinElmer/Cetus). Strains E68 (MATa ade2–1 arg4–8 leu2–3, 112 thr1–4 trp1–1 ura3–52 lys2Δ cup1–1) and E68 pol3–01 (isogenic to E68 but pol3–01) were used for construction of diploid cells. Plasmid pAC12 with the MSH2 gene under the control of the inducible GAL10 promotor has been described (34). The single-copy vector pEAA39 for expressing MSH2 by the natural promotor has been described (7). Integration vector pRS306 (35) was used to make a replacement by the msh2 point mutation in the genome of the MSH2 strain. For expression of Msh6p for immunoblot and band shift experiments, the Msh6 gene was under the control of the GAL1 promotor on a vector equivalent to pAC12 vector, except for substitution of the LEU2 gene for the URA3 gene.
Media and Reagents.
Media were prepared as described (36), and yeast cells were grown in either YPDA or selective minimal media. Synthetic selective media contained 2% galactose (Pfanstiehl Chemicals) for expressing genes from the GAL1–10 promotor. Oligonucleotides were from Genosys (The Woodlands, TX). Mutagenesis was performed by using the QuikChange kit from Stratagene.
MSH2 Allele Replacement.
The msh2-G693A allele was used to replace the wild-type MSH2 in the genome. The mutated msh2 gene, containing six histidine codons on the 5′ end, was cloned into pRS306 by using restriction enzymes XhoI and KpnI. The vector was linearized with NheI, introduced into yeast strain E134, and selected as described (37). To verify the presence of the mutation, the respective part of the msh2 gene was amplified by using primers 5′-CTGGATACGTTGCGTGATGAAATTC-3′ and 5′-CCATCATATGTACTAGTAC-3′ and Pfu polymerase (Stratagene). Sequencing reactions were performed by using the dRhodamine dye terminator cycle sequencing ready reaction kit (Perkin–Elmer) on a 377 ABI Prism sequencer. Single colonies were checked for a mutator phenotype.
Analysis of Mutators.
To study reversion in a large number of independent clones, a qualitative estimate was used based on a spot test as described (54). Briefly, the cells were plated on selective media and, after growth for 2 to 3 days, replica-plated onto media lacking lysine. After growth for 1 to 3 days, plates were analyzed for the presence of Lys revertants. Mutation rates were determined by fluctuation tests using 9–24 independent cultures, as described (31–32).
Immunoblot Analysis.
Yeast cells were grown in 6 ml of synthetic media lacking uracil overnight at 30°C (36). Cultures were diluted 1:10 with synthetic media lacking uracil, galactose was added to 2% final concentration, and cells were grown for another 8 hr to an OD600 of 0.8–0.9. Cells were harvested by using centrifugation and resuspended in 10% sucrose, 50 mM Tris⋅HCl (pH 7.5), 10 mM EDTA, 10 mM 2-mercaptoethanol, and 1 mg/ml Zymolyase (ICN). After incubation on ice for 30 min, cells were collected and resuspended in 0.5 M NaCl, 50 mM Tris⋅HCl (pH 7.5), 10 mM EDTA, 10 mM 2-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride. Cells were incubated on ice for 30 min, and debris was pelleted by using centrifugation. The supernatant was frozen at −70°C. Protein concentrations were determined by the Bradford dye method (38), using BSA as a standard. Proteins were separated by using PAGE and transferred to an Immobilon-P membrane (Millipore) by using a semi-dry blotting system (W.E.P., Seattle, WA). Polyclonal antibodies were raised in rabbits by using a multiple antigen peptide of amino acids 889–908 of yMsh2 (Research Genetics, Huntsville, AL). An alkaline phosphatase-coupled anti-rabbit antibody (Promega) was used as secondary antibody, and blots were stained by using Western Blue stabilized substrate (Promega).
DNA Mobility Shift Assay.
DNA binding assays were performed with heteroduplices as described (34). For extract preparation, cells were resuspended in 100 mM Tris⋅Cl (pH 8.0), 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, 0.5 mM DTT, and 10% glycerol. Glass beads were added, and three series of 2-min mixing followed by a 2-min incubation on ice were performed. Glass beads and cell debris were pelleted by centrifuging for 40 min. Five nanograms DNA substrate and 20 μg of extract were incubated in a reaction containing 20 mM Tris⋅HCl (pH 7.5), 5 mM MgCl2, 0.1 mM DTT, and 0.01 mM EDTA for 30 min on ice in a total volume of 25 μl. After adding 2 μl of buffer with 50% glycerol and 0.1% bromphenol blue, reactions were subjected to electrophoresis in a 6% polyacrylamide gel in 0.5× TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3). After drying, the gel was analyzed by using a Molecular Dynamics PhosphorImager.
RESULTS
Complementation and Expression of Wild-Type Msh2.
To study Msh2 function in vivo, we used a highly sensitive reporter system with the 61-bp InsE insert in the LYS2 gene, resulting in a +1 frameshift and a Lys− phenotype (31–32). A 4-bp (A/T) run in this insert was changed to a run of 14 consecutive A/T base pairs, again resulting in a +1 frameshift and yielding a Lys− phenotype. The Lys+ reversion rate was 10,000-fold higher in an msh2 mutant strain than in wild-type yeast (Table 1), and all revertants recovered from either strain had lost one A/T base pair from the run (32). Two types of plasmids carrying wild-type or mutant MSH2 genes were introduced into strains carrying this run. One contained a high-copy-number 2 μ plasmid replication origin and MSH2 under the control of the inducible GAL10 promotor. The other is a low-copy-number ARS-CEN plasmid containing the MSH2 gene under the control of its natural promotor (7). In the latter case, Msh2 protein contains a 12CA5 epitope insertion at amino acid 644.
Table 1.
Complementation of msh2 mutator phenotype by the wild-type MSH2 gene on a plasmid
| Strain | Gene in the vector | Promotor | Vector | Mutation rate × 106 | Confidence limits | Relative rate |
|---|---|---|---|---|---|---|
| MSH2* | None | — | None | 0.16 | 0.12–0.24 | 1 |
| msh2* | None | — | None | 1600 | 1,400–2,100 | 10,000 |
| msh2 | None | GAL10 | 2μ | 620 | 530–1,900 | 8,800 |
| MSH2 | GAL10 | 2μ | 0.07 | 0.03–0.2 | 1 | |
| MSH2 | MSH2 | ARS-CEN | 0.64 | 0.4–1.20 | 9 | |
| MSH2 | None | GAL10 | 2μ | 0.14 | 0.06–0.4 | 1 |
| MSH2 | GAL10 | 2μ | 0.15 | 0.12–0.25 | 1 | |
| MSH2 | MSH2 | ARS-CEN | 0.15 | 0.06–0.2 | 1 |
Mutation rates were obtained for (typically) 12 independent colonies. The respective vector construct without the MSH2 gene was not available for the ARS-CEN plasmid.
The mutation rates for these strains have already been described (32).
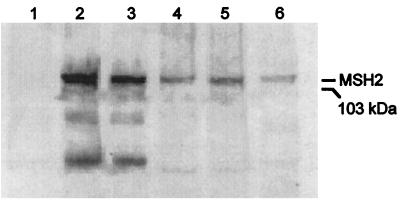
Immunoblot analysis indicated that introduction of wild-type MSH2 on the 2 μ plasmid led to high expression of wild-type Msh2p under induced conditions (Fig. 1, lane 2). Concomitantly, full complementation of the mutator phenotype is observed (Table 1). Introduction of this same vector into a wild-type yeast strain did not alter the mutation rate (Table 1), indicating that excess Msh2p does not interfere with repair of single-base deletion intermediates. Wild-type Msh2p also is detected in cells expressing Msh2p from the natural gene promotor in the ARS-CEN vector (Fig. 1, lane 4). Complementation of the mutator phenotype is observed even at this lower expression level; the mutation rate of an msh2 strain was reduced from 10,000-fold to only 9-fold above the MSH2 strain (Table 1). The fact that complementation was not complete may reflect the presence of the 12CA5 epitope within the MSH2 gene or insufficient Msh2p expression. However, expression observed from the natural promoter in the ARS-CEN vector (Fig. 1, lane 4) is similar to that detected from the natural promoter in the chromosome (Fig. 1, lane 6). The results show that the high frameshift mutation rate of an msh2-null mutant strain can be corrected by wild-type Msh2p expressed from either vector.
Figure 1.
Immunoblot analysis of wild-type and G693A mutant Msh2p. Extracts of msh2 cells were analyzed by using immunoblot analysis as described in Materials and Methods. Lane 1, 12 μg of protein extract from cells containing the control vector lacking Msh2. Lane 2, 12 μg of protein extract from cells expressing wild-type Msh2 from pAC12. Lane 3, 12 μg of protein extract from cells expressing G693A Msh2 from pAC12. Lane 4, 21 μg of protein extract from cells expressing wild-type Msh2 from the natural promoter in pEAA39. Lane 5, 21 μg of protein extract from cells expressing G693A Msh2 from the natural promoter in pEAA39. Lane 6, 21 μg of protein extract from cells expressing wild-type Msh2 from the genome. The mobility of a 103-kDa marker is indicated.
Mutator Effect of Gly693 → Ser Mutation.
We used this system to examine the effect of a yeast msh2 missense mutation analogous to that found in hMSH2 in the tumor of a patient with early onset colorectal cancer (24). Gly693 is one of four conserved residues in MutS family proteins that is in the P loop required for ATP binding and hydrolysis by Msh2p. Haploid msh2 strains expressing the G693S mutant in the Gal10 2μ plasmid or ARS-CEN plasmid have Lys+ reversion rates similar to that of the initial msh2-null mutant strain (Table 2), suggesting that the missense mutation completely inactivates MMR. In a wild-type haploid yeast strain, expression of the G693S msh2 mutant from the GAL10 promoter elevated the Lys+ reversion rate to 1,400 × 10−6, a value similar to that of a strain completely lacking Msh2p. Expression from the MSH2 promoter in the ARS-CEN plasmid produced a 7-fold increase in reversion rate over that of a wild-type strain (Table 3). Thus, the G693S mutation produces a dominant mutator phenotype in a wild-type cell when expressed at normal level.
Table 2.
Complementation of an msh2 strain by genes harboring mutations in the P loop
| MSH2 allele in the vector |
GAL10 inducible promotor 2μ plasmid
|
MSH2 promotor ARS-CEN plasmid
|
||||
|---|---|---|---|---|---|---|
| MR × 106 | CL × 106 | RR | MR × 106 | CL × 106 | RR | |
| None | 620 | 530–1,800 | 8,800 | ND | ||
| MSH2 | 0.07 | 0.03–0.2 | 1 | 0.64 | 0.4–1 | 1 |
| G693S | 2,600 | 1,800–7,300 | 37,000 | 1,100 | 990–2,200 | 1,700 |
| G688A | 14 | 4–82 | 200 | 140 | 98–220 | 220 |
| G693A | 1300 | 930–2,000 | 18,000 | 270 | 50–600 | 430 |
| K694A | 1600 | 1,100–1,900 | 22,000 | 680 | 75–940 | 1,100 |
| S695A | 1400 | 10–1,800 | 20,000 | 550 | 84–720 | 840 |
MR, median Lys+ reversion rates for (typically) 12 individual colonies; CL, 95% confidence limits; RR, relative rate, in reference to wild type; ND, not determined.
Table 3.
Dominant negative mutator effects of msh2 P-loop mutants in an MSH2 strain
| MSH2 allele in the vector |
GAL10 inducible promotor 2μ plasmid
|
MSH2 promotor ARS-CEN plasmid
|
||||
|---|---|---|---|---|---|---|
| MR × 106 | CL × 106 | RR | MR × 106 | CL × 106 | RR | |
| None | 0.14 | 0.1–0.4 | 0.9 | ND | ||
| MSH2 | 0.15 | 0.12–0.25 | 1 | 0.15 | 0.06–0.2 | 1 |
| G693S | 1,400 | 1,200–2,000 | 9,300 | 1.1 | 0.8–1.4 | 7 |
| G688A | 3 | 0.2–11 | 20 | 1.2 | 0.6–4.7 | 8 |
| G693A | 1,050 | 900–2,600 | 7,000 | 5.1 | 1.5–6.7 | 34 |
| K694A | 920 | 600–2,600 | 6,100 | 0.6 | 0.3–1.8 | 4 |
| S695A | 4,100 | 1,800–7,400 | 27,000 | 0.6 | 0.2–1.1 | 4 |
MR, median Lys+ reversion rates for (typically) 12 individual colonies; CL, 95% confidence limits; RR, relative rate, in reference to wild type; ND, not determined.
Mutator Effects of Other Missense Mutations.
To determine whether the dominant mutator effect is unique to this particular missense mutation, we examined the phenotypes of msh2 mutations in four conserved P-loop residues. Amino acids G688, G693, K694, and S695 were individually replaced with alanine. Alanine is a conservative change for glycine and is intended to reduce or eliminate interactions of the lysine and serine side chains with ATP. When expressed from the GAL10 2μ plasmid or the ARS-CEN plasmid, three of the mutants (G693A, K694A, and S695A) were unable to complement the mutator phenotype of an msh2 haploid yeast strain (Table 2). The high Lys+ reversion rates suggest complete or near-complete loss of mismatch repair function.† The strain with the G688A mutant expressed from the GAL10 promoter had a reversion rate that is lower but still 200-fold higher than the wild-type strain and 10-fold higher when expressed from the MSH2 promotor (Table 2). The data suggest that the G688A mutation is inactivating but retains some function.
When the four alanine mutant alleles were expressed from the GAL10 promoter in an MSH2 strain, all generated a mutator phenotype. The dominant mutator effect is several thousand-fold for the G693A, K694A, and S695A mutants and 20-fold for the G688A mutant.‡ Mutant proteins also show 4- to 34-fold greater effects when expressed from the natural Msh2 promotor in the ARS-CEN plasmid (Table 3). We used the G693A mutant for further study because it conferred the strongest mutator effect when expressed from the MSH2 promoter. The amount of G693A mutant protein expressed was examined by using immunoblot analysis (Fig. 1). Expression levels were similar to those for wild-type Msh2 protein, and were high with the GAL10 promoter (lane 3) and near normal with the natural promoter (compare lane 5 to lanes 4 and 6). We then determined whether the dominant mutator effect could be seen with a LYS2 allele that scores additions rather than deletions. The rate of single-base additions in an (A)12 homonucleotide run in the LYS2 gene was elevated 230-fold and 9-fold when the G693A mutation was expressed from the GAL10 or ARS-CEN plasmids, respectively (data not shown). For comparison, the reversion rate for this LYS2 allele is elevated 400-fold in an msh2-null mutant strain (32). Thus, the G693A mutation elevates the rate of addition mutations. We then examined the effect on the rate of mutation to canavanine resistance. With this gene, the mutation rate is ≈40-fold higher in an msh2 strain than in a wild-type strain, and 85% of the mutations are single-base frameshifts in shorter homonucleotide runs (39). When the G693A allele was expressed from the GAL10 promoter in a wild-type haploid yeast strain, it produced a 33-fold increase in the rate of canavanine resistance, suggesting near-complete suppression of MMR. No mutator effect was seen when the G693A mutant was expressed from the MSH2 promoter in the ARS-CEN plasmid (data not shown), likely because of the 10- to 100-fold lower sensitivity of this system.
Dominant Mutator Phenotype of G693A in an msh3 Strain.
Current evidence for MMR in yeast suggests that Msh2 forms heterodimers with either Msh3 or Msh6 and that either heterodimer can bind to single-base frameshift intermediates. To test a requirement for Msh6 and/or Msh3 for the dominant mutator phenotype of the G693A mutant, we examined reversion rates at the (A)14 run in strains expressing G693A Msh2p but disrupted for either msh3 or msh6. Disruption of MSH6 or MSH3 resulted in 140- and 15-fold increases, respectively, in reversion rates compared with a wild-type strain. This is consistent with studies demonstrating that both genes contribute to the stability of homonucleotide runs (40–41). Expression of the G693A mutant protein from the ARS-CEN plasmid in the msh6 mutant strain increased the reversion rate by <2-fold, whereas expression of the G693A mutant protein from the ARS-CEN plasmid in the msh3 strain resulted in a 6-fold increase in reversion rate. The minimal dominant mutator effect in a strain lacking Msh6p and the clear effect in a strain lacking Msh3p suggest that dominance by the G693A Msh2p mutant depends primarily on Msh6p.
G/T Mismatch Binding by Msh2–Msh6 Complexes.
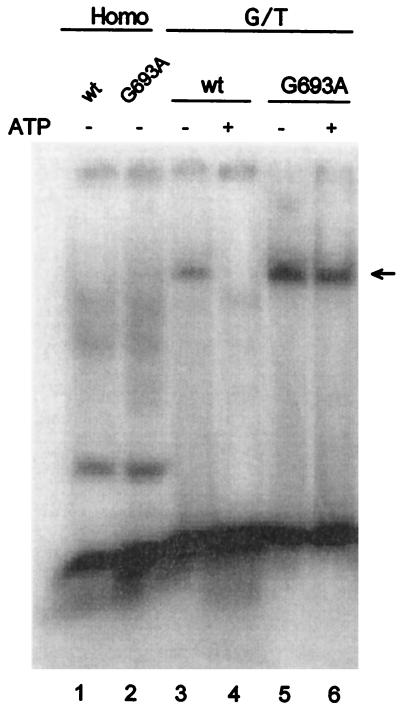
One hypothesis for the dominant mutator effect consistent with the above observations and the literature (7) is binding of an Msh2G693A–Msh6 mutant heterodimer to a mismatch with a subsequent defect in ATP-dependent dissociation from the DNA. To test this, we performed bandshift experiments with extracts of msh2 mutant cells coexpressing Msh6 protein with either wild-type or G693A mutant Msh2p. Extracts of galactose-induced yeast cells harboring an Msh6 expression vector and the wild-type or G693A Msh2 expression vector showed no specific binding to homoduplex DNA (Fig. 2 lanes 1 and 2). With DNA containing a G/T mismatch, these same extracts (Fig. 2 lanes 3 and 5) yield a novel band of slower mobility (indicated by an arrow). Immunoblot analysis confirmed that this band contains Msh2 protein, and its appearance also requires the Msh6 expression vector (data not shown), indicating that both Msh2 and Msh6 are present. When ATP is added to the binding reaction, no shifted band is detected for the wild-type complex (Fig. 2 lane 4), consistent with previous reports of ATP-dependent release of the yeast Msh2–Msh6 complex from a mispair (5, 42–43). In contrast, an extract expressing the G693A mutant protein did yield a shifted complex even in the presence of ATP (Fig. 2, lane 6).§ These data indicate that a Msh2G693A–Msh6 mutant heterodimer forms that retains mismatch binding capacity but is less sensitive to release in the presence of ATP than is wild-type MutSα.
Figure 2.
Band shifts with cell extracts overexpressing MSH2 and MSH6. Five nanograms of DNA and cell extracts were mixed in binding buffer and incubated for 30 min on ice. Samples were analyzed in a 6% polyacrylamide gel, which was then examined by using a PhosphorImager. The arrow denotes the position of the Msh2–Msh6–DNA complex. Reactions contained 20 μg of protein from cells harboring the Msh6 expression vector and overexpressing wild-type or G693A Msh2. Where indicated, ATP was added to a final concentration of 5 mM. Addition of 20- and 40-fold excesses of unlabeled heteroduplex reduced by 16-fold and 65-fold, respectively, the amount of G/T heteroduplex complex shifted by Msh2–Msh6 (data not shown). In a repeat experiment, the intensity of the band shifted by the wild-type cell extract in the absence of ATP was higher than shown here in lane 3.
Dominant Effect of G693A as a Genomic Allele.
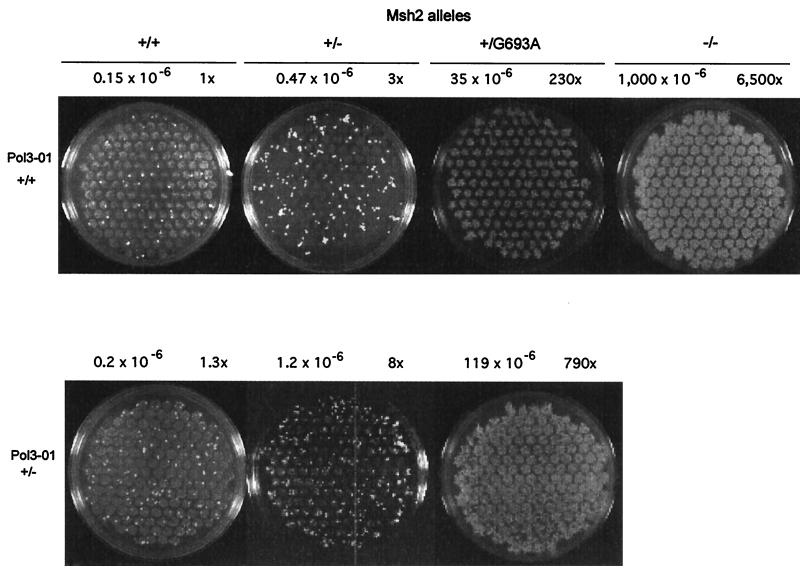
Despite the almost-normal level of expression of Msh2p from the ARS-CEN plasmid, it is possible that some cells may contain more than one copy of the plasmid, thus partly accounting for the dominant mutator phenotype (Table 3). To determine whether the dominant mutator phenotype of G693A and G693S mutants is observed when these alleles are present in its natural position in the chromosome, a genomic copy of MSH2 was replaced by the mutant alleles. As expected, a diploid strain containing two wild-type MSH2 alleles has a low reversion rate (Fig. 3) and a diploid strain disrupted for both MSH2 alleles has a high rate (Fig. 3). Both values are comparable to reversion rates for the haploid strains (Table 1). A heterozygous strain with one MSH2 allele disrupted had a 3-fold increase in mutation rate. This increase was statistically significant as determined by using Mann–Witney U test (P < 0.05) (44). A diploid strain containing one wild-type MSH2 allele and one G693A mutant allele has a mutation rate elevated 230-fold (Fig. 3). When Gly693 was replaced by a serine, a 130-fold increase in mutation rate is observed. These data indicate that the MSH2/msh2 strain has a slight spontaneous mutator phenotype, and that a strong dominant mutator phenotype is conferred by expression of the G693A or G693S msh2 mutant allele from the natural promotor on the chromosome.
Figure 3.
Dominant mutator effects in diploid yeast strains. The plates show a qualitative spot test for reversion of diploid yeast strains, performed as described (54). For strains with lower mutation rates (plates on left side), when multiple spots of cells were plated on media lacking lysine, single-revertant colonies are visible above the background spots. Strains with high reversion rates (Upper, +/G693A and −/−) have spots showing confluent growth of many Lys+ revertant colonies. Mutation rates (determined as described in ref. 32) are shown above each plate, together with the increases in rates relative to the wild-type MSH2/MSH2 strain. The respective lower and upper 95% confidence limits for these rates (×106) are MSH2/MSH2 POL3/POL3, 0.07 and 0.36; MSH2/msh2 POL3/POL3, 0.23 and 1.0; MSH2/msh2G693A POL3/POL3, 1.3 and 58; msh2/msh2 POL3/POL3, 730 and 1,300; MSH2/MSH2 POL3/pol3-01, 0.1 and 0.6; MSH2/msh2 POL3/pol3-01, 0.9 and 2.2; and MSH2/msh2G693A POL3/pol3-01, 64 and 210.
Mutator Effect in MSH2/msh2 Heterozygotes Combined with a DNA Polymerase δ Mutation.
We next examined these effects in a strain (pol3-01) altered in the 3′ → 5′ exonuclease activity of DNA polymerase δ (45). The Lys+ mutation rate in the (A)14 run in the diploid strain heterozygous for the pol3-01 mutation is similar to that of a homozygous POL3 wild-type strain when these strains harbor two wild-type copies of MSH2 (Fig. 3). Thus, the presence of the pol3-01 mutation in only one allele alone does not yield a mutator phenotype. However, the reversion rate in the double heterozygote MSH2/msh2 POL3/pol3-01 strain is 8-fold higher than for the wild-type MSH2/MSH2 POL3/POL3 strain and 6-fold higher than for the MSH2/MSH2 POL3/pol3-01 strain (Fig. 3), clearly illustrating that loss of one MSH2 allele yields a mutator phenotype. In addition, the dominant mutator effect of the G693A and the G693S allele is even more apparent in the POL3/pol3-01 background [790-fold (Fig. 3) and 230-fold, respectively].
DISCUSSION
These observations in diploid yeast heterozygous for MSH2 mutations may be relevant to the relationship between genome instability and tumorigenesis. The increased mutation rate of a heterozygous cell with a disruption of one MSH2 allele (Fig. 3) places the remaining wild-type MSH2 allele at higher risk for mutations that could lead to even greater loss of MMR activity. The mutator phenotype of MSH2/msh2Δ heterozygote is stronger in a strain that is concomitantly heterozygous for a mutation in the DNA polymerase δ gene. This situation may model subpopulations of MSH2/msh2 heterozygotes at greater than normal risk of disease by virtue of mutations in other genes that allow or even promote replication errors. The elevated mutation rates in heterozygous yeast strains may result from occasional loss of the wild-type MSH2 allele in a subpopulation of heterozygous cells, e.g., by mitotic recombination. They may also reflect a more limited MMR capacity in cells containing only one wild-type MSH2 allele, which could render heterozygous cells at greater risk of mutation under conditions of rapid proliferation (19–20) or quiescence (21–22). Heterozygosity for a defect in the 3′ → 5′ exonuclease of DNA polymerase δ may enhance the mutator effect by allowing more replication errors to escape proofreading. Alternatively, DNA polymerase δ can resynthesize DNA during MMR (46–47), and heterozygosity for a defect in its intrinsic exonuclease may directly reduce MMR if this activity participates in the excision reaction.
Given that a null mutant has a mutation rate elevated 10,000-fold (Table 1), the 3- and 8-fold mutator effects in MSH2/msh2Δ heterozygotes (Fig. 3) are small and only detected here by using a highly sensitive reporter gene. Nevertheless, as noted by Loeb (18), even small changes in mutation rates may be highly significant for initiation or promotion of tumorigenesis in humans, because many rounds of replication occur in a lifetime. Moreover, large eukaryotic genomes contain numerous repetitive coding sequences. For example, 25% of yeast gene coding sequences contain homonucleotide runs of eight or more base pairs (ref. 16, see also http://www.niehs.nih.gov/science/HTML/tablelink.html). Such sequences can be particularly susceptible to frequent replication slippage and therefore highly dependent on MMR for stability. In fact, frameshift mutations in homonucleotide runs are frequently observed in the coding sequences of genes found to be mutated in tumors (48–52).
As mentioned above (23), tumors arising in hMSH2-mutant hereditary nonpolyposis colon cancer patients often contain nonsense, frameshift, or deletion mutations in the second hMSH2 allele, resulting in complete loss of function. In contrast, the five msh2 alleles listed in Table 3 contain missense mutations that produce a dominant mutator phenotype when expressed at a normal level from the MSH2 promoter. In an early onset, sporadic colon tumor exhibiting microsatellite instability (24), serine is substituted for glycine in hMSH2 at amino acid 674, the homolog of G693 in yeast Msh2p. Our data showing dominant mutator effects for the G693S and G693A msh2 mutations (Table 3; Fig. 3) imply that a G674S mutant of hMSH2 may yield a dominant mutator phenotype when expressed with a wild-type hMSH2 allele and that this is associated with the early onset of colon cancer (24). The fact that a second mutation in hMSH2 was not found in the tumor DNA of this patient supports this hypothesis.
Yeast Msh2 and other MutS homologues share the P-loop motif with a variety of proteins with ATPase activity (25–26). The phenotypes observed here when serine or alanine is substituted for four invariant P-loop residues are interesting given that the P loop participates in the ATP binding and hydrolysis required for several MMR steps (3–8, 30, 42–43, 53). The intermediate mutator phenotype of the G688A mutant when expressed in an msh2Δ haploid yeast strain (Table 2) suggests partial retention of MMR function, despite the fact that this glycine is invariant among MutS homologues and that studies of other P-loop-containing proteins suggest that it is important for P-loop structure (27). Residual MMR function may reflect the conservative Gly → Ala substitution, which replaces a hydrogen with a methyl group. The dominant mutator effect in wild-type yeast expressing the G688A mutant (Table 3) suggests that the mutant protein does interfere with MMR, even when expressed from the MSH2 promoter in the ARS-CEN plasmid (Table 3).
The inability of the K694A and S695A missense mutants to complement an msh2 strain (Table 2) suggests complete loss of MMR function, perhaps because of alterations of important contacts with ATP (e.g., see ref. 27) or altered ability to change protein conformation. Expression of G693S or G693A also failed to complement the high mutation rate of an msh2 strain, consistent with the structural importance of this glycine inferred from studies of other P-loop-containing proteins. The G693S or G693A mutants also produce dominant mutator effects when overexpressed (Table 3), similar to results for expression of P-loop missense mutations from high-copy-number plasmids in E. coli MutS (29) and yeast Msh2 (7). However, the earlier studies did not examine (29) or failed to detect (7) a dominant mutator effect for expression of P-loop mutants from the natural promoter. Here, when the G693S or G693A mutant genes were expressed from the MSH2 promoter at a lower level or in diploid Msh2/msh2G693A/s strains, clear dominant mutator effects are still observed by using the highly sensitive lys2 (A)14 and (A)12 alleles. The bandshift data (Fig. 3) reveal that mutant Msh2G693A–Msh6 can still bind to a mismatch. This is similar to the mismatch binding capacity retained by the homologous Gly → Ala mutation in a fragment of hMSH2 (53) or by a G → D mutation in yeast Msh2 when complexed with Msh6 (7). Unlike wild-type Msh2–Msh6, the mutant Msh2G693A–Msh6 (Fig. 3) and the Msh2G693D–Msh6 complex (7) did not completely release from the mismatch in the presence of ATP. This suggests that the dominant mutator effect results from formation of a Msh2G693A–Msh6 complex that binds to mismatches but does not completely release in the presence of ATP, and therefore inhibits repair by wild-type Msh2–Msh6. The fact that the G693S and G693A produce increases in mutation rate that are smaller than the 10,000-fold effect characteristic of complete inactivation of MMR suggests that the mutant repair complex may still retain some ability to dissociate from the mismatch. Such incomplete dominance in humans may result in low penetrance for diseases such as colon cancer.
Acknowledgments
We thank Samuel E. Bennett for providing substrates for the bandshift assay and Polina Shcherbakova and Lars Pedersen for critical comments on the manuscript. K.D. is recipient of a Curt Engelhorn fellowship granted from Boehringer Mannheim and German Cancer Research Center.
ABBREVIATION
- MMR
mismatch repair
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Mutation rates were 2- to 4-fold higher when these mutants were expressed from the GAL10 promoter than when expressed from the MSH2 promoter. For G693A and K694A, the lower 95% confidence limit for the rate when expressed from the GAL10 promoter is higher than the upper 95% confidence limit for the rate when expressed from the natural promoter, indicating that these differences may be significant.
We have not observed such dominant negative effects with several other MSH2 missense mutant alleles (K.D., A.B.C., and T.A.K., unpublished data), indicating that the dominant negative effects shown in Table 3 are (thus far) specific to these alleles.
Analysis of the band intensities in Fig. 2 suggests that some ATP-dependent dissociation still occurs with the G693A mutant complex.
References
- 1.Prolla T A. Curr Opin Cell Biol. 1998;10:311–316. doi: 10.1016/s0955-0674(98)80005-7. [DOI] [PubMed] [Google Scholar]
- 2.Eisen J A. Nucleic Acids Res. 1998;26:4291–4300. doi: 10.1093/nar/26.18.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drummond J T, Li G-M, Longley M J, Modrich P. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 4.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D’Arrigo A, Truong O, Hsuan J J, Jiricny J. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 5.Iaccarino I, Palombo F, Drummond J, Totty N F, Hsuan J J, Modrich P, Jiricny J. Curr Biol. 1996;6:484–486. doi: 10.1016/s0960-9822(02)00516-x. [DOI] [PubMed] [Google Scholar]
- 6.Acharya S, Wilson T, Gradia S, Kane M F, Guerrette S, Marsischky G T, Kolodner R, Fishel R. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alani E, Sokolsky T, Studamire B, Miret J J, Lahue R S. Mol Cell Biol. 1997;17:2436–2447. doi: 10.1128/mcb.17.5.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genschel J, Littman S J, Drummond J T, Modrich P. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 9.Habraken Y, Sung P, Prakash L, Prakash S. Curr Biol. 1996;6:1185–1187. doi: 10.1016/s0960-9822(02)70686-6. [DOI] [PubMed] [Google Scholar]
- 10.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 11.Peltomäki P, Vasen H F A. Gastroenterology. 1997;113:1146–1158. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- 12.Reitmair A H, Risley R, Bristow R G, Wilson T, Ganesh A, Jang A, Peacock J, Benchimol S, Hill R P, Mak T W, et al. Cancer Res. 1997;57:3765–3771. [PubMed] [Google Scholar]
- 13.Andrew S E, Reitmair A H, Fox J, Hsiao L, Francis A, McKinnon M, Mak T W, Jirik F R. Oncogene. 1997;15:123–129. doi: 10.1038/sj.onc.1201180. [DOI] [PubMed] [Google Scholar]
- 14.DeWeese T L, Shipman J M, Larrier N A, Buckley N M, Kidd L R, Groopman J D, Cutler R G, Riele H T, Nelson W G. Proc Natl Acad Sci USA. 1998;95:11915–11920. doi: 10.1073/pnas.95.20.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunkel T A, Resnick M A, Gordenin D A. Cell. 1997;88:155–158. doi: 10.1016/s0092-8674(00)81832-2. [DOI] [PubMed] [Google Scholar]
- 16.Gordenin D A, Resnick M A. Mutat Res. 1998;400:45–58. doi: 10.1016/s0027-5107(98)00047-5. [DOI] [PubMed] [Google Scholar]
- 17.Jackson A L, Loeb L A. Genetics. 1998;148:1483–1490. doi: 10.1093/genetics/148.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loeb L A. Adv Cancer Res. 1998;72:25–56. doi: 10.1016/s0065-230x(08)60699-5. [DOI] [PubMed] [Google Scholar]
- 19.Schaaper R M. Proc Natl Acad Sci USA. 1988;85:8126–8130. doi: 10.1073/pnas.85.21.8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaaper R M. Genetics. 1989;121:205–212. doi: 10.1093/genetics/121.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg S M, Harris R S, Longerich S, Galloway A M. Mutat Res. 1996;350:69–76. doi: 10.1016/0027-5107(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 22.Richards B, Zhang H, Phear G, Meuth M. Science. 1997;277:1523–1526. doi: 10.1126/science.277.5331.1523. [DOI] [PubMed] [Google Scholar]
- 23.Knudson A G. Ann NY Acad Sci. 1997;833:58–67. doi: 10.1111/j.1749-6632.1997.tb48593.x. [DOI] [PubMed] [Google Scholar]
- 24.Børresen A-L, Lothe R A, Meling G I, Lystad S, Morrison P, Lipford J, Kane M F, Rognum T O, Kolodner R D. Hum Mol Genet. 1995;4:2065–2072. doi: 10.1093/hmg/4.11.2065. [DOI] [PubMed] [Google Scholar]
- 25.Gorbalenya A E, Koonin E V. J Mol Biol. 1990;213:583–591. doi: 10.1016/S0022-2836(05)80243-8. [DOI] [PubMed] [Google Scholar]
- 26.Walker J E, Saraste M, Runswick M J, Gay N J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pai E F, Krengel U, Petsko G A, Goody R S, Kabsch W, Wittinghofer A. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haber L T, Walker G C. EMBO J. 1991;10:2707–2715. doi: 10.1002/j.1460-2075.1991.tb07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu T-H, Marinus M G. J Bacteriol. 1994;176:5393–5400. doi: 10.1128/jb.176.17.5393-5400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iaccarino I, Marra G, Palombo F, Jiricny J. EMBO J. 1998;17:2677–2686. doi: 10.1093/emboj/17.9.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran H T, Degtyareva N P, Nadejda N K, Sugino A, Masumoto H, Gordenin D A, Resnick M A. Mol Cell Biol. 1995;15:5607–5617. doi: 10.1128/mcb.15.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran H T, Keen J D, Kricker M, Resnick M A, Gordenin D A. Mol Cell Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wach A, Brachat A, Pohlmann R, Phillipson R. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 34.Clark A B, Cook M E, Tran H T, Gordenin D A, Resnick M A, Kunkel T A. Nucleic Acids Res. 1999;27:736–742. doi: 10.1093/nar/27.3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 37.Boeke J D, LaCroute F, Fink G R. Mol Gen Genet. 1984;197:345–356. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 38.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 39.Marsischky G T, Filosi N, Kane M F, Kolodner R. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 40.Greene C N, Jinks-Robertson S. Mol Cell Biol. 1997;17:2844–2850. doi: 10.1128/mcb.17.5.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sia E A, Kokoska R J, Dominska M, Greenwell P, Petes T D. Mol Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alani E. Mol Cell Biol. 1996;16:5604–5615. doi: 10.1128/mcb.16.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Habraken Y, Sung P, Prakash L, Prakash S. J Biol Chem. 1998;273:9837–9841. doi: 10.1074/jbc.273.16.9837. [DOI] [PubMed] [Google Scholar]
- 44.Motulsky H. Intuitive Biostatistics. New York: Oxford Univ. Press; 1995. pp. 217–224. [Google Scholar]
- 45.Morrison A, Johnson A L, Johnston L H, Sugino A. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Longley M J, Pierce A J, Modrich P. J Biol Chem. 1997;272:10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 47.Gu L, Hong Y, McCulloch S, Watanabe H, Li G-M. Nucleic Acids Res. 1998;26:1173–1178. doi: 10.1093/nar/26.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J, Papadopoulos N, McKinley A J, Farrington S M, Curtis L J, Wyllie A H, Zheng S, Willson J K V, Markowitz S D, Morin P, et al. Proc Natl Acad Sci USA. 1996;93:9049–9054. doi: 10.1073/pnas.93.17.9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Markowitz S D, Wang L, Myeroff L L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, et al. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 50.Parsons R, Myeroff L L, Liu B, Willson J K V, Markowitz S D, Kinzler K W, Vogelstein B. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- 51.Wang J, Sun L, Myeroff L L, Wang X, Gentry L E, Yang J, Liang J, Zborowska E, Markowitz S D, Willson J K V, Brattain M G. J Biol Chem. 1995;270:22044–22049. doi: 10.1074/jbc.270.37.22044. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto H, Sawai H, Perucho M. Cancer Res. 1997;57:4420–4426. [PubMed] [Google Scholar]
- 53.Gradia S, Acharya S, Fishel R. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 54.Whitehouse A, Parmar R, Deeble J, Taylor G R, Phillips S E V, Meredith D M, Markham A F. Biochem Biophys Comm. 1996;229:147–153. doi: 10.1006/bbrc.1996.1771. [DOI] [PubMed] [Google Scholar]
- 55.Gordenin D A, Proscyavishus Y Y, Malkova A L, Trofimova M V, Peterzen A. Yeast. 1991;7:37–50. doi: 10.1002/yea.320070105. [DOI] [PubMed] [Google Scholar]