Abstract
Hereditary hemochromatosis (HH) is a common autosomal recessive disorder characterized by tissue iron deposition secondary to excessive dietary iron absorption. We recently reported that HFE, the protein defective in HH, was physically associated with the transferrin receptor (TfR) in duodenal crypt cells and proposed that mutations in HFE attenuate the uptake of transferrin-bound iron from plasma by duodenal crypt cells, leading to up-regulation of transporters for dietary iron. Here, we tested the hypothesis that HFE−/− mice have increased duodenal expression of the divalent metal transporter (DMT1). By 4 weeks of age, the HFE−/− mice demonstrated iron loading when compared with HFE+/+ littermates, with elevated transferrin saturations (68.4% vs. 49.8%) and elevated liver iron concentrations (985 μg/g vs. 381 μg/g). By using Northern blot analyses, we quantitated duodenal expression of both classes of DMT1 transcripts: one containing an iron responsive element (IRE), called DMT1(IRE), and one containing no IRE, called DMT1(non-IRE). The positive control for DMT1 up-regulation was a murine model of dietary iron deficiency that demonstrated greatly increased levels of duodenal DMT1(IRE) mRNA. HFE−/− mice also demonstrated an increase in duodenal DMT1(IRE) mRNA (average 7.7-fold), despite their elevated transferrin saturation and hepatic iron content. Duodenal expression of DMT1(non-IRE) was not increased, nor was hepatic expression of DMT1 increased. These data support the model for HH in which HFE mutations lead to inappropriately low crypt cell iron, with resultant stabilization of DMT1(IRE) mRNA, up-regulation of DMT1, and increased absorption of dietary iron.
Keywords: intestine, iron responsive element
Hereditary hemochromatosis (HH) is a common disorder of iron homeostasis in which the intestinal absorption of iron is excessive in relation to body iron status (1–5). The excess iron is deposited in the parenchyma of many tissues, leading to tissue damage and organ failure. Clinical consequences include liver cirrhosis, hepatocellular carcinoma, diabetes, heart failure, arthritis, and hypogonadism (6–9). The gene defective in HH, designated HFE, was found to encode a major histocompatibility complex (MHC) class I-like integral membrane protein (10) that had no obvious relationship to iron absorption. A link between HFE and iron metabolism was provided by the observations in human placenta that the HFE protein is localized on the apical surface of syncytiotrophoblast cells (the site of transferrin-mediated maternal–fetal iron transport) and is physically associated with the transferrin receptor (TfR) (11). Physical association between TfR and expressed recombinant HFE protein was also reported in cultured cells (12, 13) and in vitro (14). In cell culture, overexpressed recombinant HFE (but not HFE carrying the HH mutation) was reported to reduce the affinity of TfR for holotransferrin, suggesting a role for normal HFE in down-regulating transferrin-mediated iron uptake. Although loss of this down-regulation of transferrin-mediated iron transport might explain the excess deposition of tissue iron in HH patients, it would not explain the excess absorption of dietary iron (15). Studies on HH patients suggest that the primary defect is loss of the normal feedback mechanisms regulating absorption of dietary (nontransferrin-bound) iron across the intestinal mucosa (2–5).
Dietary iron absorption is normally tightly linked with body utilization through the sensing of body iron status in the proximal small intestine (16, 17). Several lines of evidence indicate that the body iron status is detected by the uptake of transferrin-bound iron from plasma at the basolateral surface of intestinal crypt cells (18, 19). We recently demonstrated that HFE colocalizes with and is physically associated with TfR in these cells (1). This observation suggests a mechanism by which HFE might participate in sensing body iron status by modulating transferrin-mediated uptake of plasma iron in the crypt cells. We propose that mutations of HFE in HH patients impair transferrin-mediated iron uptake and thus decrease crypt cell uptake of plasma iron. The resulting low intracellular iron pool would in turn lead to increased dietary iron absorption by daughter enterocytes. The increased absorption of dietary iron seen in response to iron deficiency has been attributed to duodenal up-regulation of the divalent metal ion transporter (DMT1) (20). We thus predicted that a relatively low intracellular iron pool in duodenal crypt cells in HH would likewise increase duodenal expression of DMT1, despite the deposition of iron in other tissues. To test this prediction, we utilized a murine model of HH in which the HFE gene has been disrupted by targeted recombination (21) and quantified expression of DMT1 mRNA in duodenal tissue. The results presented here show that the HFE−/− mice, despite elevated serum transferrin saturation and hepatic iron loading, have increased duodenal expression of DMT1 transcripts containing an iron responsive element (IRE).
MATERIALS AND METHODS
Animals.
F1 generation mice heterozygous for a targeted disruption of the HFE gene (21) were mated to produce F2 generation littermates homozygous for the wild-type (+/+) and mutant (−/−) HFE genes. Mice were weaned at 3 weeks and maintained thereafter on a standard diet [LM-485 Teklad sterilized mouse diet 7012, Harlan, which contains 0.02% (wt/wt) iron]. Fifteen HFE+/+ mice and 15 HFE−/− littermates (representing four total litters) were analyzed at 4 weeks of age. Animals were fasted for 14 h before blood sampling. After blood was obtained, the mice were sacrificed and liver tissue and duodenum (the 1.5-cm length of small intestine distal to the pylorus) were dissected for RNA isolation.
To study the effect of dietary iron deprivation on DMT1 expression in mice, duodenal tissue was obtained from adult mice with iron deficiency anemia secondary to stomach parietal cell ablation [H+/K+-ATPase β subunit (−1035 to +24)/DT-A] (22). In these experiments, nontransgenic FVB/N littermates with normal iron status were used as controls.
Genotyping.
PCR analysis of tail DNA was used to identify mice with the disrupted HFE allele. A 10-mm piece of tail was cut from each mouse and placed into 700 μl of tail digestion mix (50 mM Tris, pH 8/100 mM EDTA/100 mM NaCl/1% SDS). Proteinase K (250 μg) was added, and the samples were incubated at 55°C overnight. The samples were extracted with 700 μl of phenol/chloroform, and the aqueous phase was precipitated with isopropanol. DNA (200 ng) was utilized in 50 μl reactions which included three primers: 100 pmol of primer U1 (GACCAGAGGACCTGGGCAGACAGTGCTGAA), 5 pmol of primer U2 (TCCCCTACCCGGTAGAATTAATTCGATATC), and 50 pmol of primer D (GAGCAGACTACTCTCTCACTGCAAAGGTGA). Amplification was performed for 35 cycles of denaturing at 94°C for 45 s, annealing at 65°C for 30 s, and polymerizing at 72°C for 45 s. The PCR conditions were determined empirically to allow primers U1 and D to anneal with the wild-type (+) allele and amplify a 651-bp DNA fragment, and to allow primers U2 and D to anneal with the disrupted allele (−) and amplify a 251-bp DNA fragment. If both wild-type and disrupted alleles are present, the 651-bp and 251-bp DNA fragments are both amplified. This method of genotyping was found to give the same results as the Southern blot method previously reported (21).
Northern Blot Analyses.
Total cellular RNA was isolated from tissues of HFE+/+ and HFE−/− mice by using a guanidinium/phenol solution (RNA-Stat60, Tel-Test, Friendwood, TX), and passed over an oligo(dT) cellulose column (Poly(A) Quik, Stratagene). Three micrograms of poly(A)+ RNA from each sample was electrophoresed in 1% agarose and 2.2 M formaldehyde gels. Transcript sizes were estimated by using RNA standards (Promega). The RNA was transferred to Nytran membranes (Schleicher & Schuell) and immobilized by UV cross-linking. Blots were prehybridized, then hybridized with 32P-labeled RNA probes for murine DMT1. The template for the DMT1(total) probe consisted of nt 1323–1604 of the published murine cDNA (23), corresponding to coding sequences shared by the two classes of DMT1 transcripts: DMT1(IRE) which contain a canonical IRE in the 3′ untranslated region, and DMT1(non-IRE) which do not. Blots were prehybridized for 1 h and hybridized overnight at 65°C in 50% formamide, 5× SSPE, 5× Denhardt’s solution, 50 mM NaPO4 (pH 6.5), 200 μg/ml salmon sperm DNA, 1 mM EDTA, and 0.1% SDS. The blots were washed in 2× SSPE at room temperature (RT) for 20 min, 0.2× SSPE at RT for 20 min, twice in 0.2× SSPE/0.1% SDS at 65°C for 20 min, and subjected to autoradiography and phosphorimagery. Additional blots were hybridized with 32P end-labeled oligonucleotide probes specific for murine DMT1(IRE) or DMT1(non-IRE) transcripts. The oligonucleotide probe for DMT1(IRE) consisted of nucleotides 1821–1881 of GenBank accession no. AF029758. The oligonucleotide probe for DMT1(non-IRE) consisted of nt 1768–1828 of the published murine cDNA sequence (23). Blots were prehybridized for 1 h and hybridized overnight at 42°C in 40% formamide, 5× SSPE, 5× Denhardt’s solution, 0.05% sodium pyrophosphate, and 0.1 mg/ml yeast tRNA. The blots were washed in 6× SSPE for 15 min at RT twice, at 50°C for 1 min, and subjected to autoradiography and phosphorimagery. All blots were subsequently rehybridized with a 32P-labeled RNA probe for the rat glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript (24). Autoradiography and phosphorimagery were again performed and signal intensities quantified by using imagequant software (Molecular Dynamics). Values obtained for DMT1 transcripts were normalized to those obtained for GAPDH.
Measurement of Hepatic Iron Concentration.
Liver tissue from the HFE+/+ and HFE−/− littermates were analyzed for nonheme iron as described by Torrance and Bothwell (25), and the values were expressed as micromoles iron per gram dry weight. Tissue samples were weighed dry and digested in 3 M HCl, 10% tricholoroacetic acid at 65°C for 20 h. A total of 400 μl of each acid extract was mixed with 1.6 ml of bathophenanthroline chromagen reagent. The absorbance at 535 nm was measured in a DU-65 spectrophotometer (Beckman). Liver iron distribution was determined by light microscopic examination of formalin-fixed tissue sections stained with Perls’ Prussian blue.
Measurement of Serum Transferrin Saturation.
Blood was obtained by cardiac puncture. Serum iron and total iron binding capacity (TIBC) were measured by using the protocol of Fielding (26). Transferrin saturation was calculated as: (serum iron ÷ TIBC) × 100%.
Statistical Analysis.
Values are expressed as mean ± SEM. Differences between means were determined by using Student’s t test, with Welch’s correction for unequal variance.
RESULTS
HFE−/− Mice Demonstrate Elevated Transferrin Saturations and Excessive Liver Iron Accumulation by 4 Weeks of Age.
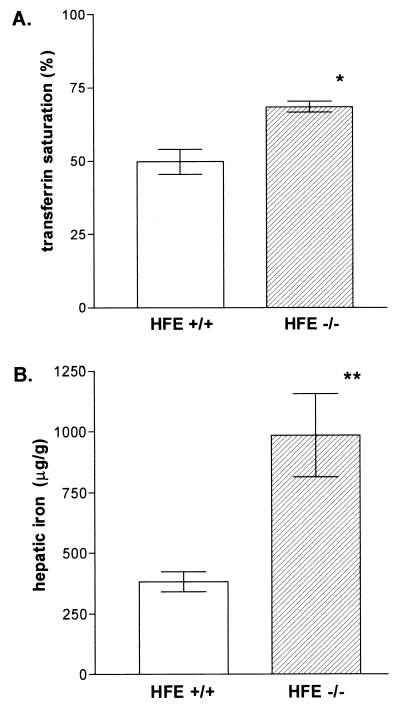
We had previously shown that serum transferrin is completely saturated in the HFE−/− mice by 10 weeks of age (21). To minimize potential down-regulatory effects of iron loading on dietary iron absorption (2, 4, 5), we analyzed the iron status of the HFE−/− animals at earlier ages. Fig. 1A shows that at 4 weeks of age serum transferrin was already more highly saturated in HFE−/− mice than in HFE+/+ littermate controls (68.4 ± 1.8% vs. 49.8 ± 4.3%), though still incompletely saturated. In addition, liver iron concentrations at this time point were significantly higher in the HFE−/− mice than the HFE+/+ controls (985 ± 171 vs. 381 ± 42 μg/g, Fig. 1B). As was reported for 10-week-old HFE−/− mice [with liver iron concentrations of 2,071 ± 450 μg/g (21)], the deposition of liver iron in the 4-week-old HFE−/− mice was predominantly in hepatocytes and demonstrated the typical zonal gradient distribution seen in patients with HH (not shown). These results indicated that the HFE−/− mice at 4 weeks of age are undergoing progressive iron loading despite incomplete transferrin saturation, a situation probably analogous to that in presymptomatic HH (27, 28). Thus, this time point was chosen for studies of duodenal expression of DMT1 in the HFE−/− mice.
Figure 1.
Serum transferrin saturations and hepatic iron concentrations and in the HFE−/− mice and HFE+/+ littermates. At 4 weeks of age the mice were sacrificed, and blood and liver specimens were obtained. (A) Serum total iron binding capacities and iron concentrations were measured. Transferrin saturations were calculated and expressed as mean ± SEM. ∗, P < 0.001. (B) Liver nonheme iron concentrations (micrograms of iron per gram dry liver) were measured and expressed as mean ± SEM. ∗∗, P < 0.005.
Iron-Deficient Mice Have Markedly Increased Duodenal Expression of DMT1(IRE) Transcripts.
We hypothesized that, despite the elevated iron status of the HFE−/− mice, the iron pool in the duodenal crypt cells would be inappropriately low and thus expected to lead to increased duodenal expression of DMT1. Before testing this hypothesis, we first characterized the regulation of DMT1 expression in mice under conditions of iron deprivation, where regulation of DMT1 has been well characterized in the rat (20). For these studies we utilized a transgenic murine model of dietary iron deficiency. Mice with targeted ablation of gastric parietal cells fail to produce the gastric acid necessary for efficient absorption of dietary iron and develop iron deficiency anemia (ref. 22, and Jeffrey Gordon, personal communication). We used Northern blot analyses to compare the duodenal expression of DMT1 in these animals with duodenal expression of DMT1 in nontransgenic littermates.
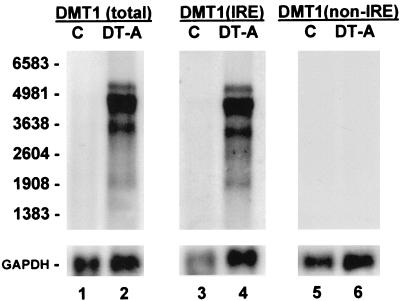
Alternative splicing of the DMT1 gene produces two distinct classes of DMT1 transcripts, which differ in nucleotide sequences encoding the amino acids at the C terminus and the 3′ untranslated region (23, 29, 30). One class, designated DMT1(IRE), includes an IRE in the 3′ untranslated region; the other class, designated DMT1(non-IRE), does not. We first quantified total DMT1 expression, which includes both DMT1(IRE) and DMT1(non-IRE) transcripts, by using a cDNA probe template containing coding sequence common to both classes. As seen in Fig. 2, a marked increase in total DMT1 transcripts was found in the transgenic iron-deficient mice over that seen in the nontransgenic littermates (≈50-fold increase for the 4.4-kb predominant transcript). We also used oligonucleotide probes specific to each of the DMT1 transcript classes on duplicate Northern blots, and found an increase only in the DMT1(IRE) transcripts. No increase in duodenal DMT1(non-IRE) expression was observed in the iron-deficient mice.
Figure 2.
RNA blot analyses of DMT1 expression in duodenum of iron-deficient mice. Three micrograms of poly(A)+ RNA from duodenum of a nontransgenic FVB/N control (C, lanes 1, 3, 5) and a transgenic H+/K+-ATPase β subunit (−1035 to +24)/DT-A iron-deficient littermate (DT-A, lanes 2, 4, 6) were electrophoresed in triplicate and blotted. Lanes 1 and 2 were hybridized with a 32P-labeled coding-sequence probe [DMT1(total)] to detect all DMT1 transcripts. Lanes 3 and 4 were hybridized with an oligonucleotide probe [DMT1(IRE)] specific to DMT1 transcripts with an IRE, and lanes 5 and 6 were hybridized with an oligonucleotide probe [DMT1(non-IRE)] specific to the DMT1 splice-variant transcripts without an IRE. Blots were exposed to film for 8 h with an intensifying screen and rehybridized with a probe for GAPDH. Positions of RNA size markers are on the left.
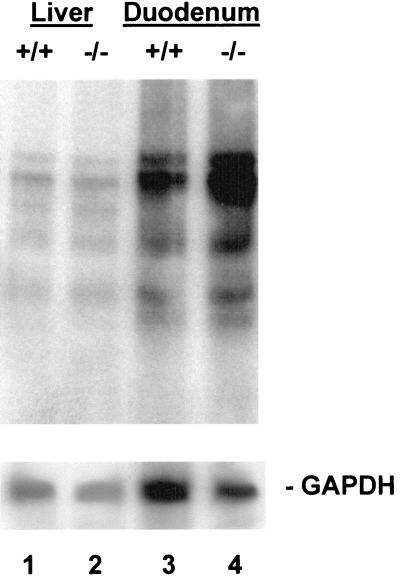
HFE−/− Mice Demonstrate Increased Duodenal Expression of DMT1(IRE) Transcripts.
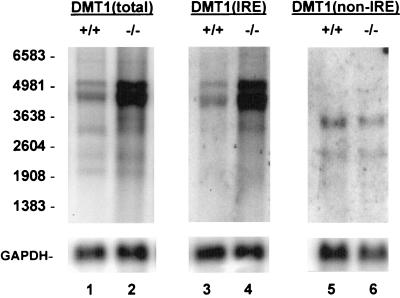
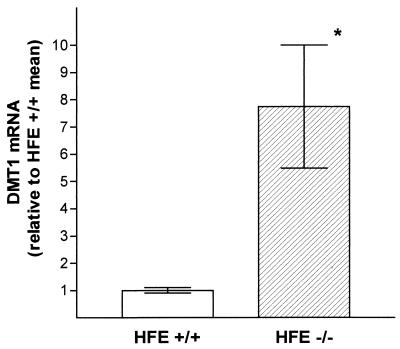
We next tested our hypothesis that the mRNA content for DMT1 would be increased in the HFE−/− mice relative to controls. In these experiments we used the 4-week-old HFE−/− mice and HFE+/+ littermates and compared the signal intensities for DMT1 transcripts by Northern blot analyses of duodenal RNA. We again used probes that allowed comparison of total DMT1 expression and of DMT1(IRE) and DMT1(non-IRE) individually. Two points are evident from Fig. 3: (i) total expression of DMT1 was indeed higher in the HFE−/− mice (lanes 1 and 2), and (ii) the increase in DMT1 expression was entirely attributable to an increase in DMT1(IRE) (lanes 3 and 4); no increase in expression of DMT1(non-IRE) transcripts was seen (lanes 5 and 6). The signal obtained from the predominant 4.4-kb DMT1 transcript in mRNA from each of 14 HFE−/− mice and 11 wild-type littermates was quantified and normalized to GAPDH mRNA expression. The level of duodenal expression of this transcript was increased 7.7-fold (on average) in the HFE−/− mice relative to HFE+/+ littermates (Fig. 4). Thus, despite elevated serum iron saturation and liver iron content in HFE−/− mice, the expression of duodenal DMT1(IRE) transcripts is up-regulated.
Figure 3.
RNA blot analysis of DMT1 expression in duodenum of HFE+/+ and HFE−/− mice. Poly(A)+ RNA (3 μg, lanes 1–4; 10 μg, lanes 5–6) from duodenum of a 4-week-old HFE+/+ mouse (lanes 1, 3, and 5) and a HFE−/− littermate (lanes 2, 4, and 6) were electrophoresed in triplicate and blotted. Lanes 1 and 2 were hybridized with a 32P-labeled coding-sequence probe [DMT1(total)] to detect all DMT1 transcripts. Lanes 3 and 4 were hybridized with an oligonucleotide probe [DMT1(IRE)] specific to DMT1 transcripts with an IRE, and lanes 5 and 6 were hybridized with an oligonucleotide probe [DMT1(non-IRE)] specific to the DMT1 splice-variant transcripts without an IRE. Blots were exposed to film for 18 h (lanes 1–4) or 48 h (lanes 5 and 6) with an intensifying screen, and rehybridized with a probe for GAPDH. Positions of RNA size markers are on the left.
Figure 4.
Quantification of duodenal DMT1 expression in HFE+/+ and HFE−/− mice. RNA blot analyses were performed on poly(A)+ RNA from duodenum of 11 4-week-old HFE+/+ mice and 14 HFE−/− littermates by using the DMT1(total) probe. Signals obtained from predominant 4.4-kb transcript were quantified by phosphorimagery and normalized to that obtained by rehybridizing the blots with a probe for GAPDH. Data are presented as mean ± SEM and expressed relative to the mean value obtained from the HFE+/+ mice. ∗, P = 0.01.
Hepatic Expression of DMT1 Is Not Increased in HFE−/− Mice.
The observation that only DMT1 transcripts which include the IRE were increased in the HFE−/− mice supports our hypothesis that the regulatory iron pool in duodenal enterocytes is low in HFE−/− mice. In iron deficiency also, the low enterocyte iron pool leads to the increased duodenal DMT1(IRE) expression (Fig. 2). However, in this setting, the systemic iron deficiency leads to increased DMT1 expression in multiple tissues in addition to duodenum (20). By contrast, in the HFE−/− mice there is systemic iron excess and iron loading of many tissues, particularly liver. We demonstrated here that the HFE−/− mice at 4 weeks of age already have an increased liver iron content (Fig. 1B). It was thus of interest to examine the level of DMT1 expression in liver. To do this, RNA blot analyses were performed on pooled poly(A)+ RNA from five HFE−/− mice and five HFE+/+ controls. The blots were hybridized with the DMT1(total) probe to examine all DMT1 transcripts. As seen in Fig. 5, no increase in DMT1 expression was seen in the pool of poly(A)+ from liver. Northern blots hybridized with oligonucleotide probes specific for DMT1(IRE) and DMT1(non-IRE) gave similar results (data not shown). On the other hand, the signal for total DMT1 transcripts in the pooled poly(A)+ RNA samples from duodenum of the same HFE−/− mice was elevated. These observations are consistent with the postulate that the increased expression of DMT1(IRE) transcripts is restricted to duodenum, where dietary iron absorption is regulated, and is not a manifestation of a generalized up-regulation of DMT1 in HFE−/− mice.
Figure 5.
RNA blot analysis of hepatic and duodenal expression of DMT1 in HFE+/+ and HFE−/− mice. Pooled poly(A)+ RNA (3 μg) from liver (lanes 1 and 2) or duodenum (lanes 3 and4) of five 4-week-old HFE+/+ mice and five HFE−/− littermates was electrophoresed, blotted, and hybridized with the DMT1(total) probe. The blot was exposed to film for 18 h with an intensifying screen and rehybridized with a probe for GAPDH.
DISCUSSION
The observation that DMT1 expression in the duodenum of the HFE−/− mice is increased provides a mechanism for the increased absorption of iron in HH, and supports the hypothesis that the regulatory iron pool of the duodenal enterocytes in HH is low. Studies on the duodenal expression of other iron-regulated proteins in patients with HH also support this hypothesis. Ferritin expression is decreased in the enterocytes of duodenal biopsy specimens of HH patients compared with controls, although increased in liver (31, 32). Likewise, TfR expression in the duodenum of HH patients does not demonstrate the down-regulation seen in the liver (33, 34). The mRNA transcripts encoding DMT1(IRE), ferritin, and TfR each include one or more IRE element(s) (35). Binding of iron regulatory proteins (IRPs) to these IREs increases as the cellular regulatory iron pool decreases. In both TfR and DMT1 transcripts the IREs are found in the 3′ untranslated region, where IRP binding leads to increased transcript stability and protein expression. The stabilization of DMT1(IRE) transcripts presumably accounts for the dramatic increase in steady-state duodenal DMT1 mRNA content seen in iron deficiency (Fig. 2). IRP binding activity has been investigated in duodenal biopsies of human patients with HH. Flanagan et al. (36) reported that, despite systemic iron loading, IRP binding activities from HH patients were similar to those from controls. Pietrangelo et al. (37) reported that duodenal IRP activities were consistently higher in HH patients than controls. Thus, most of the data suggest that the iron regulatory pool in enterocytes of HH patients is low, especially in relation to the body iron status.
The studies reported here support the hypothesis that impaired uptake of transferrin-bound iron by duodenal crypt cells (and the resulting inappropriately low crypt cell iron content) lead to dysregulation of DMT1 in the murine model of HH. It was recently shown that expressed recombinant DMT1 protein transports not only iron but other divalent metals, including cobalt, manganese, and lead (20). Earlier studies on patients with HH demonstrated increased intestinal absorption of cobalt (38), increased hepatic concentrations of manganese (39), and increased serum levels of lead (40), observations that are consistent with up-regulation of intestinal DMT1. Increased absorption of these divalent metal ions has also been shown in iron deficiency (41, 42), where up-regulation of DMT1 has been documented in the rat (20) and in the mouse (this report).
Although we attribute the proposed paradoxically low regulatory iron pool in duodenal crypt cells in patients with HH to decreased uptake of transferrin-bound iron by crypt enterocytes, an alternate explanation for the low regulatory iron pool was proposed by McClaren et al. (4). They reported an increased rate constant for transfer of mucosal iron to plasma in HH patients (4), and inferred that the primary defect in HH is enhanced transfer of dietary iron from absorptive cell to plasma at the basolateral surface. However, studies of the biology of the HFE protein which is defective in HH suggest that impaired uptake of transferrin-bound iron by crypt cells is a more likely primary defect. First, the HFE protein in duodenum is expressed predominantly in crypt cells (43), where intestinal uptake of transferrin-bound iron from plasma occurs. Second, the HFE protein colocalizes with the TfR and is physically associated with the TfR in crypt enterocytes (1). Third, there is very little HFE expressed in villus enterocytes (1, 43) where nonionic dietary iron is absorbed and transferred to plasma. Because the TfR is known to mediate the uptake of transferrin-bound iron and has never been reported to mediate transfer of intracellular iron to plasma, it seems most likely that the mutations which disrupt the function of HFE in HH impair its role in modulating transferrin-bound iron uptake. However, it is possible that an enhanced transfer of absorbed iron to plasma in the absorptive villus enterocytes is a secondary regulatory consequence of the reduced iron uptake into crypt enterocytes in the same way that the increased absorption follows the up-regulation of DMT1. If true, one would expect an up-regulation of the yet-to-be identified basolateral membrane transporter like that seen for DMT1.
Several observations suggest that the regulatory iron pool is also low in monocytes and macrophages of HH patients. Monocytes, like duodenal crypt cells, express both TfR and HFE (44). RNA bandshift assays showed a significant increase in IRP activity in monocytes of HH patients compared with controls (45). IRP activity was also increased in cultured monocyte-derived macrophages from these HH subjects, but was modulated normally by manipulation of iron levels. Furthermore, many studies of patients with HH have reported that reticulo-endothelial cells are relatively spared from iron loading until late in the disease [reviewed by McLaren (46)]. In the HFE−/− mice also, spleens were found to be relatively resistant to dietary iron loading, possibly reflecting decreased uptake of transferrin-bound iron by the HFE−/− splenic macrophages (21). These observations suggest that HFE mutations may interfere with uptake of transferrin-bound plasma iron by monocytes and macrophages in addition to crypt cells.
Finally, these studies further characterize the phenotype of the HFE−/− mouse by demonstrating iron loading of liver at an early age, when serum transferrin is elevated but incompletely saturated. These observations are in agreement with reports in human patients diagnosed early in life (27, 28) and suggest that hepatic iron loading in HH does not require an increase in plasma free (nontransferrin-bound) iron. We propose that the HFE−/− mouse at this age provides a model of presymptomatic human HH that should be a useful tool for testing additional hypotheses related to the molecular pathogenesis of HH.
Acknowledgments
We thank Dr. Jeffrey Gordon for providing the H+/K+-ATPase β subunit (−1035 to +24)/DT-A mouse tissues, Rosemary O’Neill for technical assistance, and Elizabeth Torno for editorial assistance. This work was supported by Grants DK53405 (W.S.S.), GM34182 (W.S.S.), and DK41816 (B.R.B.) from the National Institutes of Health and by support from the Fleur de Lis Foundation.
ABBREVIATIONS
- HH
hereditary hemochromatosis
- TfR
transferrin receptor
- HFE
the protein defective in hereditary hemochromatosis
- DMT1
divalent metal ion transporter (previously referred to as Nramp2 and DCT1, divalent cation transporter)
- IRE
iron responsive element
- MHC
major histocompatibility complex
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IRP
iron regulatory protein
References
- 1.Waheed A, Parkkila S, Saarnio J, Fleming R E, Zhou X-Y, Tomatsu S, Britton R S, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1999;96:1579–1584. doi: 10.1073/pnas.96.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch S R, Skikne B S, Cook J D. Blood. 1989;74:2187–2193. [PubMed] [Google Scholar]
- 3.Smith P M, Godfrey B E, Williams R. Clin Sci. 1969;37:519–531. [PubMed] [Google Scholar]
- 4.McLaren G D, Nathanson M H, Jacobs A, Trevett D, Thomson W. J Lab Clin Med. 1991;117:390–401. [PubMed] [Google Scholar]
- 5.Powell L W, Campbell C B, Wilson E. Gut. 1970;11:727–731. doi: 10.1136/gut.11.9.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartwright G E, Edwards C Q, Kravitz K, Skolnick M, Amos D B, Johnson A, Buskjaer L. N Engl J Med. 1979;301:175–179. doi: 10.1056/NEJM197907263010402. [DOI] [PubMed] [Google Scholar]
- 7.Cox T M, Lord D K. Eur J Haematol. 1989;42:113–125. doi: 10.1111/j.1600-0609.1989.tb01200.x. [DOI] [PubMed] [Google Scholar]
- 8.Edwards C Q, Griffen L M, Goldgar D, Drummond C, Skolnick M H, Kushner J P. N Engl J Med. 1988;318:1355–1362. doi: 10.1056/NEJM198805263182103. [DOI] [PubMed] [Google Scholar]
- 9.Bacon B R, Powell L W, Adams P C, Kresina T F, Hoofnagle J H. Gastroenterology. 1999;116:1–17. doi: 10.1016/s0016-5085(99)70244-1. [DOI] [PubMed] [Google Scholar]
- 10.Feder J N, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy D A, Basava A, Dormishian F, Domingo R, Jr, Ellis M C, Fullan A, et al. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 11.Parkkila S, Waheed A, Britton R S, Bacon B R, Zhou X-Y, Tomatsu S, Fleming R E, Sly W S. Proc Natl Acad Sci USA. 1997;94:13198–13202. doi: 10.1073/pnas.94.24.13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feder J N, Penny D M, Irrinki A, Lee V K, Lebron J A, Watson N, Tsuchihashi Z, Sigal E, Bjorkman P J, Schatzman R C. Proc Natl Acad Sci USA. 1998;95:1475–1477. doi: 10.1073/pnas.95.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross C N, Irrinki A, Feder J N, Enns C A. J Biol Chem. 1998;273:22068–22074. doi: 10.1074/jbc.273.34.22068. [DOI] [PubMed] [Google Scholar]
- 14.Lebrón J A, Bennett M J, Vaughn D E, Chirino A J, Snow P M, Mintier G A, Feder J N, Bjorkman P J. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- 15.Rouault T A. Hepatology. 1998;28:890–891. doi: 10.1002/hep.510280342. [DOI] [PubMed] [Google Scholar]
- 16.Powell L W, Burt M J, Halliday J W, Jazwinska E C. Semin Liver Dis. 1996;16:55–63. doi: 10.1055/s-2007-1007219. [DOI] [PubMed] [Google Scholar]
- 17.Lombard M, Chua E, O’Toole P. Gut. 1997;40:435–439. doi: 10.1136/gut.40.4.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson G J. J Gastroenterol Hepatol. 1996;11:1030–1032. doi: 10.1111/j.1440-1746.1996.tb00029.x. [DOI] [PubMed] [Google Scholar]
- 19.Conrad M E, Crosby W H. Blood. 1963;22:406–415. [PubMed] [Google Scholar]
- 20.Gunshin H, Mackenzie B, Berger U V, Gunshin Y, Romero M F, Boron W F, Nussberger S, Gollan J L, Hediger M A. Nature (London) 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 21.Zhou X-Y, Tomatsu S, Fleming R E, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt E M, Ruddy D A, Prass C E, et al. Proc Natl Acad Sci USA. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiutang L, Karam S M, Gordon J I. J Biol Chem. 1996;271:3671–3676. [PubMed] [Google Scholar]
- 23.Gruenheid S, Cellier M, Vidal S, Gros P. Genomics. 1995;25:514–525. doi: 10.1016/0888-7543(95)80053-o. [DOI] [PubMed] [Google Scholar]
- 24.Fort P, Marty L, Piechaczyk M, el Sabrouty S, Dani C, Jeanteur P, Blanchard J M. Nucleic Acids Res. 1985;13:1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torrance J D, Bothwell T H. Methods Haematol. 1980;1:90–115. [Google Scholar]
- 26.Fielding J. Methods Haematol. 1980;1:15–43. [Google Scholar]
- 27.Bassett M L, Halliday J W, Ferris R A, Powell L W. Gastroenterology. 1984;87:628–633. [PubMed] [Google Scholar]
- 28.Kaikov Y, Wadsworth L D, Hassall E, Dimmick J E, Rogers P C. Pediatrics. 1992;90:37–42. [PubMed] [Google Scholar]
- 29.Lee P L, Gelbart T, West C, Halloran C, Beutler E. Blood Cells Mol Dis. 1998;24:199–215. doi: 10.1006/bcmd.1998.0186. [DOI] [PubMed] [Google Scholar]
- 30.Fleming M D, Romano M A, Ma S, Garrick L M, Garrick M D, Andrews N C. Proc Natl Acad Sci USA. 1998;95:1148–1153. doi: 10.1073/pnas.95.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francanzani A L, Fargion S, Romano R, Piperno A, Arosio P, Ruggeri G, Ronchi G, Fiorelli G. Gastroenterology. 1989;96:1071–1078. doi: 10.1016/0016-5085(89)91625-9. [DOI] [PubMed] [Google Scholar]
- 32.Basclain K A, Shilkin K B, Withers G, Reed W D, Jeffrey G P. J Gastroenterol Hepatol. 1998;13:624–634. doi: 10.1111/j.1440-1746.1998.tb00701.x. [DOI] [PubMed] [Google Scholar]
- 33.Lombard M, Bomford A, Hynes M, Naoumov N V, Roberts S, Crowe J, Williams R. Hepatology. 1989;9:1–5. doi: 10.1002/hep.1840090102. [DOI] [PubMed] [Google Scholar]
- 34.Lombard M, Bomford A B, Polson J R, Bellingham A J, Williams R. Gastroenterology. 1990;98:976–984. doi: 10.1016/0016-5085(90)90022-s. [DOI] [PubMed] [Google Scholar]
- 35.Theil E C. Biochem J. 1994;304:1–11. doi: 10.1042/bj3040001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flanagan P R, Hajdu A, Adams P C. Hepatology. 1995;22:828–832. [PubMed] [Google Scholar]
- 37.Pietrangelo A, Casalgrandi G, Quaglino D, Gualdi R, Conte D, Milani S, Montosi G, Cesarini L, Ventura E, Cairo G. Gastroenterology. 1995;108:208–217. doi: 10.1016/0016-5085(95)90026-8. [DOI] [PubMed] [Google Scholar]
- 38.Valberg L S, Ludwig J, Olatunbosun D. Gastroenterology. 1969;56:241–251. [PubMed] [Google Scholar]
- 39.Altstatt L B, Pollack S, Feldman M H, Reba R C, Crosby W H. Proc Soc Exp Biol Med. 1967;124:353–355. doi: 10.3181/00379727-124-31741. [DOI] [PubMed] [Google Scholar]
- 40.Barton J C, Patton M A, Edwards C Q, Griffen L M, Kushner J P, Meeks R G, Leggett R W. J Lab Clin Med. 1994;124:193–198. [PubMed] [Google Scholar]
- 41.Pollack S, George J N, Reba R C, Kaufman R M, Crosby W H. J Clin Invest. 1965;44:1470–1473. doi: 10.1172/JCI105253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flanagan P R, Gaist J, Valberg L S. J Nutr. 1980;110:1754–1763. doi: 10.1093/jn/110.9.1754. [DOI] [PubMed] [Google Scholar]
- 43.Parkkila S, Waheed A, Britton R S, Feder J N, Tsuchihashi Z, Schatzman R C, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1997;94:2534–2539. doi: 10.1073/pnas.94.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saarnio, J., Parkkila, S., Parkkila, A.-K., Waheed, A., Karttunen, T. & Sly, W. S. (1999) J. Histochem. Cytochem., in press. [DOI] [PubMed]
- 45.Cairo G, Recalcati S, Montosi G, Castrusini E, Conte D, Pietrangelo A. Blood. 1997;89:2546–2553. [PubMed] [Google Scholar]
- 46.McLaren, G. D. (1989) J. Lab. Clin. Med. 137–138. [PubMed]