Abstract
Birt–Hogg–Dubé syndrome, a hamartoma disorder characterized by benign tumors of the hair follicle, lung cysts, and renal neoplasia, is caused by germ-line mutations in the BHD(FLCN) gene, which encodes a tumor-suppressor protein, folliculin (FLCN), with unknown function. The tumor-suppressor proteins encoded by genes responsible for several other hamartoma syndromes, LKB1, TSC1/2, and PTEN, have been shown to be involved in the mammalian target of rapamycin (mTOR) signaling pathway. Here, we report the identification of the FLCN-interacting protein, FNIP1, and demonstrate its interaction with 5′ AMP-activated protein kinase (AMPK), a key molecule for energy sensing that negatively regulates mTOR activity. FNIP1 was phosphorylated by AMPK, and its phosphorylation was reduced by AMPK inhibitors, which resulted in reduced FNIP1 expression. AMPK inhibitors also reduced FLCN phosphorylation. Moreover, FLCN phosphorylation was diminished by rapamycin and amino acid starvation and facilitated by FNIP1 overexpression, suggesting that FLCN may be regulated by mTOR and AMPK signaling. Our data suggest that FLCN, mutated in Birt–Hogg–Dubé syndrome, and its interacting partner FNIP1 may be involved in energy and/or nutrient sensing through the AMPK and mTOR signaling pathways.
Keywords: hamartoma syndrome, renal cancer, Birt–Hogg–Dubé, tumor suppressor
Birt–Hogg–Dubé (BHD) syndrome predisposes patients to develop hair follicle hamartomas, lung cysts, and an increased risk for renal neoplasia (1–3). BHD patients develop bilateral, multifocal renal tumors with a variety of histologies (4). We mapped the BHD locus to chromosome 17p11.2 by linkage analysis in BHD kindreds (5, 6) and identified germ-line mutations in a gene with unknown function that is highly conserved (7, 8). Twenty-two unique mutations predicted to truncate the BHD protein folliculin (FLCN), including a “hot spot” insertion/deletion in a C8 tract, were identified in 84% of BHD kindreds (9). Somatic “second-hit” mutations identified in BHD-associated renal tumors suggest a tumor-suppressor function for FLCN (10), underscored by loss of BHD mRNA expression in renal tumors from BHD patients (11).
Recent studies suggest that several hamartoma syndromes may be linked through the convergent energy/nutrient-sensing pathways involved in mammalian target of rapamycin (mTOR) regulation (12–15). These inherited syndromes are characterized by multiple hamartomas and an increased risk of cancer. Germ-line mutations have been identified in four causative genes: LKB1, responsible for Peutz–Jeghers syndrome (16–18), TSC1 and TSC2, responsible for tuberous sclerosis complex (TSC) (19), and PTEN, responsible for Cowden syndrome (20). Loss of gene function leads to dysregulation of mTOR, which regulates cell growth and size through stimulation of protein synthesis (15, 21, 22). BHD syndrome, also a hamartoma disorder, displays phenotypic similarities to TSC that have led to speculation that BHD may function in the pathway(s) signaling through mTOR (12, 23). To ascertain FLCN function, we searched for interacting proteins by coimmunoprecipitation. We identified a 130-kDa FLCN-interacting protein, FNIP1, and demonstrated its interaction with AMPK, a protein important in nutrient/energy sensing (24, 25). We found that FLCN is phosphorylated, which is facilitated by FNIP1, and suppressed by conditions that inhibit mTOR. We also found that AMPK-inhibitor treatment reduced FNIP1 phosphorylation, lowered FNIP1 expression levels, and resulted in FLCN dephosphorylation. We discuss FLCN–FNIP1–AMPK interactions and consider how dysregulation of this complex may predispose to the BHD phenotype.
Results
Identification of a 130-kDa FLCN-Interacting Protein, FNIP1, and Cloning of Human FNIP1 cDNA.
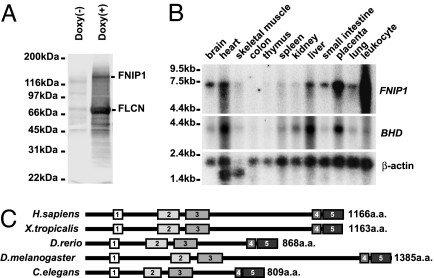
BHD encodes FLCN, a highly conserved protein with unknown function. To elucidate its biologic role, we searched for FLCN-interacting protein(s) in anti-HA immunoprecipitates from doxycycline-inducible, HA-FLCN-expressing HEK293 cells. Mass spectrometric analysis of a major 130-kDa interacting protein (Fig. 1A), designated FNIP1 (FLCN-interacting protein 1), identified peptides from two overlapping proteins, BAB85547, encoded by KIAA1961 (AB075841), and AAH01956, encoded by an uncharacterized human EST (BC001956; see peptide mass fingerprinting in Data Set 1, which is published as supporting information on the PNAS web site). Primers designed from the overlapping cDNA sequences were used to amplify and clone the full-length FNIP1 transcript (DQ145719) from a pooled tissue cDNA library. Several alternative transcripts lacking one or two of the 18 coding exons were also identified and are being verified (details in Supporting Methods, which is published as supporting information on the PNAS web site). The evolutionarily conserved 3,626-nt mRNA encoded a protein with an ORF of 1,166 aa. A comparison of FNIP1 homologs from Homo sapiens, Xenopus tropicalis, Danio rerio, Drosophila melanogaster, and Caenorhabditis elegans revealed five blocks of conserved sequence with at least 35% similarity (Fig. 1C; and see Fig. 8, which is published as supporting information on the PNAS web site), but BLAST analysis revealed no known functional domains. Northern blot analysis showed that FNIP1 was expressed in a pattern similar to BHD, with strongest expression in heart, liver, and placenta and moderate expression in tissues involved in the BHD phenotype, kidney and lung (Fig. 1B).
Fig. 1.
FNIP1, a 130-kDa FLCN-binding protein, is evolutionarily conserved and widely expressed in human tissues. (A) Stably transfected HEK293 cells expressing doxycycline-inducible HA-FLCN were lysed, and proteins were immunoprecipitated with anti-HA antibody, subjected to SDS/PAGE, transferred to PVDF membranes, and stained with colloidal gold. FNIP1 was coimmunoprecipitated with FLCN. (B) FNIP1 and BHD(FLCN) expression patterns in human adult tissues. cDNA probes were generated by PCR and hybridized to a human 12-tissue Northern blot (Clontech). β-Actin was used as loading control. (C) Schematic structure of conserved FNIP1 domains. Amino acid alignments show five blocks of conservation between FNIP1 and its homologs (Fig.8). GenBank accession nos. for FNIP1 homologs: H. sapiens (DQ145719), X. tropicalis (ENSXETP00000036209; Ensembl), D. rerio (XM_687716), D. melanogaster (NM_140686), and C. elegans (T04C4.1a; Wormbase).
FLCN and FNIP1 Interact in Vivo Through the C Terminus of FLCN and Colocalize in the Cytoplasm.
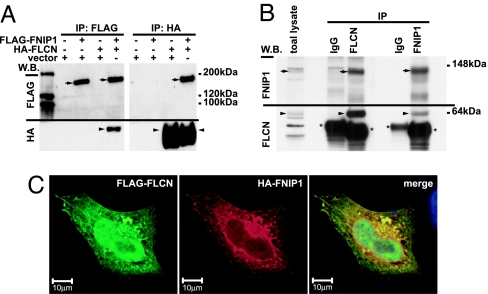
To confirm the interaction between FNIP1 and FLCN, we performed reciprocal coimmunoprecipitations in HEK293 cells transiently expressing HA-FLCN and FLAG-FNIP1 and found that HA-FLCN coimmunoprecipitated with FLAG-FNIP1 in a reciprocal fashion (Fig. 2A). Similarly, endogenous FNIP1–FLCN interaction was confirmed in HEK293 cells, blotting with polyclonal antibodies to FLCN and FNIP1 (Fig. 2B). HA-FNIP1 and FLAG-FLCN, transiently cotransfected into HeLa cells, colocalized in the cytoplasm with a reticular pattern (Fig. 2C). FLCN also localized in the nucleus.
Fig. 2.
FLCN–FNIP1 interactions. (A) HA-FLCN was cotransfected with FLAG-FNIP1 into HEK293 cells. Cell lysates were immunoprecipitated (IP) with anti-FLAG or anti-HA antibodies and blotted with both antibodies. Arrows, FLAG-FNIP1; arrowheads, HA-FLCN. (B) HEK293 lysates were immunoprecipitated with anti-FLCN and control IgG or with anti-FNIP1 and control IgG and blotted with anti-FLCN or anti-FNIP1. Arrowheads, endogenous FLCN; arrows, endogenous FNIP1; asterisks, IgG heavy-chain bands. (C) HeLa cells were cotransfected with HA-FNIP1 and FLAG-FLCN. Immunocytochemistry was performed with anti-HA and anti-FLAG antibodies. FNIP1 and FLCN colocalize with a reticular pattern in the cytoplasm. Green, FLAG-FLCN; red, HA-FNIP1.
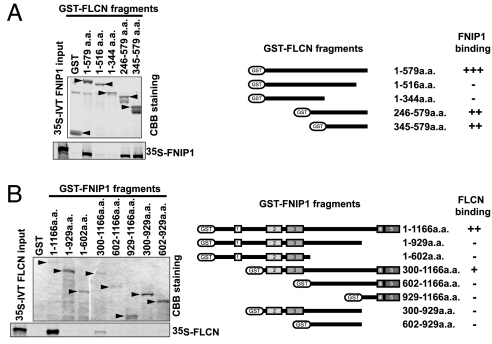
To identify the FNIP1-binding domain in FLCN, in vitro binding assays were performed with GST-FLCN mutants and 35S in vitro-translated FNIP1. FNIP1 bound to full-length GST-FLCN (1–579 aa) and mutant proteins containing the C terminus of FLCN (246–579 or 345–579 aa; Fig. 3A) but not to mutant proteins lacking the C terminus (1–516 or 1–344 aa). These interactions were confirmed in immunoprecipitates from HA-FNIP1-expressing HEK293 cells transiently transfected with FLAG-FLCN and FLAG-FLCN mutants (Fig. 9, which is published as supporting information on the PNAS web site). In support of these data, the mutant protein from the most C-terminal BHD mutation found in patients (c.2034C>T; R527X) (9) did not bind to HA-FNIP1 (data not shown). Taken together, the results support a FNIP1-binding domain in the C terminus of FLCN.
Fig. 3.
Regions of FNIP1 and FLCN necessary for binding. (A) Recombinant GST-FLCN and GST-FLCN mutants were immobilized on glutathione-Sepharose and incubated with 35S in vitro-translated (IVT) FNIP1. Binding was evaluated by SDS/PAGE and autoradiography. Coomaissee Brillant blue (CBB) staining shows relative expression of GST-FLCN proteins. FLCN mutants lacking a C terminus cannot bind FNIP1. (B) Recombinant GST-FNIP1 and FNIP1 deletion mutants were bound to glutathione-Sepharose, incubated with 35S IVT FLCN, and evaluated as in A. Deletion of conserved blocks 2/3 or 4/5 of FNIP1 disrupted the interaction with FLCN.
Optimal Binding of FLCN to FNIP1 Requires Full-Length FNIP1 Protein.
We next performed an in vitro binding assay to determine the portion of FNIP1 that was required to bind FLCN. Full-length FNIP1 protein was required for maximum binding of FLCN; only one of the deletion mutants containing residues 300-1166 (lacking conserved block 1, see Fig. 1C) bound FLCN (Fig. 3B). Both GST-FNIP1 and the FNIP1 mutant protein that bound FLCN contained four of the five conserved blocks (blocks 2–5). Mutants containing either blocks 2 and 3 or blocks 4 and 5 did not bind FLCN. The data suggest that optimal FLCN–FNIP1 interaction requires these four conserved regions of the FNIP1 protein, perhaps for proper conformation.
AMPK Was Identified as a FNIP1-Interacting Protein.
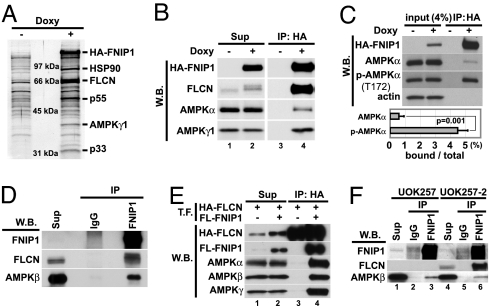
Because FNIP1 is a protein with no characteristic domains to suggest function, we searched for interacting proteins using the experimental approach described above. We identified the γ-1 subunit (40 kDa) of AMPK as well as FLCN (67 kDa) and FNIP1 in the immunoprecipitates from doxy-inducible, HA-FNIP1-expressing HEK293 cells by mass spectrometry (Fig. 4A; peptide mass fingerprinting in Data Sets 2–4, which are published as supporting information on the PNAS web site). Additional bands included HSP90 (p90) and several unverified proteins. AMPK is known to play a critical role in energy sensing by negatively regulating biosynthetic pathways and activating catabolic reactions in response to stress signals (24–26). This heterotrimeric protein kinase consists of a catalytic α-subunit and two regulatory subunits, β and γ. Anti-HA immunoprecipitates from doxy-inducible HA-FNIP1-expressing HEK293 cells were Western blotted with subunit-specific antibodies and found to contain both endogenous AMPK α- and γ1 subunits (Fig. 4B). We confirmed the interaction of endogenous FNIP1 and AMPKβ in anti-FNIP1 immunoprecipitates by Western blotting (Fig. 4D), confirming FNIP1 interaction with all subunits of AMPK. AMPK binding to FNIP1 was confirmed by an in vitro binding assay incubating HEK293 cell lysates (AMPK source) with recombinant GST-FNIP1 immobilized on glutathione-Sepharose, followed by Western analysis. Residues 602–929 were the minimum region of FNIP1 necessary for binding to AMPK, which was distinct from the FLCN-binding region. Direct binding of recombinant GST-FNIP1 to purified AMPK was also demonstrated in vitro (Fig.10 A and B, which is published as supporting information on the PNAS web site).
Fig. 4.
FNIP1 interacts with AMPK in vitro and in vivo. (A) FNIP1-interacting proteins were immunoprecipitated (IP) with anti-HA antibody from doxycycline-inducible HA-FNIP1-expressing HEK293 cells, separated by SDS/PAGE, transferred to PVDF membrane, stained with colloidal gold, and analyzed by mass spectrometry. The 40-kDa protein was identified as the γ-1 subunit of AMPK. Other interacting proteins were FLCN (67 kDa) and HSP 90 (90 kDa). (B) Cell lysates from HA-FNIP1-inducible HEK 293 cells cultured with and without doxy were immunoprecipitated with anti-HA antibody and blotted with antibodies, as indicated, showing that HA-FNIP1 and AMPK subunits interact. (C) Immunoprecipitates from HA-FNIP1-inducible HEK293 cells were blotted with antibodies as indicated. Band intensities were measured with the ImageJ program. The estimated amounts of AMPKα and phospho-AMPKα(T172) binding to FNIP1 were calculated by IP/[input (4%) × 25]. Results were from three independent experiments. Brackets, standard deviation. (D) HEK293 lysates were immunoprecipitated with anti-FNIP1 or control IgG. Immunoprecipitates were blotted with anti-FNIP1, anti-FLCN mouse monoclonal antibody, and anti-AMPKβ mouse monoclonal antibody. Endogenous FNIP1 and AMPK interact. (E) HEK293 cell lysates transfected (T.F.) with HA-FLCN alone or HA-FLCN and FLAG-FNIP1,were immunoprecipitated with anti-HA antibody and blotted with antibodies as indicated. All AMPK subunits were immunoprecipitated with FLCN in a FNIP1-dependent manner. (F) UOK257 cells (FLCN null) and FLCN-restored UOK257-2 cells were immunoprecipitated with anti-FNIP1 or control IgG and blotted with anti-FNIP1, anti-FLCNmAb, and anti-AMPKβ mAb. W.B., Western blotting; Sup, supernatant.
The catalytic α-subunit of AMPK is activated by phosphorylation on Thr-172 by AMPK kinases (27). We observed that the amount of AMPKα present in its phosphorylated form (detected by a phospho-AMPKα Thr-172-specific antibody) relative to total AMPKα was greater in HA-FNIP1 immunoprecipitates than in total cell lysates (Fig. 4C Upper). Using densitometry, we compared the total AMPKα and phospho-AMPKα in doxy-induced HA-FNIP1-expressing lysates with the amounts of these proteins in anti-HA immunoprecipitates from the lysates. We estimated that 5% of the total phospho-AMPKα but only 0.5% of the total AMPKα associated with FNIP1 in the immunoprecipitates (Fig. 4C Lower).
FLCN Binds to AMPK Through FNIP1 but Is Not Essential for FNIP1–AMPK Interaction.
To determine whether a FLCN/FNIP1/AMPK complex exists, we cotransfected HEK293 cells with an HA-FLCN expression vector with or without a FLAG-FNIP1 vector. Anti-HA immunoprecipitates were evaluated by Western blotting. All three AMPK subunits were coimmunoprecipitated with HA-FLCN in a FLAG-FNIP1-dependent manner (Fig. 4E, lane 4) confirming that the AMPK heterotrimer binds to the FLCN–FNIP1 complex in a FNIP1-dependent manner but not to FLCN alone (Fig. 4E, lane 3). We also evaluated the FNIP1–AMPK interaction in a FLCN-null renal tumor cell line, UOK257, and in UOK257-2 cells retrovirally restored with FLCN. In UOK257 cells, no FLCN was detected in total-cell lysates or anti-FNIP1 immunoprecipitates (Fig. 4F, lanes 1 and 3). However, endogenous FNIP1 was able to bind to the AMPKβ subunit in anti-FNIP1 immunoprecipitates (Fig. 4F, lane 3), and AMPK–FNIP1 binding was not affected by restoration of FLCN expression (UOK257-2; Fig. 4F, lanes 4 and 6).
FNIP1 Can Be Phosphorylated by AMPK in the FLCN/FNIP1/AMPK Complex, Which Affects Its Expression Level.
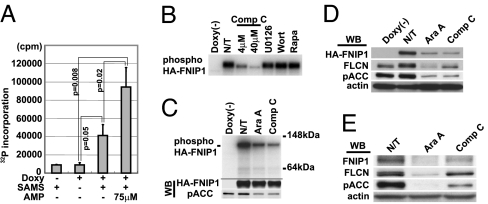
To determine whether AMPK in the FNIP1 immunoprecipitates was enzymatically active, we performed an in vitro kinase assay with anti-HA immunoprecipitates from doxycycline-induced HA-FNIP1-expressing HEK293 cells using SAMS peptide, a specific AMPK substrate (28), and [γ-32P]ATP. Immunoprecipitates from doxycycline(+) lysates showed significant AMPK activity compared with controls. The FNIP1-associated in vitro kinase activity was increased 2.5-fold by 75 μM AMP, a positive effector of AMPK (26) (Fig. 5A), supporting AMPK as the source of kinase activity.
Fig. 5.
AMPK phosphorylates FNIP1 and may regulate FNIP1 expression. (A) Cell lysates from HA-FNIP1-inducible HEK293 cells grown with and without doxy were immunoprecipitated with anti-HA antibody and assayed for AMPK in vitro kinase activity with [γ-32P]ATP and SAMS peptide with and without AMP. Radioactivity incorporated into SAMS peptide was measured by scintillation counting. Each bar represents the results of three independent experiments. Brackets, standard deviation. (B) HA-FNIP1 immunoprecipitates were incubated in AMPK kinase reaction buffer with [γ32]ATP and various kinase inhibitors (Compound C, 4 or 40 μM; UO126, 10 μM; Wortmannin, 1 μM; and rapamycin, 20 nM). Immunoprecipitates were washed and subjected to SDS/PAGE and autoradiography. (C) HA-FNIP1-inducible 293 cells were cultured for 3 h with [32P]orthophosphate with and without AMPK inhibitors (AraA, 2 mM and Compound C, 20 μM), and anti-HA immunoprecipitates were subjected to SDS/PAGE and autoradiography. Cold samples obtained under the same culture conditions were used for phosphoacetyl CoA carboxylase (pACC) and HA-FNIP1 Western blotting. (D and E) HA-FNIP1-inducible 293 cells (D) or 293 cells (E) were cultured with AraA (2 mM) or Compound C (Comp C) (30 μM) for 24 h. Exogenous or endogenous FNIP1 and endogenous FLCN were detected by Western blotting. WB, Western blotting.
We next asked whether FNIP1 might serve as a substrate for AMPK in the FNIP1–AMPK complex. When anti-HA immunoprecipitates from doxycycline-induced HA-FNIP1-expressing HEK293 cells were incubated with [γ-32P]ATP and AMPK kinase buffer, we found that HA-FNIP1 was phosphorylated, and this phosphorylation was blocked by Compound C, a specific inhibitor of AMPK (29), in a dose-dependent manner (Fig. 5B), whereas other kinase inhibitors tested (i.e., UO126, Wortmannin, and rapamycin) were unable to block HA-FNIP1 phosphorylation. Furthermore, when we labeled doxy-induced HA-FNIP1-expressing HEK293 cells in vivo with [32P]orthophosphate for 3 h, addition of Compound C or AraA (another inhibitor of AMPK) (30) to the medium reduced the amount of [32P]phosphate incorporated into HA-FNIP1 (Fig. 5C), indicating that FNIP1 may serve as a substrate for AMPK both in vitro and in vivo.
We postulated that one function of FNIP1 phosphorylation by AMPK might be regulation of protein expression. We cultured both HA-FNIP1-overexpressing HEK293 cells and 293 cells in the presence of AMPK inhibitors for 24 h and found that levels of HA-FNIP1 and endogenous FNIP1 proteins were reduced (Fig. 5 D and E, respectively). Levels of endogenous FLCN were also reduced by AMPK inhibition.
FLCN Can Exist in a Phosphorylated Form That Is Preferentially Bound to FNIP1.
In searching for a functional significance for the FLCN–FNIP1–AMPK interaction, we noted the appearance of multiple FLCN bands on Western blots and designed experiments to test their association with or regulation by FNIP1. Electrophoretic mobility shifts observed after phosphatase or calyculin A (phosphatase inhibitor) treatment suggested that FLCN existed in several phosphorylated forms (Fig. 11, which is published as supporting information on the PNAS web site). We also detected at least three electrophoretically distinct forms of endogenous FLCN in HEK293 cell lysates, which shifted to the slower migrating (phosphorylated) species in anti-FNIP1 immunoprecipitates (Fig. 4D), suggesting that FNIP1 preferentially bound phosphorylated FLCN. In addition, doxy-induced HA-FNIP1 expression produced a band shift to phospho-FLCN in total HEK293 cell lysates (Fig. 4B, lane 2), and the slower migrating phospho-FLCN preferentially immunoprecipitated with HA-FNIP1 from doxy-induced HEK293 cells (Fig. 4B, lane 4).
FLCN Phosphorylation Is Diminished by Inhibition of mTOR and AMPK.
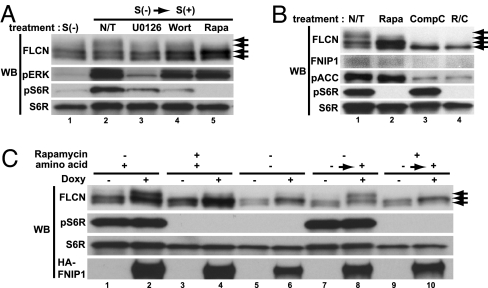
To elucidate the mechanism by which FLCN becomes phosphorylated, UOK257-2 FLCN-restored cells were grown under different culture conditions or with different kinase inhibitors. Serum withdrawal for 48 h partially decreased FLCN phosphorylation (Fig. 6A, lane 1), which was restored by serum stimulation (Fig. 6A, lane 2). Pretreatment with UO126, an inhibitor of MEK1/2, or Wortmannin, an inhibitor of PI3K, reduced FLCN phosphorylation to some extent (Fig. 6A, lanes3/4 and 2); interestingly, rapamycin, which completely inhibited mTOR activity (no pS6R), most dramatically inhibited FLCN phosphorylation by eliminating the upper phospho-FLCN band completely (Fig. 6A, lanes 5 and 2). However, rapamycin did not completely block FLCN phosphorylation, because one phospho-FLCN band was unaffected by rapamycin treatment (Fig. 6B, lane 2). Treatment of UOK257-2 with Compound C resulted in complete loss of phospho-FLCN, including the rapamycin-insensitive phospho-FLCN band (Fig. 6B, lane3) and resulted in reduced FLCN expression. The same results were obtained with AMPK inhibitor AraA (Fig. 5 D and E). In support of these data, we demonstrated phosphorylation of recombinant GST-FLCN by purified AMPK in an in vitro kinase reaction (Fig. 12, which is published as supporting information on the PNAS web site). Taken together, these data indicate that FLCN has multiple phosphorylation sites that are phosphorylated through mTOR and AMPK signaling.
Fig. 6.
FLCN phosphorylation is regulated by FNIP1 through mTOR signaling and AMPK. (A) FLCN phosphorylation is affected by mTOR activity. UOK257-2 FLCN-restored cells were cultured under different culture conditions: S(+), with serum; S(−), serum starvation for 48 h; or S(−) → S(+), 20% dialyzed serum stimulation for 30 min after 48-h serum starvation. Cells were pretreated for 2 h before stimulation with various inhibitors: nontreatment (N/T); U0126, 50 μM; Wortmannin (Wort), 1 μM; or rapamycin (Rapa), 20 nM. (B) AMPK inhibition suppresses FLCN phosphorylation. UOK 257–2 cells were cultured with rapamycin (20 nM), Compound C (CompC) (30 μM), or both for 24 h. Western blotting (WB) was performed for phospho-ACC (AMPK readout) and phospho-S6R (mTOR readout). Compound C treatment reduced FNIP1 expression and FLCN phosphorylation. (C) FLCN phosphorylation facilitated by FNIP1 expression is partially regulated by mTOR activity. HA-FNIP1-inducible HEK293 cells were cultured with and without doxy. For amino acid starvation, cells were cultured with Earl's balanced solution containing dialyzed serum, vitamins, pyruvate, and glucose for 16 h with and without doxycycline, followed by stimulation with 2× amino acids for 30 min. Rapamycin was added 2 h before stimulation.
FLCN Phosphorylation, Which Is Facilitated by FNIP1 Expression, Is Blocked by Inhibition of mTOR.
As mentioned above, FNIP1 overexpression facilitates FLCN phosphorylation (Figs. 4B and 6C, lanes 1 and 2). Interestingly, rapamycin partially inhibited FLCN phosphorylation facilitated by FNIP1 overexpression in doxycycline-induced HA-FNIP1-expressing HEK293 cells (Fig. 6C, lanes 2 and 4). Likewise, amino acid starvation, known to inhibit mTOR signaling (see pS6R readout), suppressed FLCN phosphorylation facilitated by FNIP1 overexpression (Fig. 6C, lanes 2 and 6). Amino acid stimulation restored FNIP1 overexpression-stimulated FLCN phosphorylation to amino acid-starved cells (Fig. 6C, lanes 6 and 8), and this effect was blocked by rapamycin pretreatment (Fig. 6C, lanes 8 and 10). These data suggest a role for mTOR signaling in modulating FLCN phosphorylation facilitated by FNIP1 overexpression, although it is not yet clear whether FLCN is a direct substrate of mTOR or a substrate of a downstream kinase that is activated by mTOR. Note that FNIP1 overexpression can still facilitate rapamycin-insensitive FLCN phosphorylation (lowest FLCN band shifts up, Fig. 6C, lanes 8 and 10).
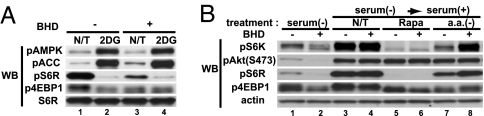
Activation of mTOR Signaling by Serum Stimulation and Its Inhibition by AMPK Activation Is Independent of FLCN Expression, but Response to Some Cell Stress Conditions Is FLCN-Dependent.
The regulation of FLCN phosphorylation by mTOR and the interaction of AMPK with FNIP1/FLCN suggested a functional relationship between FLCN/FNIP1 and mTOR. We compared the responses of the FLCN-null UOK257 and FLCN-restored UOK257-2 cell lines to physiological conditions that affect mTOR activity. We found that 2-deoxyglucose treatment activated AMPK (increased phospho-ACC), which, in turn, down-regulated mTOR in a comparable fashion in FLCN-null cells and FLCN-restored cells (Fig. 7A). mTOR signaling through PI3K and pAKT under serum-stimulated conditions and rapamycin inhibition of mTOR signaling, were unaffected by lack of FLCN expression (Fig. 7B), suggesting that FLCN may function downstream of the AMPK/mTOR signaling readouts evaluated in these experiments. On the other hand, mTOR inhibition as a result of serum starvation was significant in FLCN-restored cells but retained some signaling in FLCN-null cells (Fig. 7B). Amino acid deprivation appeared to have the opposite effect, shutting down mTOR signaling more effectively in FLCN-null cells (Fig. 7B.) Therefore, FLCN-null cells seem to respond somewhat differently to certain physiologic conditions that induce cell stress and involve the mTOR signaling pathway.
Fig. 7.
Evaluation of mTOR activity in FLCN-null and restored cells. (A) Lack of FLCN does not affect mTOR regulation by AMPK. UOK257 (FLCN-null) and UOK257-2 (FLCN-restored) cells were cultured with serum-depleted DMEM for 48 h. To activate AMPK, cells were treated with 50 mM 2-deoxyglucose (2DG) for 2 h. AMPK was activated, and mTOR activity was suppressed by 2DG in both cell lines. (B) Lack of FLCN affects mTOR activity under certain conditions. UOK257 and UOK257-2 cells were cultured without serum for 48 h, stimulated with DMEM containing 10% serum, with or without rapamycin (Rapa) (40 nM) or amino acid-free [a.a.(−)] medium with 10% dialyzed serum for 90 min. Under serum-starved conditions, FLCN-null cells did not show complete suppression of mTOR activity, whereas FLCN-null cells were more sensitive to mTOR inhibition by amino acid deprivation.
Discussion
Here, we report the identification of the FLCN-interacting protein, FNIP1, a 130-kDa protein that is moderately conserved among species, suggesting that it serves an important function in living cells. FLCN was shown to interact with FNIP1 through its C terminus in vivo and in vitro. Interestingly, nearly all mutations in BHD patients are predicted to produce a C-terminally truncated FLCN unable to bind FNIP1, which may compromise FLCN function and lead to the BHD phenotype.
We have identified AMPK as a FNIP1-interacting protein. Mutations in certain genes in the converging LKB1–AMPK–TSC1/2-mTOR and PI3K–AKT–TSC1/2-mTOR energy- and nutrient-sensing pathways lead to the development of several hamartoma syndromes. Cellular energy deficit triggers AMPK phosphorylation by LKB1 and calmodulin kinase kinase (31, 32), which negatively regulates mTOR through TSC2 phosphorylation (29, 33, 34), inhibiting protein translation and cell growth. PTEN blocks mTOR through inhibition of PI3K-activation of AKT, induced by growth factors that signal through membrane receptors (35–38). Inactivating mutations that disrupt these pathways result in dysregulated cell growth and protein synthesis, leading to a hamartoma “overgrowth” phenotype (Fig. 13, which is published as supporting information on the PNAS web site). Birt–Hogg–Dubé syndrome predisposes to hair follicle hamartomas and renal neoplasia. The interaction of FLCN with FNIP1 and AMPK suggests a possible role for FLCN and FNIP1 in the nutrient/energy-sensing pathways involving AMPK and mTOR and may provide a molecular mechanism for the BHD phenotype. Our data showing that FLCN phosphorylation facilitated by FNIP1 is mTOR- and AMPK-activity-dependent and that FNIP1 and FLCN phosphorylation and their expression were suppressed by AMPK inhibition indicate a functional interaction between FLCN/FNIP1 and mTOR/AMPK signaling pathways. Our data suggest that FLCN has multiple phosphorylation sites, and FLCN phosphorylation through AMPK may be necessary for additional phosphorylation by mTOR signaling. Furthermore, because AMPK inhibition resulted in a decreased level of FNIP1 expression, and overexpression of FNIP1 facilitated both rapamycin-sensitive and -insensitive FLCN phosphorylation, FNIP1 may be necessary for FLCN phosphorylation through both mTOR and AMPK, perhaps serving as a scaffold regulator of FLCN.
The biological mechanism explaining how FLCN functions as a tumor suppressor is still unknown. AMPK activation by 2-deoxyglucose can suppress mTOR activity in FLCN-null cells, indicating that FLCN is not necessary for mTOR suppression by AMPK. There was no difference in AMPK activity [phospho-ACC and phospho-AMPKα(T172) readouts] between FLCN-null and -restored cells, suggesting that FLCN is not an upstream regulator of AMPK. Under normal culture conditions or serum-stimulated conditions, FLCN-null cells displayed mTOR activity equal to restored cells. But FLCN-null cells exhibited different mTOR activity compared with FLCN-restored cells under certain conditions, such as serum starvation and amino acid deprivation, which is difficult to explain without further experimentation. These differences may be caused by direct or indirect effects, including possible feedback loops.
Based on these findings, we hypothesize that FNIP1 and FLCN might be downstream effectors of mTOR and AMPK and modulate energy/nutrient-sensing signaling pathways by some as-yet-unknown molecular mechanism. Further clarification of FLCN function as a tumor suppressor may contribute to a broader understanding of the molecular mechanisms responsible for other hamartoma syndromes caused by mTOR dysregulation.
Materials and Methods
Cell Lines and Cell Culture.
A FLCN-null cell line, UOK257, was established from a clear-cell renal tumor with 17p loss (10) from a BHD patient with a c.1733insC germ-line mutation (7). A FLCN-restored cell line, UOK257-2, was established by infection of UOK257 with a BHD-expressing lentiviral vector by using the ViraPower Lentiviral Expression System (Invitrogen, Carlsbad, CA). HEK293 cells expressing tetracycline-inducible HA-FLCN and FNIP1-HA were established by using the Flp-In T-Rex System (Invitrogen) according to the manufacturer's protocol.
Identification of FLCN- and FNIP-1-Interacting Proteins.
HA-FLCN- or FNIP1-HA-inducible HEK 293 cells were cultured with or without 1 μg/ml doxycycline for 36 h, lysed, immunoprecipitated with anti-HA affinity matrix, and eluted with HA peptide according to the manufacturer's protocol (Roche Applied Science, Indianapolis, IN). The eluted proteins were subjected to SDS/PAGE, transferred to PVDF membranes (ProBlott PVDF; Applied Biosystems, Foster City, CA), stained with Colloidal Gold Total Protein stain (Bio-Rad, Hercules, CA), excised and prepared as described (39), and analyzed by MALDI-TOF/MS using an Applied Biosystems Voyager-DE/STR (40) (details in Supporting Methods). Peptide mass fingerprinting and protein database searches confirmed protein identities (40).
Molecular Cloning of Human FNIP1 cDNA.
The full-length FNIP1 transcript of 3,626 nt (DQ145719) was amplified from a pooled tissue cDNA library (Clontech, Mountain View, CA) and sequenced to 6-fold coverage (details in Supporting Methods). K1AA1961 cDNA was obtained from Kazusa DNA Research Institute (Chiba, Japan).
Antibodies.
Normal rabbit IgG, HRP-labeled anti-mouse and anti-rabbit secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA); phospho-S6R, phospho-4EBP1(T37/46), phospho-ACC, phospho-AKT(S473), phospho-p70 S6 kinase(T389), p70 S6 kinase, phospho-AMPKα(T172), AMPKα, and AMPKβ1 antibodies (Cell Signaling Technology, Beverly, MA); actin antibody (Biomedical Technologies, Stoughton, MA); AMPKγ1 antibody (Zymed, San Francisco, CA); AMPKβ mouse monoclonal antibody (BD Biosciences, Franklin Lakes, NJ); FNIP-181, rabbit polyclonal antibody against a FNIP1 synthetic peptide (MAPTLFQKLFSKRTGLGAC); FLCN-102, rabbit polyclonal antibody against a FLCN peptide (QGDGNEDSPGQGEQAEE): FLCN-104, rabbit polyclonal antibody against GST-FLCN; FLCN-mAb from single-clone hybridoma cell line raised against full length GST-FLCN in the mouse.
Immunoprecipitation, Western Blotting, and Northern Blotting.
Cells were lysed in lysis or RIPA buffer and immunoprecipitated at 4°C overnight with protein G-Sepharose- (Amersham Pharmacia Biotech, Piscataway, NJ) bound antibodies. The resin was washed five times with lysis buffer, proteins were eluted with SDS sample buffer, followed by boiling, and Western blotting was performed as described (41) (details in Supporting Methods). For Northern blotting, a FNIP1 probe was PCR-generated from the unique FNIP1 sequence: forward, 5′-TGGAATCATTCAGACCCAGAA-3′; reverse, 5′-ACATCAGACACTGACGGAACC-3′. A BHD probe was generated by using exon 11 primers (7). Northern blotting was performed as described (7) by using a Human 12-lane Multiple Tissue Northern Blot (BD Biosciences).
Immunofluorescence Microscopy.
Immunofluorescence microscopy of HeLa cells transfected with FNIP1-HA and FLAG-FLCN was performed as described in Supporting Methods.
Plasmids, Transfection, Recombinant Protein Expression, and Purification.
The Gateway Protein Expression system (Invitrogen) was used to produce a variety of expression vectors encoding full-length FLCN and FNIP1, FLCN and FNIP1 deletion mutants, and GST fusion proteins produced in bacmid-infected Sf9 insect cells (details in Supporting Methods). Transfections were performed by using Lipofectamine 2000 (Invitrogen). GST-FLCN expressed in Sf9 was purified on a GST-Trap column (Amersham Pharmacia Biotech).
In Vitro Binding Assay.
GST fusion proteins were immobilized on glutathione-Sepharose beads (Amersham Biosciences, Piscataway, NJ) and incubated with 35S in vitro-translated FLCN or FNIP1 prepared by using the TNT T7 Quick kit (Promega, Madison, WI) (details in Supporting Methods). After washing, beads were boiled with SDS sample buffer and subjected to SDS/PAGE, followed by Coomassie Brilliant blue staining and autoradiography.
AMPK in Vitro Kinase Reactions.
To measure AMPK activity in anti-HA-FNIP1 immunoprecipitates, the immunoprecipitates were washed five times, and the AMPK kinase reaction was performed in reaction buffer with 100 μM SAMS substrate peptide(Upstate Biotechnology, Lake Placid, NY) and [γ-32P]ATP (Amersham Biosciences) and assayed according to the manufacturer's protocol (Upstate Biotechnology). To evaluate HA-FNIP1 phosphorylation, in vitro kinase reactions were performed on HA-FNIP1 immunoprecipitates with various kinase inhibitors.
Metabolic Whole-Cell 32P Labeling.
HA-FNIP1-inducible HEK293 cells were cultured for 24 h in standard medium containing doxycycline. Cells were washed three times with phosphate-free DMEM. Cells were labeled for 3 h with phosphate-free DMEM containing 10% dialyzed FBS and 1 mCi/ml (1 Ci = 37 GBq) [32P]orthophosphate with and without AMPK inhibitors (AraA, 2 mM and Compound C, 30 μM). AntiHA-immunoprecipitates containing [32P]HA-FNIP1 were subjected to SDS/PAGE and autoradiography.
Supplementary Material
Acknowledgments
We thank Megan Bucheimer, Kelly Esposito, Veronica Roberts, Christopher Gunning, and Peter Frank of the Protein Expression Laboratory for their excellent technical assistance; Masa-aki Nakaya for help with confocal microscopy; Debbie Morrison, Akio Yamashita, Tim Veenstra, and Ming Zhou for helpful discussions; and Len Neckers for critical reading of the manuscript. M.B. received support from the Yokohama Foundation for Advancement of Medical Science. This research was supported, in part, by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute (NCI), Center for Cancer Research. This project has been funded in whole or in part with federal funds from the NCI, NIH, under contract N01-C0-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Abbreviations
- BHD
Birt–Hogg–Dubé
- FLCN
folliculin
- mTOR
mammalian target of rapamycin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ145719).
References
- 1.Birt AR, Hogg GR, Dubé WJ. Arch Dermatol. 1977;113:1674–1677. [PubMed] [Google Scholar]
- 2.Toro JR, Glenn G, Duray P, Darling T, Weirich G, Zbar B, Linehan WM, Turner ML. Arch Dermatol. 1999;135:1195–1202. doi: 10.1001/archderm.135.10.1195. [DOI] [PubMed] [Google Scholar]
- 3.Zbar B, Alvord WG, Glenn G, Turner M, Pavlovich CP, Schmidt L, Walther M, Choyke P, Weirich G, Hewitt SM, et al. Cancer Epidemiol Biomarkers Prev. 2002;11:393–400. [PubMed] [Google Scholar]
- 4.Pavlovich CP, Walther MM, Eyler RA, Hewitt SM, Zbar B, Linehan WM, Merino MJ. Am J Surg Pathol. 2002;26:1542–1552. doi: 10.1097/00000478-200212000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt LS, Warren MB, Nickerson ML, Weirich G, Matrosova V., Toro JR, Turner ML, Duray P, Merino M, Hewitt S, et al. Am J Hum Genet. 2001;69:876–882. doi: 10.1086/323744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khoo SK, Bradley M, Wong FK, Hedblad M-A, Nordenskjold M, Teh B. Oncogene. 2001;20:5239–5242. doi: 10.1038/sj.onc.1204703. [DOI] [PubMed] [Google Scholar]
- 7.Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, Turner ML, Duray P, Merino M, Choyke P, Pavlovich CP, et al. Cancer Cell. 2002;2:157–164. doi: 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- 8.Khoo SK, Giraud S, Kahnoski K, Chen J, Motorna O, Nickolov R, Binet O, Lambert D, Friedel J, Levy R, et al. J Med Genet. 2002;39:906–912. doi: 10.1136/jmg.39.12.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt LS, Nickerson ML, Warren MB, Glenn GM, Toro JR, Merino MJ, Turner ML, Choyke PL, Sharma N, Peterson J, et al. Am J Hum Genet. 2005;76:1023–1033. doi: 10.1086/430842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vocke C, Yang Y, Pavlovich CP, Schmidt LS, Nickerson ML, Torres-Cabala CA, Merino MJ, Walther MM, Zbar B, Linehan WM. J Natl Cancer Inst. 2005;97:931–935. doi: 10.1093/jnci/dji154. [DOI] [PubMed] [Google Scholar]
- 11.Warren MS, Torres-Cabala CA, Turner ML, Merino MJ, Matrosova VY, Nickerson ML, Ma W, Linehan WM, Zbar B, Schmidt LS. Mod Pathol. 2004;17:998–1011. doi: 10.1038/modpathol.3800152. [DOI] [PubMed] [Google Scholar]
- 12.Inoki K, Corradetti MN, Guan K-L. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 13.Brugarolas J, Kaelin WG., Jr Cancer Cell. 2004;6:7–10. doi: 10.1016/j.ccr.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Martin DE, Hall MN. Curr Opin Cell Biol. 2005;17:158–166. doi: 10.1016/j.ceb.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Sarbassov DD, Ali SM, Sabatini DM. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, et al. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 17.Jenne DE, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, Muller O, Back W, Zimmer M. Nat Genet. 1998;18:38–43. doi: 10.1038/ng0198-38. [DOI] [PubMed] [Google Scholar]
- 18.Corradetti MN, Inoki K, Bardeesy N, DePinho RA, Guan KL. Genes Dev. 2004;18:1533–1538. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheadle JP, Reeve MP, Sampson JR, Kwiatkowski DJ. Hum Genet. 2000;107:97–114. doi: 10.1007/s004390000348. [DOI] [PubMed] [Google Scholar]
- 20.Eng C. Hum Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 21.Harris TE, Lawrence JC., Jr Sci STKE. 2003;2003:re15. doi: 10.1126/stke.2122003re15. [DOI] [PubMed] [Google Scholar]
- 22.Fingar DC, Blenis J. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 23.Cook JD, Walker CL. Curr Mol Med. 2004;4:813–824. doi: 10.2174/1566524043359656. [DOI] [PubMed] [Google Scholar]
- 24.Carling D. Trends Biochem Sci. 2004;29:18–24. doi: 10.1016/j.tibs.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Hardie DG. J Cell Sci. 2004;117:5479–5487. doi: 10.1242/jcs.01540. [DOI] [PubMed] [Google Scholar]
- 26.Hardie DG, Hawley SA. BioEssays. 2001;23:1112–1119. doi: 10.1002/bies.10009. [DOI] [PubMed] [Google Scholar]
- 27.Stein SC, Woods A, Jones NA, Davison MD, Carling D. Biochem J. 2000;345:437–443. [PMC free article] [PubMed] [Google Scholar]
- 28.Davies SP, Carling D, Hardie DG. Eur J Biochem. 1989;186:123–128. doi: 10.1111/j.1432-1033.1989.tb15185.x. [DOI] [PubMed] [Google Scholar]
- 29.Inoki K, Zhu T, Guan KL. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Hudson E, Chi MM, Chang AS, Moley KH, Hardie DG, Downs SM. Dev Biol. 2006;291:227–238. doi: 10.1016/j.ydbio.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 31.Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. Curr Biol. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 32.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. Proc Natl Acad Sci USA. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoki K, Li Y, Xu T, Guan KL. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Cantley LC, Neel BG. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leevers SJ, Vanhaesebroeck B, Waterfield MD. Curr Opin Cell Biol. 1999;11:219–225. doi: 10.1016/s0955-0674(99)80029-5. [DOI] [PubMed] [Google Scholar]
- 37.Inoki K, Li Y, Zhu T, Wu J, Guan KL. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 38.Potter CJ, Pedraza LG, Xu T. Nat Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 39.Iwamatsu A. Electrophoresis. 1992;13:142–147. doi: 10.1002/elps.1150130129. [DOI] [PubMed] [Google Scholar]
- 40.Jensen ON, Podtelejnikov A, Mann M. Rapid Commun Mass Spectrom. 1996;10:1371–1378. doi: 10.1002/(SICI)1097-0231(199608)10:11<1371::AID-RCM682>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 41.Baba M, Hirai S, Yamada-Okabe H, Hamada K, Tabuchi H, Kobayashi K, Kondo K, Yoshida M, Yamashita A, Kishida T, et al. Oncogene. 2003;22:2728–2738. doi: 10.1038/sj.onc.1206373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.