Abstract
Fission yeast cells belong to one of two specialized cell types, M or P. Specific environmental conditions trigger sexual differentiation, which leads to an internal program starting with pheromone signaling between M and P cells, followed by mating, meiosis, and sporulation. The initial steps of this process are controlled by Ste11p, a master transcriptional regulator that activates the expression of cell type-specific genes (only expressed in either M or P cells) as well as genes expressed in both M and P cells. Pheromone signaling is activated by Ste11p-dependent transcription and, in turn, enhances some of this transcription in a positive feedback. To obtain a genomewide view of Ste11p target genes, their cell-type specificity, and their dependence on pheromone, we used DNA microarrays along with different genetic and environmental manipulations of fission yeast cells. We identified 78 Ste11p-dependent genes, 12 and 4 of which are only expressed in M and P cells, respectively. These genes show differing grades of pheromone dependencies for Ste11p-activated transcription, ranging from complete independence to complete dependence on pheromone. We systematically deleted all novel cell type-specific genes and characterized their phenotype during sexual differentiation. A comparison with a similar data set from the distantly related budding yeast reveals striking conservation in both number and types of the proteins that define cell types. Given the divergent mechanisms regulating cell type-specific gene expression, our results highlight the plasticity of regulatory circuits, which evolve to allow adaptation to changing environments and lifestyles.
Keywords: expression profiling, master transcriptional regulator, mating type, microarray, Schizosaccharomyces pombe
Cellular differentiation is driven to a large extent by specific programs of gene expression. Yeast has distinct cell types that are required for mating, meiosis, and sporulation; this sexual differentiation provides a useful model system to understand the differentiation into specialized cell types in multicellular organisms. How the regulation of gene expression leads to specialized cell types is relatively well understood at a genomewide level in the budding yeast Saccharomyces cerevisiae (1). In this yeast, two haploid cell types, a and α, mate with each other at the first opportunity to form diploid a/α cells, which can undergo meiosis and sporulation under appropriate environmental conditions (reviewed in ref. 2).
Cells of the fission yeast Schizosaccharomyces pombe also come in two specialized cell types, called P (or h+) and M (or h-) mating types. Heterothallic strains have a fixed mating type (P or M), whereas homothallic strains (h90) can switch between both types. Unlike budding yeast, however, fission yeast cells will mate only under specific environmental conditions, most notably nitrogen starvation; the resulting diploid cells then immediately undergo meiosis and sporulation (reviewed in refs. 3 and 4). The two distantly related yeasts thus provide complementary models to identify conserved and specialized regulatory mechanisms for cell differentiation, involving both environmental and intrinsic factors.
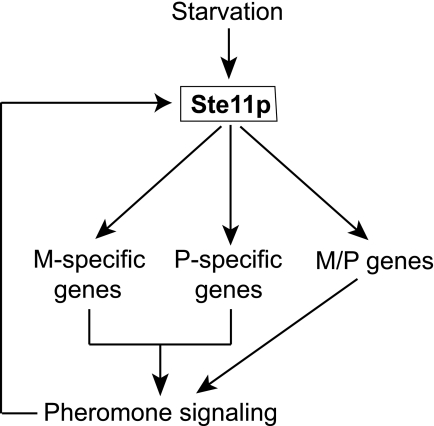
In fission yeast, starvation induces the expression of Ste11p, an HMG (high mobility group) transcription factor essential for sexual differentiation (5). Ectopic expression of Ste11p leads to sexual differentiation irrespective of nutritional conditions. Ste11p thus acts as a developmental switch, and expression of its targets induces the physiological and morphological changes that culminate in sexual differentiation (5). Some of the Ste11p target genes are cell type-specific, whereas others are induced in cells of either mating type (Fig. 1). Cell type-specific expression is mediated by cell type-specific transcription factors, Mat-Mc and Mat-Pc (5), which are encoded in the mating type locus and activated by Ste11p (6). In M cells, Mat-Mc and Ste11p physically interact and directly bind to the promoters of M specific genes (7). In P cells, Mat-Pc together with Map1p, a transcription factor of the MADS-box family, is required for activation of P specific genes. P and M cells produce specific pheromones (P and M factor, respectively) as well as receptors for the pheromones released by the opposite mating type. This system allows cells of opposite mating types to recognize each other during the mating process. The pheromone signaling activated by Ste11p-directed transcription, in turn, cooperates with Ste11p to enhance the transcription of some Ste11p-dependent genes (Fig. 1; reviewed in ref. 3).
Fig. 1.
Control of sexual differentiation by the Sc. pombe Ste11p master transcriptional regulator. Environmental changes, especially nitrogen starvation, lead to increased expression of Ste11p, which activates the transcription of cell type-specific genes as well as of genes expressed in both M and P cell types. Expression of Ste11p targets establishes pheromone signaling between cells of different mating types that, in turn, causes further activation of Ste11p activity.
Several Ste11p-dependent genes have been identified in small-scale experiments (3, 4), and we have reported global gene expression data covering later stages of sexual differentiation (8). It is not known, however, at a genomewide level what genes are directly regulated by Ste11p (rather than upon stress caused by nitrogen starvation), how many of the Ste11p-dependent genes are cell type-specific, and to what extent pheromone signaling is required for Ste11p-dependent gene expression. Here we used DNA microarrays to investigate the gene expression program during the early stages of sexual differentiation, focusing on the roles and targets of Ste11p and pheromone. We comprehensively identify cell type-specific genes and systematically delete them to assess their role in the differentiation process. We also compare our results with a similar data set from budding yeast (1) to gain insight into the evolution of the regulatory mechanisms that control cell type-specific gene expression.
Results and Discussion
Gene Expression Changes After Nitrogen Starvation.
To study the global roles of the Ste11p master transcriptional regulator in fission yeast, we used DNA microarrays to follow the gene expression response associated with depletion of nitrogen. To assess the influence of mating type and cell–cell communication on this response, we carried out time courses in heterothallic (P and M) and homothallic cells (h90). In addition, we followed h90 cells with a mutation in the ste11 gene. In homothallic cultures, pheromone communication between P and M cells leads to mating, which is immediately followed by meiosis. To separate the transcriptional response of nitrogen starvation and pheromone signaling from the meiotic gene expression program (8), we used h90 cells carrying a mutation in the fus1 gene, which specifically blocks cell–cell fusion and, therefore, entry into meiosis (9, 10).
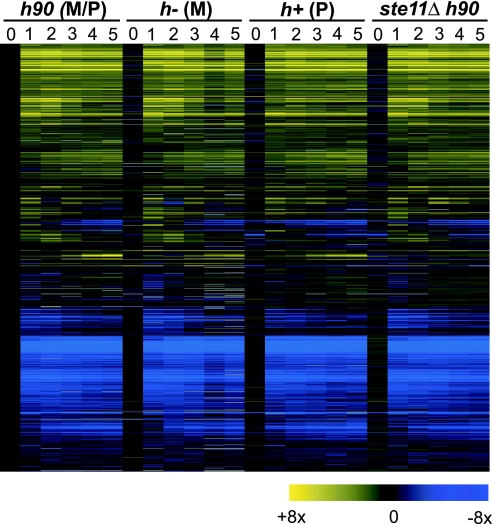
Nitrogen starvation led to extensive changes in gene expression involving almost all genes (Fig. 2). As many as 451 and 446 genes were induced or repressed 3-fold or more, respectively, compared with vegetative cells. Many of the induced (46%) and repressed (57%) genes are part of the Core Environmental Stress Response, which fission yeast cells launch in response to any stress situation (11). The majority of these changes took place within 1 h of nitrogen removal, whereas a smaller set of genes was induced only after 3 h of nitrogen starvation. This latter group contained several known Ste11p targets (see below).
Fig. 2.
Massive changes in gene expression in response to nitrogen starvation. Hierarchical clustering of all Sc. pombe genes with data for at least 50% of all conditions (4,695 genes). Rows represent genes and their expression profiles after removal of nitrogen from the medium. Columns represent hourly time points of four time course experiments (before and up to 5 h after nitrogen removal); experiments were carried out in the four genetic backgrounds indicated on top (see Materials and Methods for details). The mRNA levels at each time point relative to levels of h90 cells before nitrogen removal are color-coded as indicated at the bottom, and missing data are in gray.
Identification of Ste11p Targets.
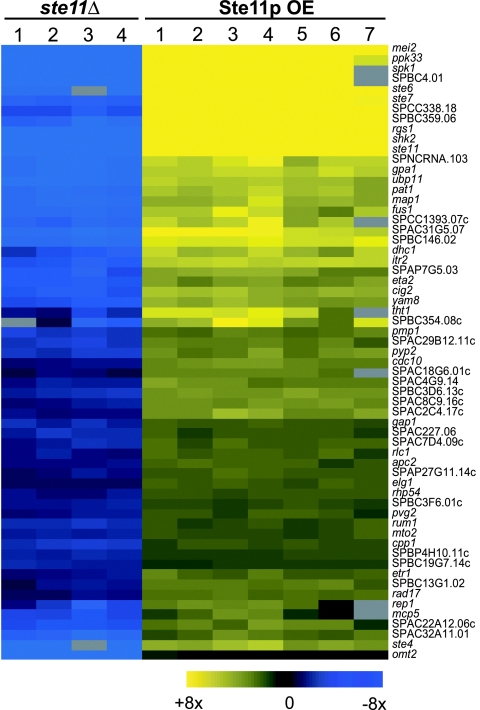
To identify Ste11p targets, we directly compared the transcriptome of homothallic WT cells (h90 fus1) with that of ste11Δ mutants under conditions that induce sexual differentiation. To allow for indirect effects of the ste11Δ mutation, we took advantage of the fact that ectopic expression of ste11 can drive cells into sexual differentiation and, therefore, is expected to cause the expression of Ste11p targets (5). We thus defined Ste11p targets as those genes whose expression was significantly reduced in a ste11Δ mutant and significantly increased when ste11 is overexpressed in vegetative cells (Fig. 3 and Fig. 4 A and B). The large number of repeated experiments and the avoidance of arbitrary thresholds allowed us to identify Ste11p targets that are only weakly regulated (Fig. 4 A and B).
Fig. 3.
Expression profiles of Ste11p target genes. (Left) Expression levels of Ste11p target genes in ste11Δ h90 deletion mutants relative to h90 WT cells, measured 5 h after nitrogen removal. Data for four independent biological repeats are shown. (Right) Expression levels of Ste11p target genes in vegetative cells overexpressing Ste11p relative to control cells containing an empty vector. Data for seven independent biological repeats are shown. The mRNA levels at each condition are color-coded as indicated at the bottom, and missing data are in gray. The corresponding genes are listed at right.
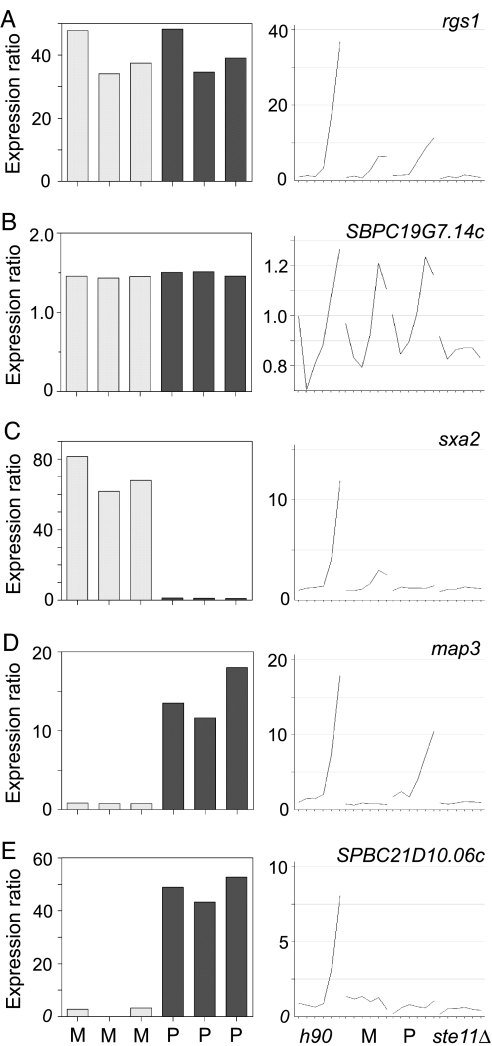
Fig. 4.
Ste11p target genes with different cell type-specificity. rgs1 (A) and SPBC19G7.14c (B) are expressed in both M and P cells; sxa2 (C) is an M specific gene; map3 (D) and SPBC21D10.06c (E) are P specific genes. (Left) Bar graphs showing expression ratios of different genes in vegetative cells overexpressing Ste11p relative to control cells containing an empty vector. Each of the six bars corresponds to an independent experiment, carried out in M or P cells as indicated at the bottom. (Right) Graphs showing expression profiles of the corresponding genes in time course experiments after nitrogen removal (as in Fig. 2). The y axes show the expression ratios of mRNA levels at each time point relative to levels of h90 cells before nitrogen removal. The experiment was carried out in four genetic backgrounds as indicated at the bottom.
A total of 62 genes behaved consistently as expected for Ste11p targets (Fig. 3; see Table 2, which is published as supporting information on the PNAS web site). We also found an additional 16 genes that were expressed at lower levels in ste11Δ mutants but were induced by ste11 overexpression in a cell type-specific manner (Fig. 4C-E; Table 3, which is published as supporting information on the PNAS web site). Our data showed good agreement with published reports on Ste11p targets (Materials and Methods). Forty-one Ste11p target genes had been studied previously, although several of them were not known to be induced or to have a function during sexual differentiation. Most of the potential Ste11p targets were induced after 3 h of nitrogen depletion, but a few genes showed a more complex profile, with a first induction within 1 h of nitrogen removal and a second increase after 3 h (data not shown). The promoters of Ste11p targets genes showed highly significant enrichment in sequence motifs related to the TR box (refs. 5 and 7; data not shown).
We have described a cluster of 40 genes that are simultaneously induced when WT diploid cells enter meiosis (“delayed genes”) (8). Most of these genes (34 of 40) were among the Ste11p targets identified here, showing that the Ste11p-dependent program launched during meiosis is similar to that of mating cells.
Functions of Genes Regulated by Ste11p.
Several Ste11p targets code for proteins required for the establishment and regulation of pheromone signaling, such as the guanine nucleotide-binding protein α-1 Gpa1p (12), the GTPase activating protein Gap1p (13), the MAP kinase Spk1p (14), the phosphatases Pyp2p and Pmp1p (15), and the regulator of G protein signaling Rgs1p (16, 17). Genes encoding both activators (Mei2p and meiRNA; ref. 18) and repressors (Pat1p) of meiosis were induced, suggesting the existence of sophisticated feedback and balancing mechanisms. The gene fus1, encoding a protein required for cell–cell fusion (10), was strongly induced, as well as two genes similar to budding yeast genes with functions in this process.
In addition, several genes with functions in early meiotic steps are induced. They encode the transcription factor Rep1p, required for premeiotic S phase (19), a protein necessary for karyogamy (Tht1p) (20), and dynein heavy chain (Dhc1p), required for nuclear movement during meiotic prophase (21). However, the induction of these genes in response to nitrogen starvation was much weaker than their induction during meiosis (8), suggesting the existence of a second step of activation specific for meiosis.
Other targets not previously studied include the gene for a novel kinase (SPCC162.10) and a gene encoding a member of the ubiquitin C-terminal hydrolase family (SPBC19C2.04C). In budding yeast, a protein of this family (Ubp3p) has a role in the modulation of pheromone transcription (22), but a similar phenomenon has not been described in fission yeast. In addition, close to 20 genes encode hypothetical proteins for which no functions can be predicted based on sequence homology. Their specific expression pattern during sexual differentiation suggests that they play a role in this process. All Ste11p targets identified are listed in Table 2.
Regulation of Ste11p Targets by Pheromone.
Pheromone cooperates with Ste11p to induce the expression of target genes (Fig. 1). To study the global extent of this effect, we compared time courses of heterothallic cells (where there is no pheromone signaling) with those of homothallic cells. The effect of pheromone on the induction of Ste11p target genes was varied: pheromone had little effect in some cases, whereas in others, it enhanced the Ste11p-dependent induction or was even absolutely required for the induction to take place (Fig. 4, compare h90 with M and P).
To directly monitor the effect of pheromone on gene expression, we used synthetic P factor (23). As P factor is rapidly degraded by the Sxa2p protease (24), we used M strains carrying a deletion in the sxa2 gene. We first starved sxa2Δ cells of nitrogen to allow the expression of Ste11p targets, including the P factor receptor and components of the pheromone pathway. Then we added synthetic P factor and followed the effects of pheromone on gene expression.
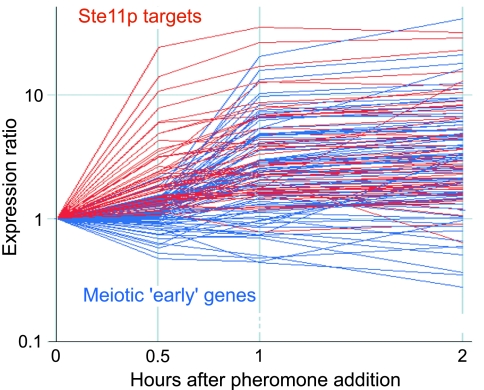
A group of genes, composed mostly of Ste11p targets, were induced within 30 min of addition of P factor and reached a plateau after 1 h (Fig. 5). Most of the Ste11p targets (90%) were induced, although the effect varied from 1.5- to 30-fold inductions.
Fig. 5.
Induction of gene expression after pheromone addition. Time course experiment after expression levels of Ste11p target genes (red) and meiotic early genes (blue; ref. 8) up to 2 h after addition of pheromone (P factor). The y axis shows the expression ratio of mRNA levels at each time point relative to the levels before addition of pheromone.
In addition, a second group of genes were induced later (between 1 and 2 h after the addition of P factor) (Fig. 5). This group was strongly enriched for meiotic “early” genes, a cluster of genes induced during meiotic prophase containing many genes required for DNA replication and recombination (8). An effect of pheromone on these genes has not been described previously. The Rep1p transcription factor (19), which is required for the expression of some early genes (refs. 19 and 25 and our unpublished observations), is a Ste11p target and strongly induced upon addition of P factor. It is likely that the high levels of Rep1p are responsible for the induction of the early genes. The physiological importance of this effect is unclear, because we did not observe a similar activation in the h90 time course (in which there is pheromone signaling, but presumably at lower levels). In addition, pheromone signaling is required for entry into meiosis but not for progression through it (26). It is possible that pheromone has a small role in enhancing the expression of the early genes during natural meiosis, thus making the process more robust. Alternatively or in addition, pheromone could have a more critical role to drive meiosis under some physiological conditions.
The mechanism by which pheromone enhances Ste11p activity is only partly understood: Ste11p is phosphorylated by the MAP kinase of the pheromone pathway (Spk1p) (27), and pheromone signaling leads to an increase in the concentration of Ste11p in the nucleus (28). Interestingly, ste11 overproduction results in the induction of all Ste11p targets, including those that are normally absolutely dependent on pheromone for their transcription (Fig. 3). This finding could mean that the overexpression of Ste11p induces an ectopic pheromone response (several components of the pathway are Ste11p targets, including the gene for the MAP kinase Spk1p), or it could be a direct consequence of the increased levels of Ste11p.
To distinguish between these two possibilities, we overexpressed ste11 in cells carrying a deletion in spk1. We expected that if the effect was indirect, Ste11p targets would not be induced in the absence of Spk1p. In fact, we observed that the effect of Ste11p overexpression was independent of the presence of Spk1p and, therefore, of the pheromone response. This finding suggests that high levels of Ste11p in the cell (and presumably in the nucleus) are sufficient to relieve the dependence of Ste11p targets on pheromone for their expression and supports the view that pheromone controls gene expression by enhancing the transport of Ste11p to the nucleus (28).
Identification of Cell Type-Specific Genes.
We initially attempted to identify cell type-specific genes by comparing the expression profiles of Ste11p-dependent genes in P and M cells. However, cells of a single mating type cannot establish pheromone communication, and the expression of many Ste11p targets is completely dependent on pheromone (Fig. 4E). This dependency prevented the identification of cell type-specific genes that require pheromone signaling for their expression.
We therefore developed an alternative strategy. While using ste11 overexpression to confirm the identity of Ste11p targets, we noticed that all known mating type-specific genes were induced to a much stronger level when Ste11p was overexpressed in cells of the mating type in which they are normally expressed (Fig. 4 C and D). We therefore looked for genes whose induction by Ste11p overexpression was cell type-dependent. In this way, we identified 12 M specific genes and 4 P specific genes (Table 3). We confirmed that these genes were indeed cell type-specific by comparing their expression profiles in time courses of P and M cells. In a few cases, their expression depended on pheromone, so it was not possible to verify their specificity (Fig. 4E). Note that this definition of cell type specificity does not necessarily mean that the genes are expressed exclusively in one mating type, but that they are induced to a higher level in a specific mating type. The promoters of M specific genes were significantly enriched in the sequence motif called an M box (7). For P specific genes, the small number of genes makes it impossible to find statistically significant enrichment of regulatory motifs (data not shown).
Six of the 12 M specific genes had been identified as such, and mutations in them cause M specific sterility. Most of them have functions related to the production of M factor, which is a small peptide modified by carboxy methylation and farnesylation. mfm1, mfm2, and mfm3 encode the M factor precursors (29), mam1 is required for its secretion (30), and mam4 has a function in the carboxy methylation (31). The latter is required in M cells for M factor maturation but has been reported not to be cell type-specific (31). We also found that the cwp1 gene, which encodes the alpha subunit of geranyl-geranyl transferase type I, is M specific. This homology suggests it may have a function in the modification of M factor, similar to the role that its Sa. cerevisiae ortholog RAM2 has in the biosynthesis of a factor. The P factor receptor is encoded by mam2 (32), and sxa2 encodes a protease that degrades extracellular P factor (33). In addition, we identified four other M specific genes of unknown function (Table 3).
Of the four P specific genes, two had previously been identified, and their mutations confer P specific sterility: map2 encodes the P factor precursor (23) and map3 the M factor receptor (34); two other genes (SPBC21D10.06c and SPAC1665.03) have not been studied.
Functional Analysis of Cell Type-Specific Genes.
The expression of cell type-specific genes confers a specific identity to a fission yeast cell in two ways: first, by allowing it to produce and secrete a particular mating pheromone; second, by allowing it to respond to pheromones of the opposite mating type. Therefore, we expected that novel genes expressed in a mating type-specific fashion have a function specific to the mating type in which they are expressed. To test this hypothesis, we used a PCR-based approach (35) to systematically delete and analyze the function of all novel mating type-specific genes (Table 3).
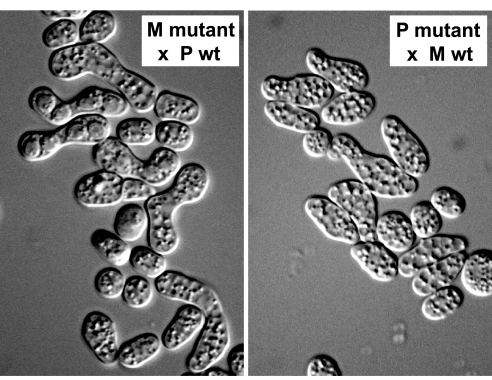
One of the two novel P specific genes behaved in the predicted way: M cells with a deletion in SPBC21D10.06c could mate and undergo meiosis, whereas P cells were completely sterile (Fig. 6). By contrast, we did not observe any phenotype in cells carrying a deletion in SPAC1665.03. One of the five novel M specific genes showed M specific partial sterility (SPAP11E10.02c), whereas deletion of cwp1 and SPAPB1A10.02 resulted in lethality in both cell types, and deletion of SPAC11H11.03c and SPAC11H11.03c did not cause any observable phenotype. The fact that some essential genes are cell type-specific may reflect an elevated requirement for them in a specific cell type during differentiation. For instance, it is likely that Cwp1p has an essential function in protein farnesylation (in both mating types) and a specific function in the production of M factor in M cells.
Fig. 6.
Deletion of the P specific gene SPBC21D10.06c causes P specific sterility. (Left) SPBC21D10.06cΔ M cells crossed to WT P cells are able to mate and form asci with four spores. (Right) SPBC21D10.06cΔ P cells cannot mate with WT M cells.
The two genes that caused cell type-specific sterility (SPBC21D10.06c in P cells and SPAP11E10.02c in M cells) encode potential cell surface proteins that are similar to each other (43% identity over 825 aa) and to yeast proteins involved in cell adhesion. Their deletion causes strikingly similar phenotypes in which cells produce long and thin mating projections but fail to attach to each other (Fig. 6). Our results suggest that SPBC21D10.06c and SPAP11E10.02c encode cell type-specific agglutinins, which allow specific aggregation between cells of opposite mating types. SPAP11E10.02c and SPBC21D10.06c have been independently identified and named mam3 and map4, respectively (M. Yamamoto, personal communication).
We believe that we have comprehensively identified and characterized the genes that establish cell type-specific identity in fission yeast. Most of these genes (11 of 16) show cell type-specific phenotypes and are involved in the production of molecules that confer cell identity (pheromones and agglutinins) or that allow the recognition of the opposite mating type (pheromone reception and agglutinins).
Comparison with Budding Yeast.
Budding and fission yeasts probably separated >1 billion years ago (36). The programs of sexual differentiation in the two yeasts show both similarities and differences (see Introduction). All ascomycetes, including fission and budding yeasts, have a bipolar mating system, consisting of a single mating-type locus with two alleles (P and M in Sc. pombe; a and α in Sa. cerevisiae) (37). The budding yeast mating-type loci also encode transcription factors that regulate the expression of target genes. Despite these similarities, the molecular mechanisms that control expression of cell type-specific genes are not conserved. For instance, the system in budding yeast involves both induction and repression of target genes, whereas in fission yeast, it appears that only activation is used.
A recent study has systematically identified cell type-specific genes in budding yeast (1). We compared the nature of the products of cell type-specific genes in both yeasts (Table 1). Sa. cerevisiae a cells specifically express genes encoding a pheromone (a-factor), a transporter required for pheromone secretion, a receptor for the pheromone of the opposite mating type, a protease that degrades α-factor, and a specific agglutinin. This set is highly reminiscent of the genes specifically expressed in Sc. pombe M cells. α cells, on the other hand, express genes encoding α-factor pheromone, a receptor for a-factor and a specific agglutinin. These genes are similar to P specific genes from fission yeast. Note that although the Sc. pombe and Sa. cerevisiae cell type-specific genes encode products with similar functions, they are not necessarily orthologs.
Table 1.
Comparison between Sc. pombe and Sa. cerevisiae cell type-specific genes
| Protein function | Sc. pombe | Sa. cerevisiae | % identity(BLAST e value) |
|---|---|---|---|
| M cell/a cell | |||
| Pheromone (farnesylated peptide) | mfm1/mfm2/mfm3 | MFA1/MFA2 | No sequence similarity, structurally similar |
| Pheromone transporter | mam1 | STE6 | 30% (1e−148) |
| Protease for pheromone of opposite cell type | sxa2 | BAR1 | No significant similarity |
| Agglutinin | SPAP11E10.02c | AGA2 | No significant similarity |
| Pheromone receptor | mam2 | STE2 | 25% (4e−11) |
| Others (not similar functions) | mam4/cwp1/SPAPB1A10.02 SPAC11H11.03c/SPAC11H11.05c | DPS1/DPS2/ASG7 | Not applicable |
| P cell/α cell | |||
| Pheromone (peptide) | map2 | MFα1, MFα2 | No sequence similarity, structurally similar |
| Agglutinin | SPBC21D10.06c | SAG1 | No significant similarity |
| Pheromone receptor | map3 | STE3 | 26% (7e−23) |
| Others (not similar functions) | SPAC1665.03 | YLR040C | Not applicable |
Cell type-specific genes from Sa. cerevisiae are described in ref. 1, and those from Sc. pombe are listed in Table 3. Note that the comparisons are based on biological function and the genes are not necessarily orthologs.
Our results show that, despite a profound lack of conservation of the regulatory mechanism that control cell type-specific gene expression, there is a strong conservation of the target genes. Similarly, a conserved set of genes is modulated in response to a wide variety of stresses in both fission and budding yeast, although the mechanisms regulating these genes are diverged (11, 38). These results support the view that regulatory circuits are highly plastic and may evolve faster to allow cells to adapt to changing environmental conditions and to different lifestyles (39, 40).
Materials and Methods
Experimental Design.
Time courses of h90 fus1-B20, h-, h+, and h90 ste11Δ were carried out once each. Cells were grown in Edinburgh minimal medium (EMM) containing NH4Cl as a nitrogen source, washed with minimal medium without NH4Cl (EMM-N), and resuspended in EMM-N. Cells then were grown at 28°C and samples taken every hour for 5 h. For direct comparisons of h90 fus1 and h90 ste11Δ, cells were treated as described above and samples were taken after 5 h at 28°C. This experiment was carried out four times. For overexpression experiments, cells carrying a plasmid containing ste11 under the control of the nmt1 thiamine-repressible promoter (41) (pREP1-ste11) or an empty vector (pREP1) were grown in EMM containing thiamine. Cells were washed with EMM without thiamine (EMM-T), resuspended in EMM-T, and grown at 32°C for 18 h. The experiment was performed three times in P cells and three times in M cells. P factor (a gift from Paul Nurse, The Rockefeller University, New York, NY) was synthesized as described in ref. 42. For the pheromone experiments, sxa2Δ h- cells were grown in EMM, washed with EMM-N, resuspended in EMM-N, and grown for 5 h at 28°C. The culture was split in half, and P factor (at a final concentration of 1.5 μg/ml) or a corresponding amount of ethanol was added to the cells. Samples were taken at 0, 0.5, 1, and 2 h.
Microarrays and Data Analysis.
RNA preparation, labeling, microarray production, and analysis are described in ref. 43. Microarrays were scanned with a Genepix 4000B scanner and analyzed with Genepix software (Axon Instruments, Union City, CA). Clustering and visualization was done with GeneSpring (Agilent Technologies, Palo Alto, CA). Data were hierarchically clustered by using the standard (Fig. 2) or distance (Fig. 3) correlation. We determined statistical significance by using SAM (Significance Analysis of Microarrays) (44), with the false discovery rate adjusted to <0.1%.
Validation of Results.
We compared our list of potential Ste11p targets with published data (Table 2 and data not shown). Of 17 genes whose expression had been directly tested for Ste11p dependency, we identified 15 genes (byr2; ref. 45) that do not behave as a Ste11p target in our experiments, and ste11 cannot be tested with our experimental setup). We also compared our lists of cell type-specific genes with published results. We identified all eight genes previously reported to be cell type-specific (Table 3). Mam4, which has been reported to be expressed in both mating types (31), behaved as M specific in our experiments.
Supplementary Material
Acknowledgments
We thank Sergio Moreno (Centro del Investigacion del Cances, Salamanca, Spain), Iain Hagan (Paterson Institute, Manchester, U.K.), and Paul Nurse for strains and reagents; Masayuki Yamamoto for communicating unpublished results; and Samuel Marguerat and Luis López Maury for comments on the manuscript. Richard Browning and Benjamin Schuster-Böckler constructed the deletions of M specific genes. Work in our laboratory is funded by Cancer Research UK Grant C9546/A5262.
Abbreviation
- EMM
Edinburgh minimal medium.
Footnotes
Conflict of interest statement: No conflicts declared.
This article is a PNAS direct submission.
Data deposition: The microarray data have been deposited in the ArrayExpress database, www.ebi.ac.uk/arrayexpress (accession no. E-TABM-139).
References
- 1.Galgoczy DJ, Cassidy-Stone A, Llinas M, O'Rourke SM, Herskowitz I, DeRisi JL, Johnson AD. Proc Natl Acad Sci USA. 2004;101:18069–18074. doi: 10.1073/pnas.0407611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sprague GF, Blair LC, Thorner J. Annu Rev Microbiol. 1983;52:536–553. doi: 10.1146/annurev.mi.37.100183.003203. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen O. In: The Molecular Biology of Schizosaccharomyces pombe. Egel R, editor. Heidelberg, Germany: Springer; 2004. pp. 281–296. [Google Scholar]
- 4.Yamamoto M, Imai I, Watanabe Y. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Life Cycle and Cell Biology. Pringle JR, Broach JR, Jones EW, editors. Plainview, NY: Cold Spring Harbor Lab Press; 1997. pp. 1035–1106. [Google Scholar]
- 5.Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M. Genes Dev. 1991;5:1990–1999. doi: 10.1101/gad.5.11.1990. [DOI] [PubMed] [Google Scholar]
- 6.Kelly M, Burke J, Smith M, Klar A, Beach D. EMBO J. 1988;7:1537–1547. doi: 10.1002/j.1460-2075.1988.tb02973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kjaerulff S, Dooijes D, Clevers H, Nielsen O. EMBO J. 1997;16:4021–4033. doi: 10.1093/emboj/16.13.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mata J, Lyne R, Burns G, Bähler J. Nat Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- 9.Bresch C, Muller G, Egel R. Mol Gen Genet. 1968;102:301–306. doi: 10.1007/BF00433721. [DOI] [PubMed] [Google Scholar]
- 10.Petersen J, Weilguny D, Egel R, Nielsen O. Mol Cell Biol. 1995;15:3697–3707. doi: 10.1128/mcb.15.7.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bähler J. Mol Biol Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obara T, Nakafuku M, Yamamoto M, Kaziro Y. Proc Natl Acad Sci USA. 1991;88:5877–5881. doi: 10.1073/pnas.88.13.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imai Y, Miyake S, Hughes DA, Yamamoto M. Mol Cell Biol. 1991;11:3088–3094. doi: 10.1128/mcb.11.6.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gotoh Y, Nishida E, Shimanuki M, Toda T, Imai Y, Yamamoto M. Mol Cell Biol. 1993;13:6427–6434. doi: 10.1128/mcb.13.10.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Didmon M, Davis K, Watson P, Ladds G, Broad P, Davey J. Curr Genet. 2002;41:241–253. doi: 10.1007/s00294-002-0301-3. [DOI] [PubMed] [Google Scholar]
- 16.Pereira PS, Jones NC. Genes Cells. 2001;6:789–802. doi: 10.1046/j.1365-2443.2001.00465.x. [DOI] [PubMed] [Google Scholar]
- 17.Watson P, Davis K, Didmon M, Broad P, Davey J. Mol Microbiol. 1999;33:623–634. doi: 10.1046/j.1365-2958.1999.01510.x. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe Y, Yamamoto M. Cell. 1994;78:487–498. doi: 10.1016/0092-8674(94)90426-x. [DOI] [PubMed] [Google Scholar]
- 19.Sugiyama A, Tanaka K, Okazaki K, Nojima H, Okayama H. EMBO J. 1994;13:1881–1887. doi: 10.1002/j.1460-2075.1994.tb06457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tange Y, Horio T, Shimanuki M, Ding DQ, Hiraoka Y, Niwa O. J Cell Biol. 1998;140:247–258. doi: 10.1083/jcb.140.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto A, West RR, McIntosh JR, Hiraoka Y. J Cell Biol. 1999;145:1233–1249. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Dohlman HG. J Biol Chem. 2002;277:15766–15772. doi: 10.1074/jbc.M111733200. [DOI] [PubMed] [Google Scholar]
- 23.Imai Y, Yamamoto M. Genes Dev. 1994;8:328–338. doi: 10.1101/gad.8.3.328. [DOI] [PubMed] [Google Scholar]
- 24.Ladds G, Rasmussen EM, Young T, Nielsen O, Davey J. Mol Microbiol. 1996;20:35–42. doi: 10.1111/j.1365-2958.1996.tb02486.x. [DOI] [PubMed] [Google Scholar]
- 25.Li YF, Smith GR. Genetics. 1997;146:57–67. doi: 10.1093/genetics/146.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willer M, Hoffmann L, Styrkarsdottir U, Egel R, Davey J, Nielsen O. Mol Cell Biol. 1995;15:4964–4970. doi: 10.1128/mcb.15.9.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kjaerulff S, Lautrup-Larsen I, Truelsen S, Pedersen M, Nielsen O. Mol Cell Biol. 2005;25:2045–2059. doi: 10.1128/MCB.25.5.2045-2059.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin J, Kang W, Leung B, McLeod M. Mol Cell Biol. 2003;23:3253–3264. doi: 10.1128/MCB.23.9.3253-3264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davey J. EMBO J. 1992;11:951–960. doi: 10.1002/j.1460-2075.1992.tb05134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christensen PU, Davey J, Nielsen O. Mol Gen Genet. 1997;255:226–236. doi: 10.1007/s004380050493. [DOI] [PubMed] [Google Scholar]
- 31.Imai Y, Davey J, Kawagishi-Kobayashi M, Yamamoto M. Mol Cell Biol. 1997;17:1543–1551. doi: 10.1128/mcb.17.3.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitamura K, Shimoda C. EMBO J. 1991;10:3743–3751. doi: 10.1002/j.1460-2075.1991.tb04943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imai Y, Yamamoto M. Mol Cell Biol. 1992;12:1827–1834. doi: 10.1128/mcb.12.4.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka K, Davey J, Imai Y, Yamamoto M. Mol Cell Biol. 1993;13:80–88. doi: 10.1128/mcb.13.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 36.Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB. Science. 2001;293:1129–1133. doi: 10.1126/science.1061457. [DOI] [PubMed] [Google Scholar]
- 37.Souza CA, Silva CC, Ferreira AV. Genet Mol Res. 2003;2:136–147. [PubMed] [Google Scholar]
- 38.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scannell DR, Wolfe K. Genome Biol. 2004;5:206. doi: 10.1186/gb-2004-5-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsong AE, Miller MG, Raisner RM, Johnson AD. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- 41.Maundrell K. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 42.Stern B, Nurse P. EMBO J. 1997;16:534–544. doi: 10.1093/emboj/16.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lyne R, Burns G, Mata J, Penkett CJ, Rustici G, Chen D, Langford C, Vetrie D, Bähler J. BMC Genomics. 2003;4:27. doi: 10.1186/1471-2164-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tusher VG, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Styrkarsdottir U, Egel R, Nielsen O. Mol Gen Genet. 1992;235:122–130. doi: 10.1007/BF00286189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.