Abstract
Monitoring genome-wide, cell-specific responses to human disease, although challenging, holds great promise for the future of medicine. Patients with injuries severe enough to develop multiple organ dysfunction syndrome have multiple immune derangements, including T cell apoptosis and anergy combined with depressed monocyte antigen presentation. Genome-wide expression analysis of highly enriched circulating leukocyte subpopulations, combined with cell-specific pathway analyses, offers an opportunity to discover leukocyte regulatory networks in critically injured patients. Severe injury induced significant changes in T cell (5,693 genes), monocyte (2,801 genes), and total leukocyte (3,437 genes) transcriptomes, with only 911 of these genes common to all three cell populations (12%). T cell-specific pathway analyses identified increased gene expression of several inhibitory receptors (PD-1, CD152, NRP-1, and Lag3) and concomitant decreases in stimulatory receptors (CD28, CD4, and IL-2Rα). Functional analysis of T cells and monocytes confirmed reduced T cell proliferation and increased cell surface expression of negative signaling receptors paired with decreased monocyte costimulation ligands. Thus, genome-wide expression from highly enriched cell populations combined with knowledge-based pathway analyses leads to the identification of regulatory networks differentially expressed in injured patients. Importantly, application of cell separation, genome-wide expression, and cell-specific pathway analyses can be used to discover pathway alterations in human disease.
Keywords: anergy, apoptosis, costimulatory receptors, immunosuppression, network analysis
An emerging opportunity exists to greatly enhance the accuracy and content of molecular information from human cells and/or tissues through the introduction of high-throughput technologies, such as oligonucleotide microarrays and cellular pathway analyses (1). Such molecular information should lead to a better understanding of disease states in the asymptomatic and progressive, symptomatic phases of disease. These new insights promise to provide multiple benefits to clinical medicine, including improved diagnosis and prevention, earlier interventions before symptomatic disease, and individualized therapies. For example, genome-wide expression analyses may provide the foundation for future diagnostics to optimize resource utilization and improve outcome in the severely injured patient. Furthermore, this “discovery science” may lead to a greater understanding of the specific mechanisms responsible for the multiple pathological alterations in the host's inflammatory and immune responses, ultimately resulting in individualized therapies for these patients (2).
The clinical application of molecular information about human disease is often stymied by an inadequate understanding of the functional integration of complex interactive cellular processes. This is particularly problematic for the study of inflammation, which is an integrated host response, characterized by numerous interactions among multiple cell types (e.g., T cells and monocytes). Although genome-wide expression analysis offers an unbiased opportunity to dissect these processes, such analyses are complicated by cellular heterogeneity. For example, blood leukocytes represent a composite mixture of several cellular subpopulations (3). Unfortunately, cellular heterogeneity makes it difficult to distinguish changes in gene expression that represent true biological responses from variations arising simply from changes in the distribution of individual cell populations. A major challenge to the clinical application of these approaches, therefore, is first to integrate high-throughput technologies using cell subpopulations that have been purified to near homogeneity, then to extract meaningful biological knowledge with appropriate cell-specific pathway analyses, and, finally, to validate this biological information at the functional protein level.
Here, we isolated highly enriched cell populations from peripheral blood leukocytes of severely injured patients and demonstrated how genome-wide expression integrated with system-level network analyses can both identify signaling pathways and facilitate the understanding of the host immunological response to severe injury. T cell apoptosis and anergy and decreased antigen presentation by blood monocytes are considered pivotal in the development of postinjury multiple organ dysfunction syndrome (MODS) (4–10). Defects in T cell and monocyte function, commonly seen in severely injured patients, may be associated with the differential expression of several recently described receptors and signaling molecules (11–14). By focusing on two specific peripheral blood leukocyte populations known to interact (CD2+CD3+ T cells and CD14+ monocytes), we identified concordant perturbations of ligands and receptors for several pathways that can lead to cellular anergy in T cells and failure of antigen presentation in monocytes. Equally important, however, this combined experimental and computational approach can be readily applied to address multiple similar hypotheses in other human diseases, and it represents an important strategic approach for future clinical research.
Results
Isolation of Enriched Cell Populations.
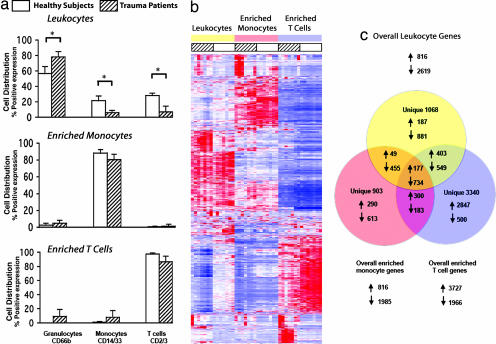
Peripheral venous blood was obtained from seven subjects with defined MODS after experiencing severe traumatic injury (15) (Table 1). Blood was also obtained from seven age-, sex-, and ethnicity-matched healthy subjects. Recognizing that reciprocal alterations in monocyte and T cell receptor expression and signaling could be examined simultaneously in individual cell populations, total blood leukocytes and enriched T cell and monocyte populations were isolated from the same peripheral blood samples by using a two-step negative immunoselection protocol. In general, neutrophil numbers were significantly increased in the trauma patients with MODS (median 80% vs. 60%; P < 0.05), whereas the proportions of T cells (8% vs. 28%; P < 0.05) and monocytes (6% vs. 21%; P < 0.05) were reduced compared with healthy subjects (Fig. 1a). Disparate cell population representation between patients and controls was corrected in the isolated T cells and monocytes.
Table 1.
Summary of patients' or matched healthy volunteers' demographics for both genomic as well as functional and protein expression analysis
| All male subjects | Age, yr (range) | Initial ISS (range) | MODS(range) | Sample postinjury day(range) |
|---|---|---|---|---|
| Genomic studies | ||||
| Patients (n = 7) | 48 (19–75) | 41 (17–59) | 9 (5–14) | 10 (4–14) |
| Controls (n = 7) | 48 (22–75) | N/A | N/A | N/A |
| Proteomic analyses | ||||
| Patients (n = 11) | 51 (20–78) | 25 (11–50) | 9 (6–19) | 10 (4–30) |
| Controls (n = 15) | 51 (27–78) | N/A | N/A | N/A |
The injury severity score (ISS) is a standard injury grading designed by the Association of Automotive Medicine (15). The MODS score of >6 indicates multiple organ failure (15). MODS scores were taken on the day of blood draw. “Sample postinjury day” refers to the day of sample collection. N/A, not applicable.
Fig. 1.
Genomic analysis of highly enriched T cells identifies genes whose expression differs between trauma patients and healthy subjects. (a) T cell, monocyte, and granulocyte distribution in the whole-blood leukocyte fraction from trauma patients and healthy subjects. Cell isolation protocols (see Methods) generated highly enriched cell populations. ∗, P < 0.02. (b) Apparent gene expression of leukocytes, T cells, and monocytes reveals different patterns between trauma patients and healthy subjects. Red indicates increased gene expression, and blue indicates reduced expression. (c) Venn diagram of genes whose expression is significantly different (5% false discovery rate) between trauma patients and healthy subjects.
Genomic Expression Analysis of Isolated Cell Subpopulations Reveals Unique Gene Expression Patterns.
Genome-wide expression analysis was performed on whole-blood leukocytes and the enriched populations of T cells and monocytes (Fig. 1b). With a false discovery rate of 5%, the mRNA abundance of 7,643 genes was significantly different in at least one cell population (either total leukocytes, highly enriched T cells, or monocytes) between trauma patients and matched healthy subjects. Alterations in apparent gene expression were cell-population-dependent (Fig. 1 b and c). In the enriched T cell population, severe trauma altered the apparent expression of 5,693 genes (3,340 uniquely), whereas the expression of 2,801 genes was altered in the monocytes (903 uniquely). There were only 911 genes (11.9% of 7,643) whose expression changed in common among all three cell populations (Fig. 1c). Severe trauma produced dramatic cell-specific changes in the human transcriptome, with expression of ≈15–20% of the human genome significantly altered in either T cells or monocytes.
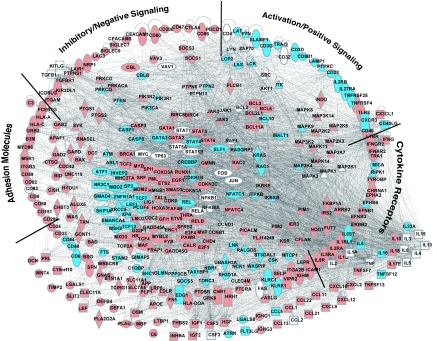
The biological relationships among these cell-specific changes in gene expression were explored by using a knowledge-based network analysis (16). Specialized blood leukocytes, such as monocytes and T cells, perform critical but distinct physiological functions. T cell apoptosis and anergy as well as blood monocyte hyporesponsiveness contribute to the immunopathology in trauma and sepsis as well as in cancer and some chronic infections (4–7, 17–31). To elucidate any genomic-based contribution to these complex functional alterations, we began with a systematic identification from a knowledge base of mammalian biology (16). As an example, among the 750 genes that have been annotated as having T lymphocyte-specific functions, 338 genes were perturbed significantly in the trauma patients (see Table 2, which is published as supporting information on the PNAS web site). Focusing on these 338 genes, we constructed a complex T cell network that identified all known interactions in the knowledge-based database (Fig. 2; for greater detail, see Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 2.
Overall network of perturbed T cell-specific gene interactions. Of 749 genes with specific T cell functions identified in the knowledge base, 338 were significantly perturbed in the trauma patients vs. healthy subjects' T cells. A comprehensive interaction network was constructed among these 338 genes plus 40 additional T cell-specific genes that closely interacted with genes in the pathway.
Specific Pathway Analysis Comparing T Cell, Monocyte, and Total Leukocyte Gene Expression.
Inspection and localization of the complex pathways of blood monocyte and T cell gene expression revealed several distinct transcriptional alterations in multiple functional networks, including those involved in cellular energetics, cell cycling, chemokine signaling, protein synthesis and degradation, and signal transduction, as well as in the targeted alterations in cellular apoptosis and T cell activation. In particular, this inspection revealed a concerted decrease in the expression of multiple genes involved in T cell activation pathways coupled with an increase in the expression of genes involved in T cell inhibitory pathways. A comparison between these genes' expression in T cells, monocytes, and leukocytes (Table 3, which is published as supporting information on the PNAS web site) showed substantial differences.
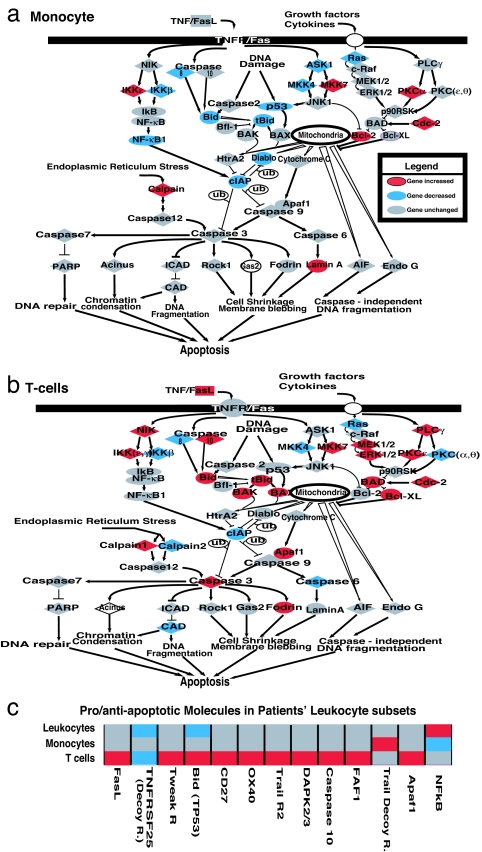
The specific alterations in expression of T cell genes characterized as involved in apoptosis and anergy were of particular interest in this patient population (18–31). Increased T cell, but not blood monocyte, apoptosis has been reported in severely injured or septic patients with MODS (4, 5, 10). Apparent gene expression for proteins involved in proapoptotic and antiapoptotic pathways was simultaneously examined in the enriched T cell and monocyte populations (Fig. 3a and b). Increased expression of a number of proapoptotic proteins was observed in the patients' enriched T cell population but not in either their total leukocyte or enriched monocyte populations (Fig. 3 b and c). Uniquely detected proapoptotic gene expression in the patients' T cells included increased TRAIL receptor 2 (DR5), increased TWEAKR, and increases in a proapoptotic molecule (FAF1), which was identified as blocking NF-kβ activation (18) (Fig. 3c). Simultaneously, gene expression of several antiapoptotic proteins was either suppressed or unchanged in the patients' enriched T cells but differentially increased or unchanged in either their monocytes or total leukocytes.
Fig. 3.
An altered balance of proapoptotic and antiapoptotic molecules in trauma patients' T cells could indicate specifically increased vulnerability to apoptotic depletion. A general apoptosis pathway was overlaid with patient genes altered at the 5% false discovery rate. (a) Expression of monocyte proapoptotic genes were unchanged or reduced (caspase 3, caspase 10, BAD, and Apa11). (b) Specific T cell expression of proapoptotic molecules was increased (caspase 3, caspase 10, Apaf1, Faf1, TWEAKR, and TRAIL2), with decreased decoy receptor signaling (TNFRSF25). (c) Analysis of the total leukocyte population failed to reveal T cell-specific changes or increases in proapoptotic molecules. Selected relevant apoptotic genes not in the standard apoptosis pathway were included for comparison.
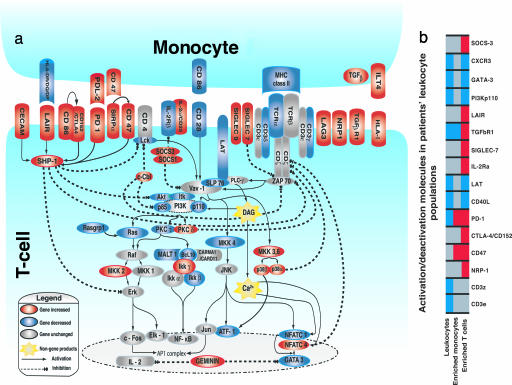
T cell anergy also is a characteristic of severe injury pathology (7–9). We therefore examined gene expression in several recently characterized T cell activation/inhibitory pathways that are suggested from in vitro and murine studies to be involved in anergy or hyporesponsiveness (11–14, 18–25, 27–31). As summarized schematically in Fig. 4a, the apparent gene expression of several immunoinhibitory signaling molecules, like SHP-1, c-Cbl, SOCS 1/SOCS3, and PKC-δ, was increased in T cells from the severe trauma patients. In addition, increased gene expression of a cohort of recently described inhibitory receptors was also seen in the enriched T cell population from trauma patients. In particular, gene expression of SIGLEC7/9, SIRPα-CD47, CD86, CTLA4, PD-1, CECAM (CD66a), LAIR, NRP-1, and Lag3, all of which are described as inhibitory signaling receptors in T cells, was significantly increased (11–14, 19, 21–23). Importantly, many of these unique perturbations were only observed in the patients' enriched T cell populations but not in either their total leukocytes or enriched monocytes, further supporting the necessity to examine enriched leukocyte subsets (Fig. 4b).
Fig. 4.
Interface between altered patient monocyte costimulatory/inhibitory gene expression and T cell activation/inhibition pathways. (a) Increased gene expression (red) of inhibitory costimulation receptor/ligand combinations in monocytes and T cells and concordant decreased expression (blue) of stimulatory receptor/ligand combinations. Concordant increases in expression of inhibitory signal transduction pathways (dotted lines) and decreased or unchanged (gray) gene expression in T cell activation pathways (solid lines) are shown. (b) Examples of cell-specific gene expression alternations involved in T cell inhibition/stimulation. T cell depletion in total leukocytes leads to misdetection of some T cell-specific gene expression as depressed or unchanged.
Monocytes from these same patients also demonstrated reduced expression of several genes required for T cell costimulation, such as HLA-DR and CD86, whereas expression of juxtacrine monocyte ligands with potential monocyte and T cell reciprocal inhibitory activation function, such as PD-L2, CD47, and ILT-4, was increased (14, 23–25) (Fig. 4a). Thus, a molecular choreography was evident between monocytes and T cells, associating trauma-induced T cell apoptosis, reduced T cell proliferation, and inhibition of T cell and monocyte activation with concomitant decreases in monocyte antigen presentation.
Functional Validation of Selected Receptor Alterations Identified by Genomic Analysis.
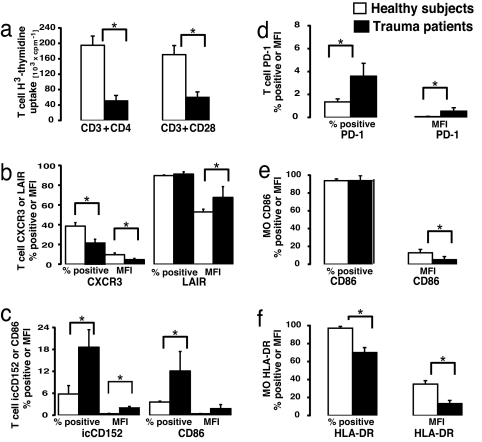
To confirm that some of the above observations in T cell and monocyte gene expression reflect alterations in protein abundance as well as T cell dysfunction, we matched an additional 11 trauma patients by age, gender, ethnicity, and organ dysfunction with an additional 15 healthy subjects. Protein expression and functional proteomic analysis could not be performed on samples from the original seven patients in whom gene expression analyses were conducted, because of limitations on blood sample volumes. Isolated T cells from this second cohort of severely traumatized patients with MODS were identified as being immunologically dysfunctional by their inability to proliferate in response to ex vivo immobilized anti-CD3CD4 or anti-CD3CD28 ligation (Fig. 5a). Flow cytometric analyses corroborate the mRNA abundance findings, because T cell surface protein expression of CD86, PD-1, CXCR3, CD152, and LAIR protein was increased significantly in the severely injured patients (Fig. 5 b–d). Similarly, monocyte CD86 (B7-2) and HLA-DR surface protein abundance were significantly reduced (Fig. 5 e and f).
Fig. 5.
Selective validation of patients' gene expression alterations by functional and protein expression analysis. (a) T cells from a matched cohort of 11 trauma patients with MODS and 15 matched healthy subjects were cultured with immobilized anti-CD3/anti-CD4 or anti-CD3/anti-CD28, and proliferation was assessed as a measure of T cell functional competence (P = 0.0037 and 0.0025, respectively). (b and c) Flow-cytometry-assessed expression of proteins presented as the total percentage of T cells (CD2) expressing marker and mean fluorescent intensity (MFI) involved in T cell migration (CXCR3). ∗, P = 0.003 for percent positive and 0.013 for MFI. T cell activation/inhibition markers were assessed. ∗, intracellular CD152, P = 0.021; CD86, P = 0.04; and LAIR, P = 0.015. (d) Comparison of T cells from trauma patients and healthy subjects for expression of inhibitory receptor PD-1, whose mRNA abundance was increased by microarray. Data are presented as the percentage T cells expressing the receptor and MFI. ∗, P = 0.047 and 0.001, respectively. (e and f) Decreased monocyte cell surface expression of costimulatory receptors, CD86 (∗, P = 0.009 for MFI), and HLA-DR as the percentage of monocytes (P = 0.009 expressing the receptor; P = 0.003 for MFI).
Discussion
Application of high-throughput technologies, such as oligonucleotide microarrays, poses significant challenges to sample collection, processing, and data interpretation in the clinical setting (32). This study demonstrates how cell-specific transcriptome and pathway analyses can reveal signaling pathways that lead to immunological dysfunction in hospitalized patients with severe trauma. Perturbations in the in vivo expression of genes involved in inhibitory and apoptotic pathways from human T cells and monocytes were uncovered by this discovery science approach and then validated with functional proteomics.
Here, we have identified a number of human T cell signaling pathways involved in apoptosis and anergy in which gene expression was dramatically altered by severe trauma. Furthermore, we validated differential expression of a number of these proteins by using functional proteomics. These results could be achieved only by applying the discovery power of genome-wide expression analyses to highly enriched cell populations and by exploring the resultant information analyzed with pathway-specific analytical tools.
We previously demonstrated that human endotoxicosis is associated with remarkably diverse changes in the human transcriptome affecting a number of previously unanticipated pathways and functional modules (16). We revealed unexpected complexity in human responses to this prototypical microbial challenge. However, a more thorough exploration of the human inflammatory response required understanding the genomic response in more enriched, individualized cell populations. We could not have identified that trauma-related genomic responses in two interacting cell populations are concordant and aimed at inducing anergy and tolerance without combining genome-wide expression analysis of enriched cell populations with cell-specific pathway analysis. The resulting identification of molecules in these alternative pathways provides insights into the augmented T cell apoptosis and anergy seen in human trauma. These unique changes in gene expression suggest targets for further exploration as mediators of postinjury, T cell immune dysfunctions.
These findings strikingly reveal that interactions among signaling pathways can be deconstructed in hospitalized patient populations by combining genome-wide expression on highly enriched cell populations and cell specific pathway analyses. Application of these approaches to human disease is practically unlimited. Our studies employ enriched cell subpopulations to provide a cell-specific, time-course analysis of a large hospitalized patient cohort. A variety of other cell–cell interactions that have been implicated in host response to injury also can be explored with these experimental approaches by focusing on cell–cell interactions with other subpopulations (PMN, NK lymphocytes, B lymphocytes, and dendritic cells). Importantly, these analytical strategies are generally applicable to the evaluation of high-throughput analyses in human disease states that involve significant systemic innate and specific immune dysfunctions. These strategies can be used simultaneously to uncover reciprocal alterations in cell-type-specific activation/inhibition pathways relevant to divergent clinical disease states.
Methods
Subjects.
Informed consent was obtained from 18 severely injured patients and 22 healthy subjects under a protocol approved by the University of Rochester School of Medicine Institutional Review Board. All patients experienced severe blunt trauma and developed MODS (15, 33) (Table 1; see also Table 4, which is published as supporting information on the PNAS web site). No trauma patient had undiagnosed or untreated infections at the time of blood sampling. Patients were treated with prophylactic antibiotics as clinically indicated. None of the patients received blood products in the previous 48 h.
Blood Sampling.
For the genomic analyses, seven severely traumatized patients and seven matched healthy subjects provided 60 and 90 ml of venous blood, respectively, which was collected and divided into three aliquots. One aliquot was diluted in 20 volumes of bicarbonate-buffered ammonium chloride solution (0.826% NH4Cl/0.1% KHCO3/0.0037% Na4EDTA in H2O) for the isolation of total leukocytes. Cells were recovered by centrifugation (400 × g at 4°C), washed once in ice-cold PBS, and resuspended in RLT buffer (600 μl per every 107 cells) (Qiagen, Valencia, CA). Samples were sheared by passage through shredder columns (Qiagen), and the eluate was immediately frozen at −70°C until the RNA was extracted (16).
Isolation of T cells and Monocytes.
Enriched T cells and monocytes from two simultaneous aliquots were obtained with a two-step sequential negative-depletion procedure (see Supporting Methods, which is published as supporting information on the PNAS web site). The initial step involved rosetting unwanted cells with antibody and density centrifugation removal, followed by a second enrichment step using commercially available antibody-bound bead columns (34). Cell purity was in excess of 95% in the controls but surprisingly showed good concordant gene expression to cells enriched only with the first step rosetting isolation (Table 5, which is published as supporting information on the PNAS web site). The distribution of cell phenotypes in the original total leukocyte and subsequent enriched T cells or monocytes was determined by flow cytometry with conjugated antibodies specific for T cells (CD2+ and CD3+), monocytes (C14+ and CD33+), neutrophils (CD66b+), B-cells (CD19+), or NK cells (CD56+) (see Fig. 7, which is published as supporting information on the PNAS web site). Expression of monocyte markers CD14 and CD33 decreases in trauma, so monocyte purity was calculated by subtracting the quantities of identified contaminating cells contained in the total enriched monocyte population.
Flow Cytometric Analysis of Monocyte and T Cell Functional Proteomics.
For the receptor/ligand expression and functional analysis of the additional 11 severely injured patients and 15 healthy subjects, 30 ml of venous blood was collected, anticoagulated with EDTA, and processed as described above. T cells and monocytes were stained for four-color flow cytometry as previously described (35). Analysis of T cell functional responses to T cell receptor (CD3CD28) stimulation was as described (9).
RNA Isolation, Processing, and Oligonucleotide Microarray Hybridization.
Total cellular RNA was isolated from the leukocytes, T cells, or monocytes by using a commercial kit (RNAeasy; Qiagen) with on-column DNase treatment. RNA purity and integrity was confirmed by spectrophotometry (A260/A280 ratio) and capillary electrophoresis (2100 Bioanalyzer; Agilent Technologies, Palo Alto, CA). cRNA synthesis was performed with 2 μg of total cellular RNA by using Affymetrix (Santa Clara, CA) protocol. cRNA was hybridized onto Hu133A oligonucleotide arrays (Affymetrix) and processed by the manufacturer's protocol.
Microarray Data Analysis.
We analyzed 22,411 probe sets on the U133A arrays as described (32). Briefly, normalization was performed with dChip, and the expression level was modeled by using the perfect match only. Probe sets significantly different between individual leukocyte populations from patients and healthy subjects were identified by using Significance Analysis of Microarrays (36), with an estimated false discovery rate of <5% based on 1,000 permutations of the data set. Probe sets were then mapped to unique genes based on Entrez GeneIDs (www.ncbi.nih.gov/Entrez).
Pathway Analysis.
The Ingenuity Pathways Knowledge Base, a comprehensive knowledge base of biological findings for genes of human, mouse, and rat, was used to construct pathways and functional modules. Details of the pathway analysis method are described in ref. 16. To identify focus genes in isolated T cells, the database was systematically queried, and 750 genes were identified as having T lymphocyte-specific functions. Among these genes, 338 were perturbed significantly in trauma patients (see Table 2). All other genes were ranked according to the specificity of connections, i.e., the percentage of interactions in the 338 genes that are significantly perturbed. A comprehensive interaction network was constructed among these 338 genes along with the top 40 additional genes most closely connected to this set. The resulting network had high connectivity (P < 10−15; Fisher's exact test).
Statistical Analysis of Flow Cytometry Data.
Distribution of each analyzed variable within studied groups was tested for homogeneity and normality (Brown–Forsyth and Shapiro–Wilk W tests), respectively, as a check for parametric data distribution. For parametric data, a Student's t test or ANOVA was used, whereas a Mann–Whitney U test was calculated when the sample sizes were smaller than 25 per group for each of the calculated contrasts (95% confidence level using a two-tailed model).
Supplementary Material
Acknowledgments
We thank S. Fisher for clinical assistance; J. Wilhelmy, S. MacMillan, and J. Strickland for technical assistance; and C. Graziano for graphics. This work was supported by National Institute of General Medical Sciences Awards U54 GM-62119 (to R.G.T.) and 5R01 GM-65237 (to C.M.-G.).
Abbreviation
- MODS
multiple organ dysfunction syndrome.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE5580).
References
- 1.Hood L, Heath JR, Phelps ME, Lin B. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 2.Berg J. J Am Med Assoc. 2005;294:2685–2686. [Google Scholar]
- 3.Murray JI, Whitfield ML, Trinklein ND, Myers RM, Brown PO, Botstein D. Mol Biol Cell. 2004;15:2361–2374. doi: 10.1091/mbc.E03-11-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hotchkiss RS, Swanson PE, Freeman BD, Tinsley KW, Cobb JP, Matuschak GM, Buchman TG, Karl IE. Crit Care Med. 1999;27:1230–1251. doi: 10.1097/00003246-199907000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Wesche DE, Lomas-Neira JL, Perl M, Chung CS, Ayala A. J Leukocyte Biol. 2005;78:325–337. doi: 10.1189/jlb.0105017. [DOI] [PubMed] [Google Scholar]
- 6.De AK, Laudanski K, Miller-Graziano CL. J Immunol. 2003;170:6355–6362. doi: 10.4049/jimmunol.170.12.6355. [DOI] [PubMed] [Google Scholar]
- 7.Murphy TJ, Choileain NN, Zang Y, Mannick JA, Lederer JA. J Immunol. 2005;174:2957–2963. doi: 10.4049/jimmunol.174.5.2957. [DOI] [PubMed] [Google Scholar]
- 8.Knoferl MW, Angele MK, Catania RA, Diodato MD, Bland KI, Chaudry IH. J Appl Physiol. 2003;95:529–535. doi: 10.1152/japplphysiol.01201.2002. [DOI] [PubMed] [Google Scholar]
- 9.De AK, Kodys KM, Pellegrini J, Yeh B, Furse RK, Bankey P, Miller-Graziano CL. Clin Immunol. 2000;96:52–66. doi: 10.1006/clim.2000.4879. [DOI] [PubMed] [Google Scholar]
- 10.Hotchkiss RS, Tinsley KW, Swanson PE, Schmieg RE, Jr, Hui JJ, Chang KC, Osborne DF, Freeman BD, Cobb JP, Buchman TG, et al. J Immunol. 2001;166:6952–6963. doi: 10.4049/jimmunol.166.11.6952. [DOI] [PubMed] [Google Scholar]
- 11.Krishnan S, Farber DL, Tsokos GC. J Immunol. 2003;171:3325–3331. doi: 10.4049/jimmunol.171.7.3325. [DOI] [PubMed] [Google Scholar]
- 12.Acuto O, Mise-Omata S, Mangino G, Michel F. Immunol Rev. 2003;192:21–31. doi: 10.1034/j.1600-065x.2003.00034.x. [DOI] [PubMed] [Google Scholar]
- 13.Saito T, Yamasaki S. Immunol Rev. 2003;192:143–160. doi: 10.1034/j.1600-065x.2003.00022.x. [DOI] [PubMed] [Google Scholar]
- 14.Latour S, Tanaka H, Demeure C, Mateo V, Rubio M, Brown EJ, Maliszewski C, Lindberg FP, Oldenborg A, Ullrich A, et al. J Immunol. 2001;167:2547–2554. doi: 10.4049/jimmunol.167.5.2547. [DOI] [PubMed] [Google Scholar]
- 15.Marshall JC, Cook DJ, Christou NV, Bernard GR, Sprung CL, Sibbald WJ. Crit Care Med. 1995;23:1638–1652. doi: 10.1097/00003246-199510000-00007. [DOI] [PubMed] [Google Scholar]
- 16.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, et al. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 17.Matsutani T, Samy TS, Rue LW III, Bland KI, Chaudry IH. Am J Physiol Cell Physiol. 2005;288:C1109–C1116. doi: 10.1152/ajpcell.00478.2004. [DOI] [PubMed] [Google Scholar]
- 18.Park MY, Jang HD, Lee SY, Lee KJ, Kim E. J Biol Chem. 2004;279:2544–2549. doi: 10.1074/jbc.M304565200. [DOI] [PubMed] [Google Scholar]
- 19.Jang IK, Gu H. Curr Opin Immunol. 2003;15:315–320. doi: 10.1016/s0952-7915(03)00048-7. [DOI] [PubMed] [Google Scholar]
- 20.Spitaler M, Cantrell DA. Nat Immunol. 2004;5:785–790. doi: 10.1038/ni1097. [DOI] [PubMed] [Google Scholar]
- 21.Ikehara Y, Ikehara SK, Paulson JC. J Biol Chem. 2004;279:43117–43125. doi: 10.1074/jbc.M403538200. [DOI] [PubMed] [Google Scholar]
- 22.Khoury SJ, Sayegh MH. Immunity. 2004;20:529–538. doi: 10.1016/s1074-7613(04)00116-5. [DOI] [PubMed] [Google Scholar]
- 23.Chen L. Nat Rev Immunol. 2004;4:336–347. doi: 10.1038/nri1349. [DOI] [PubMed] [Google Scholar]
- 24.Macian F, Im SH, Garcia-Cozar FJ, Rao A. Curr Opin Immunol. 2004;16:209–216. doi: 10.1016/j.coi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Papanikolaou NA, Vasilescu ER, Suciu-Foca N. Hum Immunol. 2004;65:700–705. doi: 10.1016/j.humimm.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Blattman JN, Greenberg PD. Nat Immunol. 2006;7:227–228. doi: 10.1038/ni0306-227. [DOI] [PubMed] [Google Scholar]
- 27.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 28.Greenwald RJ, Freeman GJ, Sharpe AH. Annu Rev Immunol. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611. [DOI] [PubMed] [Google Scholar]
- 29.Rieux-Laucat F, Fischer A, Deist F. Curr Opin Immunol. 2003;15:325–331. doi: 10.1016/s0952-7915(03)00042-6. [DOI] [PubMed] [Google Scholar]
- 30.Lichtner M, Maranon C, Vidalain PO, Azocar O, Hanau D, Lebon P, Burgard M, Rouzioux C, Vullo V, Yagita H, et al. AIDS Res Hum Retroviruses. 2004;20:175–182. doi: 10.1089/088922204773004897. [DOI] [PubMed] [Google Scholar]
- 31.Bosque A, Pardo J, Martinez-Lorenzo MJ, Iturralde M, Marzo I, Pineiro A, Alava MA, Naval J, Anel A. J Leukocyte Biol. 2005;77:568–578. doi: 10.1189/jlb.0904514. [DOI] [PubMed] [Google Scholar]
- 32.Cobb JP, Mindrinos MN, Miller-Graziano C, Calvano SE, Baker HV, Xiao W, Laudanski K, Brownstein BH, Elson CM, Hayden DL, et al. Proc Natl Acad Sci USA. 2005;102:4801–4806. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chawda MN, Hildebrand F, Pape HC, Giannoudis PV. Injury. 2004;35:347–358. doi: 10.1016/S0020-1383(03)00140-2. [DOI] [PubMed] [Google Scholar]
- 34.Asea A, Kraeft SK, Kurt-Jones EA, Stevenson MA, Chen LB, Finberg RW, Koo GC, Calderwood SK. Nat Med. 2000;6:435–442. doi: 10.1038/74697. [DOI] [PubMed] [Google Scholar]
- 35.De AK, Miller-Graziano CL, Calvano SE, Laudanski K, Lowry SF, Moldawer LL, Remick DG, Jr, Rajicic N, Schoenfeld D, Tompkins RG. J Immunol. 2005;175:6155–6162. doi: 10.4049/jimmunol.175.9.6155. [DOI] [PubMed] [Google Scholar]
- 36.Tusher VG, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.