Abstract
Histone deacetylases (HDACs) are enzymes that regulate the functions of histones as well as nonhistones by catalyzing the removal of acetyl groups from lysine residues. HDACs regulate many biological processes, including the cell division cycle and tumorigenesis. Although recent studies have implicated HDAC8 in tumor cell proliferation, the molecular mechanisms linking HDAC8 to cell growth remain unknown. Here, we report that the human ortholog of the yeast ever-shorter telomeres 1B (EST1B) binds HDAC8. This interaction is regulated by protein kinase A-mediated HDAC8 phosphorylation and protects human EST1B (hEST1B) from ubiquitin-mediated degradation. Phosphorylated HDAC8 preferentially recruits Hsp70 to a complex that inhibits the CHIP (C-terminal heat shock protein interacting protein) E3 ligase-mediated degradation of hEST1B. Importantly, HDAC8 regulation of hEST1B protein stability modulates total telomerase enzymatic activity. Our findings reveal a novel mechanism by which HDAC8 contributes to tumorigenesis by regulating telomerase activity.
Histone acetylation and deacetylation are dynamic posttranslational modifications mediated by the opposing enzymatic activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively. These reversible modifications regulate diverse biological functions ranging from cell cycle control to tumor formation. Increasing evidence suggests the need for a balance between HAT and HDAC activities for appropriate gene expression. Many studies have demonstrated the adverse consequences of abnormal HAT and HDAC expressions. For example, the inappropriate recruitment of HDACs has been implicated in both developmental defects and human diseases such as promyelocytic leukemia and lymphoma (34). HDAC inhibitors display antitumor activity by promoting either tumor cell apoptosis or normal cell differentiation (37).
Based on similarity to yeast proteins, human HDACs can be divided into three different groups. Class I HDACs (HDAC1, 2, 3, and 8) share extensive amino acid sequence homology with the yeast RPD3 protein. Class II HDACs (HDAC4, 5, 6, 7, 9, and 10) are homologous to the yeast HDA1 protein. Class III HDACs (SIRT1, 2, 3, 4, 5, 6, and 7) share significant homology with the yeast SIR2 protein and require NAD+ for enzymatic activity. A new human HDAC, HDAC11, was reported to exhibit properties of both class I and II HDACs (16, 18).
The least studied and understood class I HDAC is HDAC8. The HDAC8 cDNA was identified initially by three independent groups using sequence homology database searches for class I HDACs (3, 23, 54). The HDAC8 gene encodes a 377-amino-acid protein with a predicted molecular mass of 45 kDa. HDAC8 mRNA is expressed in multiple human tissues, including liver, heart, brain, lung, pancreas, placenta, prostate, and kidney. While HDAC8 can deacetylate all core histones in vitro, it preferentially deacetylates histones H3 and H4. Consistent with the presence of a putative nuclear localization signal, HDAC8 is localized predominantly in the nucleus (23), although recent studies have demonstrated that it also may be present in the cytoplasm of human smooth muscle cells (11, 58, 59). Durst et al. (13) showed that HDAC8 specifically associates with the inv (16) fusion protein found in acute myeloid leukemia. A recent study found that HDAC8 influences beta interferon gene expression (41).
The structure of HDAC8 consists of a single α/β domain that includes an eight-stranded parallel β sheet sandwiched between 13 α helices (48, 55). Using different HDAC inhibitors, Vannini et al. (55) showed that HDAC8 has a restrictive inhibition pattern different than that of other class I HDACs. In addition, they showed that knockdown of HDAC8 by RNA interference inhibits the growth of human lung, colon, and cervical cancer cell lines. The latter finding underscores the important role of HDAC8 in regulating tumor cell proliferation.
Like most HDACs, HDAC8 is a target for posttranslational modification, most notably phosphorylation. HDAC8 is phosphorylated by cyclic AMP-dependent protein kinase A (PKA) both in vitro and in vivo (33). The phospho-acceptor site of HDAC8 is Ser39, and interestingly, HDAC8 enzymatic activity is negatively regulated by this PKA-mediated phosphorylation. Although we have demonstrated that the phosphorylation of HDAC8 results in the hyperacetylation of histones H3 and H4, the precise biological and functional consequences of HDAC8 phosphorylation remain to be elucidated.
In the present study, we used a modified bacterial two-hybrid system (29) to identify proteins that specifically interact with phosphorylated HDAC8. We found that the human ever-shorter telomeres 1B (hEST1B) interacts with HDAC8 in a PKA-mediated, phosphorylation-dependent manner. Furthermore, the phosphorylation of HDAC8 protects hEST1B from degradation mediated by the E3 ubiquitin ligase CHIP (C-terminal heat shock protein interacting protein) via the recruitment of Hsp70. Our results demonstrate a novel mechanism by which a telomerase-associated protein is regulated by HDAC8. These findings support the hypothesis that HDAC8 is critically involved in telomerase regulation and provide a possible explanation for the involvement of HDAC8 in tumor cell proliferation.
MATERIALS AND METHODS
Plasmids, antibodies, and proteins.
Expression plasmids for human HDAC8 were constructed using the p3XFlag-CMV-10 vector (Sigma), and the HDAC8(S39A) mutant was generated as described previously (33). cDNAs encoding full-length human EST1B and EST1A were amplified by PCR using Image Clone (Open Biosystem) and inserted into pcDNA3.1-HA. Full-length human CHIP was cloned by PCR amplification using a HeLa cDNA library as a template and the following primers: 5′-CGGGATCCCCATGAAGGGCAAGGAGGAG and 3′-GGAATTCCTCAGTAGTCCTCCACCC. The PCR product was digested with BamHI and EcoRI and inserted into the myc-tag-containing vector pCMV-tag3C (Stratagene). To generate the U box deletion mutant of CHIP (amino acids 1 to 189), the U box domain was excised by ApaI digestion and isolated and the remaining DNA, which lacks the U box domain, then was religated. For the His-tagged fusion protein, the 5′ end of the CHIP open reading frame was digested first with BamHI and blunt ended and then digested with XhoI to release the insert from pCMV-tag3C. This fragment was inserted into EcoRV- and XhoI-digested pET-30a (Novagen) to create pET-CHIP. pET-EST1B was generated by PCR using the following primers: 5′-AGCTTTGTTTAAACTGATGAGCCAAGGCCCCCCCACAG and 3′-CCGGAATTCTTATCAACCAATTTCCTTCCACTGCTTG. Amplified cDNA was cloned into EcoRV- and EcoRI-digested pET-30a. pCMV-PKAcat was purchased from Clonetech. HDAC8 short interfering RNA (siRNA) and controls were purchased from Santa Cruz Biotechnology and Dharmacon. The anti-HDAC8 antibody was described previously (33). Rabbit polyclonal anti-EST1B antibody was raised against a peptide corresponding to the N terminus of hEST1B (SQESRSDLEDMEEEEGTRSC). Anti-Ub and anti-AcK antibodies were purchased from Upstate Biotechnology. Anti-Flag M2, antihemagglutinin (anti-HA), and anti-poly-His antibodies and Flag-ubiquitin were purchased from Sigma. Anti-myc, anti-GAPDH, anti-β-actin, anti-PKAcat, anti-Hsp70, and anti-Hsp90 antibodies were purchased from Santa Cruz Biotechnology. E1 and Ubc-H5 were purchased from Calbiochem.
Cell culture and transfection.
HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% calf serum, 100 IU of penicillin per ml, and 100 μg of streptomycin per ml. Cells were transfected with DNA using Lipofectamine 2000 (Invitrogen) for 48 h and harvested for each assay as described in the figure legends.
Immunoblotting and coimmunoprecipitation.
Cells were lysed in modified radioimmunoprecipitation assay buffer as described previously (33). For the coimmunoprecipitation assays, lysates were precleared with 20 μl of anti-immunoglobulin G-agarose and immunoprecipitated with primary antibody for 3 h. The immune complexes were washed extensively with lysis buffer, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred onto nitrocellulose membranes. The membranes were probed with the appropriate antibodies, and proteins were detected using the chemiluminescent detection kit (Pierce). To detect the endogenous interaction of HDAC8 with hEST1B, immunoprecipitation was performed using TrueBlot (eBioscience) according to the manufacturer's instructions.
Bacterial two-hybrid assay.
Escherichia coli two-hybrid assays were performed according to the manufacturer's instructions (Stratagene). Briefly, to generate inducible bacterial expression vectors, the region encoding the PKA catalytic subunit was fused to full-length HDAC8 and cloned into the pBT vector, which contained the bacteriophage λ cI protein (pBT-HDAC8.PKA). This plasmid was cotransformed with a HeLa cDNA library fused to the α subunit of RNA polymerase into a reporter strain that contained the lacZ promoter. Positive clones were selected based on carbenicillin resistance and β-galactosidase expression. To confirm a specific interaction with phosphorylated HDAC8, positive clones were retransformed into a reporter strain, together with pBT-HDAC8.PKA, and subjected to a second round of selection. To examine fusion protein expression levels and the extent of PKA-mediated HDAC8 phosphorylation in the E. coli cells, an aliquot of bacterial culture harvested after IPTG (isopropyl-β-d-thiogalactopyranoside) induction was subjected to direct immunoblotting and in vitro kinase assays.
Establishment of MC5HD8 cell line.
To generate a clone stably expressing HDAC8, p3XFlag-CMV-HDAC8 was transfected into HeLa cells with Lipofectamine. One day later, cells were subcultured and grown in the presence of 400 μg/ml of G418 (Invitrogen). Cells were then maintained in selection medium for about 2 weeks until G418-resistant colonies appeared. Single colonies were picked using cloning cylinders (Bellco) and transferred to 24-well plates. Individual clones were then maintained in medium containing 200 μg/ml of G418. A colony with high Flag-HDAC8 expression, MC5HD8, was selected for further analysis.
Luciferase activity assay.
Reporter and effector plasmids were cotransfected into HeLa cells grown in six-well plates at 50 to 70% confluence (seeded at 5 × 105 cells/well one day before). Forty-eight hours posttransfection, cells were harvested and luciferase activities were measured using a Berthold Lumat model LB 9501 luminometer.
Assays of endogenous hEST1B and β-actin mRNA.
Total RNA was isolated using TRIzol reagent (Invitrogen) as described by the manufacturer, quantified, and subjected to reverse transcription using Moloney murine leukemia virus reverse transcriptase (RT) (Promega) and specific antisense oligodeoxynucleotides (TTGTCATACAGCCGCTTGAG for hEST1B and CAAACATGATCTGGGTCATCTTCT for β-actin). After reverse transcription, cDNA was amplified by PCR using the same antisense oligodeoxynucleotides plus sense oligodeoxynucleotides (TCAGGGAAGGAGATGGATTG for hEST1B and GCTCGTCGTCGACAACGGCTC for β-actin).
Protein expression and purification.
His-EST1B and His-CHIP were produced in the E. coli strain BL21(DE3)pLysS (Novagen). After IPTG induction for 4 h, cells were lysed by sonication. His-tagged recombinant proteins were purified by Ni2+ chelation chromatography. Protein expression was verified by Coomassie blue staining.
In vitro ubiquitination assay.
In vitro ubiquitination assays were performed using bacterially expressed recombinant hEST1B and CHIP proteins. Briefly, hEST1B protein immobilized on Ni-nitrilotriacetic acid agarose was incubated at 30°C for 120 min in 50 μl of reaction solution (50 mM Tris-HCl, pH 7.4, 120 mM NaCl, 5 mM MgCl2, 4 mM ATP, and 0.5 mM dithiothreitol) containing 10 μg Flag-ubiquitin, 50 ng E1, 0.5 μg Ubc-H5, and 2 μg CHIP. After incubation, bead-bound proteins were washed three times with reaction buffer, resolved on SDS-PAGE, and subjected to direct immunoblotting.
TRAP assay.
Detection of telomerase activity in HeLa cells was performed according to the manufacturer's recommendations (Chemicon International) with minor modifications. Briefly, hEST1B immunoprecipitates used in telomeric-repeat amplification protocol (TRAP) reactions contained 2 μCi of [α-32P]dCTP (3,000 Ci/mmol; Amersham). The reaction mixture was incubated at 30°C for 30 min and then cycled 30 times at 94°C for 30 s and 59°C for 30 s. The PCR products were separated on 12% nondenaturing polyacrylamide gels and visualized by autoradiography.
RESULTS
EST1B interacts specifically with phosphorylated HDAC8.
To identify proteins that interact with phosphorylated HDAC8, we used a modified bacterial two-hybrid system (29). Briefly, we engineered a “bait and kinase” expression plasmid that expressed the catalytic portion of PKA (the kinase) and HDAC8 (the bait) fused to λ cI. We confirmed that the HDAC8 portion of the fusion protein was phosphorylated by PKA in this context (data not shown). The “bait and kinase” expression plasmid was transformed into E. coli, together with plasmids expressing a library of HeLa cell cDNAs fused to the α subunit of bacterial RNA polymerase (Stratagene). Target proteins that interact with HDAC8 facilitate the cI and α subunit interactions that drive the λ cI-dependent transcription of reporter genes encoding β-lactamase and β-galactosidase. Of the 4 × 106 transformants screened, 15 were positive for β-galactosidase activity, carbenicillin resistance, and the inability to activate the system in the absence of HDAC8. Out of these 15 phospho-HDAC8-interacting clones, 9 encoded proteins that interacted with HDAC8 in the absence of PKA, indicating that they did not require HDAC8 phosphorylation for interaction. DNA sequencing of the remaining six clones revealed that two clones contained the gene encoding the human Est1p-like protein B, a telomerase-associated factor (44, 47). This interaction was chosen for further characterization.
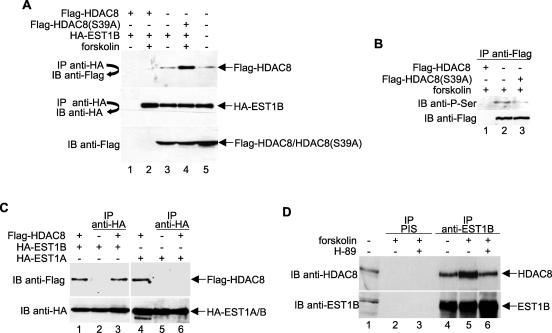
To confirm the hEST1B-HDAC8 interaction, HeLa cells were transfected with plasmids that expressed Flag-HDAC8 and HA-EST1B. Lysates were prepared from transfected cells, and after normalizing for equal amounts of HA-EST1B, immunoprecipitations were performed with anti-HA antibodies followed by Western blot analysis with anti-Flag antibodies. As shown in Fig. 1A, Flag-HDAC8 protein coprecipitated with HA-EST1B and, consistent with our results from bacterial two-hybrid experiments, the interaction was enhanced by forskolin, which induces the PKA-mediated phosphorylation of HDAC8 (Fig. 1A, top panel, compare lanes 3 and 4). Flag-HDAC8(S39A), which is refractory to phosphorylation by PKA (33), bound HA-EST1B at the same level as the non-forskolin-treated sample did, confirming that the association of hEST1B with HDAC8 is regulated by HDAC8 phosphorylation (Fig. 1A, top panel, lane 5). Figure 1B shows the forskolin-induced phosphorylation of wild-type HDAC8 but not the S39A mutant.
FIG. 1.
hEST1B interacts with HDAC8 in a phosphorylation-dependent manner. (A) PKA-mediated HDAC8 phosphorylation increases its association with hEST1B. HA-EST1B and Flag-HDAC8, either wild type or mutant, were coexpressed in HeLa cells in the absence or presence of forskolin. Following immunoprecipitation with an anti-HA antibody, the samples were resolved by SDS-PAGE and the presence of coimmunoprecipitated HDAC8 was detected by immunoblotting with an anti-Flag antibody. (B) Foskolin induces the phosphorylation of wild-type but not S39A mutant HDAC8. Flag-tagged wild-type or S39A mutant HDAC8 was transfected into HeLa cells in the presence or absence of forskolin. Following immunoprecipitation with anti-Flag antibodies, the samples were resolved by SDS-PAGE and phosphorylated HDAC8 was detected with an anti-phospho-serine antibody. (C) Phosphorylated HDAC8 preferentially interacts with hEST1B, but not hEST1A. Plasmids that express the indicated EST1 were transfected into cells, together with a plasmid that expresses HDAC8 in the presence of forskolin. (D) Endogenous hEST1B and HDAC8 interact. HeLa cells were treated with either forskolin or H-89 as indicated, and lysates were immunoprecipitated with anti-hEST1B antibodies. The immunoprecipitates were washed, and the proteins were separated by SDS-PAGE. Coimmunoprecipitated HDAC8 was detected by immunoblotting using TrueBlot to reduce the background due to the immunoglobulin G heavy chain. All protein concentrations were adjusted according to the expression levels of HA-EST1B. For example, in panel A, the reaction displayed in lane 2 contains three times more lysate than that in lane 3. −, absence of; +, presence of; IP, immunoprecipitate; IB, immunoblot; PIS, preimmune serum.
Unlike the Saccharomyces cerevisiae genome, which codes for only one Est1 protein, the human genome contains at least three genes which encode Est1p orthologs. Two of these genes (hEST1A and hEST1B) are known to associate with telomerase (44, 47). To determine whether hEST1A also binds phosphorylated HDAC8, we performed coimmunoprecipitation experiments using extracts prepared from cells transfected with a plasmid expressing hEST1A. The results clearly demonstrate that phosphorylated HDAC8 specifically interacts with hEST1B, but not hEST1A (Fig. 1C, top panels, compare lanes 3 and 6).
Next, we investigated whether endogenous HDAC8 and hEST1B proteins interact in vivo. A coimmunoprecipitation was performed using HeLa whole-cell extracts with anti-EST1B antibodies, followed by immunoblotting with anti-HDAC8 antibodies. As shown in Fig. 1D, HDAC8 coimmunoprecipitated with hEST1B and the level of coimmunoprecipitation was increased in the presence of forskolin (top panel, lanes 4 and 5). The PKA inhibitor H-89 reversed the effect of forskolin (lane 6), confirming the importance of HDAC8 phosphorylation in the hEST1B-HDAC8 interaction.
HDAC8 phosphorylation protects hEST1B from ubiquitin-mediated degradation.
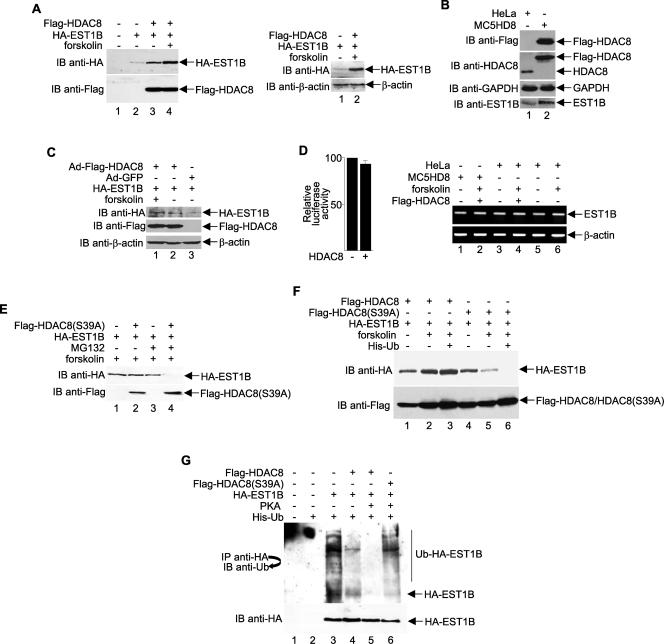
During our hEST1B-HDAC8 interaction studies, we repeatedly found that hEST1B protein levels were elevated in the presence of HDAC8 (Fig. 2A, top left panel, compare lanes 2 and 3). Furthermore, we found that forskolin-mediated activation of PKA, which increases HDAC8 phosphorylation, caused an additional increase in hEST1B protein levels (Fig. 2A, top left panel, lane 4), whereas actin levels remain unchanged (Fig. 2A, top right panel).
FIG. 2.
HDAC8 phosphorylation protects hEST1B protein from ubiquitin-mediated degradation. (A) HA-EST1B and Flag-HDAC8 were expressed in HeLa cells in the absence or presence of forskolin and analyzed by Western blotting. Western blotting with anti-β-actin (right panel) was used as a control. (B) Western blot analyses of extracts derived from HeLa cells or a HeLa cell line that overexpresses HDAC8 (MC5HD8). (C) Western blot analyses from extracts of adenovirus-infected cells to determine the effects of overexpression of HDAC8 on hEST1B. (D) Left panel, luciferase reporter assays showing that overexpression of HDAC8 does not alter the transcription of the cytomegalovirus promoter (pCMV-Luc). Transfections were normalized to equal amounts of DNA with parental expression vector. The results are the means ± standard deviations from three separate transfections. Right panel, semiquantitative RT-PCR was performed to analyze expression of the hEST1B gene. PCR products of cDNA samples are shown. (E) MG132 blocks hEST1B degradation. Cells were treated with 10 μM MG132 for 18 h before harvesting. hEST1B protein was detected by an anti-HA antibody. (F) HDAC8 phosphorylation protects hEST1B from degradation. Cells were transfected with His-ubiquitin, together with wild-type or mutant HDAC8, in the presence of HA-EST1B as indicated. (G) Inhibition of hEST1B ubiquitination by phosphorylated HDAC8. The level of hEST1B ubiquitination was determined by using an anti-ubiquitin antibody in the presence of His-ubiquitin after treatment with MG132. The amount of protein used in this experiment was adjusted according to the expression level shown in panel A. Ub, ubiquitin; −, absence of; +, presence of; IP, immunoprecipitate; IB, immunoblot.
A stable HeLa cell line, MC5HD8, that overexpresses Flag-HDAC8 was established, and Western blot analysis with anti-EST1B confirmed the up-regulation of hEST1B by HDAC8 (Fig. 2B, bottom panel, compare lanes 1 and 2). Similarly, the overexpression of HDAC8 using an adenovirus vector (33), followed by Western blot analysis, confirmed that HDAC8 increases the level of hEST1B (Fig. 2C).
Because the cytomegalovirus promoter used to drive expression of the HA-EST1B plasmid was not activated by HDAC8 (Fig. 2D, left panel), we ruled out the possibility that HDAC8 simply increased the transcription of HA-EST1B in the transfected or adenovirus-infected cells. Semiquantitative RT-PCR analysis was performed to confirm that HDAC8 has no effect on endogenous hEST1B mRNA (Fig. 2D, right panel). Therefore, we hypothesized that phosphorylated HDAC8 regulates hEST1B protein degradation.
To determine whether hEST1B protein levels are regulated by proteasome-dependent pathways, constructs expressing hEST1B were cotransfected with the HDAC8(S39A) mutant in the presence or absence of MG132, a specific inhibitor of proteasome-dependent pathways. As shown in Fig. 2E, hEST1B degradation in cells expressing the HDAC8(S39A) mutant was inhibited by MG132 treatment (top panel, compare lanes 2 and 4). In the same experiment, control cells without HDAC8(S39A) expression had slightly increased levels of hEST1B in the presence of MG132, probably due to endogenous HDAC8 protein (lanes 1 and 3). Therefore, we concluded that hEST1B degradation is mediated by a proteasome-dependent pathway that is regulated by phosphorylated HDAC8.
To determine whether HDAC8 phosphorylation protects hEST1B from degradation, we transfected cells with plasmids that express HA-EST1B, Flag-HDAC8, or Flag-HDAC8(S39A) and histidine-tagged ubiquitin. To induce HDAC8 phosphorylation, cells were either treated with forskolin or transfected with the catalytic subunit of PKA. As determined by Western blot analyses, hEST1B protein levels were consistently decreased in HDAC8(S39A)-transfected cells (Fig. 2F, compare lanes 4 and 5 with lane 2) and completely diminished in cells overexpressing ubiquitin (lane 6). In contrast, in the presence of phosphorylated HDAC8, hEST1B was not degraded even when ubiquitin was overexpressed (Fig. 2F, compare lanes 2 and 3 with lanes 5 and 6), suggesting that HDAC8 phosphorylation protects hEST1B from ubiquitin-mediated degradation. To examine the protective role of HDAC8 in hEST1B degradation, cells were treated with MG132 after transfection. As shown in Fig. 2G, hEST1B is highly ubiquitinated (lane 3), and coexpression of hEST1B with HDAC8 inhibited this ubiquitination (compare lanes 3 and 4). Strikingly, in cells overexpressing PKA, which induces HDAC8 phosphorylation, hEST1B ubiquitination was diminished (lane 5). As expected, the HDAC8(S39A) mutant did not protect hEST1B from ubiquitination (compare lanes 5 and 6), providing strong evidence that hEST1B protein is protected from ubiquitination by the PKA-mediated phosphorylation of HDAC8.
Ubiquitination of hEST1B is not regulated by acetylation.
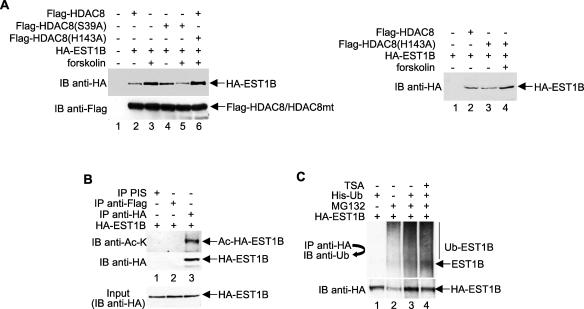
The modulation of protein stability by ubiquitination is often affected by subsequent additional posttranslational modifications, such as phosphorylation and acetylation. For example, the p53 protein is specifically acetylated at multiple lysine residues by p300/CBP, thereby inhibiting its ubiquitination-mediated degradation by Mdm2 (26). Previously, we reported that HDAC8 deacetylase activity was negatively regulated by PKA-mediated phosphorylation (33). Therefore, it is possible that the ability of phosphorylated HDAC8 to protect hEST1B from ubiquitin-mediated degradation is due to its decreased enzymatic activity and possibly the hyperacetylation of hEST1B. To investigate this possibility, we examined the requirement of HDAC8 enzymatic activity for protection of hEST1B degradation by transfecting cells with various plasmids expressing HDAC8 mutants and hEST1B. The level of hEST1B protein expression was altered consistently by the phosphorylation status of HDAC8 (Fig. 3A, left panel, lanes 2 and 3). In addition, the catalytically inactive but phosphorylation-competent HDAC8 mutant H143A regulated hEST1B expression to nearly the same extent as phosphorylated HDAC8 did (lane 6), indicating that HDAC8 enzymatic activity is not required to protect hEST1B from degradation. As a control, we show that the H143A mutant does not have the same stabilizing effect in the absence of forskolin (Fig. 3A, right panel).
FIG. 3.
hEST1B ubiquitination is not affected by its acetylation. (A) HA-EST1B and Flag-tagged wild-type, S39A mutant, or H143A mutant HDAC8 were coexpressed in HeLa cells. After the lysate was resolved by SDS-PAGE, the hEST1B protein level was determined by immunoblotting using an anti-HA antibody. (B) hEST1B is acetylated in vivo. Proteins expressing HA-EST1B were immunoprecipitated using an anti-HA antibody. For negative controls, preimmune serum (PIS) and anti-Flag antibodies were used. Acetylated hEST1B proteins were detected by immunoblotting with an anti-acetyl lysine (anti-Ac-K) antibody. To confirm equal amounts of HA-EST1B in the lysates, a small portion was removed before antibody incubation and resolved by SDS-PAGE. (C) hEST1B ubiquitination is not affected by TSA treatment. Cells were treated with 400 ng/ml TSA for 18 h and MG132 for 5 h before harvesting. Ub, ubiquitin; −, absence of; +, presence of; IP, immunoprecipitate; IB, immunoblot.
To confirm that the inhibition of HDAC8 activity does not cause increased hEST1B acetylation but does protect it from degradation, hEST1B was overexpressed in cells, and the hEST1B acetylation level was examined by anti-acetyl lysine antibodies in vivo. As shown in Fig. 3B, hEST1B was acetylated in vivo (lane 3). Additionally, these hEST1B-overexpressing cells were treated with trichostatin A (TSA) to induce the hyperacetylation of hEST1B and MG132 in the presence of ubiquitin. As expected, the hEST1B ubiquitination level was not affected by its acetylation state (Fig. 3C, lanes 3 and 4). These results demonstrate that, while hEST1B is acetylated, its ubiquitination is not affected by its acetylation state. Moreover, although hEST1B ubiquitination is regulated by HDAC8 phosphorylation, it is likely that this regulation may require another cofactor whose function is indirectly affected by HDAC8 phosphorylation.
Hsp70/90 forms a complex with hEST1B and HDAC8.
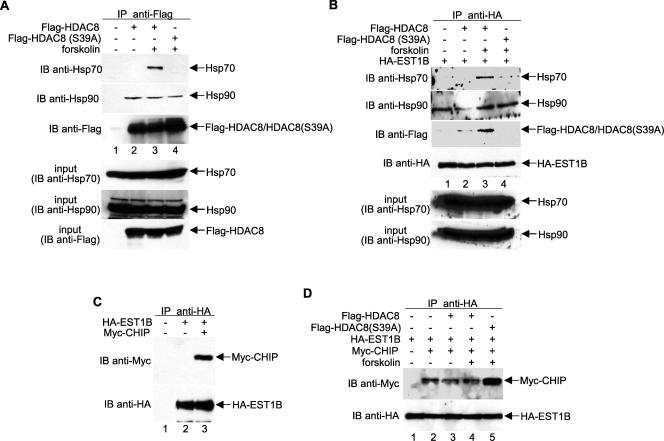
The bacterial two-hybrid assay also identified STIP1 (also known as HOP1) as an interacting partner of PKA-phosphorylated HDAC8. HOP1 is a bridging protein that links Hsp70 and Hsp90 and is required for some receptor-Hsp90 heterocomplex assembly (7, 12, 32). Therefore, HOP1 may also serve as an adaptor protein which recruits chaperone proteins, such as Hsp70 and Hsp90, to the HDAC8-EST1B complex. To investigate whether chaperone proteins interact with HDAC8, we transfected HeLa cells with plasmids expressing Flag-tagged wild-type or mutant HDAC8, immunoprecipitated the proteins with anti-Flag antibodies, and examined coimmunoprecipitated proteins with anti-Hsp70 and anti-Hsp90 antibodies. The results, presented in Fig. 4A, demonstrate that both Hsp70 and Hsp90 are HDAC8-interacting proteins. However, only the Hsp70-HDAC8 interaction was regulated by HDAC8 phosphorylation, such that the association was dramatically increased by PKA-mediated phosphorylation of HDAC8 (Fig. 4A, top panel, lane 3). In agreement with this observation, Hsp70, but not Hsp90, preferentially interacts with the phosphorylated HDAC8-hEST1B complex (Fig. 4B, top panel, lane 3). Therefore, we hypothesize that Hsp70/90 binding regulates hEST1B ubiquitination, and Hsp70, but not Hsp90, is an important determinant of the mechanism underlying the regulation of HDAC8-mediated hEST1B protein stability.
FIG. 4.
HDAC8 recruits Hsp70 and CHIP to hEST1B in a phosphorylation-dependent manner. (A) HeLa cells were transfected with either wild-type or S39A HDAC8, and lysates were immunoprecipitated with an anti-Flag antibody. The presence of Hsp proteins was determined using anti-Hsp70 or anti-Hsp90 antibodies. (B) HDAC8-Hsp protein complexes were immunoprecipitated as described above. The amount of protein used was adjusted according to the expression level shown in Fig. 2A. (C) hEST1B is a CHIP ubiquitin ligase-interacting protein. HA-EST1B and Myc-CHIP were expressed in HeLa cells and harvested after 48 h for analysis. (D) CHIP recruitment to hEST1B is dependent on HDAC8 phosphorylation. Myc-CHIP constructs were transfected with HA-EST1B and Flag-HDAC8 in the presence or absence of forskolin. The ability of CHIP to interact with hEST1B was determined with an anti-Myc antibody. −, absence of; +, presence of; IP, immunoprecipitate; IB, immunoblot.
HDAC8 regulates the binding of hEST1B to CHIP.
Recent reports have suggested CHIP as a possible link between chaperones and the degradation system (2, 39, 43). CHIP is a chaperone-interacting protein that binds to Hsp/Hsc70 and Hsp90. It contains an amino-terminal tetratricopeptide domain and a carboxyl-terminal U box that possesses E3 ubiquitin ligase activity (2, 10). To examine whether CHIP is a hEST1B-binding partner, cells were transfected with plasmids expressing HA-EST1B and Myc-CHIP. Coimmunoprecipitations were performed using anti-HA antibodies followed by immunoblotting with anti-Myc antibodies. The results indicate that CHIP interacts with hEST1B (Fig. 4C, upper panel, lane 3). In addition, the inhibition of HDAC8 phosphorylation significantly enhances the CHIP-hEST1B interaction (Fig. 4D, upper panel, lane 5), suggesting that HDAC8 phosphorylation regulates the interaction between CHIP and hEST1B.
CHIP ubiquitinates hEST1B, and HDAC8 phosphorylation protects hEST1B from CHIP-mediated degradation.
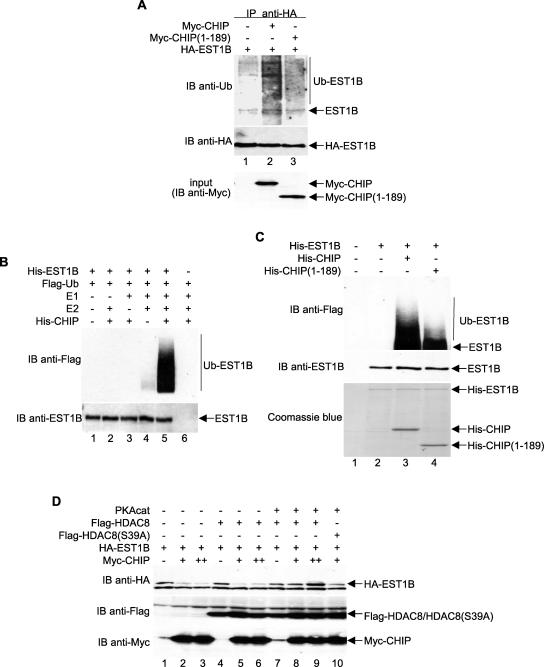
To examine the possible role of CHIP as an E3 ligase involved in hEST1B ubiquitination, cells were transfected with wild-type and catalytically inactive CHIP and treated with MG132 in the presence of ubiquitin. Interestingly, overexpression of the wild-type CHIP, but not the CHIP U box deletion mutant (amino acids 1 to 189), significantly enhanced hEST1B ubiquitination (Fig. 5A), suggesting that hEST1B ubiquitination is mediated through the U box domain of CHIP.
FIG. 5.
CHIP is an E3 ubiquitin ligase specific for hEST1B, and HDAC8 phosphorylation regulates CHIP-mediated degradation of hEST1B. (A) Ubiquitinated hEST1B proteins were visualized with an anti-ubiquitin antibody in the lysates from cells expressing HA-EST1B and various Myc-CHIP constructs. (B) An in vitro ubiquitination assay was performed using bacterially expressed proteins as indicated. After the reaction, Ni-nitrilotriacetic acid agarose-bound proteins were washed extensively and resolved by SDS-PAGE. Ubiquitin conjugation on hEST1B proteins was detected with an anti-Flag antibody. (C) An in vitro ubiquitination assay was performed using the bacterially expressed CHIP deletion mutant. (D) cDNAs expressing HA-EST1B, Flag-HDAC8, and Myc-CHIP were transfected into HeLa cells, and immunoblots were performed as indicated. Ub, ubiquitin; −, absence of; +, presence of; IP, immunoprecipitate; IB, immunoblot.
To determine whether hEST1B is a direct target of CHIP-mediated ubiquitination, His-EST1B was used in an in vitro ubiquitination assay with various enzymes involved in the ubiquitin pathway. UbcH5 was used as the E2 enzyme since it is the E2 ubiquitin-conjugating enzyme that specifically interacts with CHIP (39). The results clearly demonstrated that hEST1B was ubiquitinated directly by the enzymatic activity of UbcH5-CHIP (Fig. 5B, lane 5). The expression of a mutant with a U box deletion greatly diminished hEST1B ubiquitination (Fig. 5C, lane 4), an effect which demonstrates the role of U box in mediating CHIP enzymatic activity. Together, our data demonstrate that hEST1B ubiquitination is mediated by the CHIP E3 ligase whose activity is regulated by Hsp70 in an HDAC8 phosphorylation-dependent manner. To examine whether HDAC8 phosphorylation modulates CHIP-mediated hEST1B degradation, hEST1B was expressed in the presence or absence of phosphorylated HDAC8 together with CHIP. The levels of hEST1B were decreased in response to CHIP expression (Fig. 5D, compare lanes 1 to 3). In contrast, hEST1B was protected against this CHIP-dependent decrease when coexpressed with wild-type phosphorylated HDAC8 (lanes 7 to 9) but not the HDAC8(S39A) mutant (lane 10). These results demonstrate that hEST1B is targeted by CHIP-mediated ubiquitination and that HDAC8 phosphorylation prevents this modification by stimulating the association of Hsp70 with the hEST1B-CHIP complex.
Silencing of HDAC8 results in hEST1B degradation and reduction of total telomerase activity.
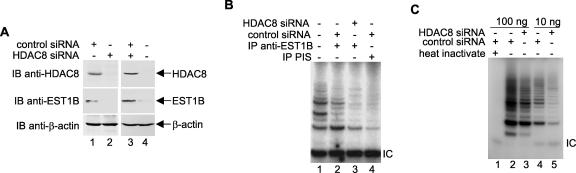
Both hEST1A and hEST1B are associated with telomerase activity and bind human telomerase reverse transcriptase (hTERT) in vitro (47). To determine whether HDAC8-mediated hEST1B regulation affects its telomerase activity, we performed TRAP assays using lysates prepared from HDAC8 knockdown cells. As shown in Fig. 6A, two different pools of HDAC8 siRNA effectively depleted HDAC8 from HeLa cells and consequently caused a dramatic decrease in hEST1B (compare lanes 1 and 2 and lanes 3 and 4). Accordingly, hEST1B-associated telomerase activity was diminished in HDAC8 knockdown cells (Fig. 6B, lanes 2 and 3). Importantly, total telomerase activity was significantly decreased in the absence of HDAC8 (Fig. 6C, compare lanes 3 and 5 with lanes 2 and 4). However, we were unable to detect any change in the average telomere length in the HDAC8 knockdown cells (data not shown). These data suggest a direct in vivo contribution of hEST1B as well as an indirect HDAC8 effect on total telomerase catalytic activity.
FIG. 6.
Silencing of HDAC8 results in hEST1B degradation and the down-regulation of total telomerase activity. A TRAP assay was performed to detect hEST1B-associated telomerase and total telomerase activity in HDAC8 knockdown cells. (A) Two different pools of HDAC8 siRNA oligonucleotides purchased from Santa Cruz Biotechnology (sc-35548, lane 2) and Dharmacon (M-003500-02, lane 4) were used to knock down HDAC8 expression, and the effects on hEST1B protein levels were examined. (B) The TRAP assay was performed using 2 μl of immunoprecipitates. Telomerase activity was visualized by autoradiography following resolution on nondenaturing gels. As a negative control for hEST1B-specific telomerase interaction, immunoprecipitates with preimmune serum (PIS) were used (lane 4). A positive control for telomerase activity is shown in lane 1. IC, internal control. (C) Total telomerase activity was measured using a different amount of lysates. For a negative control, heat-inactivated lysate was prepared from control siRNA-transfected cells (lane 1). −, absence of; +, presence of; IP, immunoprecipitate; IB, immunoblot.
DISCUSSION
In normal human cells, telomeres are shortened with each successive round of cell division due to repressed telomerase activity. This shortening ultimately leads to senescence. However, cancer cells escape such senescence by stimulating telomerase activity (for a review, see references 1 and 4). Telomerase activation requires the induction of its catalytic subunit, hTERT. Many studies have revealed the presence of a TSA response element in the hTERT promoter and a possible cofactor involved in the recruitment of HDAC to this region (9, 22, 61). Consistent with these reports, the treatment of cells with TSA abolished the interaction between HDAC1 and Sp1, which would be recruited to the hTERT promoter, thereby derepressing hTERT promoter activity (53). However, the contribution of telomerase to tumorigenesis extends beyond maintaining telomere length. In fact, recent findings have implicated the involvement of hTERT in processes like stem cell proliferation, cell survival, and chromosomal healing (15, 21, 45, 49, 57, 63). In this study, we identified a novel transcription-independent regulation of telomerase activity by the histone deacetylase HDAC8. Our results showed that HDAC8 is a regulator of the telomerase-associated protein hEST1B, the human homolog of yeast Est1p. HDAC8 interacted with hEST1B in a PKA-mediated, phosphorylation-dependent manner and protected hEST1B from ubiquitin-mediated degradation. In addition, we demonstrated that the ubiquitin E3 ligase CHIP was responsible for hEST1B degradation. HDAC8 phosphorylation at Ser39 recruits Hsp70 to a complex that suppresses the CHIP-mediated hEST1B degradation, thus providing a mechanism for the HDAC8 phosphorylation-mediated protection of hEST1B from degradation. Furthermore, we found that knockdown of HDAC8 leads to a significant reduction in telomerase activity without affecting average telomere length.
In yeast, telomeres exist as multiprotein complexes consisting of Est1, Est2, Est3, cdc13, and Tlc1, all of which are essential for telomerase activity (52). Deficiencies in any of the genes encoding these proteins result in the same est phenotype, a progressive telomere shortening leading to eventual cell death. Est1 is a cell cycle-regulated activator of telomerase that binds to an inactive, telomere-bound Est2-Tlc1 RNA complex in late S phase and then interacts with one or more cdc13 molecules arrayed on the G tail (51). The human Est1p homologs, hEST1A and hEST1B, have been shown to associate with telomerase activity in vivo and bind hTERT in vitro independently of telomerase RNA (44, 47). Interestingly, overexpression of hEST1A, but not hEST1B, was found to cooperate with hTERT to lengthen telomeres in vivo (47). Other than associating with telomerase, hEST1B (also known as hSMG5) has been shown to be a critical component of an mRNA surveillance complex which eliminates premature mRNAs by nonsense-mediated mRNA decay (8, 42). In the present study, our findings strongly argue for a significant contribution of hEST1B to total telomerase enzymatic activity independently of telomere length maintenance, as evidenced by the HDAC8 knockdown experiment. In addition, we demonstrate that PKA-phosphorylated HDAC8 preferentially interacts with hEST1B, but not hEST1A, supporting the functional differences between the two homologs. We propose that human telomere length is maintained by hEST1A-associated telomerase activity, while hEST1B-associated telomerase activity plays a distinct role in promoting tumorigenesis. In fact, using an hTERT mutant, Stewart et al. (50) have shown functional dissection of telomerase and the telomerase enzymatic activity, specifically that telomere elongation activity was not critical for tumorigenesis. Additionally, conditional TERT activation recently has been shown to induce stem cell proliferation in hair follicles and, interestingly, this function is independent of its telomere synthesizing activity (45). However, further investigation is needed to clarify the exact role of hEST1B.
The link between acetylation/deacetylation and ubiquitination comes from numerous studies demonstrating that acetylation generally stabilizes protein levels by inhibiting ubiquitination, whereas deacetylation has the opposite effect. The HDAC6 complex contains proteins that are involved in the ubiquitin signaling pathway (46). In addition, HDAC1 and SIRT1 are known to regulate the p53 protein, a well-known target of the ubiquitin pathway (26, 35, 36, 40, 56). However, in the present study, the ubiquitination of hEST1B was found to be dependent neither on its acetylation status nor on the deacetylase activity of HDAC8. Instead, our findings suggest an alternate and distinct mechanism of hEST1B regulation that requires multiple cofactors. One cofactor required for hEST1B ubiquitination is CHIP, an E3 ubiquitin ligase. CHIP was originally identified as an Hsp/Hsc70 binding protein, and its U box domain was characterized as an ubiquitin chain formation-catalyzing domain (2). CHIP has been implicated in the ubiquitination of many other proteins, including unfolded CFTR (38), glucocorticoid receptor (60), and E2A (24). In this case, the E3 function of CHIP requires the help of chaperone proteins, such as members of the heat shock protein family (27, 62). Our study supports the roles of CHIP and chaperone proteins in the ubiquitin pathway. As shown in the in vitro ubiquitination assay, wild-type CHIP, but not a U box deletion mutant of CHIP, facilitated hEST1B ubiquitination, suggesting that CHIP is a de novo E3 ligase responsible for hEST1B ubiquitination. Consistent with the role of CHIP as a chaperone-dependent E3 ligase, hEST1B ubiquitination also was regulated by interaction with the molecular chaperone Hsp70.
The mechanism by which Hsp proteins regulate CHIP ligase activity is controversial. In the case of E2A and c-ErbB2 ubiquitination, CHIP activity was increased upon the inhibition of chaperone binding to CHIP (62). In contrast, CHIP enhanced the ubiquitination of Pael-R in the absence of Hsp70 (25). The binding of Hsp70 to the Parkin-CHIP complex negatively regulated its ubiquitin ligase activity. Like HDAC1, HDAC2, and HDAC3, HDAC8 coimmunoprecipitated with the ATP-dependent chaperone protein Hsp70 (28). In our current model, we propose that phosphorylated HDAC8 preferentially recruits Hsp70 protein to the hEST1B-CHIP complex. In this context, Hsp70 binding inhibits CHIP ligase activity, thereby stabilizing hEST1B by preventing its ubiquitin-mediated degradation. It is possible that HDAC8 may regulate the recruitment of Hsp70 to the complex by deacetylating the protein, which could contribute to its inhibitory effect on CHIP. Further studies are necessary to determine whether the enzymatic activity of HDAC8 is responsible for regulating the binding of Hsp70 to CHIP.
Protein ubiquitination is involved in several processes that are important in controlling cellular events in both normal and tumor cells. Recent work has suggested a link between ubiquitination and telomere regulation. UbcD1, an ubiquitin-conjugating enzyme in Drosophila, is needed to ensure proper telomere structure, suggesting that at least one of the telomere-associated proteins may be a target of the ubiquitin pathway (5). Pof3, a yeast F box protein required for telomere function, contains substrate specificity for SCF ubiquitin ligase (30). TRF1, a sequence-specific telomere DNA binding protein that serves as a negative regulator of telomere length, is subject to ubiquitination and degradation by the proteasome (6). However, how TRF1 activity is regulated by ubiquitination remains unclear. The current study provides a novel mechanism for the regulation of the telomerase-associated protein hEST1B by ubiquitination-mediated degradation via the CHIP pathway.
A previous study suggested that cells derived from SUV39DN mice, which are deficient in H3-Lys9 methylation, contain abnormally long telomeres due to reduced di- and trimethylation at that site (17). Since the N-terminal tails of histones undergo many different modifications that are regulated by combinatorial events, it is highly possible that telomere length modulation requires several histone modification enzymes. While the role of telomerase in tumor cell growth and progression to malignancy has been well documented in human cancers, such as malignant glioma and breast cancers (19, 31), ways in which telomerase could be targeted for therapeutic applications remain to be developed. It is tempting to speculate that HDAC8-regulated tumor cell proliferation is mediated primarily through its interaction with hEST1B, which provides a regulatory scaffold complex that, in turn, regulates its associated partner, telomerase activity. In this respect, specific HDAC8 inhibitors might be excellent candidates for therapeutic purposes.
It is noteworthy that the molecular chaperone Hsp90 is required for proper telomerase function and is stably associated with telomerase, while the interaction of Hsp70 with telomerase is transient (14, 20). We found that hEST1B interacted with Hsp90 directly, while the hEST1B-Hsp70 association required phosphorylated HDAC8. Therefore, HDAC8 phosphorylation appears to regulate chaperone/telomerase association via the hEST1B protein, an effect which can alter either the assembly of the telomerase complex or its proper function. While the functions of HDAC8 remain poorly understood, our work clearly demonstrates a unique role for this enzyme and shows that HDAC8 possesses functions beyond histone modification. Since telomerase appears to be a governing factor in growth promotion and tumor progression, our results suggest that the regulation of the hEST1B-telomerase complex by HDAC8 may have far-reaching implications in the understanding and treatment of human cancers.
Acknowledgments
We thank Jian-Dong Chen for plasmids and many helpful discussions, Michele Glozak and Mary Zhang for critical reading of the manuscript, and the Moffitt Cancer Center Core Facility for their assistance.
This work was supported by grants to E.S. from the NIH (GM58486 and CA109699) and the Kaul Foundation and postdoctoral fellowships from the American Heart Association to H.L. and N.S.
REFERENCES
- 1.Aisner, D. L., W. E. Wright, and J. W. Shay. 2002. Telomerase regulation: not just flipping the switch. Curr. Opin. Genet. Dev. 12:80-85. [DOI] [PubMed] [Google Scholar]
- 2.Ballinger, C. A., P. Connell, Y. Wu, Z. Hu, L. J. Thompson, L. Y. Yin, and C. Patterson. 1999. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19:4535-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buggy, J. J., M. L. Sideris, P. Mak, D. D. Lorimer, B. McIntosh, and J. M. Clark. 2000. Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem. J. 350:199-205. [PMC free article] [PubMed] [Google Scholar]
- 4.Cech, T. R. 2004. Beginning to understand the end of the chromosome. Cell 116:273-279. [DOI] [PubMed] [Google Scholar]
- 5.Cenci, G., R. B. Rawson, G. Belloni, D. H. Castrillon, M. Tudor, R. Petrucci, M. L. Goldberg, S. A. Wasserman, and M. Gatti. 1997. UbcD1, a Drosophilan ubiquitin-conjugating enzyme required for proper telomere behavior. Genes Dev. 11:863-875. [DOI] [PubMed] [Google Scholar]
- 6.Chang, W., J. N. Dynek, and S. Smith. 2003. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 17:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S., V. Prapapanich, R. A. Rimerman, B. Honore, and D. F. Smith. 1996. Interactions of p60, a mediator of progesterone receptor assembly, with heat shock proteins hsp90 and hsp70. Mol. Endocrinol. 10:682-693. [DOI] [PubMed] [Google Scholar]
- 8.Chiu, S. Y., G. Serin, O. Ohara, and L. E. Maquat. 2003. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9:77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong, Y. S., and S. Bacchetti. 2000. Histone deacetylation is involved in the transcriptional repression of hTERT in normal human cells. J. Biol. Chem. 275:35665-35668. [DOI] [PubMed] [Google Scholar]
- 10.Connell, P., C. A. Ballinger, J. Jiang, Y. Wu, L. J. Thompson, J. Hohfeld, and C. Patterson. 2001. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3:93-96. [DOI] [PubMed] [Google Scholar]
- 11.de Leval, L., D. Waltregny, J. Boniver, R. H. Young, V. Castronovo, and E. Oliva. 2006. Use of histone deacetylase (HDAC8), a new marker of smooth muscle differentiation, in the classification of mesenchymal tumors of the uterus. Am. J. Surg. Pathol. 30:319-327. [DOI] [PubMed] [Google Scholar]
- 12.Dittmar, K. D., K. A. Hutchison, J. K. Owens-Grillo, and W. B. Pratt. 1996. Reconstitution of the steroid receptor · hsp90 heterocomplex assembly system of rabbit reticulocyte lysate. J. Biol. Chem. 271:12833-12839. [DOI] [PubMed] [Google Scholar]
- 13.Durst, K. L., B. Lutterbach, T. Kummalue, A. D. Friedman, and S. W. Hiebert. 2003. The inv(16) fusion protein associates with corepressors via a smooth muscle myosin heavy-chain domain. Mol. Cell. Biol. 23:607-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsythe, H. L., J. L. Jarvis, J. W. Turner, L. W. Elmore, and S. E. Holt. 2001. Stable association of hsp90 and p23, but not hsp70, with active human telomerase. J. Biol. Chem. 276:15571-15574. [DOI] [PubMed] [Google Scholar]
- 15.Fu, W., J. G. Begley, M. W. Killen, and M. P. Mattson. 1999. Anti-apoptotic role of telomerase in pheochromocytoma cells. J. Biol. Chem. 274:7264-7271. [DOI] [PubMed] [Google Scholar]
- 16.Gao, L., M. A. Cueto, F. Asselbergs, and P. Atadja. 2002. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J. Biol. Chem. 277:25748-25755. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Cao, M., R. O'Sullivan, A. H. Peters, T. Jenuwein, and M. A. Blasco. 2004. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 36:94-99. [DOI] [PubMed] [Google Scholar]
- 18.Gregoretti, I. V., Y. M. Lee, and H. V. Goodson. 2004. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338:17-31. [DOI] [PubMed] [Google Scholar]
- 19.Herbert, B. S., W. E. Wright, and J. W. Shay. 2001. Telomerase and breast cancer. Breast Cancer Res. 3:146-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holt, S. E., D. L. Aisner, J. Baur, V. M. Tesmer, M. Dy, M. Ouellette, J. B. Trager, G. B. Morin, D. O. Toft, J. W. Shay, W. E. Wright, and M. A. White. 1999. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 13:817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holt, S. E., V. V. Glinsky, A. B. Ivanova, and G. V. Glinsky. 1999. Resistance to apoptosis in human cells conferred by telomerase function and telomere stability. Mol. Carcinog. 25:241-248. [PubMed] [Google Scholar]
- 22.Hou, M., X. Wang, N. Popov, A. Zhang, X. Zhao, R. Zhou, A. Zetterberg, M. Bjorkholm, M. Henriksson, A. Gruber, and D. Xu. 2002. The histone deacetylase inhibitor trichostatin A derepresses the telomerase reverse transcriptase (hTERT) gene in human cells. Exp. Cell Res. 274:25-34. [DOI] [PubMed] [Google Scholar]
- 23.Hu, E., Z. Chen, T. Fredrickson, Y. Zhu, R. Kirkpatrick, G. F. Zhang, K. Johanson, C. M. Sung, R. Liu, and J. Winkler. 2000. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J. Biol. Chem. 275:15254-15264. [DOI] [PubMed] [Google Scholar]
- 24.Huang, Z., L. Nie, M. Xu, and X. H. Sun. 2004. Notch-induced E2A degradation requires CHIP and Hsc70 as novel facilitators of ubiquitination. Mol. Cell. Biol. 24:8951-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imai, Y., M. Soda, S. Hatakeyama, T. Akagi, T. Hashikawa, K. I. Nakayama, and R. Takahashi. 2002. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol. Cell 10:55-67. [DOI] [PubMed] [Google Scholar]
- 26.Ito, A., Y. Kawaguchi, C. H. Lai, J. J. Kovacs, Y. Higashimoto, E. Appella, and T. P. Yao. 2002. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21:6236-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, J., C. A. Ballinger, Y. Wu, Q. Dai, D. M. Cyr, J. Hohfeld, and C. Patterson. 2001. CHIP is a U box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276:42938-42944. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, C. A., D. A. White, J. S. Lavender, L. P. O'Neill, and B. M. Turner. 2002. Human class I histone deacetylase complexes show enhanced catalytic activity in the presence of ATP and co-immunoprecipitate with the ATP-dependent chaperone protein Hsp70. J. Biol. Chem. 277:9590-9597. [DOI] [PubMed] [Google Scholar]
- 29.Joung, J. K., E. I. Ramm, and C. O. Pabo. 2000. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc. Natl. Acad. Sci. USA 97:7382-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katayama, S., K. Kitamura, A. Lehmann, O. Nikaido, and T. Toda. 2002. Fission yeast F-box protein Pof3 is required for genome integrity and telomere function. Mol. Biol. Cell 13:211-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komata, T., T. Kanzawa, Y. Kondo, and S. Kondo. 2002. Telomerase as a therapeutic target for malignant gliomas. Oncogene 21:656-663. [DOI] [PubMed] [Google Scholar]
- 32.Kosano, H., B. Stensgard, M. C. Charlesworth, N. McMahon, and D. Toft. 1998. The assembly of progesterone receptor-hsp90 complexes using purified proteins. J. Biol. Chem. 273:32973-32979. [DOI] [PubMed] [Google Scholar]
- 33.Lee, H., N. Rezai-Zadeh, and E. Seto. 2004. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol. Cell. Biol. 24:765-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, R. J., L. Nagy, S. Inoue, W. Shao, W. H. Miller, Jr., and R. M. Evans. 1998. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 391:811-814. [DOI] [PubMed] [Google Scholar]
- 35.Luo, J., A. Y. Nikolaev, S. Imai, D. Chen, F. Su, A. Shiloh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107:137-148. [DOI] [PubMed] [Google Scholar]
- 36.Luo, J., F. Su, D. Chen, A. Shiloh, and W. Gu. 2000. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408:377-381. [DOI] [PubMed] [Google Scholar]
- 37.Marks, P., R. A. Rifkind, V. M. Richon, R. Breslow, T. Miller, and W. K. Kelly. 2001. Histone deacetylases and cancer: causes and therapies. Nat. Rev. Cancer 1:194-202. [DOI] [PubMed] [Google Scholar]
- 38.Meacham, G. C., C. Patterson, W. Zhang, J. M. Younger, and D. M. Cyr. 2001. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3:100-105. [DOI] [PubMed] [Google Scholar]
- 39.Murata, S., Y. Minami, M. Minami, T. Chiba, and K. Tanaka. 2001. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2:1133-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy, M., J. Ahn, K. K. Walker, W. H. Hoffman, R. M. Evans, A. J. Levine, and D. L. George. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13:2490-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nusinzon, I., and C. M. Horvath. 2006. Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation. Mol. Cell. Biol. 26:3106-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohnishi, T., A. Yamashita, I. Kashima, T. Schell, K. R. Anders, A. Grimson, T. Hachiya, M. W. Hentze, P. Anderson, and S. Ohno. 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell 12:1187-1200. [DOI] [PubMed] [Google Scholar]
- 43.Qian, S.-B., H. McDonough, F. Boellmann, D. M. Cyr, and C. Patterson. 2006. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature 440:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reichenbach, P., M. Hoss, C. M. Azzalin, M. Nabholz, P. Bucher, and J. Lingner. 2003. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr. Biol. 13:568-574. [DOI] [PubMed] [Google Scholar]
- 45.Sarin, K. Y., P. Cheung, D. Gilison, E. Lee, R. I. Tennen, E. Wang, M. K. Artandi, A. E. Oro, and S. E. Artandi. 2005. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature 436:1048-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seigneurin-Berny, D., A. Verdel, S. Curtet, C. Lemercier, J. Garin, S. Rousseaux, and S. Khochbin. 2001. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol. 21:8035-8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snow, B. E., N. Erdmann, J. Cruickshank, H. Goldman, R. M. Gill, M. O. Robinson, and L. Harrington. 2003. Functional conservation of the telomerase protein Est1p in humans. Curr. Biol. 13:698-704. [DOI] [PubMed] [Google Scholar]
- 48.Somoza, J. R., R. J. Skene, B. A. Katz, C. Mol, J. D. Ho, A. J. Jennings, C. Luong, A. Arvai, J. J. Buggy, E. Chi, J. Tang, B. C. Sang, E. Verner, R. Wynands, E. M. Leahy, D. R. Dougan, G. Snell, M. Navre, M. W. Knuth, R. V. Swanson, D. E. McRee, and L. W. Tari. 2004. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure 12:1325-1334. [DOI] [PubMed] [Google Scholar]
- 49.Sprung, C. N., G. E. Reynolds, M. Jasin, and J. P. Murnane. 1999. Chromosome healing in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 96:6781-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart, S. A., W. C. Hahn, B. F. O'Connor, E. N. Banner, A. S. Lundberg, P. Modha, H. Mizuno, M. W. Brooks, M. Fleming, D. B. Zimonjic, N. C. Popescu, and R. A. Weinberg. 2002. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl. Acad. Sci. USA 99:12606-12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taggart, A. K., S. C. Teng, and V. A. Zakian. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297:1023-1026. [DOI] [PubMed] [Google Scholar]
- 52.Taggart, A. K., and V. A. Zakian. 2003. Telomerase: what are the Est proteins doing? Curr. Opin. Cell Biol. 15:275-280. [DOI] [PubMed] [Google Scholar]
- 53.Takakura, M., S. Kyo, Y. Sowa, Z. Wang, N. Yatabe, Y. Maida, M. Tanaka, and M. Inoue. 2001. Telomerase activation by histone deacetylase inhibitor in normal cells. Nucleic Acids Res. 29:3006-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van den Wyngaert, I., W. de Vries, A. Kremer, J. Neefs, P. Verhasselt, W. H. Luyten, and S. U. Kass. 2000. Cloning and characterization of human histone deacetylase 8. FEBS Lett. 478:77-83. [DOI] [PubMed] [Google Scholar]
- 55.Vannini, A., C. Volpari, G. Filocamo, E. C. Casavola, M. Brunetti, D. Renzoni, P. Chakravarty, C. Paolini, R. De Francesco, P. Gallinari, C. Steinkuhler, and S. Di Marco. 2004. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl. Acad. Sci. USA 101:15064-15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaziri, H., S. K. Dessain, E. Ng Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]
- 57.Vermeesch, J. R., D. Falzetti, G. Van Buggenhout, J. P. Fryns, and P. Marynen. 1998. Chromosome healing of constitutional chromosome deletions studied by microdissection. Cytogenet. Cell Genet. 81:68-72. [DOI] [PubMed] [Google Scholar]
- 58.Waltregny, D., L. De Leval, W. Glenisson, S. Ly Tran, B. J. North, A. Bellahcene, U. Weidle, E. Verdin, and V. Castronovo. 2004. Expression of histone deacetylase 8, a class I histone deacetylase, is restricted to cells showing smooth muscle differentiation in normal human tissues. Am. J. Pathol. 165:553-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waltregny, D., W. Glenisson, S. L. Tran, B. J. North, E. Verdin, A. Colige, and V. Castronovo. 2005. Histone deacetylase HDAC8 associates with smooth muscle alpha-actin and is essential for smooth muscle cell contractility. FASEB J. 19:966-968. [DOI] [PubMed] [Google Scholar]
- 60.Wang, X., and D. B. DeFranco. 2005. Alternative effects of the ubiquitin-proteasome pathway on glucocorticoid receptor down-regulation and transactivation are mediated by CHIP, an E3 ligase. Mol. Endocrinol. 19:1474-1482. [DOI] [PubMed] [Google Scholar]
- 61.Won, J., J. Yim, and T. K. Kim. 2002. Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells. J. Biol. Chem. 277:38230-38238. [DOI] [PubMed] [Google Scholar]
- 62.Xu, W., M. Marcu, X. Yuan, E. Mimnaugh, C. Patterson, and L. Neckers. 2002. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. USA 99:12847-12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, H., W. Fu, and M. P. Mattson. 2000. The catalytic subunit of telomerase protects neurons against amyloid beta-peptide-induced apoptosis. J. Neurochem. 75:117-124. [DOI] [PubMed] [Google Scholar]