Abstract
The binding of the prereplication complex proteins Orc1, Orc2, Mcm3, Mcm7, and Cdc6 and the novel DNA unwinding element (DUE) binding protein DUE-B to the endogenous human c-myc replicator was studied by chromatin immunoprecipitation. In G1-arrested HeLa cells, Mcm3, Mcm7, and DUE-B were prominent near the DUE, while Orc1 and Orc2 were least abundant near the DUE and more abundant at flanking sites. Cdc6 binding mirrored that of Orc2 in G1-arrested cells but decreased in asynchronous or M-phase cells. Similarly, the signals from Orc1, Mcm3, and Mcm7 were at background levels in cells arrested in M phase, whereas Orc2 retained the distribution seen in G1-phase cells. Previously shown to cause histone hyperacetylation and delocalization of replication initiation, trichostatin A treatment of cells led to a parallel qualitative change in the distribution of Mcm3, but not Orc2, across the c-myc replicator. Orc2, Mcm3, and DUE-B were also bound at an ectopic c-myc replicator, where deletion of sequences essential for origin activity was associated with the loss of DUE-B binding or the alteration of chromatin structure and loss of Mcm3 binding. These results show that proteins implicated in replication initiation are selectively and differentially bound across the c-myc replicator, dependent on discrete structural elements in DNA or chromatin.
Eukaryotic DNA replication initiates at a large number of chromosomal origins, controlled by the ordered assembly of multiprotein replication complexes and the cell cycle-dependent activity of kinases that phosphorylate them (49). In cases where origins have been transposed to other chromosomal locations, they have been found to colocalize with genetically defined replicators, i.e., sequences capable of promoting DNA replication at ectopic genomic sites (3, 6, 7, 9, 16, 22, 47, 48, 50, 61). In metazoan systems, replication origins or replicators are bound by homologues of proteins first characterized for the yeast Saccharomyces cerevisiae (5, 34, 38, 40, 61, 64, 66, 73, 76), suggesting that the basic mechanisms controlling replication initiation are conserved among eukaryotes. In S. cerevisiae, replicators typically comprise a binding site for the hexameric origin recognition complex ORC and a DNA unwinding element (DUE) (45, 56). ORC enables the Cdc6-, Cdt1-dependent recruitment of the MCM helicase complex to replication origins, forming a prereplication complex (pre-RC) (18, 69) early during the G1 phase of the cell cycle. Cyclin-dependent kinase and DDK activities promote the binding of Mcm10, Cdc45, and RPA to form preinitiation complexes and unwind the DNA template in advance of replication (76). The effect of kinase activity on the pre-RC is partially to disassemble ORC and release MCMs and Cdc6 from chromatin (1, 37, 38, 65).
The 2.4-kb 5′ region of the human c-myc gene contains multiple transcription factor binding sites and a DUE that is unwound in vivo (10, 25). The DUE is situated in a 100-bp zone containing three 10/11 matches to the S. cerevisiae ARS consensus sequence. Our laboratory initially reported that replication initiates in this region (43, 51, 52), and Vassilev and Johnson first used quantitative PCR (qPCR) to define the replication initiation zone (74). Subsequent work has confirmed that replication initiates in the 5′ flanking DNA of the c-myc gene in multiple species (19, 23, 57, 62, 71, 77). The 2.4-kb c-myc core origin endowed plasmids with ARS activity in vitro (12) and in vivo (51, 52, 71) and showed replicator activity when moved to an ectopic chromosomal location (47, 50). This region displays an ordered chromatin structure stable to chromosomal translocation (39), and mutational analyses have identified regions of the replicator essential for replication initiation, including the DUE (47).
In the present work we used chromatin immunoprecipitation (ChIP) to show that the human analogs of the yeast ORC, MCM, and Cdc6 proteins bind preferentially and selectively to the c-myc replicator. The distributions of Mcm3 and Mcm7 are similar in asynchronous cells, with the greatest ChIP signal at, and upstream of, the DNA unwinding element. These distributions change in parallel in cells synchronized in G1 or M phases. By contrast, Orc1, Orc2, and Cdc6 appear to be least abundant at the DUE and each displays a different temporal pattern of replicator binding. We show also that the DNA unwinding element binding protein DUE-B, identified using the c-myc DUE as bait in a yeast one-hybrid assay (17), preferentially binds near the c-myc DUE in a pattern comparable to that of the MCMs in asynchronous and G1-phase cells. Furthermore, at an ectopic locus, c-myc replicator deletions that removed the DUE or altered chromatin structure suppressed DUE-B or Mcm3 binding, respectively, and eliminated origin activity. The relationship between chromatin structure, MCM binding, and origin activity was supported by the demonstration that inhibition of histone deacetylase activity by trichostatin A (TSA) caused a redistribution of Mcm3 binding similar to the broadening of the c-myc replication initiation zone. These results suggest that pre-RC proteins bind nonrandomly to the c-myc replicator and that c-myc origin activity is a function of ORC, MCM, Cdc6, and DUE-B binding to c-myc chromatin.
MATERIALS AND METHODS
Cell lines and synchronization.
HeLa cells were grown at 37°C in 5% CO2 (47). The generation of isogenic HeLa cell lines carrying either a single copy of the 2.4-kb c-myc replicator core region or specific deletions of the c-myc replicator at an ectopic location was described previously (47). HeLa cells were synchronized at M phase by 20 h of incubation in the presence of 50 ng/ml of nocodazole in complete medium. To accumulate cells at G1 phase, HeLa cells were treated with 300 μM mimosine for 20 h. Daudi Burkitt lymphoma cells were obtained from the ATCC and were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10 mM HEPES, 1 mM sodium pyruvate, 10% fetal bovine serum, and 50 μg/ml gentamicin.
ChIP.
Polyclonal antibodies against human Orc1, Orc2, Mcm3, and Mcm7 were raised and purified as described previously (65). Affinity-purified antibody against human Cdc6 protein was generously provided by Nicholas Heintz (University of Vermont), and antibody used against Orc1 in chromatin immunoblotting was generously provided by C. Obuse (Kyoto University). Rabbit antiserum against acetylated histone H4 and normal rabbit serum were obtained from Upstate Biotechnology. Polyclonal antibody against DUE-B protein was generated in rabbits by using bacterially expressed recombinant DUE-B (17).
Cross-linked chromatin was prepared as described previously (65), with the exception that the sonicated chromatin was also subjected to micrococcal nuclease (MNase) digestion. To study the endogenous c-myc locus and the lamin B2 replicator, chromatin was digested with 0.05 U micrococcal nuclease (Sigma) per 100 μg of chromatin at 37°C for 5 min to limit the maximum length of each fragment to 200 to 400 bp. To study the ectopic c-myc locus, 0.01 U micrococcal nuclease per 100 μg of chromatin was used at room temperature for 5 min to generate fragments of ∼2 kb. Digestions were stopped by adding EGTA to 5 mM. Equal amounts of cross-linked chromatin (250 μg) were used for each ChIP. Chromatin was diluted with 11× NET (550 mM Tris-HCl, pH 7.4, 1.65 M NaCl, 5.5 mM EDTA, 5.5% NP-40) to a final concentration of 1× NET. Fifteen micrograms of DUE-B antiserum, 15 μg of Mcm3, Mcm7, Orc2, or Cdc6 antibody, 15 μl of polyclonal Orc1 antiserum, or an equivalent amount of normal rabbit serum was used for ChIP. Antibodies were allowed to bind the chromatin complex for 2 h at room temperature. The antibody:chromatin complex was mixed with protein A-agarose beads-salmon sperm DNA (Upstate Biotechnology) and incubated for 2 h at room temperature. Antibody complex washing and purification of coprecipitated DNA were carried out according to the method of Schepers et al. (67).
PCR.
Real-time PCR was performed with an ABI Prism 7000 system using SYBR green fluorescence. One-sixtieth or 1/120 aliquot of ChIP or input DNA, respectively, was used for each qPCR. Primer sequences are listed in Table 1. Data were compiled from at least three qPCR repeats from each of three independent ChIP experiments. Serially diluted HeLa genomic DNA was used to generate a standard curve for each primer set (r2 > 0.99) and to determine the sequence-tagged site (STS) copy number in ChIP DNA. Based on the slopes of the standard curves, the efficiency of amplification for each primer set was greater than 89%. The relative enrichment at each STS is the percentage of input chromatin DNA precipitated with specific antibody divided by the percentage of input chromatin DNA precipitated with normal rabbit serum. For all experiments, error bars indicate standard deviations.
TABLE 1.
Oligonucleotide primers used in this work
| STS or primer | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| 5′ | GCCACGGCTCAAGCAATC | GCCTCATGGCTGTAATCCTATCTACT |
| A | GGCTCCTGCGGGAAGG | CCTGACGGTGTCTGATCACTTAGA |
| B | TGCCATTACCGGTTCTCCA | TTCAACCGCATAAGAGATGGTG |
| C | GGGAAAGACGCTTTGCAGC | TTTGCCGCAAACGCG |
| D | TTCACAAGGGTCTCTGCTGACTC | GCGGGACCGGACTTCCTA |
| E | CGCGCCCATTAATACCCTT | AGGGCCGCGCTTTGA |
| F | CACTTTGCACTGGAACTTACAACA | GAATAGCCTCCCCGCGTC |
| G | TTGTGTGCCCCGCTCC | TTCCTGTTGGTGAAGCTAACGTT |
| H | TGGTCTTCCCCTACCCTCTCA | TGGAGTCTTGCGAGGCG |
| 3′ | GGAGACTATGATAACAGCCAGAGTTG | TCCTTTGCCTACCTCTCACCTT |
| LB2A | ATGAAGCGGATGTCTAAGAAAG | CGCCTGGGTCCTGTTTACAC |
| LB nonorigin | GAAAACACGCCTCTGGAGACA | TCAGGGACCCTGTTACCACC |
| 2q20 | GTGACCAGCCACCAACTAAGC | CACCGCCAAAGACTGCACTA |
| Nested-PCR primer no. 1 | AATCCAGTGTCTTGCTTTCA | |
| Nested-PCR primer no. 2 | AGCCAGGTTTCAGAAGAGAC | |
| Nested-PCR primer no. 3 | GCATTGTTAGATTTCATACA |
For nested PCR, primer sets specific for the endogenous (primers 1 and 2) and ectopic (primers 2 and 3) loci were used to amplify aliquots of input DNA and ChIP DNA. First-round PCR was carried out for 25 cycles when the products were at the logarithmic phase of amplification. PCR conditions were 94°C for 10 min; 25 cycles of 94°C for 40 s, 56°C for 40 s, and 72°C for 2 min; and then 72°C for 7 min. Diluted products of these reactions were used for qPCR with primer sets STS-B and STS-C. Protein binding at the ectopic c-myc locus was normalized using protein binding at the endogenous c-myc locus as an internal standard, i.e., enrichment at the ectopic locus is shown as the ratio of DNA copies at the ectopic locus versus DNA copies at the endogenous c-myc locus.
Isolation of a chromosomal site depleted for DUE-B protein binding.
DNA from total chromatin (input) or from ChIP DNA with DUE-B antibody was amplified with random amplified polymorphic DNA primer 50-01 from Genosys, Inc. (5′ GTGCAATGAG 3′), as follows: 94°C for 10 min; 45 cycles each of 94°C for 40 s, 35°C for 1 min, and 72°C for 2 min; 72°C for 7 min. A 225-bp product that was amplified in the input DNA but not in the ChIP DNA was gel purified, cloned, and sequenced. The 2p20 chromosomal location (GenBank accession no. AC098853) was determined by BLAST analysis and designated the DUE-B depleted site. Replication origin activity is expressed as the abundance of 1- to 2-kb nascent DNA at the DUE-B depleted 2q20 site or the c-myc origin (STS-myc2) relative to that at a site in the human β-globin locus (BG3) (35).
RESULTS
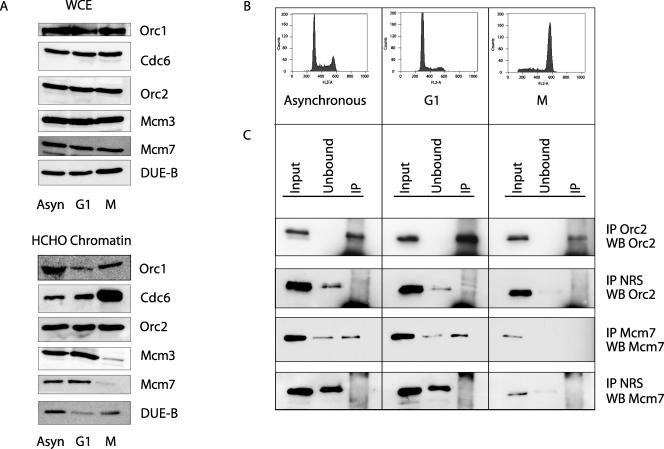
The structures of the endogenous c-myc replicator and of deletion mutants of the replicator integrated at an ectopic site in HeLa cells (47, 50) are schematized in Fig. 1. Highlighted are the c-myc promoters, the DUE, the segments deleted from the ectopic-site integrants, and the positions of the primer sets used for real-time quantitative PCR of immunoprecipitated (IP) DNA. Formaldehyde-cross-linked chromatin from asynchronously growing cells or populations enriched in G1-phase cells with mimosine or in mitotic cells with nocodazole was immunoprecipitated using polyclonal antibodies previously employed to examine the binding of pre-RC proteins to other regions of chromatin (30, 38, 64, 65, 67). In the HeLa cells used here, the levels of abundance of pre-RC proteins and DUE-B did not vary substantially in whole-cell extracts of asynchronous cells and cells synchronized by mimosine or nocodazole (Fig. 2A, top). These results agree with previous reports, except in the case of Orc1, where twofold- to fourfold-lower levels have been reported for mitotic cells (53, 54, 59, 70). This may be due to differences in the synchronizing reagents or the greater stringency of the mitotic selection used previously. The association of Orc1 with cross-linked chromatin was lower in G1-phase- and M-phase-synchronized cells than in asynchronous cells (53, 54, 59, 70), while the increased levels of Cdc6 on chromatin in nocodazole-arrested cells (53, 54) were enhanced by HCHO cross-linking (Fig. 2A, bottom).
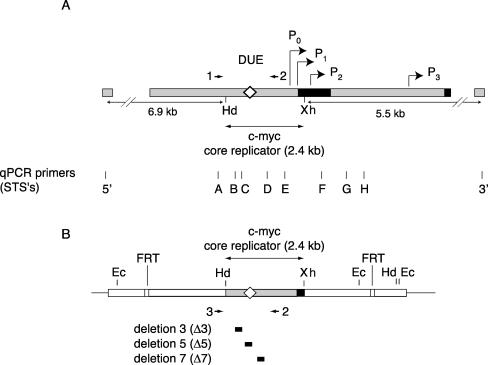
FIG. 1.
Maps of DNA sequences used in this study. (A) Endogenous human c-myc locus. The first c-myc exon and the 5′ part of the second exon are shown as black boxes. The HindIII/XhoI fragment constitutes the wild-type c-myc core replicator (Hd, HindIII; Xh, XhoI). The diamond denotes the DUE region. The c-myc promoters (bent arrows) are indicated as P0, P1, P2, and P3. Arrowheads indicate primers 1 and 2 used in nested PCR (see text). 5′, 3′, and A through H denote qPCR STS primer sets. (B) Structure of the ectopic c-myc replicator locus in cell lines containing the integrated wild-type (map) or deleted (black boxes) core replicator constructs (45). Arrowheads indicate primers 2 and 3 used in nested PCR (see text). The two qPCR primer sites STS-B and STS-C used in nested PCR are indicated. Ec, EcoRI; FRT, FLP recombination target.
FIG. 2.
Cell synchrony. (A) Whole-cell extract (WCE) or formaldehyde-cross-linked (HCHO) chromatin was prepared from asynchronous (Asyn) cells or cells arrested with mimosine (G1) or nocodazole (M) and immunoblotted for the indicated proteins. Protein from equal numbers of asynchronous or G1- or M-phase cells was loaded. (B) Asynchronous HeLa cells, HeLa cells synchronized in G1 with mimosine, or HeLa cells synchronized in M with nocodazole were stained with propidium iodide and analyzed by flow cytometry. (C) ChIP chromatin from asynchronous or G1- or M-phase HeLa cultures analyzed by Western blotting (WB), using the indicated antibodies. Input, cross-linked chromatin from asynchronous, G1, and M cells before ChIP (5% aliquot); unbound, supernatant fraction after ChIP (5% aliquot); IP, immunoprecipitate (50% aliquot). NRS, normal rabbit serum.
Figure 2B shows the degree of cell synchrony in these cultures. To confirm that the chromosomal abundance of pre-RC proteins in cells synchronized by mimosine or nocodazole was similar to that in cells synchronized by thymidine block (37, 38), immunoblotting was performed on chromatin precipitated with control normal rabbit serum or antibodies against Orc2 or Mcm7 from equal amounts of input chromatin. Orc2 was present at roughly equal amounts in the input from asynchronous cells or cells in G1 or M phase (Fig. 2C) and was efficiently immunoprecipitated on chromatin by Orc2-specific antibody but not by control antiserum. In contrast, Mcm7 was found in both chromatin-bound and unbound fractions of asynchronous and G1 cells but was significantly reduced in cultures arrested in mitosis by nocodazole and not detectably bound to mitotic chromatin.
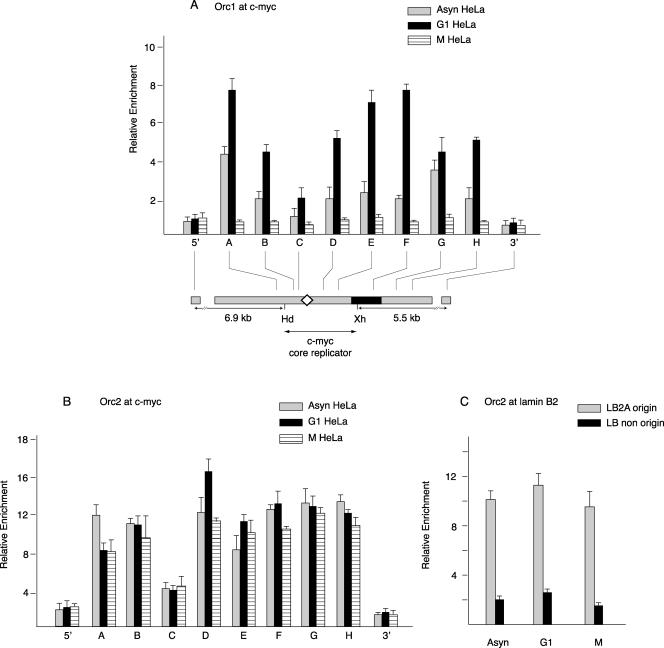
To test whether ORC proteins are localized to the c-myc replicator, cross-linked chromatin was prepared from HeLa cells in asynchronous growth or arrested in G1 phase or in M phase. qPCR was used to quantitate the amount of each of 10 STSs (A to H, 5′, and 3′) (Fig. 1) along the c-myc replicator. The relative enrichment for each STS is the amount of that STS in the DNA IP by the specific antibody relative to that precipitated by the control antiserum from an equal input amount of chromatin. The highest level of Orc1 binding was seen in the chromatin from cells in G1 phase, whereas cells blocked in mitosis showed only low abundance, comparable to the signals at the 5′ and 3′ distal sites, which we define as background. We interpret the signal from the immunoprecipitated DNA to indicate the presence of Orc1 on DNA, although the nature of immunoprecipitation makes it formally possible that low Orc1 signals reflect epitope masking, e.g., at the 5′ and 3′ distal sites and during M phase, rather than the absence of protein binding. Chromatin from asynchronous cells showed an intermediate level of Orc1 abundance (Fig. 3A). In both asynchronous and G1-phase cells, the pattern of Orc1 binding was nonrandom across the c-myc replicator, showing the lowest enrichment at STS-C, which reports on binding at the DUE/ARS consensus sequence region. In contrast to Orc1, Orc2 showed similar distributions in chromatin from asynchronous, G1, and M cells (Fig. 3B), although like Orc1, Orc2 revealed a significantly lower signal near the DUE than at other sites across the replicator.
FIG. 3.
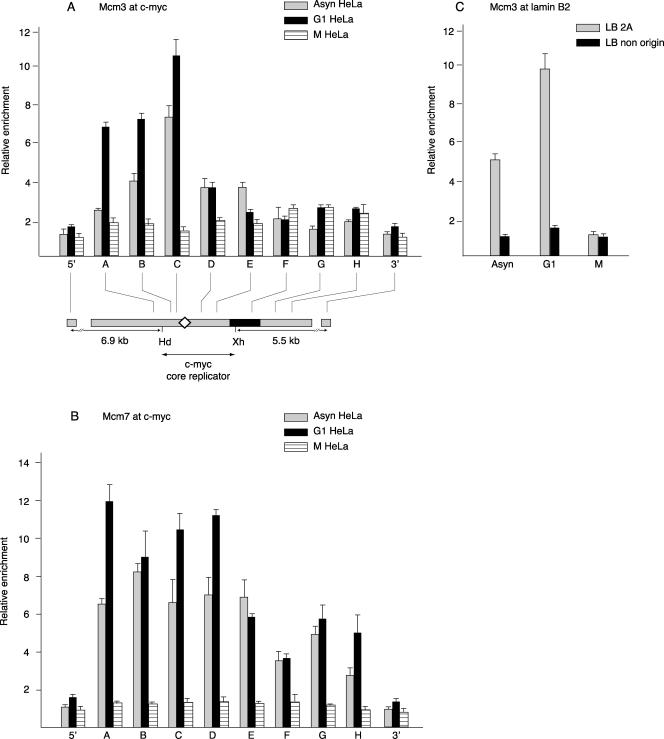
Orc1 and Orc2 binding. Shown are results from ChIP using chromatin from asynchronous (Asyn), mimosine-arrested G1-phase, or nocodazole-arrested M-phase HeLa cells with antibodies against (A) Orc1 or (B) Orc2 proteins. The ChIP DNA was used for qPCR of STSs in the c-myc replicator. The relative enrichment (ordinate) at each STS is the copy number of the DNA precipitated with the specific antibody divided by the copy number of the DNA precipitated by normal rabbit serum at the same STS. Map details are described in the legend for Fig. 1. (C) Orc2 protein binding at the lamin B2 replicator. ChIP DNA was used to measure the abundance of STS LB2A sequences (110 bp from the transition point at the lamin B2 origin) and lamin B2 nonorigin sequences (5 kb downstream of the lamin B2 origin) by qPCR.
To verify the ability of the Orc2 antibody to detect differences in Orc2 binding at a characterized origin, we assayed the same ChIP DNA for lamin B2 origin sequences. The lamin B2A (LB2A) primer site is 110 bp downstream of the lamin B2 origin of bidirectional replication, and the lamin B2 nonorigin site is 5 kb downstream of the lamin B2 origin (1). ChIP revealed a four- to fivefold enrichment for Orc2 at the LB2A site compared to that at the nonorigin site (Fig. 3C), confirming the preferential binding of Orc2 at the lamin B2 origin site observed previously (1), despite differences in cross-linking agent.
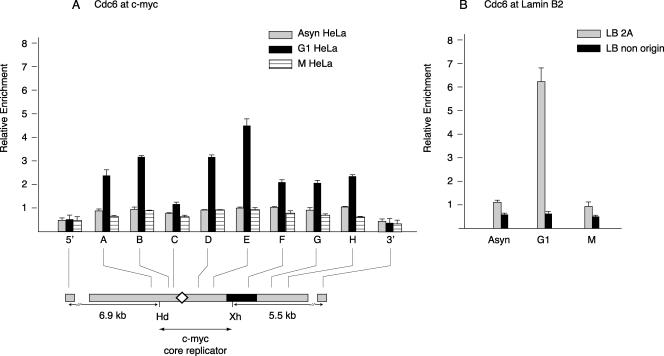
During the formation of the pre-RCs in G1 phase, Cdc6 is recruited by the origin recognition complex (32, 72). To determine whether Cdc6 recruitment is observed at the c-myc replicator, cross-linked chromatin was IP with anti-Cdc6 antibody (a generous gift of I. Sharon and N. Heintz). Cdc6 levels were low in c-myc-associated chromatin from mitotic cells or asynchronous cells but were significantly elevated in chromatin from cells arrested in G1 (Fig. 4). The pattern of Cdc6 binding was similar to that of Orc1, particularly with respect to the minimal abundance near the DUE in the ChIP chromatin. Despite the similarity in distribution of Orc1, Orc2, and Cdc6, the STS copy number was nominally lower in Orc1 or Cdc6 ChIP than in Orc2 ChIP. This could be due to the greater stability of Orc2 or to the less efficient precipitation of Orc1 and Cdc6. To validate these measurements, the binding of Cdc6 was examined at the lamin B2 origin (Fig. 4B). The highest level of Cdc6 binding to the origin and the greatest selectivity of binding was again observed to occur in G1-arrested cells, in quantitative agreement with results obtained using UV cross-linking (1). Moreover, Cdc6 and Orc2 ChIP showed similar preferential binding to the lamin B2 origin site versus the nonorigin site. These results support the view that two constituents of the pre-RC, Orc1 and Cdc6, are nonrandomly bound to the c-myc replicator in a cell cycle-dependent pattern.
FIG. 4.
Cdc6 binding. Cross-linked chromatin from asynchronous (Asyn), mimosine-arrested G1-phase, or nocodazole-arrested M-phase HeLa cells was precipitated with affinity-purified antibodies against Cdc6 protein, and the ChIP DNA was used for qPCR of STSs in the (A) c-myc or (B) lamin B2 replicator locus. Map details are described in the legend for Fig. 1.
The hexameric Mcm2-7 complex and two subcomplexes, the Mcm3/Mcm5 dimer and the Mcm4/Mcm6/Mcm7 trimer, have been reported to play important roles in DNA replication initiation (15, 31, 42, 60, 79). Antibodies against Mcm3 or Mcm7 were used to examine MCM complex loading at the c-myc replicator. A previous study of the nuclear abundance of MCM proteins suggested that the highest level of chromatin association would occur during G1 phase (53). As anticipated, the highest levels of Mcm3 and Mcm7 binding to the c-myc replicator were observed with G1-phase cells and the lowest with M-phase cells (Fig. 5A and B). Surprisingly, in contrast to the patterns of Orc1, Orc2, and Cdc6 binding, which were relatively low near the c-myc DUE, Mcm3 and Mcm7 showed high levels of binding at STS-C, near the DUE. To confirm that the ChIP chromatin mirrored the abundance of sequences reported for another origin, the same chromatin preparations were analyzed for Mcm3 binding to the lamin B2 origin and nonorigin sequences. As shown in Fig. 5C, Mcm3 binding was most apparent and selective in G1-phase cells. These data are quantitatively comparable to those reported by Abdurashidova et al. (1). Earlier reports have suggested that MCM proteins move from ORC-dependent sites of pre-RC loading (21, 64, 66). The present observation that Mcm3 and Mcm7 are enriched near the c-myc DUE, while Orc1, Orc2, and Cdc6 are relatively depleted there, may be a reflection of this phenomenon.
FIG. 5.
Mcm3 and Mcm7 binding. ChIP on chromatin from asynchronous (Asyn), mimosine-arrested G1-phase, or nocodazole-arrested M-phase HeLa cells with antibodies against (A) Mcm3 or (B) Mcm7 proteins, quantitated at the c-myc replicator by qPCR. Map details are described in the legend for Fig. 1. (C) Mcm3 protein binding at the lamin B2 replicator.
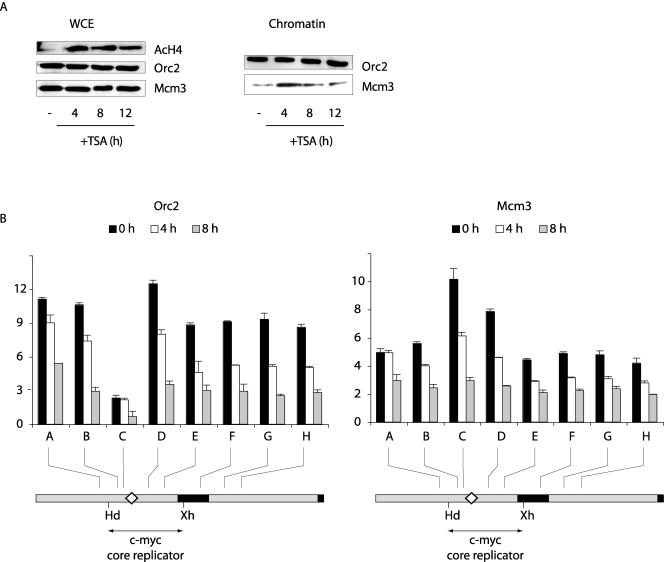
It was previously shown that treatment of cells with a single dose of the histone deacetylase inhibitor TSA reversibly decreases the selectivity of replication initiation at the human c-myc origin, in parallel with the time course of histone hyperacetylation (35) (Fig. 6A). When ChIP was used to examine the distribution of Orc2 and Mcm3 at the c-myc origin in cells treated with TSA, a quantitative decrease in Orc2 immunoprecipitation of c-myc DNA sequences was observed, but the qualitative pattern of Orc2 binding, particularly the low abundance of Orc2 near the DUE, was nominally unchanged (Fig. 6B). The decrease in Orc2 ChIP may be a consequence of Orc2 lysine acetylation (28), since similar amounts of Orc2 were associated with chromatin over this time course (Fig. 6A). In contrast, the hyperacetylation of histones correlated with an increase in chromatin-bound Mcm3, a more uniform pattern of Mcm3 binding across the origin, and the loss of preferential Mcm3 binding at the DUE (Fig. 6B). Hence, the patterns of Mcm3 binding and replication initiation change in parallel upon hyperacetylation of chromatin.
FIG. 6.
Redistribution of Mcm3 following TSA treatment. (A) Whole-cell extract (WCE) or chromatin (51) was prepared from cells treated with TSA for 0 h, 4 h, 8 h, or 12 h and immunoblotted for the indicated proteins. (B) ChIP on chromatin from HeLa cells treated with TSA for 0 h, 4 h, or 8 h, using antibodies against (left) Orc2 or (right) Mcm3. The ChIP DNA was used for qPCR of STSs in the c-myc replicator. Map details are described in the legend for Fig. 1. AcH4, acetylated histone H4.
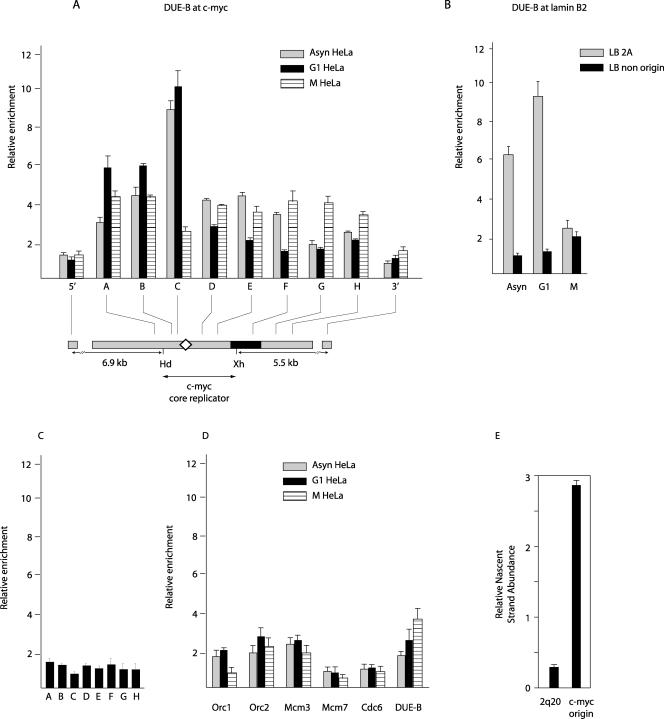
A DNA unwinding element binding protein, DUE-B, was identified using the c-myc DUE as bait in a yeast one-hybrid screen and was shown by competition electrophoretic mobility shift assays to bind the c-myc DUE bait in a sequence-dependent manner in the presence of HeLa nuclear proteins (17). ChIP with anti-DUE-B antibody revealed that DUE-B is preferentially bound at or near the c-myc DUE in asynchronous cells and in G1-arrested cells (Fig. 7A) in a pattern similar to that of Mcm3 (cf. Fig. 5A). In contrast, in nocodazole-blocked cells DUE-B binding to the replicator region (STS-A to STS-H) remained above background levels (STS-5′ and STS-3′). This observation is consistent with the increased association of DUE-B with cross-linked chromatin in M-phase- versus G1-phase-arrested cells (Fig. 2) and implies that DUE-B may redistribute or associate with additional chromosomal sites after the G1 phase. Like Orc2, Cdc6, and Mcm3, DUE-B is more abundant and shows greater binding selectivity at the LB2A origin site than at a nonorigin site in asynchronous cells and cells arrested in G1 (Fig. 7B). In contrast to the patterns of ORC, Cdc6, and MCM binding at the lamin B2 nonorigin site, DUE-B levels increased nominally in nocodazole-blocked cells (Fig. 7B). To confirm that the uniform level of DUE-B binding to the c-myc replicator was different from the level of nonspecific background binding, we used the anti-DUE-B antibody to IP chromatin from an asynchronous population of Daudi Burkitt lymphoma cells that do not express DUE-B protein or mRNA (M. Kemp, unpublished data). In these cells, one copy of the c-myc gene has been translocated into the immunoglobulin heavy chain locus from a breakpoint more than 170 kb 5′ to the c-myc core replicator (33). Anti-DUE-B ChIP with Daudi cross-linked chromatin showed only background-level binding across the c-myc replicator (Fig. 7C).
FIG. 7.
DUE-B binding and origin activity. Shown are results of ChIP on chromatin from asynchronous (Asyn), G1-phase, or M-phase HeLa cells with polyclonal antibodies against DUE-B protein. The ChIP DNA was used for qPCR at STSs in the (A) c-myc or (B) lamin B2 replicator. Map details are described in the legend for Fig. 1. (C) Cross-linked chromatin was isolated from G1-arrested Daudi cells, which do not express DUE-B, and immunoprecipitated with polyclonal antibodies against DUE-B, and the precipitated DNA was used for qPCR of STSs in the c-myc replicator. (D) Decreased pre-RC protein binding at a site selected for low DUE-B binding. The chromosome 2p20 DNA sequence underrepresented in the anti-DUE-B antibody ChIP was isolated from the input HeLa chromatin pool (see Materials and Methods). qPCR analysis of ChIP DNA with Orc1, Orc2, Mcm3, Mcm7, Cdc6, or DUE-B antibody was carried out at the 2p20 site. (E) Replication origin activity (1- to 2-kb nascent-strand abundance) at the DUE-B depleted 2q20 site compared to that at the endogenous c-myc origin.
The preceding results demonstrate that DUE-B and pre-RC proteins bind to replicator regions in the genome. To test the converse hypothesis, we identified a site that does not efficiently bind DUE-B in asynchronous HeLa cells by comparison of the products of random PCR amplification of DNA from total cross-linked chromatin or anti-DUE-B ChIP chromatin. A 225-bp fragment present in the amplified product of total chromatin but absent in the PCR product of anti-DUE-B ChIP DNA was cloned, sequenced, and localized to chromosome band 2q20 (not shown). Real-time PCR primers were designed to monitor the binding of DUE-B and pre-RC proteins at the DUE-B depleted 2q20 site in asynchronous, G1-phase, and M-phase cells. No significant enrichment was observed at this chromosomal site for any of the pre-RC proteins tested, and there was no significant fluctuation of Orc2, Mcm3, Mcm7, or Cdc6 abundance between G1- and M-phase cells (Fig. 7D). The decrease in Orc1 abundance at the 2q20 site in nocodazole-arrested cells may reflect the decreased chromatin binding of Orc1 during M phase (44). In contrast, DUE-B levels increase moderately at the 2q20 site in mitotic cells, as at the lamin B2 nonorigin site and at c-myc replicator sites distal to the DUE. Since DUE-B levels do not vary over the cell cycle, these results support the idea that DUE-B redistribution occurs prior to or during mitosis. The low levels of pre-RC protein binding at the 2q20 site suggested that the DUE-B depleted 2q20 site would not display origin activity. Consistent with this expectation, the abundance of short nascent DNA strands at the 2q20 site was ∼10-fold lower than at the c-myc origin (Fig. 7E). Combined with the results of ChIP at the c-myc, lamin B2, and 2q20 loci, these data indicate that pre-RC components and DUE-B are preferentially bound to replicator elements during G1.
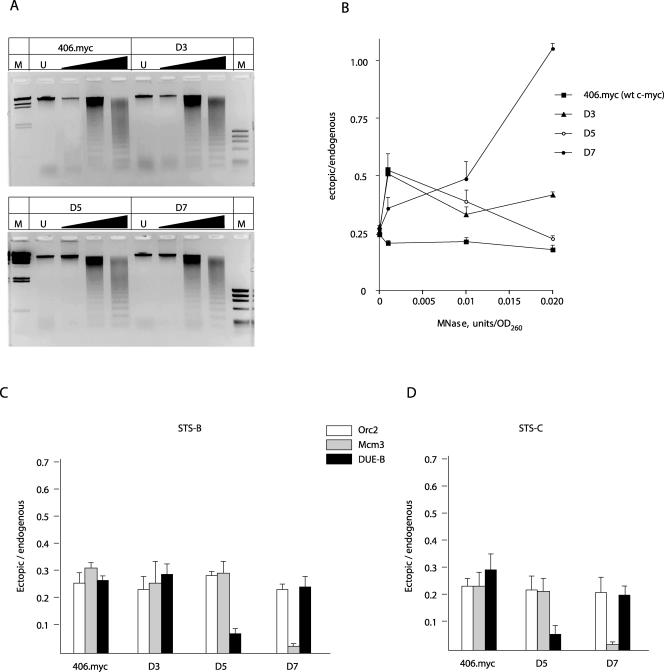
The relationship between DNA structure, origin activity, and replication protein binding was probed using c-myc replicator deletion mutants integrated at an ectopic chromosomal site. The 2.4-kb 5′ flanking DNA of the c-myc gene acts as a replicator when integrated at the Flp recombinase target site in HeLa/406 cells (chromosome band 18p11.22; 406.myc cells). Nascent-strand quantitation of a series of c-myc replicator deletion mutants integrated at this ectopic site showed that nucleotides 384 to 533 (removed in deletion 3; Δ3 cells [Fig. 1]) were not essential for full origin activity, while deletion of the DUE (deletion 5; Δ5 cells [Fig. 1]) or a downstream segment containing a positioned nucleosome (deletion 7; Δ7 cells [Fig. 1]) eliminated origin activity (47). To correlate replicator activity and protein binding, a nested-PCR strategy that included the endogenous c-myc replicator as an internal standard was used. ChIP DNA was isolated, and preparative PCR of a 1.45-kb fragment from the endogenous or ectopic replicator was carried out using locus-specific primer pairs 1 and 2 or 2 and 3 (Fig. 1), respectively. PCR was performed for 25 cycles so as not to exceed the logarithmic phase of amplification. The preparative PCR products were then diluted and quantitated by real-time PCR at STS-B or STS-C. We note that this procedure reports on the presence of the entire 1.45-kb template in the qPCR analyses that follow. First, MNase digestion was used to compare the levels of chromatin accessibility at the endogenous and ectopic sites in 406.myc, Δ3, Δ5, and Δ7 cells. Figure 8A shows that the rates of digestion of bulk chromatin are similar for the four cell lines. When the abundances of the c-myc replicator templates were compared in undigested DNA, the ratio was close to 0.25, as expected of pseudotetraploid cells containing one ectopic copy of the replicator (Fig. 8B). The sensitivities of the ectopic and endogenous sequences to MNase digestion were similar in 406.myc cells containing the wild type c-myc core replicator at the ectopic locus, indicating that there is not a gross change of chromatin structure that accounts for origin activity at the ectopic site, since the nature of the nested PCR is such that the assay is sensitive to the first nuclease cleavage within the c-myc PCR template. In the Δ3 and Δ5 cell lines, the ectopic replicator was slightly less sensitive to digestion at very low MNase concentrations. However, the Δ7 cell line showed the greatest resistance to nuclease digestion of the ectopic replicators, where the ectopic locus was approximately fourfold less sensitive to digestion than the endogenous locus at the MNase concentration of 0.02 U/unit of optical density at 260 nm.
FIG. 8.
Effect of sequence deletion on protein binding to the c-myc replicator. The 406.myc cell line containing the ectopic wild-type (wt) c-myc replicator and the three cell lines containing c-myc replicator deletion mutant constructs (Δ3, Δ5, and Δ7 [Fig. 1]) (45) were compared. (A) MNase digestion dose response. Gels were stained with ethidium bromide. M, markers of HindIII-digested λ DNA (left) and HaeIII-digested ΦX174 DNA (right). (B) Nested-PCR quantitation at STS-B of MNase resistance at the ectopic and endogenous replicators. OD260, optical density at 260 nm. (C and D) Orc2, Mcm3, and DUE-B binding at the ectopic and endogenous c-myc replicators assayed by ChIP at (C) STS-B and (D) STS-C (Fig. 1).
We next compared the abundances of ectopic and endogenous replicator sequences by Orc2, Mcm3, or DUE-B ChIP. Orc2, Mcm3, and DUE-B were cross-linked as efficiently (on a per-copy basis) to the endogenous c-myc replicator as to the wild-type c-myc core replicator at the ectopic locus in 406.myc cells (Fig. 8C), and the Δ3 cell line mutation did not significantly affect Orc2, Mcm3, or DUE-B binding. In contrast, deletion of the DUE in the Δ5 cell line resulted in a selective decrease in DUE-B association with the ectopic replicator, whereas Mcm3 and Orc2 binding were not detectably altered. These data argue that the DUE is necessary for DUE-B binding to the c-myc replicator in vivo and suggest that while ORC and MCM binding do not require the presence of DUE-B, they are not sufficient for replicator activity in the absence of the DUE region.
In the Δ7 cell line, a segment of DNA harboring a positioned nucleosome was deleted from the ectopic replicator. Orc2 and DUE-B proteins remained efficiently cross-linked to the replicator, but a large decrease in Mcm3 binding was detected. The loss of Mcm3 binding thus correlated with the change in nuclease accessibility of the replicator chromatin. These results are similar to those of Lipford and Bell (46), where removal of a positioned nucleosome at the S. cerevisiae ARS1 locus resulted in the loss of MCM binding and origin activity without a decrease in ORC binding. Similar to the conclusions derived from the Δ5 cell line, these data suggest that DUE-B and ORC binding are not sufficient for replicator activity.
DISCUSSION
Using chromatin immunoprecipitation, we showed that the human homologues of the yeast pre-RC proteins Orc1, Orc2, Mcm3, Mcm7, and Cdc6 and the human DNA unwinding element binding protein DUE-B bind nonrandomly to the human c-myc replicator and that the levels of binding of Orc1, Mcm3, Mcm7, Cdc6, and DUE-B vary over the cell cycle. Examined over 10 sequence-tagged sites spanning the c-myc replicator, Orc1 and Orc2 are most abundant in asynchronous and G1-arrested cells at sites flanking the DUE and ARS consensus region. While we interpret low ChIP signals as indicative of low protein association, we cannot rule out the possibility that local modification of proteins reduces the efficiency of cross-linking or immunoprecipitation. This caveat notwithstanding, analysis of ORC binding in cells arrested in M phase suggested that Orc1 is not stably bound to the c-myc or lamin B2 replicators, whereas Orc2 was present at similar levels in asynchronous, G1-phase, and M-phase cells. Orc1, Orc2, and Cdc6 were depleted near the DUE (STS-C) compared to the neighboring sites, whereas Mcm3, Mcm7, and DUE-B revealed considerably higher abundance at STS-C. An MCM-enriched region flanked by ORC bound regions has been reported in a detailed ChIP scan of ori2004 in fission yeast (68). This study suggested that cooperative binding of ORC to AT-rich regions I and III promotes MCM loading in the flanking region II, consistent with a model in which ORC binds to the replicator and recruits MCM proteins to flanking chromatin (21, 24, 41, 64). The behavior of Orc1 and Orc2 at the c-myc replicator mimics that observed at the PRKDC-MCM4 (40, 66) and lamin B2 (1) origins, and a similar pattern of stable Orc2 association with chromatin has been described in several other studies (53, 66). The elevated Orc2 binding which we observed at the LB2A site in M-phase chromatin differs from that reported previously (1). Possibly a change in ORC composition or conformation during M phase makes the complex less susceptible to the zero-length UV cross-linking used previously (1), whereas Orc2 may be cross-linked to DNA by formaldehyde through protein-protein bridges.
The similarity in ChIP patterns of Cdc6 and ORC at the c-myc replicator is consistent with the recruitment of Cdc6 by ORC; however, the efficiency of Cdc6 cross-linking to the c-myc replicator in asynchronous cells was low, possibly because Cdc6 is bound transiently (13, 58, 63) or because the antibody used in our ChIP experiments does not recognize the phosphorylated form of Cdc6 reported to reside on S-phase chromatin (4). That complete pre-RCs are assembled at the c-myc replicator is supported by the observation that Orc1, Orc2, Mcm3, Mcm7, and Cdc6 levels significantly above background are found at the c-myc and lamin B2 replicators during G1 phase, but only low levels are found at the 5′ and 3′ sites distal to the c-myc replicator and at the chromosome 2p20 site selected for depleted DUE-B binding. On the other hand, we have consistently observed a very low level of Cdc6 cross-linking to the c-myc replicator in nocodazole-blocked HeLa cells. Cdc6 levels are also greatly reduced at the LB2A origin site in M-phase HeLa cells, comparable to the levels at the LB2 nonorigin site, similar to the observations made earlier (1). The presence of Orc1 and Cdc6 on cross-linked mitotic chromatin, but their low appearance at the c-myc replicator, suggests that these protein epitopes are masked on M-phase chromatin or that Orc1 and Cdc6 are removed from origin sites to other chromosomal locations before mitosis. A recent immunofluorescence study posited that a fraction of Cdc6 phosphorylated by cyclin A-Cdk2 remains associated with chromatin throughout S phase (4), whereas the present experiments find a small residual amount of Cdc6 by ChIP of asynchronous cells. Possible quantitative differences between these results may arise from inefficient immunoprecipitation of low levels of Cdc6 or phosphorylated Cdc6.
DUE-B is preferentially cross-linked to the c-myc DUE region in asynchronous cells and G1-arrested HeLa cells but is distributed more evenly over the replicator in cells arrested in mitosis. Taken with the demonstration that binding of DUE-B is dependent on the presence of a segment of the replicator essential for c-myc origin activity and that DUE-B is enriched at the lamin B2 origin site, we suggest that DUE-B is involved in the initiation of replication. In this light, the temporal and spatial similarities in binding of DUE-B, Mcm3, and Mcm7 and the contrast to the binding of Orc1, Orc2, and Cdc6 are striking. DUE-B is not essential for DNA replication in Daudi cells, and yet small interfering RNA knockdown of DUE-B slows entry of HeLa cells into S phase and efficient immunodepletion of DUE-B from Xenopus laevis egg extracts inhibits sperm chromatin replication, which can be rescued by affinity-purified DUE-B expressed in HeLa cells (17). From HeLa or Xenopus egg extracts, DUE-B coisolates with the major maintenance DNA methyltransferase Dnmt1 (Kemp, unpublished), which has also been implicated in the process of DNA repair (27, 36, 55, 78). We propose, therefore, that DUE-B or a coisolating protein is involved in the recruitment of DNA repair or modifying proteins to replicating DNA and that the effect of DUE-B depletion on S-phase progression is linked to the integrity of the cellular checkpoint system.
The nuclear abundance of DUE-B protein remains the same in G1- and M-phase cells (17), although the redistribution of DUE-B in mitotic chromatin implies that the protein binds to previously depleted sites. Notably more DUE-B protein binding was observed downstream of c-myc STS-C in mitotic cells than in G1 cells. Similarly, there were appreciable increases in DUE-B association at the lamin B2 nonorigin and chromosome 2p20 sites in M-phase cells but not at the c-myc 5′ and 3′ distal sites. An explanation of this observation must await identification of the determinants of DUE-B binding. DUE-B expressed in insect cells is able to inhibit RPA loading and replication of sperm chromatin in Xenopus egg extracts (17). Considered with the colocalization of DUE-B and MCM proteins at the c-myc and lamin B2 replicators, we propose that DUE-B associates with the pre-RC during G1 at or near the DNA unwinding element. Once the MCMs are loaded and unwinding is complete, DUE-B may relocate to participate in additional nuclear processes.
Multiple structures contribute to eukaryotic replicator activity (3, 6, 7, 45, 47), suggesting that both DNA sequence and chromatin packaging influence replication initiation. To investigate how the structure of the c-myc replicator affects pre-RC formation, we made use of isogenic HeLa cell lines harboring either the 2.4-kb core c-myc replicator or deletion mutants integrated at the same ectopic chromosomal location. The ratio of Orc2 immunoprecipitable sequences at the wild-type ectopic and endogenous c-myc replicators was close to 0.25, similar to the copy number ratio of the ectopic and endogenous c-myc loci in these cell lines. The data imply that Orc2 binds to the ectopic and endogenous replicators with similar efficiencies. In two of the mutant cell lines, Δ5, in which the DUE region is deleted, and Δ7, in which the replicator chromatin structure was altered, origin activity was eliminated (47). The presence of Orc2 at the nonfunctional c-myc replicators shows that in human cells, as in yeast, ORC binding is not sufficient for replicator activity. The DUE region contains a zone of easily unwound DNA and three matches to the S. cerevisiae ACS. This region was originally identified as the c-myc far-upstream element (FUSE) and binding site for the FUSE binding protein FBP (20, 26). FBP binding is sensitive to the extent of DNA unwinding, and we speculate that FBP and DUE-B may interact through binding to this region. In the Δ5 cell line, deletion of the DUE/FUSE is correlated with a decrease in DUE-B binding and the loss of c-myc replicator activity. These results show that DUE-B binding is dependent on the DUE/FUSE in vivo but that neither the DUE nor DUE-B binding is essential for Orc2 or Mcm3 binding. These observations are consistent with the finding that DUE-B interacts with the pre-RC after MCM binding and before RPA binding in Xenopus egg extracts (17).
At the ectopic c-myc locus in the Δ7 cell line, Mcm3 association was significantly impaired, whereas neither Orc2 nor DUE-B protein association was affected. To our knowledge, these are the first demonstrations that mutation of a specific region of a metazoan replicator influences the binding of replication proteins in vivo. The correlation between an altered chromatin structure and decreased Mcm3 binding at the ectopic c-myc replicator is reminiscent of the loss of Mcm3 binding but not Orc1 binding at ARS1 of S. cerevisiae following loss of a nucleosome positioning signal (46). The ordered nucleosome arrangement of the c-myc replicator is stable to translocation (39), suggesting that the c-myc chromatin structure is a function of the underlying DNA sequence. It has recently been shown that recruitment of the CREB transcription factor or an open chromatin structure associated with basal transcription is compatible with c-myc replicator activation (22). That chromatin structure can influence origin activity has been shown by the demonstration that trichostatin A treatment leads to the exposure of novel sites of replication initiation (35) and that histone acetyltransferase recruitment to the Drosophila melanogaster ACE3/ori-β chorion origin locally stimulates DNA replication (2). The present ChIP data provide a possible mechanism for this effect, namely, the redistribution of MCM proteins. In response to a single dose of TSA, histone H4 acetylation occurs within 4 h; however, the change in the distribution of Mcm3 and replication initiation sites (35) continues until at least 8 h after TSA treatment. These observations suggest that histone acetylation is temporally upstream of events leading to pre-RC formation or that pre-RC formation responds to other effects of TSA. In conjunction with reports that multiple pre-RC constituents interact with the histone acetyltransferase HBO1 (14, 28, 29) and that deletion of the histone deacetylase Rpd3 from the budding yeast genome allows many late-firing replication origins to initiate DNA synthesis earlier in S phase (8, 75), these data imply that modulations of chromatin structure that affect transcription may also influence pre-RC formation and replicator activity.
The region 5′ to the c-myc gene acts as a replication origin in human (47, 57), chicken (62), frog, and mouse cells (23), indicating conservation of origin function. A recent report (37) focused on the temporal pattern of protein binding to the c-myc replicator to show that pre-RC formation follows the models derived from studies of yeast and frog systems (reviewed in reference 11). The present results are consistent with those of Kinoshita and Johnson (37), showing the preferential binding of pre-RC proteins near the c-myc DUE, but extend the spatial resolution of pre-RC protein binding across the c-myc replicator by using shorter chromatin fragments to demonstrate that ORC and MCM binding sites are separable (cf. reference 64). The observation that ORC, MCM, and DUE-B binding show differential sensitivities to replicator sequence deletions strengthens the view that the binding of these proteins depends on different aspects of replicator structure.
Finally, a striking outcome of this study is the similarity in association of DUE-B and Mcm3 at the c-myc replicator during G1 phase and the preferential binding of DUE-B and Mcm3 near the lamin B2 origin. Whereas analysis of the Δ5 and Δ7 cell lines shows that these proteins bind the c-myc replicator independently of one another, their colocalization near the c-myc and lamin B2 DUEs may reflect a functional interaction that is either direct or codependent on the structure of the replicator. Future studies will examine the implications of this interaction.
Acknowledgments
We are very grateful to I. Sharon and N. Heintz for providing affinity-purified Cdc6 antibody and to C. Obuse for providing Orc1 antibody for immunoblotting.
M.G. and G.L. were supported by the WSU School of Medicine postdoctoral program, and M.K. was supported by the WSU Biomedical Sciences Ph.D. program. This work was funded by NIH grant GM53819 to M.L.
REFERENCES
- 1.Abdurashidova, G., M. B. Danailov, A. Ochem, G. Triolo, V. Djeliova, S. Radulescu, A. Vindigni, S. Riva, and A. Falaschi. 2003. Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J. 22:4294-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aggarwal, B. D., and B. R. Calvi. 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430:372-376. [DOI] [PubMed] [Google Scholar]
- 3.Aladjem, M. I., L. W. Rodewald, J. L. Kolman, and G. M. Wahl. 1998. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science 281:1005-1009. [DOI] [PubMed] [Google Scholar]
- 4.Alexandrow, M. G., and J. L. Hamlin. 2004. Cdc6 chromatin affinity is unaffected by serine-54 phosphorylation, S-phase progression, and overexpression of cyclin A. Mol. Cell. Biol. 24:1614-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexandrow, M. G., M. Ritzi, A. Pemov, and J. L. Hamlin. 2002. A potential role for mini-chromosome maintenance (MCM) proteins in initiation at the dihydrofolate reductase replication origin. J. Biol. Chem. 277:2702-2708. [DOI] [PubMed] [Google Scholar]
- 6.Altman, A. L., and E. Fanning. 2001. The Chinese hamster dihydrofolate reductase replication origin beta is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity. Mol. Cell. Biol. 21:1098-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altman, A. L., and E. Fanning. 2004. Defined sequence modules and an architectural element cooperate to promote initiation at an ectopic mammalian chromosomal replication origin. Mol. Cell. Biol. 24:4138-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aparicio, J. G., C. J. Viggiani, D. G. Gibson, and O. M. Aparicio. 2004. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:4769-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Austin, R. J., T. L. Orr-Weaver, and S. P. Bell. 1999. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 13:2639-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bazar, L., D. Meighen, V. Harris, R. Duncan, D. Levens, and M. Avigan. 1995. Targeted melting and binding of a DNA regulatory element by a transactivator of c-myc. J. Biol. Chem. 270:8241-8248. [DOI] [PubMed] [Google Scholar]
- 11.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 12.Berberich, S., A. Trivedi, D. C. Daniel, E. M. Johnson, and M. Leffak. 1995. In vitro replication of plasmids containing human c-myc DNA. J. Mol. Biol. 245:92-109. [DOI] [PubMed] [Google Scholar]
- 13.Bowers, J. L., J. C. Randell, S. Chen, and S. P. Bell. 2004. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol. Cell 16:967-978. [DOI] [PubMed] [Google Scholar]
- 14.Burke, T. W., J. G. Cook, M. Asano, and J. R. Nevins. 2001. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem. 276:15397-15408. [DOI] [PubMed] [Google Scholar]
- 15.Burkhart, R., D. Schulte, D. Hu, C. Musahl, F. Gohring, and R. Knippers. 1995. Interactions of human nuclear proteins P1Mcm3 and P1Cdc46. Eur. J. Biochem. 228:431-438. [PubMed] [Google Scholar]
- 16.Carminati, J. L., C. G. Johnston, and T. L. Orr-Weaver. 1992. The Drosophila ACE3 chorion element autonomously induces amplification. Mol. Cell. Biol. 12:2444-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casper, J. M., M. G. Kemp, M. Ghosh, G. M. Randall, A. Vaillant, and M. Leffak. 2005. The c-myc DNA-unwinding element-binding protein modulates the assembly of DNA replication complexes in vitro. J. Biol. Chem. 280:13071-13083. [DOI] [PubMed] [Google Scholar]
- 18.Devault, A., E. A. Vallen, T. Yuan, S. Green, A. Bensimon, and E. Schwob. 2002. Identification of Tah11/Sid2 as the ortholog of the replication licensing factor Cdt1 in Saccharomyces cerevisiae. Curr. Biol. 12:689-694. [DOI] [PubMed] [Google Scholar]
- 19.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 20.Duncan, R., L. Bazar, G. Michelotti, T. Tomonaga, H. Krutzsch, M. Avigan, and D. Levens. 1994. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 8:465-480. [DOI] [PubMed] [Google Scholar]
- 21.Edwards, M. C., A. V. Tutter, C. Cvetic, C. H. Gilbert, T. A. Prokhorova, and J. C. Walter. 2002. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 277:33049-33057. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh, M., G. Liu, G. Randall, J. Bevington, and M. Leffak. 2004. Transcription factor binding and induced transcription alter chromosomal c-myc replicator activity. Mol. Cell. Biol. 24:10193-10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girard-Reydet, C., D. Gregoire, Y. Vassetzky, and M. Mechali. 2004. DNA replication initiates at domains overlapping with nuclear matrix attachment regions in the Xenopus and mouse c-myc promoter. Gene 332:129-138. [DOI] [PubMed] [Google Scholar]
- 24.Harvey, K. J., and J. Newport. 2003. CpG methylation of DNA restricts prereplication complex assembly in Xenopus egg extracts. Mol. Cell. Biol. 23:6769-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hay, N., J. M. Bishop, and D. Levens. 1987. Regulatory elements that modulate expression of human c-myc. Genes Dev. 1:659-671. [DOI] [PubMed] [Google Scholar]
- 26.He, L., J. Liu, I. Collins, S. Sanford, B. O'Connell, C. J. Benham, and D. Levens. 2000. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 19:1034-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermann, A., H. Gowher, and A. Jeltsch. 2004. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 61:2571-2587. [DOI] [PubMed] [Google Scholar]
- 28.Iizuka, M., T. Matsui, H. Takisawa, and M. M. Smith. 2006. Regulation of replication licensing by acetyltransferase Hbo1. Mol. Cell. Biol. 26:1098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iizuka, M., and B. Stillman. 1999. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 274:23027-23034. [DOI] [PubMed] [Google Scholar]
- 30.Illenye, S., and N. H. Heintz. 2004. Functional analysis of bacterial artificial chromosomes in mammalian cells: mouse Cdc6 is associated with the mitotic spindle apparatus. Genomics 83:66-75. [DOI] [PubMed] [Google Scholar]
- 31.Ishimi, Y. 1997. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 272:24508-24513. [DOI] [PubMed] [Google Scholar]
- 32.Jares, P., and J. J. Blow. 2000. Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev. 14:1528-1540. [PMC free article] [PubMed] [Google Scholar]
- 33.Joos, S., F. G. Haluska, M. H. Falk, B. Henglein, H. Hameister, C. M. Croce, and G. W. Bornkamm. 1992. Mapping chromosomal breakpoints of Burkitt's t(8;14) translocations far upstream of c-myc. Cancer Res. 52:6547-6552. [PubMed] [Google Scholar]
- 34.Keller, C., E. M. Ladenburger, M. Kremer, and R. Knippers. 2002. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J. Biol. Chem. 277:31430-31440. [DOI] [PubMed] [Google Scholar]
- 35.Kemp, M. G., M. Ghosh, G. Liu, and M. Leffak. 2005. The histone deacetylase inhibitor trichostatin A alters the pattern of DNA replication origin activity in human cells. Nucleic Acids Res. 33:325-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, M., B. N. Trinh, T. I. Long, S. Oghamian, and P. W. Laird. 2004. Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res. 32:5742-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinoshita, Y., and E. M. Johnson. 2004. Site-specific loading of an MCM protein complex in a DNA replication initiation zone upstream of the c-MYC gene in the HeLa cell cycle. J. Biol. Chem. 279:35879-35889. [DOI] [PubMed] [Google Scholar]
- 38.Kreitz, S., M. Ritzi, M. Baack, and R. Knippers. 2001. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem. 276:6337-6342. [DOI] [PubMed] [Google Scholar]
- 39.Kumar, S., and M. Leffak. 1991. Conserved chromatin structure in c-myc 5′ flanking DNA after viral transduction. J. Mol. Biol. 222:45-57. [DOI] [PubMed] [Google Scholar]
- 40.Ladenburger, E. M., C. Keller, and R. Knippers. 2002. Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol. Cell. Biol. 22:1036-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laskey, R. A., and M. A. Madine. 2003. A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. EMBO Rep. 4:26-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, J. K., and J. Hurwitz. 2001. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl. Acad. Sci. USA 98:54-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leffak, M., and C. D. James. 1989. Opposite replication polarity of the germ line c-myc gene in HeLa cells compared with that of two Burkitt lymphoma cell lines. Mol. Cell. Biol. 9:586-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, C. J., and M. L. DePamphilis. 2002. Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol. 22:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin, S., and D. Kowalski. 1997. Functional equivalency and diversity of cis-acting elements among yeast replication origins. Mol. Cell. Biol. 17:5473-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lipford, J. R., and S. P. Bell. 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell 7:21-30. [DOI] [PubMed] [Google Scholar]
- 47.Liu, G., M. Malott, and M. Leffak. 2003. Multiple functional elements comprise a mammalian chromosomal replicator. Mol. Cell. Biol. 23:1832-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu, L., H. Zhang, and J. Tower. 2001. Functionally distinct, sequence-specific replicator and origin elements are required for Drosophila chorion gene amplification. Genes Dev. 15:134-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Machida, Y. J., J. L. Hamlin, and A. Dutta. 2005. Right place, right time, and only once: replication initiation in metazoans. Cell 123:13-24. [DOI] [PubMed] [Google Scholar]
- 50.Malott, M., and M. Leffak. 1999. Activity of the c-myc replicator at an ectopic chromosomal location. Mol. Cell. Biol. 19:5685-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McWhinney, C., and M. Leffak. 1990. Autonomous replication of a DNA fragment containing the chromosomal replication origin of the human c-myc gene. Nucleic Acids Res. 18:1233-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McWhinney, C., and M. Leffak. 1988. Episomal persistence of a plasmid containing human c-myc DNA, p. 467-471. In B. Stillman and T. Kelly (ed.), Cancer cells, vol. 6. CSH Laboratory Press, New York, N.Y. [Google Scholar]
- 53.Méndez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendez, J., X. H. Zou-Yang, S. Y. Kim, M. Hidaka, W. P. Tansey, and B. Stillman. 2002. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell 9:481-491. [DOI] [PubMed] [Google Scholar]
- 55.Mortusewicz, O., L. Schermelleh, J. Walter, M. C. Cardoso, and H. Leonhardt. 2005. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl. Acad. Sci. USA 102:8905-8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Natale, D. A., R. M. Umek, and D. Kowalski. 1993. Ease of DNA unwinding is a conserved property of yeast replication origins. Nucleic Acids Res. 21:555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nenguke, T., M. I. Aladjem, J. F. Gusella, N. S. Wexler, and N. Arnheim. 2003. Candidate DNA replication initiation regions at human trinucleotide repeat disease loci. Hum. Mol. Genet. 12:1021-1028. [DOI] [PubMed] [Google Scholar]
- 58.Oehlmann, M., A. J. Score, and J. J. Blow. 2004. The role of Cdc6 in ensuring complete genome licensing and S phase checkpoint activation. J. Cell Biol. 165:181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohta, S., Y. Tatsumi, M. Fujita, T. Tsurimoto, and C. Obuse. 2003. The ORC1 cycle in human cells. II. Dynamic changes in the human ORC complex during the cell cycle. J. Biol. Chem. 278:41535-41540. [DOI] [PubMed] [Google Scholar]
- 60.Pacek, M., and J. C. Walter. 2004. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 23:3667-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paixao, S., I. N. Colaluca, M. Cubells, F. A. Peverali, A. Destro, S. Giadrossi, M. Giacca, A. Falaschi, S. Riva, and G. Biamonti. 2004. Modular structure of the human lamin B2 replicator. Mol. Cell. Biol. 24:2958-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phi-van, L., C. Sellke, A. von Bodenhausen, and W. H. Stratling. 1998. An initiation zone of chromosomal DNA replication at the chicken lysozyme gene locus. J. Biol. Chem. 273:18300-18307. [DOI] [PubMed] [Google Scholar]
- 63.Randell, J. C., J. L. Bowers, H. K. Rodriguez, and S. P. Bell. 2006. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol. Cell 21:29-39. [DOI] [PubMed] [Google Scholar]
- 64.Ritzi, M., M. Baack, C. Musahl, P. Romanowski, R. A. Laskey, and R. Knippers. 1998. Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J. Biol. Chem. 273:24543-24549. [DOI] [PubMed] [Google Scholar]
- 65.Ritzi, M., K. Tillack, J. Gerhardt, E. Ott, S. Humme, E. Kremmer, W. Hammerschmidt, and A. Schepers. 2003. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 116:3971-3984. [DOI] [PubMed] [Google Scholar]
- 66.Schaarschmidt, D., E. M. Ladenburger, C. Keller, and R. Knippers. 2002. Human Mcm proteins at a replication origin during the G1 to S phase transition. Nucleic Acids Res. 30:4176-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takahashi, T., E. Ohara, H. Nishitani, and H. Masukata. 2003. Multiple ORC-binding sites are required for efficient MCM loading and origin firing in fission yeast. EMBO J. 22:964-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tanaka, S., and J. F. Diffley. 2002. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat. Cell Biol. 4:198-207. [DOI] [PubMed] [Google Scholar]
- 70.Tatsumi, Y., S. Ohta, H. Kimura, T. Tsurimoto, and C. Obuse. 2003. The ORC1 cycle in human cells. I. Cell cycle-regulated oscillation of human ORC1. J. Biol. Chem. 278:41528-41534. [DOI] [PubMed] [Google Scholar]
- 71.Trivedi, A., S. E. Waltz, S. Kamath, and M. Leffak. 1998. Multiple initiations in the c-myc replication origin independent of chromosomal location. DNA Cell Biol. 17:885-896. [DOI] [PubMed] [Google Scholar]
- 72.Van Hatten, R. A., A. V. Tutter, A. H. Holway, A. M. Khederian, J. C. Walter, and W. M. Michael. 2002. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J. Cell Biol. 159:541-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vashee, S., C. Cvetic, W. Lu, P. Simancek, T. J. Kelly, and J. C. Walter. 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 17:1894-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vassilev, L., and E. M. Johnson. 1990. An initiation zone of chromosomal DNA replication located upstream of the c-myc gene in proliferating HeLa cells. Mol. Cell. Biol. 10:4899-4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vogelauer, M., L. Rubbi, I. Lucas, B. J. Brewer, and M. Grunstein. 2002. Histone acetylation regulates the time of replication origin firing. Mol. Cell 10:1223-1233. [DOI] [PubMed] [Google Scholar]
- 76.Walter, J., and J. Newport. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell 5:617-627. [DOI] [PubMed] [Google Scholar]
- 77.Waltz, S. E., A. A. Trivedi, and M. Leffak. 1996. DNA replication initiates non-randomly at multiple sites near the c-myc gene in HeLa cells. Nucleic Acids Res. 24:1887-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang, K. Y., and C. K. James Shen. 2004. DNA methyltransferase Dnmt1 and mismatch repair. Oncogene 23:7898-7902. [DOI] [PubMed] [Google Scholar]
- 79.You, Z., Y. Ishimi, H. Masai, and F. Hanaoka. 2002. Roles of Mcm7 and Mcm4 subunits in the DNA helicase activity of the mouse Mcm4/6/7 complex. J. Biol. Chem. 277:42471-42479. [DOI] [PubMed] [Google Scholar]