Abstract
Nucleoside diphosphate kinase (NDPK) (nm23/awd) belongs to a multifunctional family of highly conserved proteins (∼16 to 20 kDa) including two well-characterized isoforms (NDPK-A and -B). NDPK catalyzes the conversion of nucleoside diphosphates to nucleoside triphosphates, regulates a diverse array of cellular events, and can act as a protein histidine kinase. AMP-activated protein kinase (AMPK) is a heterotrimeric protein complex that responds to the cellular energy status by switching off ATP-consuming pathways and switching on ATP-generating pathways when ATP is limiting. AMPK was first discovered as an activity that inhibited preparations of acetyl coenzyme A carboxylase 1 (ACC1), a regulator of cellular fatty acid synthesis. We recently reported that NDPK-A (but not NDPK-B) selectively regulates the α1 isoform of AMPK independently of the AMP concentration such that the manipulation of NDPK-A nucleotide trans-phosphorylation activity to generate ATP enhanced the activity of AMPK. This regulation occurred irrespective of the surrounding ATP concentration, suggesting that “substrate channeling” was occurring with the shielding of NDPK-generated ATP from the surrounding medium. We speculated that AMPK α1 phosphorylated NDPK-A during their interaction, and here, we identify two residues on NDPK-A targeted by AMPK α1 in vivo. We find that NDPK-A S122 and S144 are phosphorylated by AMPK α1 and that the phosphorylation status of S122, but not S144, determines whether substrate channeling can occur. We report the cellular effects of the S122 mutation on ACC1 phosphorylation and demonstrate that the presence of E124 (absent in NDPK-B) is necessary and sufficient to permit both AMPK α1 binding and substrate channeling.
Nucleoside diphosphate kinase (NDPK) (nm23/awd) has been recognized for over 40 years as a ubiquitous enzyme that catalyzes the transfer of the γ-phosphate of a nucleoside triphosphate to a different nucleoside diphosphate (14). For example, the addition of ADP to GTP generates ATP plus GDP. This versatility allows NDPK to function as a nucleotide converter, balancing different cytosolic nucleotide pools, but cannot explain the necessity for nine human isoforms (Nm23-H1 to Nm23-H9) transcribed from different nm23 genes, constituting a family of structurally and functionally conserved proteins with differing functions (23). NDPK was also reported to be a protein histidine kinase (9), and the most widely expressed isoforms of NDPK (NDPK-A and -B) regulate a diverse array of cellular events including growth and development, tumor metastasis, and transcriptional regulation (9, 25). Despite sharing 88% sequence identity, these two isoforms have distinct cellular functions (3), and accumulating evidence indicates that protein-protein interactions can modulate the specific molecular actions of NDPK, with new binding partners being identified at an increasing rate (25). One such example is our recent report that an NDPK-A/AMP-activated protein kinase (AMPK) α1 complex associates with the cystic fibrosis transmembrane conductance regulator (7), providing a mechanistic link between ion transport and energy metabolism (17, 18, 27).
AMPK is a heterotrimeric protein with a 63-kDa catalytic α subunit and two regulatory β and γ subunits (38 and 36 kDa, respectively), each of which is encoded by distinct genes (α1, α2, β1, β2, γ1, γ2, and γ3) (22, 28). AMPK is a sensor of cellular energy status, responding to the cytosolic AMP/ATP ratio that itself varies as the square of the ADP/ATP ratio, with the whole process being equilibrated by the adenylate kinase reaction (11). Once activated by a rise in the cellular AMP concentration, AMPK phosphorylates several downstream substrates. The net effect switches off ATP-consuming pathways (e.g., fatty acid synthesis and cholesterol synthesis) and switches on ATP-generating pathways (e.g., fatty acid oxidation and glycolysis) (12).
Our previous study demonstrated that the AMPK α1 (but not α2) catalytic subunit is associated with NDPK-A (but not NDPK-B). We also provided in vitro evidence that local channeling of ATP (produced from GTP plus ADP) within the complex by an NDPK-catalyzed reaction is capable of altering the activity of AMPK in an AMP-independent manner (6). This association and the consequent substrate-channeling function provide a novel explanation for the paradox that AMPK needs ATP to function and yet must remain active when the ATP supply is limiting. We found that NDPK-generated ATP is specifically channeled to AMPK α1 irrespective of the surrounding ATP concentration for subsequent use as a phosphate donor to a recognized downstream in vivo target, acetyl coenzyme A carboxylase (ACC). Thus, substrate channeling altered the phosphorylation of this regulator of fat metabolism. Importantly, basal AMPK α1 activity towards ACC was also inhibited by ∼50% when nucleotides were manipulated to “steal” ATP from the complex, i.e., by adding GDP to generate complex-local GTP.
In this work, we report that two residues, S122 and S144, on NDPK-A are targeted by AMPK α1 in vitro and in vivo and that one local residue, E124, is both necessary and sufficient for the interaction of AMPK α1 with NDPK-A. Using recombinant and transfected NDPK-A proteins containing single point mutations of these residues, we find that the phosphorylation status of S122 (but not S144) determines whether NDPK-A is able to channel substrate; i.e., when AMPK phosphorylates S122, substrate channeling occurs, and when S122 cannot be phosphorylated, substrate channeling does not occur. We also provide in vivo phosphorylation data demonstrating the physiological relevance of this mechanism using ACC1 phosphorylation status as a downstream indicator. These data provide further insights into the mechanism relevance and specificity of the interaction between NDPK-A and AMPK α1.
MATERIALS AND METHODS
NDPK/AMPK cDNA constructs and purification.
pcDNA3.1 constructs containing cDNA encoding human AMPK α1, AMPK α2, AMPK γ1, AMPK γ2, and AMPK β (gifts of D. Carling) as well as NDPK-A and NDPK-B (gifts of M. L. Lacombe) were grown in bacterial cultures using ampicillin resistance selection. Mutation of specific residues on NDPK-A (S122A, S122D, S144A, S144D, and E124K) was accomplished using the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions and using gene-specific primers encoding each point mutation.
Affinity purification of the expressed cDNA was achieved using the incorporated poly-His tag according to the Lebendiker method (Protein Purification Facility, Hebrew University of Jerusalem). Briefly, 1 liter of LB broth was inoculated with pcDNA3.1-His+ plasmid cDNA containing sequence-verified AMPK α1, AMPK α2, NDPK-A, or NDPK-B and grown for 10 to 12 h in the presence of ampicillin. The bacteria were pelleted for 10 min at 4,000 rpm, resuspended in 10 ml of lysis buffer (50 mM NaPO4, 0.3 M NaCl, 8 M urea, and 10 mM imidazole, pH 8, with 2% Tween 20 and protease inhibitors [Boehringer] added fresh), and sonicated for three 15-s bursts. Lysate was then passed through a high-gauge needle, and DNA/insoluble proteins were pelleted by centrifugation at 13,000 rpm for 5 min. Supernatant from this spin was added to 5 ml of start buffer (0.2 M NaPO4, 0.5 M NaCl, and 10 mM imidazole, pH 7.4) and applied to a 5-ml Ni2+ Sepharose affinity purification column (Bio-Rad) preequilibrated in start buffer. The sample was passed through the column three times and then washed with a further 10 column volumes of start buffer prior to the elution of His-tagged proteins with 7 ml of elution buffer (0.2 M NaPO4, 0.5 M NaCl, and 500 mM imidazole, pH 7.4) collected in 0.5-ml fractions by gravity flow. Column fractions were screened for target protein by Bradford analysis (3a) and Western blotting using isoform-specific antibodies. The purity of the eluted protein was verified by both Coomassie blue and silver staining and determined to be over 90% pure.
NDPK-A-, AMPK α1-, and AMPK α2-null mice.
NDPK-A-null mouse liver (and skeletal muscle) tissue (and tissues from littermate wild-type control mice) (see Fig. 4B) was obtained as detailed previously (1). AMPK α1- and AMPK α2-null murine liver tissue was generated as detailed previously (16). Tissues were snap-frozen, and membranes/cytosol was extracted as described below.
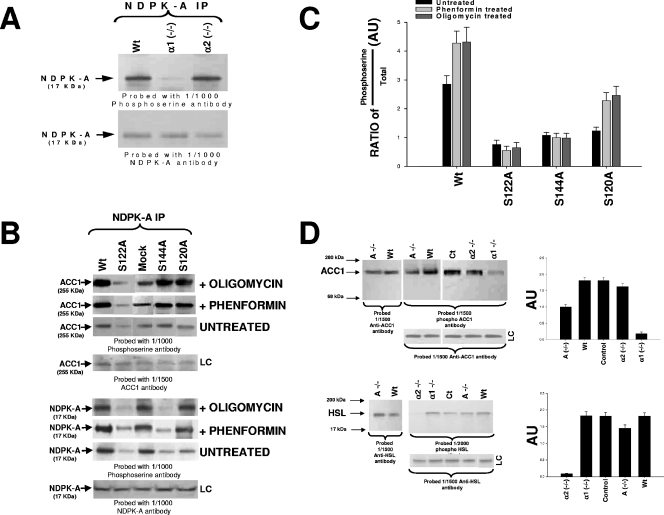
FIG. 4.
In vivo phosphorylation studies. (A) NDPK-A precipitations (IP) from 10 μg of liver cytosolic protein from wild-type littermate control (Wt), AMPK α1-null (α1 −/−), or AMPK α2-null (α2 −/−) mice run on 4 to 12% SDS-PAGE gels and probed with either a 1/1,000 dilution of anti-phosphoserine antibody or anti-NDPK-A antibody, as labeled. (B) HepG2 human-derived liver cell line transfected with various NDPK-A constructs: wild type, S122A, S144A, S120A, or empty vector control (mock). Transfected cells were either left untreated or incubated with 2 mM phenformin or 5 μg/ml oligomycin for 60 min. Ten micrograms of the cytosolic fraction from each construct was precipitated using an NDPK-A antibody as indicated, run on 4 to 12% SDS-PAGE gels, blotted, and probed using either a 1/1,000 dilution of phosphoserine or NDPK-A or ACC1 antibodies, as labeled. (C) Densitometry (histogram) data from four independent experiments were expressed in arbitrary units (AU) as a ratio of the phosphoserine amount over total protein amount ± standard error of the mean. (D) In the upper left panel, 10 μg of liver cytosolic protein from either wild-type (Wt), NDPK-A-null (A −/−), wild-type littermate control (Ct), AMPK α1-null (α1 −/−), or AMPK α2-null (α2 −/−) mice was run on 4 to 12% SDS-PAGE gels and probed with a 1/1,500 dilution of either anti-ACC1 antibody (leftmost blot) or anti-phospho-ACC1 antibody (center and right blots). In the lower left panel, 10 μg of the cytosolic fraction from mouse skeletal muscle from either wild-type (Wt), NDPK-A-null (A −/−), wild-type littermate control (Ct), AMPK α1-null (α1 −/−), or AMPK α2-null (α2 −/−) mice was run on an SDS-PAGE gel and probed with either a 1/1,500 dilution of anti-HSL antibody or a 1/2,000 dilution of phospho-HSL antibody. In each case, loading controls (LC) are included. Densitometry (D, right histograms) data from four independent experiments were calculated as described above for panel C.
Antibodies used in this study.
Anti-NDPK-H1 (nm301) mouse monoclonal and anti-NDPK-H2 (L-15) goat polyclonal antibodies were supplied by Santa Cruz. AMPK α1-, AMPK α2-, ACC1-, and phospho-ACC1-specific antibodies were kindly provided by D. G. Hardie, University of Dundee, and D. Carling, Imperial College London. The specificities of the antibodies to their targets have been validated elsewhere previously (3, 6, 33). Phosphoserine, hormone-sensitive lipase (HSL), and phospho-HSL antibodies were purchased from Abcam.
Cytosolic protein extraction.
The fractionation of tissue/cells was carried out as previously reported (24). Briefly, homogenization of tissue or cell extracts was achieved using mechanical lysis in 3 volumes of ice-cold buffer I (10 mM Tris, 20 mM NaH2PO4, and 1 mM EDTA, pH 7.8, containing a protease inhibitor cocktail). Osmolarity was restored by adding a solution containing a 1/20 volume of 2.4 M KCl, a 1/40 volume of 1.2 M NaCl, and a 1/5 volume of 1.25 M sucrose. Mixtures were centrifuged for 5 min at 500 × g. The pellets contained nuclei. Supernatants were diluted in buffer II (30 mM imidazole, 120 mM KCl, 30 mM NaH2PO4, and 250 mM sucrose, pH 6.8, with a protease inhibitor as described above) to a protein concentration of ∼0.7 mg/ml. Samples were then spun down at 7,000 × g for 15 min to remove mitochondria. The supernatant was then subjected to a spin at 30,000 × g for 30 min. The resultant pellets contained the remaining membrane proteins, and the supernatants contained cytosolic proteins.
Immunoprecipitation.
Prior to precipitation, dimethyl pimelimidate linking of antibody to Sepharose beads was performed as previously described (6). We used a 7-μl slurry of dimethyl pimelimidate-antibody-linked protein A/protein G beads as required and added cytosolic protein as detailed in each figure legend. Samples were then incubated with shaking for 60 min at 4°C and pelleted in a desk-top centrifuge, followed by three 1-ml washes with standard assay buffer containing 1 M NaCl and three 1-ml washes into standard assay buffer. Precipitation pellets were resuspended in 20 μl of assay buffer and assayed/probed as described above.
Western blot analysis.
Protein concentrations were determined using the method of Bradford (3a). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using the Novex (Invitrogen) system with 4 to 12% Bis-Tris polyacrylamide gels and MES (morpholineethanesulfonic acid) buffer, which allowed good separation of proteins between 17 and 180 kDa. Western blotting analysis was performed as detailed previously (5, 6). Briefly, nitrocellulose membranes were blocked in Tris-buffered saline-Tween (TBS-Tween) (0.5% Tween 20) plus 5% milk powder for 30 min, followed by four 15-min washes in TBS-Tween. Primary antibodies were incubated for 90 min, followed by four 15-min washes. Species-specific secondary antibodies were used according to the manufacturer's instructions and incubated for 45 min, followed by four 15-min washes. Enhanced chemiluminescent reagent (Amersham) was used for visualization.
Overlay binding analysis.
Purified bacterial recombinant NDPK-A and its various mutants or four NDPK regional peptides were immobilized on nitrocellulose membranes by spotting. The membranes were then blocked and washed as detailed above, a TBS-Tween solution containing either 10 or 15 μg of purified recombinant protein (as labeled in figure legends as either AMPK α1, AMPK α2, or CK2α) was overlaid onto the membrane and incubated for 1 h with shaking followed by four 15-min washes in TBS-Tween (as described above), and a 1/1,000 dilution of isoform-specific probing antibody against the overlaid protein was incubated as described above for the Western blotting protocol, followed by detection using secondary antibody and enhanced chemiluminescence visualization.
AMPK activity (SAMS) assay.
AMPK activity was assayed by measuring the incorporation of 32P from 500 nM [γ32P]ATP into 1 mM of the synthetic peptide substrate “SAMS” (HMRSAMSGLHLVKRR) exactly as detailed previously by Sullivan et al. (29). In detail, samples were suspended to a final volume of 25 μl in standard assay buffer (50 mM HEPES, pH 7.0, 50 mM NaF, 1 mM EGTA, 1 mM EDTA, 0.2% Tween 20, 10% glycerol). Assay mixtures were incubated at 30°C for 10 min, and the assays were terminated by spotting a 15-μl aliquot onto a 1-cm2 piece of P-81 phosphocellulose paper and washing them three times for 5 min in 1% phosphoric acid. Samples were then air dried, and the incorporation of labeled phosphate was quantified using a Packard Instant Imager (21). Slight variations of this assay were used throughout the experiments by substituting the SAMS peptide with various substrates (NDPK peptides or NDPK mutants).
RESULTS
The novel NDPK-A/AMPK α1 interaction.
Previous work from this laboratory identified a novel, bidirectional, and functionally relevant interaction between NPDK-A and the AMPK α1 catalytic subunit in both liver and lung cytosol (6). This interaction occurred in both reciprocal immunoprecipitation and recombinant assays. We therefore obtained NDPK-A-null (1), AMPK α1-null, and AMPK α2-null mouse (30) liver cytosolic extracts to investigate the specificity and functional relevance of this interaction.
Western blot analysis of NDPK-A-null tissue.
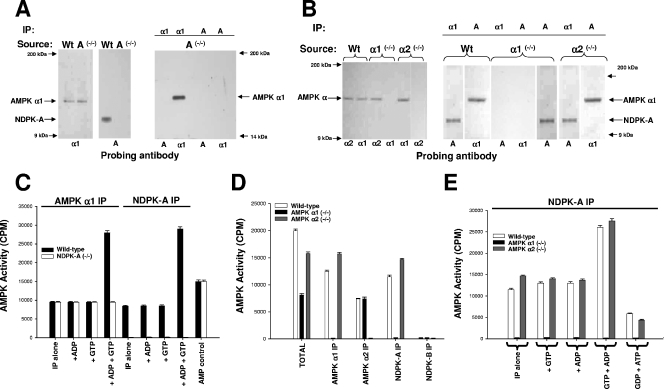
Where relevant, in the figures, the covalently bead-linked precipitating antibody used in a given experiment is shown at the top of each lane, and the isoform-specific probing antibody is shown at the bottom. To confirm the isoform specificity of the interaction with respect to NDPK, NDPK-A wild-type and knockout mouse liver extracts were probed for AMPK α1 and NDPK-A as indicated (Fig. 1A, left and middle panels). The data in the left panels confirm that NDPK-A has been knocked out without altering AMPK α1 and, furthermore, that an AMPK α1 antibody, while able to precipitate AMPK itself, was unable to coprecipitate any detectable NDPK-A (right panel). Conversely, an NDPK-A antibody was unable to precipitate either NDPK-A or AMPK α1, indicating that the interaction is absent under NDPK-A-null conditions (right panel). The positive controls for wild-type liver are not shown but have been described previously (6).
FIG. 1.
Investigation of AMPK α1, AMPK α2, and NDPK-A knockout tissues. The precipitating tissue source is shown above each blot together (where relevant) with the precipitating antibody. The lower part of each blot denotes a 1/1,000 dilution of probing antibody as indicated. (A) Ten micrograms of extracted NDPK-A wild-type (Wt) or NDPK-A-null (−/−) cytosol probed with sheep anti-AMPK α1 or mouse anti-NDPK-A (left and middle panels). The right panel shows anti-AMPK α1 or mouse anti-NDPK-A antibodies used to precipitate (IP) 10 μg of extracted cytosol and probed with either an AMPK α1- or NDPK-A-specific antibody, as indicated. (B) The procedure used was the same as that described above (A), except that the tissue source was extracted liver cytosol from murine wild-type (Wt), AMPK α1 knockout (−/−), or AMPK α2 knockout (−/−) tissue. (C) Ten micrograms of extracted NDPK-A wild-type or NDPK-A-null (−/−) cytosol precipitated using either an AMPK α1- or NDPK-A specific antibody and assayed for AMPK-SAMS activity in the presence/absence of 500 nM ADP, GTP, and ADP plus GTP, as indicated. Error bars (n = 3) on all assay histograms indicate the ranges and not the standard errors. (D) Ten micrograms of extracted AMPK wild-type, AMPK α1-null (−/−), or AMPK α2-null (−/−) cytosol precipitated using either an AMPK α1-, AMPK α2-, NDPK-A-, or NDPK-B-specific antibody and assayed for AMPK-SAMS activity relative to the input (total). (E) Ten micrograms of extracted AMPK wild-type, AMPK α1-null (−/−), or AMPK α2-null (−/−) cytosol precipitated using an NDPK-A-specific antibody and assayed for AMPK-SAMS activity in the presence/absence of 500 nM ADP, GTP, and ADP plus GTP, as indicated; the middle bars show no activity in each triplet.
Western blot analysis of AMPK α1- and α2-null tissue.
To confirm the specificity of the interaction with respect to the two isoforms of AMPK, cytosolic liver extracts from wild-type and AMPK α1- and AMPK α2-null mice were probed for AMPK α1, AMPK α2, and NDPK-A as indicated (Fig. 1B, left panel). This confirms that AMPK α1 and α2 have been selectively knocked out, as both isoforms are present in the wild-type tissue extract, and only the alternate isoform (AMPK α2 in the case of α1-null tissue and vice versa) is present in each knockout tissue extract. Furthermore, in the right-hand panel, we show that an AMPK α1-specific antibody is able to precipitate itself and coprecipitate NDPK-A from both wild-type and AMPK α2-null extracts but not from the AMPK α1-null extract, thus confirming the specificity of the interaction of NDPK-A with AMPK α1 in a native tissue.
AMPK activity in NDPK-A-null tissue.
To test the functional specificity of the AMPK/NDPK-A interaction, we used the well-established AMPK-SAMS assay, which is based on a peptide containing a consensus motif based around the AMPK target serine on acetyl coenzyme A carboxylase, an in vivo AMPK substrate (29). Figure 1C shows AMPK activity in a series of immunoprecipitates of AMPK α1 and NDPK-A. Previously, we were able to use an NDPK-A antibody to quantitatively precipitate >95% of the cytosolic AMPK α1 activity (6). Under the same conditions (Fig. 1C), both an AMPK α1 antibody and an NDPK-A antibody are able to precipitate similar amounts of AMPK activity from wild-type mouse liver cytosolic extracts (filled bars), and only the AMPK α1 antibody is able to precipitate this level of AMPK activity under NDPK-A-null conditions (compare open bars). Importantly, and unlike wild-type cytosol, the basal, AMP-stimulated AMPK α1 activity that is precipitated from NDPK-A-null tissue cannot be augmented in the presence of ADP plus GTP (Fig. 1C). These data are consistent with our recent hypothesis that NDPK-A function is critical for augmented AMPK α1 activity above levels found with AMP alone (Fig. 1C).
AMPK activity in AMPK α1 and α2 knockout tissue.
As a reciprocal check for specificity, we investigated mouse liver tissue extracts from AMPK α1 and AMPK α2 knockout mice (30). As shown in Fig. 1D, precipitations were performed by using AMPK α1-, AMPK α2-, NDPK-A-, or NDPK-B-specific antibodies (as indicated), and the precipitated AMPK-SAMS activity from cytosolic extracts of wild-type, AMPK α1-null, or AMPK α2-null mouse liver was measured. We observed that AMPK α1 antibody precipitated AMPK-SAMS activity in all cases except under the AMPK α1-null condition (likewise for AMPK α2). NDPK-A was able to precipitate AMPK-SAMS activity only from extracts where AMPK α1 was present, while no AMPK activity coprecipitated with NDPK-B, consistent with our previous observation that NDPK-B does not associate with AMPK α1 or α2. We will return to this difference below. Furthermore, in the NDPK-A precipitates shown in Fig. 1E, we never observed AMPK α1 activity under α1-null conditions (middle bar of each triplet), indicating that NDPK-A is able to precipitate AMPK activity only when AMPK α1 is present and that the presence or absence of AMPK α2 in the extract has no effect on this interaction. Additionally, Fig. 1E confirms that substrate channeling (i.e., NDPK-A-mediated ATP generation from GTP plus ADP local to the AMPK α1 catalytic subunit) is unable to occur when AMPK α1 is not present. As expected, the data shown in Fig. 1E confirm our previous finding that AMPK α1 activity can also be decreased below basal control levels by ATP sequestration when the nucleotide generation profile favors NDPK-dependent substrate stealing (i.e., ATP is “stolen” by NDPK-A by creating GTP from ATP plus GDP, despite millimolar concentrations of ATP in the medium).
What part of NDPK-A is phosphorylated by AMPK α1?
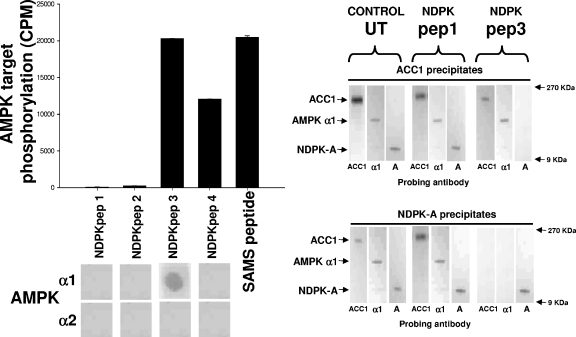
Next, we determined the specific area(s) of NDPK-A that either interacts with or potentially becomes phosphorylated by AMPK α1 in vitro. First, we generated four peptides spanning most of the NDPK-A primary sequence and adopted an overlay binding protocol to investigate potential interacting regions. We then performed “AMPK assays” but substituting the peptides as substrate to determine whether phosphorylation occurred. The left panel of Fig. 2 shows a screen of four different NDPK-A peptides used as substrates for purified rat liver AMPK in vitro. The first four peptides (NDPKpep1, 2, 3, and 4) correspond to exposed regions of NDPK-A (amino acids 49 to 62, 92 to 105, 111 to 125, and 139 to 152, respectively), as determined by crystal structure; alongside these, a SAMS peptide positive control is shown. We observed that purified rat liver AMPK was able to phosphorylate NDPKpep3, NDPKpep4, and SAMS peptide but not NDPKpep1 or 2. Furthermore, although AMPK was able to phosphorylate both NDPKpep3 and NDPKpep4, the AMPK α1 catalytic subunit binds only to NDPKpep3 on the overlay assay (Fig. 2, lower left panel).
FIG. 2.
NDPK peptides. The left histogram shows pure, active rat liver AMPK used to phosphorylate a selection of peptides as follows. Four peptides (NDPKpep1, 2, 3 and 4) corresponding to regions of NDPK-A (amino acids 49 to 62, 92 to 105, 111 to 125 and 139 to 152, respectively) were used, and the SAMS peptide was used as a positive control, each at 1 mM concentration. For the left overlay, the overlay protocol involves a slight variation of the above-described Western blotting protocol in that each of the NDPK-A peptides was immobilized on a nitrocellulose membrane and then blocked/washed as described above for the Western blot protocol. Furthermore, 10 μg of pure, recombinant AMPK α1 or α2 protein was overlaid onto the peptides, with binding being detected using isoform-specific antibodies as described above for Western blotting. In the right blots, 10 μg of rat liver cytosol either left untreated (UT) or incubated with a molar excess of NDPKpep1 (pep1) or NDPKpep3 (pep3) was precipitated using an ACC1 (upper right) or NDPK-A (lower right) antibody and probed with either ACC1-, AMPK α1 (α1)-, or NDPK-A (A)-specific antibodies as labeled at the bottom of each lane. NDPKpep3 specifically interferes with the ACC1/AMPK α1/NDPK-A complex association.
We previously demonstrated that NDPK-A and AMPK α1 were present in a precipitate of ACC1 from cytosol (10). To further confirm this interaction in a functional context with respect to ACC1, we incubated 10 μg of liver cytosol that was either untreated or treated with a molar excess of either NDPKpep1 or NDPKpep3, as labeled in Fig. 2 (right panels), and precipitated the cytosol using an ACC1 antibody (6) or an NDPK-A antibody. We probed these precipitates with ACC1, AMPK α1, or NDPK-A antibodies. As described above, the covalently bead-linked precipitating antibody used is shown at the top of each panel, and for each lane, the isoform-specific probing antibody is indicated at the bottom. The untreated control and NDPKpep1-incubated cytosol had no effect on the tripartite complex of ACC1, AMPK α1, and NDPK-A. We observed that the NDPKpep3 peptide alone is able to interfere with complex composition (Fig. 2, right panel, top and bottom final three lanes), confirming the region of interaction of the AMPK α1/NDPK-A complex shown in the left panel of Fig. 2. Specifically, NDPKpep3 (but not NDPKpep1) eliminates NDPK-A from an ACC1 precipitate, and conversely, both ACC1 and AMPK α1 no longer coprecipitate with NDPK-A.
Identification of the AMPK target residues on NDPK.
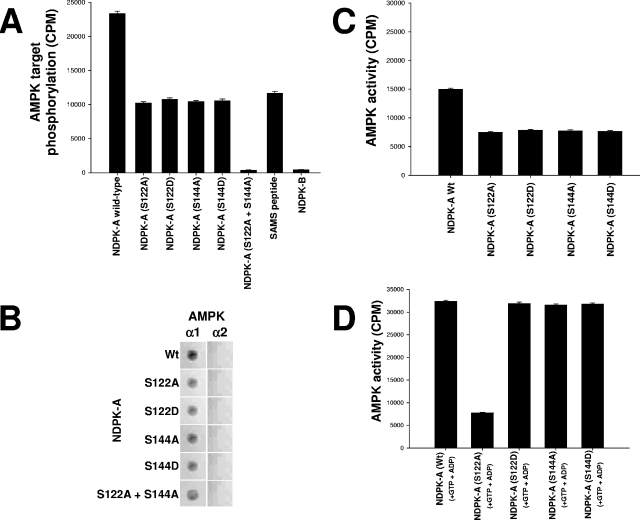
Inspection of the primary amino acid sequence of NDPK corresponding to the above-described phosphorylated regions at amino acids 111 to 125 and 139 to 152 revealed two candidate serine residues, S122 and S144, which at least partially conform to the established minimal consensus recognition motif for AMPK (8). They were individually point mutated in full-length NDPK-A to either alanine (A) or aspartic acid (D) (AMPK site phosphomimic) residues as indicated. We tested the hypothesis that AMPK α1 could phosphorylate these NDPK-A mutants. Figure 3A presents a series of AMPK assays performed using wild-type NDPK-A, the S122A, S122D, S144A, or S144D mutant, or the S122A-plus-S144A double mutant as a substrate alongside SAMS (positive control) and NDPK-B (negative control) (note that both serine 122 and serine 144 residues are conserved between NDPK-A and -B). We observed that wild-type NDPK-A incorporates twice as much phosphate in the presence of AMPK as any of the single point mutations, as might be expected due to the loss of one target site (the SAMS peptide contains only one site). Furthermore, AMPK was unable to phosphorylate the NDPK-A S122A-plus-S144A double mutant. These data confirm that S122 and S144 residues are in vitro targets for AMPK in NDPK-A. The data also show that despite serine conservation at both sites in NDPK-B, no phosphorylation occurs, consistent with our previous data showing no interaction between NDPK-B and AMPK. This difference suggests that a motif(s) exists within the AMPK α1 interaction region on NDPK-A (NDPKpep3) but not NDPK-B, which specifies the interaction with AMPK α1.
FIG. 3.
Specific NDPK-A amino acids targeted by AMPK and substrate-channeling ability of NDPK-A mutants. (A) Pure, active rat liver AMPK was used to phosphorylate a selection of substrates as follows. Fifteen micrograms (each) of bacterial recombinant purified proteins, wild-type NDPK-A, NDPK-A S122A, NDPK-A S122D, NDPK-A S144A, NDPK-A S144D, NDPK-A S122A-plus-S144A double mutant, and NDPK-B (negative control), was used. SAMS peptide (1 mM) was included as a positive control. (B) Overlay binding analysis where each of the NDPK-A mutants and wild-type recombinant proteins was immobilized on a nitrocellulose membrane and then blocked/washed as described above. Ten micrograms of pure, recombinant AMPK α1 or α2 protein was overlaid onto the membrane, and binding was detected using isoform-specific AMPK antibodies. (C) Histogram measuring the capacity of pure, active rat liver AMPK to phosphorylate as a substrate (10 μg of wild-type NDPK compared to mutant proteins [S122A/D and S144A/D]). (D) Histogram measuring the ability of 10 μg of bacterial recombinant wild-type/mutant NDPK-A protein, as labeled, to channel labeled ATP (generated from labeled GTP plus ADP) to 1 mM SAMS peptide.
Binding interaction of AMPK with NDPK mutants.
Next, we adopted an overlay binding protocol to investigate binding interactions between regions of NDPK-A and AMPK α1. Figure 3B shows a dot blot matrix of each NDPK mutant spotted onto nitrocellulose membranes and then overlaid with recombinant AMPK α1 or AMPK α2 protein. Isoform-specific AMPK antibodies were then used to probe for binding to each AMPK isoform. We show that whereas a mutation of S122 and S144 can abolish AMPK phosphorylation of NDPK, singly or in combination, these mutations do not abolish (but only attenuate) binding of AMPK α1 to NDPK-A.
Phosphorylation status of S122 on NDPK-A determines whether substrate channeling can occur.
Next, we tested whether these mutations had any effect on the ability of NDPK-A to channel ATP to AMPK α1. We incubated purified rat liver AMPK α1 with different mutants of recombinant wild-type NDPK-A, S122A, S122D, S144A, or S144D, which were precipitated using a specific NDPK-A antibody (Fig. 3C and D). We measured the coimmunoprecipitated AMPK activity by standard SAMS assay either alone (Fig. 3C) or in the presence of GTP plus ADP to promote substrate channeling (Fig. 3D). We observed that wild-type NDPK-A was able to precipitate AMPK activity and channel substrate as expected (first bars in Fig. 3C and D, respectively). NDPK-A S122A was able to precipitate a reduced amount of AMPK activity (not unexpectedly, as the binding efficacy is partially reduced, as shown in Fig. 3B), but this S122A mutant was unable to channel substrate despite the presence of GTP plus ADP (compare second bars in 3C and D). In contrast, NDPK-A S122D (AMPK site phosphomimic), S144A, and S144D each precipitated comparable amounts of AMPK activity (i.e., similar to the S122A mutant) (Fig. 3B) but were all observed to channel substrate normally (Fig. 3D, third to fifth bars). We interpret these data to indicate that firstly, the phosphorylation status of S122 on NDPK-A critically determines whether NDPK-A can channel ATP to AMPK α1 and that secondly, the phosphorylation status of S144 has no effect on substrate channeling.
In vivo phosphorylation studies.
As shown in Fig. 4A, we tested the in vivo phosphorylation status of NDPK-A under AMPK α1 and α2 knockout conditions using total mouse liver extracts from wild-type, AMPK α1-null, and AMPK α2-null tissue. We observed that the deletion of AMPK α1, but not AMPK α2, resulted in a significant reduction in NDPK-A phosphoserine content relative to that of the wild-type littermate control, with no detectable effect on the overall cellular quantity of NDPK-A (Fig. 4A, lower blot). Next, we investigated whether S122 and S144 are in vivo NDPK-A targets for AMPK. We transfected HepG2 human-derived liver cells with wild-type or the S122A, S144A, or S120A NDPK-A mutation using a mock (empty vector) transfection as a control. Residue S120 was chosen as a positive control, as this NDPK residue is a known target for protein kinase CK2 (see below). Each set of transfected cells was then either left untreated or exposed to 2 mM phenformin or 5 μg/ml oligomycin to stress cells and activate AMPK (Fig. 4B). Cytosolic cell extracts were immunoprecipitated using an NDPK-A antibody and probed using the phosphoserine-specific antibody as described above. The lower panel of Fig. 4B shows that untreated cells manifest a reduction in baseline phosphoserine content with respect to wild-type levels when S122, S144, or S120 residues were each singly mutated to alanine (top upper panel). In the phenformin- and oligomycin-treated cell extracts where AMPK was stimulated (confirmed by AMPK assay) (data not shown), we observed an increase in NDPK-A phosphoserine band intensity in wild-type, mock-transfected, and S120A mutated samples alone, with no increment under S122A and S144A mutated conditions. The histogram shown in Fig. 4C quantitates these data as the ratio of phosphoserine to total protein band intensity (LC lanes), as measured by densitometry from four independent experiments. In the upper panel of Fig. 4B, we demonstrate that only the mutation of serine 122 functionally correlates with ACC1 phosphorylation. Thus, S122A (but not S144A) fails to augment ACC1 phosphorylation relative to all other mutants after cell stress with either phenformin or oligomycin. This selectivity for S122 is consistent with our proposal that substrate channeling regulates ACC1 function.
For further in vivo confirmation, we studied the role of NDPK-A in the regulation of AMPK function, with the AMPK- and NDPK-A-null tissues probed using phosphorylation-specific antibodies against two established AMPK substrates, ACC1 and HSL. The former substrate regulates fatty acid fate, and the latter substrate provides the rate-limiting step in the release of fatty acids from triacylglycerol. The deletion of NDPK-A reduced the amount of phospho-ACC1 by approximately 45%, whereas the total amount of protein remained unchanged (see the histograms in Fig. 4D, lower right panels, which present the ratio of phosphoserine intensity to total protein as described above). The data confirm that whereas the AMPK α2 deletion has no significant effect on the phospho-ACC1 status, the deletion of AMPK α1 almost abolished phospho-ACC1 (by approximately 90%) (Fig. 4D, upper left and right panels). Conversely, when phospho-HSL levels were measured in mouse skeletal muscle cell cytosol, the deletion of AMPK α1 had little effect on phospho-HSL content with respect to the control, but the deletion of AMPK α2 almost abolished phosphorylation (approximately 95% with respect to the control). Deletion of NDPK-A resulted in a smaller reduction (approximately 20% with respect to the control) (Fig. 4D, lower left and right panels). The combined data are consistent with the notion that NDPK-A plays a functional role in AMPK α1, but not AMPK α2, function towards important physiological target proteins.
A single amino acid difference between NDPK-A and NDPK-B determines the interaction with AMPK α1.
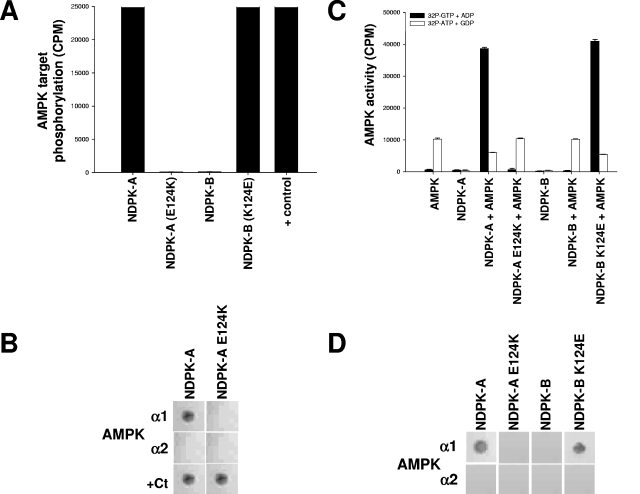
As described above, we found that AMPK α1 binds NDPK-A in the region of amino acids 111 to 125 (Fig. 2). Since this interaction is specific to NDPK-A (but not NDPK-B) with respect to AMPK α1, we reasoned that the specificity for NDPK-A might be dependent on sequence differences in this area relative to NDPK-B. A comparison of the relevant sequences showed identity apart from residue 124, which is a glutamic acid (E124) in NDPK-A but a lysine in NDPK-B (K124). The significant difference in charge prompted us to test the effect of a reciprocal mutation of NDPK-A and NDPK-B to their equivalent respective residues (E124K for NDPK-A and K124E for NDPK-B) on the observed interaction with AMPK α1. Figure 5A shows AMPK-dependent phosphorylation incorporation into NDPK-A, NDPK-A E124K (pseudo-NDPK-B), NDPK-B, and NDPK-B K124E (pseudo-NDPK-A) recombinant proteins and a positive phosphorylation control for the NDPK 124 mutants (E124K is shown, and K124E is not shown, but both were identical). This positive control was another known NDPK-targeting kinase, CK2, which was previously reported to target S120 on NDPK (2). Figure 5B shows an overlay binding matrix testing the ability of pure, recombinant AMPK α1 and AMPK α2 catalytic subunits to bind the NDPK-A or E124K mutant protein immobilized on nitrocellulose membranes. We observed that the E124K mutant abrogates the binding of AMPK α1 to NDPK-A. These mutations had no effect on binding to the positive control enzyme, CK2 (bottom panels). As shown in Fig. 5C, we tested the ability of the NDPK 124 mutants to channel substrate to or steal substrate from AMPK-directed SAMS phosphorylation using either [32P]GTP plus ADP (resulting in labeled, NDPK-generated ATP) or [32P]ATP plus GDP (resulting in labeled NDPK-generated GTP), respectively. We observe that whereas the NDPK-A E124K (pseudo-NDPK-B) mutant lost the ability to channel or steal substrate, the NDPK-B K124E (pseudo-NDPK-A) mutant has gained this ability. Importantly, the pseudo-NDPK-A mutant also manifests the ability to steal ATP away from AMPK α1 (Fig. 5C, last open bar) when GDP is present (6).
FIG. 5.
A single amino acid on NDPK-A determines its association with AMPK α1. (A) Pure, active rat liver AMPK was used to phosphorylate a selection of substrates, 15 μg (each) of NDPK-A, NDPK-A(E124K), NDPK-B, and NDPK-B(K124E). In the final lane, CK2α holoenzyme (+ control) was used to phosphorylate NDPK-A (shown) and NDPK-B (not shown) mutant recombinant proteins, and both were identical. (B) NDPK-A and NDPK-A E124K mutant recombinant proteins were immobilized on nitrocellulose membranes and then blocked/washed as described above for the Western blot protocol. Furthermore, 10 μg of purified, recombinant AMPK α1, AMPK α2, or CK2α protein was overlaid onto the membrane, with binding detected using isoform-specific antibodies for each protein, as described above for the Western blot protocol. (C) Substrate-channeling ability of NDPK mutants was tested by adding, where indicated, active rat liver AMPK to 15 μg of either NDPK-A, NDPK-A E124K, NDPK-B, or NDPK-B K124E recombinant protein. Either [32P]GTP plus ADP (resulting in labeled NDPK-generated ATP) or [32P]ATP plus GDP (resulting in labeled NDPK-generated GTP) was used to demonstrate substrate channeling or substrate stealing, respectively, and also to act as cross-contamination control for each kinase individually. (D) NDPK-A, NDPK-A E124K, NDPK-B, and NDPK-B K124E proteins were immobilized on nitrocellulose membranes and then blocked/washed as described above for the Western blot protocol. Furthermore, 10 μg of purified, recombinant AMPK α1 or AMPK α2 protein was overlaid onto the membrane, with binding detected using isoform-specific antibodies for each protein, as described above for the Western blot protocol.
Figure 5D shows an overlay binding matrix testing the ability of pure, recombinant AMPK α1 and AMPK α2 catalytic subunits to bind the NDPK-A, NDPK-A E124K, NDPK-B, or NDPK-B K124E protein immobilized on nitrocellulose membranes. Thus, we observed that whereas the NDPK-A E124K mutant abrogates both binding and phosphorylation by AMPK α1, the NDPK-B K124E mutant is able to both bind and channel substrate to AMPK α1.
DISCUSSION
The principal findings of this study are that for NDPK-A, the phosphorylation status of a single amino acid, S122, determines whether NDPK-A channels ATP to AMPK α1, thereby regulating AMPK α1 activity towards an important regulator of fatty acid metabolism, ACC1. Additionally, we demonstrate that the presence of a glutamic acid at position 124 is both necessary and sufficient for this interaction with AMPK α1. The absence of E124 in NDPK-B explains our recent demonstration that NDPK-A (but not NDPK-B) specifically interacts with AMPK α1 and that this interaction allows NDPK-A-generated ATP (from GTP plus ADP) to be preferentially used by AMPK α1 as a phosphate donor in downstream AMPK mechanisms (6). Importantly, we use the term “preferentially” to describe our finding that this locally generated ATP does not interact with ATP in bulk medium, suggesting that the NDPK-A/AMPK α1 complex partitions local ATP in some manner. Crucially, within this region, we find that NDPK-A only channels substrate and regulates ACC1, provided that S122 is capable of being phosphorylated or exists as a phosphomimic (Fig. 5). Thus, AMPK α1 can be regulated by another protein in the absence of AMP.
Parallel data using null tissue further confirm the specificity of the NDPK-A/AMPK α1 interaction when liver cytosols from wild-type, NDPK-null, AMPK α1-null, and AMPK α2-null mouse tissue were compared (1, 30). We demonstrate that in the absence of AMPK α1 (but not AMPK α2), NDPK is unable to coprecipitate AMPK-SAMS activity. Conversely, in the absence of NDPK-A, substrate channeling does not occur, and only basal AMPK α1 activity (autophosphorylation) is present. Since gross amounts of AMPK α1 proteins are unaffected by the loss of NDPK-A, the absence of substrate channeling is likely to be functionally significant.
Functional relevance was investigated in two ways. First, to isolate the specific regions/residues of NDPK-A targeted by AMPK, we used four peptides corresponding to exposed regions of NDPK-A and found that AMPK was able to phosphorylate NDPK-A in only two regions, NDPKpep3 and NDPKpep4 (Fig. 2 and 5). Overlay binding analysis of these peptides revealed that AMPK α1 binds only the NDPKpep3 peptide. Second, the functional relevance of this interacting region was confirmed using ACC1 and NDPK-A precipitations (Fig. 2, right panel) to show that incubation with a molar excess of NDPKpep3 could disrupt the complex.
As shown in Fig. 3, we observed that within NDPKpep3, S122 and S144 on NDPK-A are AMPK targets in vitro. This notion was extended in vivo by immunoprecipitating NDPK-A from a human-derived liver cell line (HepG2) transfected with wild-type and AMPK-targeted point-mutated variants of NDPK-A (Fig. 4). These cells were stressed with two different stimuli (phenformin and oligomycin) to activate AMPK. We observed that for untreated samples, the loss of S122, S144, and S120 resulted in the expected reduction in phosphoserine band intensity should they be bona fide in vivo targets for the relevant kinases. Analysis of the phenformin/oligomycin-treated cells demonstrated an increase in NDPK-A phosphoserine band intensity under wild-type, mock-transfected, and S120A conditions, but crucially, there was no change in the S122A and S144A mutants. Thus, we propose that S122 and S144 in NDPK-A are AMPK targets in vivo. Although further work will have to establish the role of S144 phosphorylation, the phosphorylation status of S122 may be important for the function of AMPK towards the in vivo target ACC1. This notion is supported by the differential change in the phosphorylation status of two established AMPK substrates (ACC1 and HSL) by comparing wild-type and NDPK-A-null tissues. We observed that the loss of NDPK-A from liver cytosol results in a reduction (∼45%) in phospho-ACC1 compared with the wild type (Fig. 4D). Conversely, we observed that an AMPK α1 knockout results in an almost total loss of ACC1 phosphorylation (Fig. 4D, upper left panel). The combined data indicate that while AMPK α1 is the primary driver of ACC1 phosphorylation in liver cytosol, NDPK-A is also responsible for a significant portion of that phosphorylation (approximately 45% compared to that of the wild type). Interestingly, the phosphorylation status of HSL in skeletal muscle is predominantly AMPK α2 dependent (32), and as such, the loss of NDPK-A might not be expected to have any major effect on phospho-HSL levels. Nevertheless, the loss of NDPK-A reduced phospho-HSL by ∼15 to 20% (Fig. 4D, lower panels). The significance of this unexpected finding is beyond the scope of the current investigation.
The charge on the amino acid at position 124 on NDPK-A is negative (but is positive in NDPK-B), and we find this to be a critical determinant of the interaction with AMPK α1 (Fig. 5). The glutamic acid at position 124 is highly conserved among NDPK isoforms, apart from NDPK-B. Thus, further work will have to determine whether NDPK-C, NDPK-D, etc., can also channel substrate via AMPK α1. Point mutation of residue 124 reciprocally (i.e., NDPK-A E124K and NDPK-B K124E) resulted in an NDPK-A mutant that could no longer bind or channel substrate to AMPK α1 and, conversely, an NDPK-B “pseudo-A” mutant that could now bind and channel substrate to AMPK α1. The generation of reciprocal pseudomutants of NDPK-A and NDPK-B allows us to speculate that NDPK-B may be channeling substrate in vivo to unknown associated proteins (similar to our reported NDPK-A/AMPK α1 interaction) by virtue of the fact that the core channeling mechanism remains intact (manifest by the ability of NDPK-B to channel substrate) (Fig. 5C). Such latent binding partners could be crucial for our understanding of unexplained differences between NDPK-A and NDPK-B functions. Further work will have to establish the significance of the finding that S122 is also present in NDPK-B but that this form fails to bind AMPK α1.
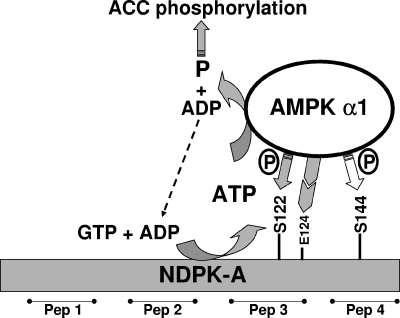
Figure 6 crystallizes our observations and presents a working model showing that AMPK α1-dependent phosphorylation at S122 on NDPK-A determines whether NDPK-A is able to channel ATP to AMPK α1 and that E124 is required before this isoform-specific protein-protein interaction can proceed.
FIG. 6.
Model of interaction between AMPK α1 and NDPK-A. The phosphorylation status of residue S122 on NDPK-A determines whether NDPK-A channels ATP derived from GTP plus ADP to AMPK α1 as the substrate in the phosphorylation of downstream AMPK targets, for example, ACC1. Mutation of residue E124K on NDPK-A eliminates the specific interaction of NDPK-A with AMPK α1 (not shown). Serine 144 was also identified as an AMPK α1 target, but experiments revealed that its phosphorylation status has no bearing on substrate channeling or ACC1 phosphorylation. Peptides (Pep) 1 to 4 refer to surface regions of NDPK-A.
Historically, the previously reported actions of AMPK have been described after pharmacological activation by 5-amino-4-imidazolecarboxamide riboside (13, 20), an AMP-mimetic agent that activates both α1 and α2 catalytic isoforms. In order to address this limitation, AMPK α1 and α2 knockout mice were recently generated (30). Mice lacking the α1 isoform present no gross defect in glucose homeostasis, whereas mice lacking the α2 isoform exhibit glucose intolerance and reduced insulin sensitivity (31). Therefore, investigation into the α2 knockout has proceeded apace, resulting in the establishment of a role for AMPK α2 in glucose homeostasis. This, taken alongside the known nuclear preponderance and greater AMP dependence of the α2 isoform, suggests that AMPK α2 functions in both short-term ATP conservation and long-term energy homeostasis and gene regulation (26). Adding to the emerging variance in function between AMPK catalytic subunit isoforms, our data reveal a specific dynamic for AMPK α1 as part of a novel complex with NDPK-A and, crucially, the critical controller of fatty acid fat, ACC1. We speculate that this complex can rationalize and respond to levels of GTP additively to ATP (and possibly other triphosphates, although this remains to be tested), altering AMPK α1 activity without changes in AMP and irrespective of bulk cytosol ATP. We also report a downstream in vivo consequence in the control of cellular fatty acid metabolism. This finding is consistent with our previous finding that phosphate can be transferred from GTP to ACC1 via substrate channeling through NDPK-A (6). This report also establishes that the AMPK α1 isoform is functionally linked to NDPK-A in a native tissue and that this complex is able to channel substrate towards only one of the two isoforms of AMPK that are involved in energy conservation. We suggest that substrate channeling could maintain AMPK activity under conditions of local ATP depletion (6). It is therefore not unreasonable to propose that the two catalytic isoforms of AMPK might differentially address both long- and short-term energy homeostasis by distinct mechanisms.
Mice carrying a homozygous germ line-null mutation in the NM23-M1 gene (equivalent murine gene product to NDPK-A) have an unexplained phenotype. NDPK-A-null mice are smaller (20 to 25% lower weight than wild-type controls), display a high rate of neonatal mortality, and manifest altered mammary gland function in surviving adult females. Our previous work showed that ACC, a key regulator of fat metabolism, was part of the NDPK-A/AMPK α1 complex and that the substrate-channeling phenomenon described here altered the phosphorylation of ACC bidirectionally (6). Our present work demonstrates that this complex can be disrupted by peptides corresponding to a small part of the NDPK protein (Fig. 2). Since fat metabolism is essential for weight control and milk formation, we hypothesize that the weight reduction in NDPK-A-null mice might be linked to the loss of the NDPK-A/AMPK α1 complex mediated by the aberrant control of ACC and thus fat metabolism. Furthermore, since NDPK function is related to membrane turnover (15), fatty acid synthesis (10), breast cancer metastasis (19), and mammary development (4), this might explain defective mammary gland function in pregnant females who fail to feed their pups. Thus, our findings have a wide biological applicability given that NDPK-A function is also critical for the metastatic potential of breast cancer cells.
In Fig. 6, we model our understanding of the interaction of AMPK α1 with NDPK-A. We have incorporated our observation that an initial AMPK phosphorylation of S122 primes the NDPK-A/AMPK α1 complex towards substrate channeling and thus ATP conservation. Our findings shed light on molecular interactions of two critical cellular energy regulators, which could contribute to our understanding of the mechanism by which AMPK protects against cellular stress by decreasing fatty acid synthesis and increasing fatty acid oxidation. We propose that this is accomplished via NDPK-regulated AMPK-mediated phosphorylation of ACC (6), creating a novel “fat controller.” Additionally, we propose that AMPK, the cellular fuel gauge, acquires an alternative fuel source in GTP via NDPK-A when cells are under stress.
Acknowledgments
We are grateful to Grahame Hardie, David Carling, Angela Woods, and Marie-Lise Lacombe for constructs, antibodies, and reagents.
We thank the Wellcome Trust (075237/Z/04/Z) and the Cystic Fibrosis Trust (PJ439) for their support. Local university support was provided for K.J.T.
There are no conflicts of interest.
REFERENCES
- 1.Arnaud-Dabernat, S., P. M. Bourbon, A. Dierich, M. Le Meur, and J. Y. Daniel. 2003. Knockout mice as model systems for studying nm23/NDP kinase gene functions. Application to the nm23-M1 gene. J. Bioenerg. Biomembr. 35:19-30. [DOI] [PubMed] [Google Scholar]
- 2.Biondi, R. M., M. Engel, M. Sauane, C. Welter, O. G. Issinger, L. Jimenez de Asua, and S. Passeron. 1996. Inhibition of nucleoside diphosphate kinase activity by in vitro phosphorylation by protein kinase CK2. Differential phosphorylation of NDP kinases in HeLa cells in culture. FEBS Lett. 399:183-187. [DOI] [PubMed] [Google Scholar]
- 3.Bosnar, M. H., J. De Gunzburg, R. Bago, L. Brecevic, I. Weber, and J. Pavelic. 2004. Subcellular localization of A and B Nm23/NDPK subunits. Exp. Cell Res. 298:275-284. [DOI] [PubMed] [Google Scholar]
- 3a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Caligo, M. A., G. Cipollini, L. Fiore, S. Calvo, F. Basolo, P. Collecchi, F. Ciardiello, S. Pepe, M. Petrini, and G. Bevilacqua. 1995. NM23 gene expression correlates with cell growth rate and S-phase. Int. J. Cancer 60:837-842. [DOI] [PubMed] [Google Scholar]
- 5.Crawford, R. M., H. J. Ranki, C. H. Botting, G. R. Budas, and A. Jovanovic. 2002. Creatine kinase is physically associated with the cardiac ATP-sensitive K+ channel in vivo. FASEB J. 16:102-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford, R. M., K. J. Treharne, O. G. Best, R. Muimo, C. E. Riemen, and A. Mehta. 2005. A novel physical and functional association between nucleoside diphosphate kinase H1 and AMP-activated protein kinase alpha1 in liver and lung. Biochem. J. 392:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Crawford, R. M., K. J. Treharne, O. G. Best, C. E. Riemen, R. Muimo, D. C. Gruenert, S. Arnaud-Dabernat, J. Y. Daniel, and A. Mehta. 8February2006, posting date. NDPK-A (but not NDPK-B) and AMPK α1 (but not AMPK α2) bind the cystic fibrosis transmembrane conductance regulator in epithelial cell membranes. Cell. Signal. [Online.] doi: 10.1016/j.cellsig.2006.01.001. [DOI] [PubMed]
- 8.Dale, S., W. A. Wilson, A. M. Edelman, and D. G. Hardie. 1995. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 361:191-195. [DOI] [PubMed] [Google Scholar]
- 9.Engel, M., M. Veron, B. Theisinger, M. L. Lacombe, T. Seib, S. Dooley, and C. Welter. 1995. A novel serine/threonine-specific protein phosphotransferase activity of Nm23/nucleoside-diphosphate kinase. Eur. J. Biochem. 234:200-207. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda, M., A. Ishii, Y. Yasutomo, N. Shimada, N. Ishikawa, N. Hanai, N. Nagata, T. Irimura, G. L. Nicolson, and N. Kimura. 1996. Decreased expression of nucleoside diphosphate kinase alpha isoform, an nm23-H2 gene homolog, is associated with metastatic potential of rat mammary-adenocarcinoma cells. Int. J. Cancer 65:531-537. [DOI] [PubMed] [Google Scholar]
- 11.Hardie, D. G., and D. Carling. 1997. The AMP-activated protein kinase—fuel gauge of the mammalian cell? Eur. J. Biochem. 246:259-273. [DOI] [PubMed] [Google Scholar]
- 12.Hardie, D. G., and S. A. Hawley. 2001. AMP-activated protein kinase: the energy charge hypothesis revisited. Bioessays 23:1112-1119. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi, T., M. F. Hirshman, E. J. Kurth, W. W. Winder, and L. J. Goodyear. 1998. Evidence for 5′ AMP-activated protein kinase mediation of the effect of muscle contraction on glucose transport. Diabetes 47:1369-1373. [DOI] [PubMed] [Google Scholar]
- 14.Heidbuchel, H., G. Callewaert, J. Vereecke, and E. Carmeliet. 1992. Membrane-bound nucleoside diphosphate kinase activity in atrial cells of frog, guinea pig, and human. Circ. Res. 71:808-820. [DOI] [PubMed] [Google Scholar]
- 15.Ishijima, Y., N. Shimada, M. Fukuda, H. Miyazaki, N. Y. Orlov, T. G. Orlova, T. Yamada, and N. Kimura. 1999. Overexpression of nucleoside diphosphate kinases induces neurite outgrowth and their substitution to inactive forms leads to suppression of nerve growth factor- and dibutyryl cyclic AMP-induced effects in PC12D cells. FEBS Lett. 445:155-159. [DOI] [PubMed] [Google Scholar]
- 16.Jorgensen, S. B., B. Viollet, F. Andreelli, C. Frosig, J. B. Birk, P. Schjerling, S. Vaulont, E. A. Richter, and J. F. Wojtaszewski. 2004. Knockout of the alpha2 but not alpha1 5′-AMP-activated protein kinase isoform abolishes 5-aminoimidazole-4-carboxamide-1-beta-4-ribofuranosidebut not contraction-induced glucose uptake in skeletal muscle. J. Biol. Chem. 279:1070-1079. [DOI] [PubMed] [Google Scholar]
- 17.Kahn, B. B., T. Alquier, D. Carling, and D. G. Hardie. 2005. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 1:15-25. [DOI] [PubMed] [Google Scholar]
- 18.Konig, J., R. Schreiber, M. Mall, and K. Kunzelmann. 2002. No evidence for inhibition of ENaC through CFTR-mediated release of ATP. Biochim. Biophys. Acta 1565:17-28. [DOI] [PubMed] [Google Scholar]
- 19.MacDonald, N. J., A. De la Rosa, M. A. Benedict, J. M. Freije, H. Krutsch, and P. S. Steeg. 1993. A serine phosphorylation of Nm23, and not its nucleoside diphosphate kinase activity, correlates with suppression of tumor metastatic potential. J. Biol. Chem. 268:25780-25789. [PubMed] [Google Scholar]
- 20.Merrill, G. F., E. J. Kurth, D. G. Hardie, and W. W. Winder. 1997. AICA riboside increases AMP-activated protein kinase, fatty acid oxidation, and glucose uptake in rat muscle. Am. J. Physiol. 273:E1107-E1112. [DOI] [PubMed] [Google Scholar]
- 21.Muimo, R., Z. Hornickova, C. E. Riemen, V. Gerke, H. Matthews, and A. Mehta. 2000. Histidine phosphorylation of annexin I in airway epithelia. J. Biol. Chem. 275:36632-36636. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen, J. N., K. J. Mustard, D. A. Graham, H. Yu, C. S. MacDonald, H. Pilegaard, L. J. Goodyear, D. G. Hardie, E. A. Richter, and J. F. Wojtaszewski. 2003. 5′-AMP-activated protein kinase activity and subunit expression in exercise-trained human skeletal muscle. J. Appl. Physiol. 94:631-641. [DOI] [PubMed] [Google Scholar]
- 23.Postel, E. H. 1998. NM23-NDP kinase. Int. J. Biochem. Cell Biol. 30:1291-1295. [DOI] [PubMed] [Google Scholar]
- 24.Ranki, H. J., R. M. Crawford, G. R. Budas, and A. Jovanovic. 2002. Ageing is associated with a decrease in the number of sarcolemmal ATP-sensitive K+ channels in a gender-dependent manner. Mech. Ageing Dev. 123:695-705. [DOI] [PubMed] [Google Scholar]
- 25.Roymans, D., R. Willems, D. R. Van Blockstaele, and H. Slegers. 2002. Nucleoside diphosphate kinase (NDPK/NM23) and the waltz with multiple partners: possible consequences in tumor metastasis. Clin. Exp. Metastasis 19:465-476. [DOI] [PubMed] [Google Scholar]
- 26.Salt, I., J. W. Celler, S. A. Hawley, A. Prescott, A. Woods, D. Carling, and D. G. Hardie. 1998. AMP-activated protein kinase: greater AMP dependence, and preferential nuclear localization, of complexes containing the alpha2 isoform. Biochem. J. 334:177-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schreiber, R., A. Hopf, M. Mall, R. Greger, and K. Kunzelmann. 1999. The first-nucleotide binding domain of the cystic-fibrosis transmembrane conductance regulator is important for inhibition of the epithelial Na+ channel. Proc. Natl. Acad. Sci. USA 96:5310-5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stapleton, D., K. I. Mitchelhill, G. Gao, J. Widmer, B. J. Michell, T. Teh, C. M. House, C. S. Fernandez, T. Cox, L. A. Witters, and B. E. Kemp. 1996. Mammalian AMP-activated protein kinase subfamily. J. Biol. Chem. 271:611-614. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan, J. E., F. Carey, D. Carling, and R. K. Beri. 1994. Characterisation of 5′-AMP-activated protein kinase in human liver using specific peptide substrates and the effects of 5′-AMP analogues on enzyme activity. Biochem. Biophys. Res. Commun. 200:1551-1556. [DOI] [PubMed] [Google Scholar]
- 30.Viollet, B., F. Andreelli, S. B. Jorgensen, C. Perrin, D. Flamez, J. Mu, J. F. Wojtaszewski, F. C. Schuit, M. Birnbaum, E. Richter, R. Burcelin, and S. Vaulont. 2003. Physiological role of AMP-activated protein kinase (AMPK): insights from knockout mouse models. Biochem. Soc. Trans. 31:216-219. [DOI] [PubMed] [Google Scholar]
- 31.Viollet, B., F. Andreelli, S. B. Jorgensen, C. Perrin, A. Geloen, D. Flamez, J. Mu, C. Lenzner, O. Baud, M. Bennoun, E. Gomas, G. Nicolas, J. F. Wojtaszewski, A. Kahn, D. Carling, F. C. Schuit, M. J. Birnbaum, E. A. Richter, R. Burcelin, and S. Vaulont. 2003. The AMP-activated protein kinase alpha2 catalytic subunit controls whole-body insulin sensitivity. J. Clin. Investig. 111:91-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watt, M. J., A. G. Holmes, S. K. Pinnamaneni, A. P. Garnham, G. R. Steinberg, B. E. Kemp, and M. A. Febbraio. 2006. Regulation of HSL serine phosphorylation in skeletal muscle and adipose tissue. Am. J. Physiol. Endocrinol. Metab. 290:E500-E508. [DOI] [PubMed] [Google Scholar]
- 33.Woods, A., I. Salt, J. Scott, D. G. Hardie, and D. Carling. 1996. The alpha1 and alpha2 isoforms of the AMP-activated protein kinase have similar activities in rat liver but exhibit differences in substrate specificity in vitro. FEBS Lett. 397:347-351. [DOI] [PubMed] [Google Scholar]