Abstract
Eukaryotic genomes are divided into independent transcriptional domains by DNA elements known as insulators. The gypsy insulator, a 350-bp element isolated from the Drosophila gypsy retrovirus, contains twelve degenerate binding sites for the Suppressor of Hairy-wing [Su(Hw)] protein. Su(Hw) associates with over 500 non-gypsy genomic sites, the functions of which are largely unknown. Using a bioinformatics approach, we identified 37 putative Su(Hw) insulators (pSIs) that represent regions containing clustered matches to the gypsy insulator Su(Hw) consensus binding sequence. The majority of these pSIs contain fewer than four Su(Hw) binding sites, with only seven showing in vivo Su(Hw) association, as demonstrated by chromatin immunoprecipitation. To understand the properties of the pSIs, these elements were tested for enhancer-blocking capabilities using a transgene assay system. In a complementary set of experiments, effects of the pSIs on transcriptional regulation of genes at the natural genomic location were determined. Our data suggest that pSIs have complex genomic functions and, in some cases, establish insulators. These studies provide the first direct evidence that the Su(Hw) protein contributes to the regulation of gene expression in the Drosophila genome through the establishment of endogenous insulators.
Genes with similar expression profiles are clustered in eukaryotic genomes (5, 38, 39, 43, 73). These observations suggest that these genomes are divided into discrete domains that facilitate the regulation of gene expression. Functional domains may be established by assembly of higher-order chromatin structures that constrain the activity of enhancers and silencers to ensure that these transcriptional elements interact with the appropriate target promoter. The demarcation of chromatin domains is associated with a conserved class of DNA elements known as insulators (6, 13, 17, 41, 61, 75, 79). Insulators possess two functional properties. First, they block the action of enhancers and silencers when placed between these regulatory elements and a promoter. Second, they protect gene expression from chromosomal position effects that result from ectopic placement of genes in the genome. These blocking effects are accomplished without inactivation of the intrinsic properties of any of the regulatory elements, implying that insulators disrupt signaling between enhancers, silencers, and promoters (10, 66). The mechanism by which insulators establish independent functional domains is unclear, but it might depend upon interactions between insulators and/or nuclear substructures to form loop domains that limit the action of transcriptional regulatory elements (9, 23, 78).
One of the best-characterized insulators is the Drosophila gypsy insulator. This element resides within the 5′-untranslated region of the gypsy retrotransposon (33). The gypsy insulator contains twelve clustered, degenerate repeats of a consensus sequence, 5′-YRYTGCATAYYY-3′, where Y represents a pyrimidine and R represents a purine residue, that bind the Suppressor of Hairy-wing [Su(Hw)] protein (72). The number and spacing of these Su(Hw) sites are determinants of enhancer blocking, with a minimum of four clustered sites required to reconstitute an insulator (28, 31, 65). At least twenty different enhancers have been tested and found to be blocked by the gypsy insulator, indicating that this insulator is active at different times and in distinct tissues throughout Drosophila development (24, 29, 31, 36, 37). The gypsy insulator also blocks repressive effects from silencers, such as the Polycomb group of proteins (45, 62, 69). Taken together, these findings suggest that the gypsy insulator has a general capacity to prevent communication between long-range transcriptional regulatory elements and promoters.
At least three proteins are required for gypsy insulator function. The Su(Hw) protein binds the gypsy insulator through a twelve-zinc-finger DNA binding domain. This protein recruits two BTB/POZ (broad-complex, tramtrack, and bric-a-brac/poxvirus and zinc finger proteins) domain proteins, Modifier of mdg4 (Mod) 67.2 and Centrosomal protein (CP) 190, through direct protein interactions (20, 27, 53). The gypsy insulator proteins localize to over five hundred genomic sites within the Drosophila polytene chromosomes, with <1% of these sites corresponding to sites of gypsy retrotransposon insertion. Su(Hw) and Mod67.2 colocalize at nearly all of these sites, whereas CP190 is found at ∼70% of the Su(Hw) sites (53). These observations suggest that the non-gypsy Su(Hw) binding sites represent endogenous insulators.
At present, little is known about the properties of genomic Su(Hw) binding sites that are outside of the gypsy retrotransposon. The first identified site, named 1A2 for its cytological location, was determined to reside within an intergenic region located between the independently regulated yellow and achaete genes (28, 54). Transgene assays demonstrated that 1A2 has enhancer-blocking activity. These observations led to the proposal that 1A2 represents an endogenous Su(Hw) insulator that defines the regulatory independence of the yellow and achaete genes (54), a postulate that has not been directly tested.
To identify additional non-gypsy Su(Hw) binding sites, we used a bioinformatic approach to search for genomic regions that contain clusters of a Su(Hw) binding site that matched the gypsy consensus. We identified 37 putative Su(Hw) insulators (pSIs). Of these, 31 contained fewer than four Su(Hw) binding sites, indicating that the arrangement of endogenous Su(Hw) sites is not similar to that found in the gypsy insulator. The in vivo and in vitro properties of the pSIs were characterized. Through chromatin immunoprecipitation (ChIP) studies, we determined that a subset of pSIs were associated with the Su(Hw) protein in vivo. Electrophoretic mobility shift assay (EMSA) analyses demonstrated that the in vivo Su(Hw) occupancy did not correlate with the apparent in vitro DNA binding affinity of the Su(Hw) protein, implying that determinants in addition to the presence of a Su(Hw) binding site influence Su(Hw) localization to chromosomes. The insulator effects of the pSIs were tested using two assays. First, we used transgene analyses to determine whether the endogenous sequences conferred enhancer blocking, employing the yellow gene as a reporter. Second, we determined the role of the pSIs within the natural location, by examining effects of the loss of the Su(Hw) and Mod67.2 insulator proteins on expression of genes adjacent to the pSIs. Our studies demonstrate that the nature of endogenous Su(Hw) binding sites is complex, with some, but not all, establishing endogenous insulators.
MATERIALS AND METHODS
Computer search and analysis of Su(Hw) binding sites.
The FLY ENHANCER program was used to screen the Drosophila melanogaster genome (release 1) for clusters of Su(Hw) binding sites (47). Gene models surrounding identified clusters were updated to release 4 of the genome using Flybase and GenePalette software (University of California) (58). Neighboring binding sites that deviated from the gypsy consensus sequence were identified using GenePalette and the European Molecular Biology Open Software Suite. Regions containing clusters were PCR amplified out of genomic DNA from Canton S flies, cloned into the Topo II vector (Invitrogen), and sequenced. The pSIs ranged in size from 364 to 889 bp. Matrix attachment region (MAR) analysis was performed using MAR-Wiz (Futuresoft) (70).
Analysis of in vitro Su(Hw) protein binding.
Full-length Su(Hw) protein was expressed and purified from Escherichia coli DE3 cells. The su(Hw) cDNA was cloned into a modified pET21a expression vector (Novagen) that contained an amino-terminal T7 tag and a carboxy-terminal FLAG tag, followed by a six-His tag. Expression of Su(Hw) was induced with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and cells were grown at 18°C overnight. After harvesting, cells were lysed by sonication, and the lysate was cleared by centrifugation at 45,000 rpm. Purification of the Su(Hw) protein involved two chromatography steps, first with a Ni-nitrilotriacetic acid resin (QIAGEN) and then by salt elution from a Mono Q column.
Apparent DNA binding affinities were determined using an EMSA. pSI DNA fragments were PCR amplified from Canton S genomic DNA, cloned into Topo vectors (Invitrogen), EcoRI digested to release the pSI, and end labeled using 32P-dATP and Klenow enzyme. For each reaction, 2 fmol of labeled DNA (∼1,000 to 10,000 cpm) were incubated with 0, 0.003, 0.01, 0.03, 0.1, 0.3, and 1 μg of Su(Hw) protein in 25 mM HEPES, 200 mM KCl, 10 μM ZnCl2, 5 mM dithiothreitol, 1 μg poly(dI-dC) and 15% glycerol. After 15 min of incubation at room temperature, reaction products were separated by electrophoresis overnight on a 1% agarose 0.1× Tris-borate-EDTA gel at 4°C. Gels were dried and analyzed by autoradiography. Counts in bound and unbound bands were measured using an Instant Imager (Packard). The apparent association constants (M−1) were determined by nonlinear least-squares analysis of a Langmuir binding equation for noncooperative binding using Kaleidagraph (Synergy Software) (40).
Chromatin immunoprecipitation.
Chromatin was prepared from either third-instar larvae or 0- to 12-h embryos. Larval chromatin was prepared as described previously (54). For embryonic chromatin, embryos were homogenized in grinding buffer (15 mM HEPES, pH 7.6, 10 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 350 mM sucrose, 1 mM dithiothreitol, 1× protease inhibitor cocktail [Roche], and 0.1 mg/ml phenylmethylsulfonyl fluoride) and centrifuged twice at 5,000 × g to isolate nuclei. The nuclear suspension (∼109 nuclei/ml) was cross-linked with 1% formaldehyde at room temperature for 5 min. Nuclei were washed and lysed, and the chromatin was sheared to an average length of ∼700 bp by sonication. In each ChIP experiment, a chromatin solution containing ∼20 μg of genomic DNA was incubated with either specific or nonspecific antibody. Immunoprecipitation and washing were performed as described previously (54). The precipitated products were analyzed by PCR followed by polyacrylamide gel electrophoresis, ethidium bromide staining, and quantitation of digital fluorescent images. For input PCRs, 0.1% of DNA applied to a ChIP reaction was used as template in a 50-μl reaction volume. PCRs were kept within the linear range of amplification as determined by a 2 (±0.3)-fold increase in product amount between duplicate reactions offset by one cycle difference. To determine the percentage of input (Fig. 1), PCR products were amplified from at least three separate immunoprecipitation products from at least two different chromatin preparations. The Su(Hw) ChIPs used a rabbit anti-Su(Hw) antibody described previously (54). For the Mod67.2 ChIP, an affinity-purified chicken antibody directed against residues 403 to 610 of the Mod67.2 was utilized. Normal rabbit immunoglobulin G (IgG; Sigma) and chicken preimmune IgG were used as nonspecific antibody controls. Antibody chromatin complexes were captured with either protein A-Sepharose beads (Sigma) or anti-IgY conjugated agarose beads (PrecipHen; Aves Labs). Primer sequences used in PCR for the ChIP analysis will be provided upon request.
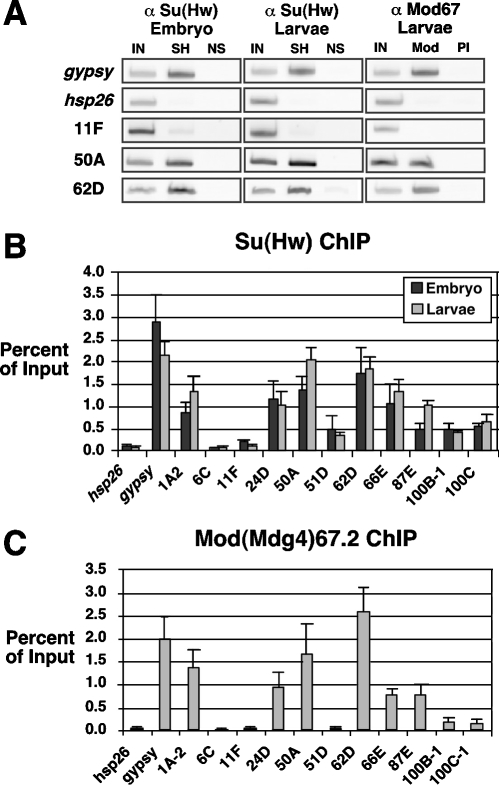
FIG. 1.
ChIP analysis of pSI-binding sites. (A) Representative examples of PCR products obtained from chromatin material that was directly purified (input [IN]) or immunoprecipitated with Su(Hw) (SH), nonspecific rabbit antibody (NS), Mod(mdg4)67.2 antibody (Mod), or preimmune antibody (PI). PCR products were obtained using primers specific to the gypsy insulator, the hsp26 coding region, and the pSIs 11F, 50A, and 62D. (B and C) The percentage of input was determined by quantifying the intensity of the PCR product in the antibody-enriched fraction relative to the input. Averages of at least three ChIP experiments are shown. Enrichment resulting from the Su(Hw) antibody is shown in panel B, using chromatin isolated from embryos (black bars) and third-instar larvae (gray bars). Enrichment resulting from the Mod(mdg4) antibody is shown in panel C, using chromatin isolated from larvae.
Enhancer-blocking assay.
The yellow gene was used as a reporter of the enhancer-blocking capability of the pSIs. pSIs isolated from Canton S genomic DNA were cloned between direct repeats of loxP sites and inserted into the yellow gene at position −900 bp relative to the start site of transcription. In the case of the pSI at cytological location 62D, the pSI was directly cloned into the yellow gene. P element vectors carrying a modified yellow gene and the mini-white gene were injected into the host y1w67c23 strain. Only transgenic lines with single transposon insertions were established and analyzed.
Phenotypes were determined by crossing transgenic males to y1w67c23 females and scoring pigmentation in the wing and body cuticle of 3-day-old female progeny. Flies were raised on standard cornmeal and agar medium at 25°C with 70% humidity. Pigmentation scores were determined by comparison with progeny from parallel standard control crosses as previously described, where 1 represents the null phenotype and 5 represents the wild-type phenotype (49). Insulator activity of the pSI was confirmed by comparing the cuticle coloration of flies that had a pSI-containing transposon with that of flies carrying a derivative transposon inserted at the same genomic location that was deleted for the pSI. These derivative lines were obtained by crossing the transgenic line to a stock expressing the Cre recombinase as described previously (14). Confirmation of excision was verified by either Southern blot analysis or PCR on genomic DNA.
Real-time PCR.
RNA for real-time PCR experiments was isolated from larvae and pupae of three genotypes. First, the control flies carried the y2 allele, in which a gypsy retrotransposon was located between the wing and body enhancers and the promoter of the yellow gene. Second, one set of experimental flies were homozygous for the su(Hw)v allele, in which the promoters of the divergent su(Hw) and rpII15 genes are deleted (34). As rpII15 is an essential gene, a P element rescue construct containing rpII15 was integrated onto the su(Hw)v chromosome to permit recovery of su(Hw)v homozygotes. To generate a stock in which homozygous su(Hw)v animals could be identified phenotypically in the larval and pupal stages, the su(Hw)f chromosome in the su(Hw)v/su(Hw)f stock was replaced with the TM6B balancer chromosome that carries the dominant Tubby (Tb1) allele. Tb1 larvae and pupae have a shortened, fatter (tubby) phenotype than those of the wild type. Virgin females of the genotype y2 v f1; su(Hw)v, P[v+, rpII15]/TM6B, Tb1 were crossed to y2 v f1; su(Hw)v/TM6B, Tb1 males. Larval and pupal y2 v f1; su(Hw)v, P[v+, rpII15]/su(Hw)v progeny were selected by their nontubby phenotype. The su(Hw)v, P[v+, rpII15] chromosome is homozygous lethal, necessitating the cross to a stock with a different su(Hw)v chromosome. Third, the second set of experimental flies were y− ac− w1118; mod(mdg4)u1/mod(mdg4)u1, in which a Stalker retrotransposon insertion into the mod(mdg4) locus introduces a premature stop codon, eliminating production of the Mod67.2 insulator-specific isoform (20, 22).
Total RNA was isolated from larvae that were collected 6 days after egg laying, and pupae that were collected on 8 and 10 days after egg laying. RNA was isolated from 120 larvae or pupae per genotype using TRIzol reagent (GibcoBRL), according to the manufacturer's instructions. Genomic DNA was removed by treatment with amplification grade DNase I (1 U per 10 μg; Invitrogen) followed by purification with a QIAGEN RNeasy kit. RNA was reverse transcribed into cDNA with a QIAGEN Omniscript kit. Each 20-μl reaction volume included 2 μg of RNA, 1 μM oligo-dT primer, 0.5 mM deoxynucleoside triphosphates, 10 U of RNase-out RNase inhibitor (Invitrogen), and 4 U of Omniscript. Reactions were performed for 50 min at 45°C followed by 10 min at 65°C to inactivate the reverse transcriptase.
Real-time PCR was performed using an iCycler Thermal Cycler and iQ SYBR green Supermix (Bio-Rad Laboratories). Reactions were performed in a 20-μl volume, using 0.05 μg of cDNA template and 200 nM forward and reverse primers. Due to the low abundance of the Or24a and CG32853 transcripts, 0.2 μg of cDNA was used per reaction. Primer sequences will be provided upon request. Thermocycler conditions were as follows: 95°C for 2.5 min and then 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds for 40 cycles. Following the amplification process, a melt curve was generated between 55°C and 95°C, with a reading every 0.5°C to confirm that a single PCR product was obtained. For each experiment, duplicate or triplicate reactions were performed and averaged, with the standard deviation among the replicates being no greater than 0.5 cycle threshold (CT). At least three independent experiments were performed for each primer set, using at least two independent RNA samples.
The fold change in expression of each gene in su(Hw) and mod(mdg4) mutants relative to the y2 control was determined with the ΔΔCT method (Applied Biosystems; Prism 7700 Users Bulletin no. 2), using Ras2 as an internal control for data normalization. A two-tailed Student t test determined whether the fold change in expression of each gene was statistically different than that of the RpL32 negative control, assumed to be unchanged in all stages of development and mutant backgrounds. Changes in gene expression (increase or decrease) were considered significant if greater than twofold with a P value of <0.05 compared to RpL32. Results were also calculated using a method that incorporates the efficiency of each primer set (55). Values obtained differed from those obtained by the ΔΔCT method by less than 10%, and genes that displayed a significant change (>2-fold, with P values of <0.05) were identical to those determined by the ΔΔCT method.
RESULTS
Bioinformatic approaches have been used to identify cis-regulatory sequences, such as enhancers, using the principle that these control elements are compact and include closely spaced binding sites for the same or multiple transcription factors (2, 3, 18, 46, 47, 59, 67). While less is known about the organization of insulators, several, including the gypsy insulator, are 0.5 to 1 kb in length and contain clusters of factor binding sites, implying that insulator properties parallel those of enhancers (1, 19, 35, 60). These observations suggested that non-gypsy Su(Hw) sites could be identified by searching the Drosophila genome for clusters of the gypsy insulator Su(Hw) consensus site. In this analysis, we used the FLY ENHANCER program (47) to query release 1 of the genomic sequence for clusters of two or more consensus sites within a 250-bp window. This screen identified 37 clusters of sites (pSIs). This is far fewer than the 500 sites predicted by immunolocalization of the Su(Hw) protein within polytene chromosomes, supporting our previous finding that endogenous sites of Su(Hw) association differ from the gypsy insulator (54).
Genomic DNA sequences encompassing each Su(Hw) cluster were obtained, and each pSI was assigned a name based on the predicted cytological position of the genomic region. The properties of the pSIs are summarized in Table 1. Examination of the location of the Su(Hw) cluster relative to the annotation of the genome showed that all but one of the pSIs reside in an intronic or intergenic region. While the number of pSIs is small, these data indicate a bias away from coding regions, as exonic regions represent nearly 25% of the euchromatic genome. Next, we determined the number and organization of Su(Hw) binding sites present in each pSI. We found that both parameters differed from the gypsy insulator. Surprisingly, 27 pSIs contain only two Su(Hw) consensus sequences. Of the remaining pSIs, only 6C has more than four gypsy consensus sites, with pSIs 24D and 62D containing four sites, if imperfect consensus sites are also considered. Furthermore, the Su(Hw) sites within many pSIs are not closely spaced, with nearly a third containing sites separated by more than 60 nucleotides, a distance of separation previously associated with diminished enhancer blocking (65). These properties raised the possibility that most pSIs may not represent robust insulators. These data further enforce the conclusion that genomic sites of Su(Hw) association do not closely resemble the gypsy insulator.
TABLE 1.
List of pSIsa
| pSI | No. of sitesb
|
Spacingc | Relative location | Apparent Ka (M−1)d | ChIP resulte | |
|---|---|---|---|---|---|---|
| Full match | Mismatch | |||||
| 3A | 2 | 0 | 24 | Intergenic | ND | <0.2 |
| 4C | 2 | 0 | 48 | Intronic | ND | <0.2 |
| 6C | 14 (15) | 8 (9) | 2 | Intronic | 8.4 × 106 | <0.2 |
| 7B | 2 | 0 | 169 | Intergenic | ND | <0.2 |
| 8E | 2 (1) | 0 (1) | 58 | Intronic | ND | <0.2 |
| 11A | 2 | 1 | 0, 172 | Intergenic | ND | <0.2 |
| 11F | 3 | 0 | 43, 42 | Intergenic | 4.6 × 107 | <0.2 |
| 13C | 2 (1) | 0 | 47 | Intronic | ND | <0.2 |
| 16B | 2 | 0 | 44 | Intronic | ND | <0.2 |
| 18A-2 | 2 | 0 | 37 | Intergenic | ND | <0.2 |
| 18A-3 | 2 | 0 | 13 | Intergenic | ND | <0.2 |
| 18C | 2 | 1 | 58, 53 | Intronic | ND | <0.2 |
| 23A | 3 (2) | 0 (1) | 14, 206 | Exonic | 1.5 × 107 | <0.2 |
| 24D | 2 (1) | 2 | 23, 23, 183 | Intergenic | 4.4 × 107 | 1.02 |
| 26A | 2 | 0 | 213 | Intronic | ND | <0.2 |
| 34A | 2 | 0 | 26 | Intergenic | 2.1 × 107 | <0.2 |
| 36C | 2 | 0 | 91 | Intergenic | ND | <0.2 |
| 42A | 2 | 0 | 246 | Intergenic | ND | <0.2 |
| 44C | 2 | 2 | 160, 74, 15 | Intergenic | ND | <0.2 |
| 50A | 2 | 0 | 38 | Intergenic | 4.6 × 107 | 2.02 |
| 50B | 2 | 0 | 32 | Intronic | ND | <0.2 |
| 51D | 2 | 0 | 214 | Intergenic | ND | 0.34 |
| 54B | 2 | 0 | 211 | Intronic | ND | <0.2 |
| 59E | 2 (1) | 0 (1) | 135 | Intronic | ND | <0.2 |
| 61C | 2 | 0 | 9 | Intronic | 1.9 × 106 | <0.2 |
| 62D | 3 | 1 | 34, 73, 33 | Intergenic | 2.6 × 107 | 1.85 |
| 66E | 2 | 0 | 41 | Intronic | 2.0 × 107 | 1.33 |
| 87E | 2 (1) | 0 | 29 | Intergenic | 2.7 × 107 | 1.02 |
| 90C | 2 | 0 | 175 | Intergenic | ND | <0.2 |
| 90D | 2 | 0 | 21 | Intronic | ND | <0.2 |
| 94D | 2 | 0 | 36 | Intergenic | ND | <0.2 |
| 98B | 2 | 0 | 38 | Intergenic | 2.1 × 106 | <0.2 |
| 100B-1 | 2 | 0 | 49 | Intergenic | 1.2 × 107 | 0.40 |
| 100B-2 | 2 | 0 | 87 | Intergenic | ND | <0.2 |
| 100C | 2 | 0 | 188 | Intronic | ND | 0.66 |
Boldface indicates pSIs that bind the Su(Hw) protein in vivo.
Number of predicted Su(Hw) sites in Berkeley Drosophila Genome Project release 1 of the genome sequence. Observed numbers of sites in cloned sequences isolated from Canton S DNA are reported in parentheses. Full sites represent a 12/12 match to the gypsy consensus, while mismatch sites represent an 11/12 match.
Distance in base pairs between sites.
Ka, apparent association constant. ND, not determined.
Percentage of input recovered in Su(Hw) ChIP in Canton S third-instar larval chromatin.
Previous studies of the 1A2 insulator indicated that enhancer blocking depended upon the two Su(Hw) sites and additional flanking sequences (28). These observations suggest that at endogenous genomic locations, the Su(Hw) protein may cooperate with other insulator proteins to define regulatory isolation. As a first step to determine whether these newly identified pSIs were composite elements, we searched the genomic region encompassing each pSI for the presence of binding sites for other Drosophila insulator proteins, such as BEAF, ZW5/SBP, and GAGA (16, 19, 30, 35, 64). In all cases, binding sites for these proteins were not identified.
Analyses of in vivo pSI association with gypsy insulator proteins.
To gain insights into the properties of the pSIs, each region was isolated by PCR amplification of genomic DNA isolated from the standard Drosophila strain Canton S. Two of the pSIs were located in a large ∼180-bp repeat cluster and were not further analyzed. Sequence analysis of the remaining 35 pSIs, showed that several sequence variations existed between the published genomic database and our isolated pSIs, consistent with a high level of polymorphism associated with intronic and intergenic regions (Table 1).
To evaluate the potential insulator function of the pSIs, we used ChIP to determine whether these sequences were associated with the Su(Hw) protein in vivo. Chromatin was isolated from third-instar larvae, a stage known to have robust Su(Hw) association with the gypsy and 1A2 insulators (54). Chromatin was immunoprecipitated with a Su(Hw) antibody, and the degree of DNA enrichment was analyzed by PCR using primers that flanked each pSI. Primers flanking the gypsy insulator and the hsp26 coding region were used as positive and negative controls, respectively (Fig. 1). Of the 35 pSIs tested, 7 showed an increased level of amplification in the α-Su(Hw) chromatin immunoprecipitated material relative to the negative controls (Table 1). A range of pSI enrichment was obtained, with two pSIs (50A and 62D) showing levels similar to that of the gypsy insulator, three (24D, 66E, and 87E) showing intermediate levels, and two (100B-1 and 100C) showing a low level of enrichment. Surprisingly, pSI 6C, which had the highest number of Su(Hw) consensus sites, was not amplified in chromatin precipitated with the Su(Hw) antibody. These data indicate that the majority of pSIs are not bound by the Su(Hw) protein in vivo. Further, Su(Hw) association does not correlate with the number of binding sites.
The lack of Su(Hw) association with most pSIs might be due to the developmental stage used in the ChIP analysis. While we did not favor this possibility because the gypsy insulator blocks enhancers active at multiple stages of development, we addressed this issue by repeating the ChIP experiments using chromatin isolated from early embryos. Importantly, data obtained from these studies demonstrate that the same seven pSIs that bound the Su(Hw) protein in larval chromatin were associated with this protein in chromatin isolated from embryos. Further, the overall level of enrichment of each pSI in the two developmental stages was similar (Fig. 1; Table 1). These findings suggest that a factor(s) in addition to a consensus Su(Hw) binding site influence(s) chromosomal association of the Su(Hw) protein.
Analyses of in vitro pSI association with gypsy insulator proteins.
To investigate factors that contribute to in vivo Su(Hw) pSI occupancy, we determined whether the absence of in vivo Su(Hw) association reflected an inability of these sequences to bind the Su(Hw) protein in vitro. Previous studies indicated that DNA recognition by the Su(Hw) protein depends, in part, upon sequences flanking the Su(Hw) consensus site that direct DNA bending (68, 71). These observations imply that for some pSIs, the Su(Hw) consensus sites might reside in an unfavorable environment that decreases the DNA binding affinity of the Su(Hw) protein. To test this possibility, we purified the Su(Hw) protein from E. coli and performed the EMSA to test in vitro binding. In these studies, we determined apparent DNA association constants for Su(Hw) binding to the gypsy insulator, six pSIs that were associated with the Su(Hw) protein in vivo, and six that were not (Fig. 2; Table 1).
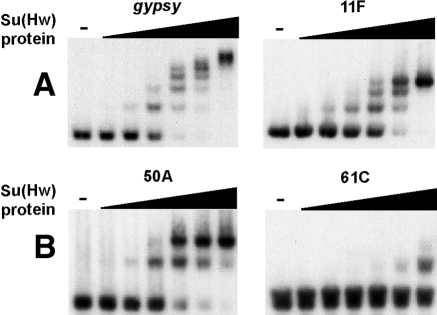
FIG. 2.
Analysis of in vitro Su(Hw) association with pSIs. (A) EMSA results are shown for 32P-labeled probes corresponding to the gypsy insulator and pSIs 11F, 50A, and 61C. Either no Su(Hw) protein (−) or increasing amounts of purified recombinant Su(Hw) protein were combined with labeled probes. The amount of Su(Hw) protein was increased threefold in each lane, beginning at 0.003 μg.
The gypsy insulator has 12 Su(Hw) binding sites. EMSA analysis demonstrated that the Su(Hw) protein produced up to six distinct migrating species (Fig. 2), with an apparent association constant for Su(Hw) binding to the gypsy insulator of 8.9 × 107 M−1. That we observed 6 instead of 12 independent species may reflect either incomplete occupancy of sites in the gypsy insulator or an inability to resolve species with more than six bound proteins.
The binding properties of the Su(Hw) protein to in vivo associated pSIs were determined. These experiments demonstrated that increasing amounts of the Su(Hw) protein produced slower-migrating bands, with the number of shifted species corresponding to the number of gypsy consensus sequences contained in the pSI. The apparent Su(Hw) association constants for this set of pSIs were two- to fourfold lower than that determined for the gypsy insulator (Table 1).
We tested six pSIs that failed to bind in vivo. Our EMSA studies demonstrated that three did not bind the Su(Hw) protein in vitro (Table 1; Fig. 2). We note that the spacing of the Su(Hw) sites might influence in vitro DNA association, as two of the nonbinding pSIs (6C and 61C) contained Su(Hw) sites separated by less than 10 bp. Interestingly, we found that three of the pSIs that were not associated with the Su(Hw) protein in vivo did bind this protein in vitro, with apparent association constants similar to those of in vivo associated pSIs (Table 1). These data imply that differences in the intrinsic Su(Hw) binding affinity do not account for the distinctions in Su(Hw) association in vivo. For example, 11F (not bound in vivo) and 50A (bound in vivo), carry three or two copies of an identical Su(Hw) consensus sequence, TGTTGCATACTT, and bind the Su(Hw) protein with the same apparent affinity in vitro (Fig. 2; Table 1). We postulate that the sequence context of the Su(Hw) sites influences protein occupancy in vivo, with sequences outside of the Su(Hw) sites contributing to Su(Hw) association. These data imply that extrinsic factors may be an important determinant of in vivo association of the Su(Hw) protein, limiting the effectiveness of applying only bioinformatics approaches to identify sites of in vivo association.
Mod67.2 associates with the pSIs.
The Su(Hw) and Mod67.2 proteins colocalize extensively at non-gypsy genomic sites in polytene chromosomes. These findings imply that pSIs associated with the Su(Hw) protein in vivo should be bound by Mod67.2. This prediction was confirmed by ChIP analysis of chromatin isolated from third-instar larvae (Fig. 1). The pSIs that showed strong Su(Hw) association were similarly enriched in chromatin immunoprecipitated with the Mod67.2 antibody, while pSIs with lower Su(Hw) association showed weaker precipitation with this antibody. The recruitment of Mod67.2 to the pSIs supports the postulate that these sequences represent endogenous insulators.
Multiple pSIs block enhancer-activated transcription.
To test whether the pSIs had insulator properties, an enhancer-blocking assay was used. This assay provides a direct assessment of the ability of a sequence to prevent regulatory interactions. In these experiments, we used the yellow gene as a reporter, as this gene has been extensively used to test the properties of the gypsy insulator (24, 28, 51, 54, 63). The yellow gene encodes a protein responsible for dark pigmentation of larval and adult cuticle structures. Several independent tissue-specific enhancers are required for yellow transcription, including the upstream wing and body enhancers and the intronic bristle enhancer (4, 15, 25). P element transposons were generated that carried each pSI, flanked by loxP sites, inserted between the wing and body enhancers and the yellow promoter. If the inserted pSI functioned to set up an insulator, then pigmentation should be decreased specifically in the adult wing and body cuticle but not in the bristles. To validate whether reduced yellow expression was due to the pSI, transgenic flies carrying the pSI reporter gene were crossed to flies expressing Cre recombinase, and derivative lines that carried a deletion of the pSI were established. In this way, enhancer blocking was evaluated by direct comparison of pigmentation levels in adult flies carrying yellow transgenes inserted at the same genomic location that either contained or lacked the pSI.
A subset of the pSIs was chosen for enhancer-blocking analysis (Table 2). These included pSIs that failed to bind (6C, 11F, 34A, and 61C) and bound (24D, 50A, 62D, 66E, and 87E) the Su(Hw) protein in vivo. We predicted that transgenic flies carrying transposons with the nonbinding pSIs would not change yellow expression and should serve as a negative control. This postulate was supported by analysis of three of the four nonbinding pSIs, where transgenic flies displayed levels of wing and body pigmentation that were near wild type (scores of 4 and 3+) (42, 54). The outlier among these pSIs was 61C, in which transgenic flies had intermediate pigmentation levels (scores of 3+ and 3). As pSI 61C showed Su(Hw) binding that was weak in vitro and undetectable in vivo, it is likely that this reduction in yellow expression is Su(Hw) independent.
TABLE 2.
Pigmentation scores of transformed lines carrying pSI-yellow transgenes and Cre recombinase derivativesa
| pSI | Binds Su(Hw) in vivo | Cuticle pigmentation scores for wing, body (no. of lines)
|
|
|---|---|---|---|
| With pSIb | Without pSIc | ||
| gypsy | Yes | 2+, 2 (26) | NA |
| 6C | No | 4, 3+ (6) | ND |
| 11F | No | 4, 3+ (11) | ND |
| 24D | Yes | 4, 3 (13) | 3+, 3 (4) |
| 34A | No | 4, 3+ (7) | ND |
| 50A | Yes | 3+, 3 (6) | 4+, 4 (6) |
| 61C | No | 3+, 3 (3) | ND |
| 62D | Yes | 2+, 2 (10) | NA |
| 66E | Yes | 3, 2+ (6) | 4, 3+ (6) |
| 87E | Yes | 3, 2+ (6) | 4, 3+ (6) |
Boldface indicates pSIs that show enhancer-blocking ability.
Insertion of the pSI between the wing-body enhancer and yellow promoter.
Deletion of the pSI using Cre recombinase. Boldface indicates lines showing a pigmentation increase in the deletion derivative lines. A score of 1 represents null or nearly null pigmentation, and a score of 5 represents wild-type pigmentation. NA, not applicable; ND, not determined.
Analysis of the in vivo binding pSIs showed that four of the five tested displayed enhancer blocking. Transgenic lines carrying yellow transposons with 50A had intermediate levels of pigmentation in the wing and body cuticle (scores of 3+ and 3), whereas low levels of cuticle pigmentation were observed for 62D (scores of 2+ and 2) and 66E and 87E (scores of 3 and 2+). In all cases, bristle pigmentation was at a wild-type level in these transgenic flies. These results indicate that 50A, 62D, and 87E blocked the yellow wing and body enhancers. This conclusion was confirmed by side-by-side comparison of the cuticle coloration in flies carrying yellow transposons, with or without the pSI, located at the same genomic site. We found that for three of six transgenic pSI lines for 50A and 87E and four of six lines for 66E, cuticle pigmentation was lighter in the presence of the pSI than in its absence (Table 2), indicating that the pSI was responsible for the lowered level of yellow gene expression. That 62D provided the strongest block among the in vivo binding pSIs of the yellow enhancers supports previous findings of a correlation between the number of Su(Hw) binding sites and the strength of enhancer blocking (31, 65).
In our enhancer-blocking assay, the exceptional in vivo binding pSI was 24D. Transgenic lines carrying the yellow 24D transposon had average wing and body pigmentation scores similar to those of transgenic flies carrying the nonbinding pSI transposons (scores of 4 and 3 compared to 4 and 3+ [Table 2]). Further, in side-by-side comparisons of flies carrying the transposons that contained or lacked 24D, the cuticle color in the wing and body did not significantly change (scores of 3+ and 3). The in vitro and in vivo binding data for 24D were similar to those for 87E, suggesting that the absence of enhancer blocking by 24D does not correlate with an ability to associate with the Su(Hw) protein (Tables 1 and 2). These observations indicate that the properties of endogenous regions associated with the Su(Hw) protein depend upon the context of the Su(Hw) sites and that at some genomic locations, the formation of an insulator may require cooperation with other nearby factors.
Effects of pSIs on endogenous gene expression.
Insulators are proposed to divide eukaryotic chromosomes into independent transcriptional domains. As such, if the pSIs established a genomic insulator, then a loss of the associated insulator proteins, Su(Hw) and Mod67.2, should change the expression of genes located adjacent to the pSI. To test this possibility, the level of RNA accumulation of genes next to the pSIs was determined in y2 control flies and in su(Hw) and mod(mdg4) mutant backgrounds. We studied effects in su(Hw)v and mod(mdg4)u1 mutant backgrounds. The su(Hw)v allele is a null allele caused by a deletion that encompasses the su(Hw) promoter (34). The mod(mdg4)u1 allele is caused by insertion of the Stalker transposon, resulting in the production of a truncated Mod67.2 protein lacking the Su(Hw) interaction domain (20, 22). We reasoned that if a pSI represented an endogenous insulator, then expression of genes adjacent to the pSIs should be altered in the su(Hw) null background, as the Su(Hw) protein is an essential component of the gypsy insulator (48). However, prediction of the outcome caused by the loss of Mod67.2 was more complicated, as in the absence of this protein, the gypsy insulator has been shown to have gene-specific effects, in some cases showing retention of enhancer blocking, loss of enhancer blocking, or promoter-specific silencing (11, 21, 22).
Most of the genes that are adjacent to the pSIs are predicted genes. As such, molecular information about the expression profile and the transcriptional regulatory elements in these regions is lacking. These limitations made it impossible to predict the direction or degree of the change in gene expression. To study expression from these predicted genes, RNA was isolated from three collections (6, 8, and 10 days after egg laying). Late stages of development were chosen to ensure that the maternally supplied insulator proteins were depleted. RNA accumulation for genes adjacent to each pSI was determined using real-time PCR and normalized to the level of Ras2 RNA, as previous studies indicated that expression of this gene is constant throughout development (50). The change in gene expression was determined by dividing normalized levels of RNA accumulation in the su(Hw) and mod(mdg4) mutant backgrounds by normalized values determined for RNA accumulation in y2 flies. Several controls were included in our expression analysis (Fig. 3). First, we studied accumulation of su(Hw) mRNA in the su(Hw) and mod(mdg4) mutants, which carry deleted and intact su(Hw) genes, respectively. As predicted, su(Hw) mRNA was undetectable in su(Hw)v flies and changed less than twofold in the mod(mdg4)u1 background. Second, we determined the change in accumulation of yellow mRNA, as all strains carried the extant y2 allele. Loss of the Su(Hw) protein reverses the enhancer-blocking effects of the gypsy insulator, restoring yellow transcription, whereas loss of the Mod67.2 protein causes promoter-specific silencing and a reduction in yellow transcription (21, 22). As predicted, yellow expression increased in the su(Hw) mutant background (Fig. 3), with the largest change observed in day 8 RNA, a time point coincident with the known temporal pattern of yellow transcription (26). Further, we observed a decreased change in mod(mdg4) mutants relative to the wild-type (y2) control (Fig. 3). Third, we determined levels of RNA accumulation for several housekeeping genes (Gapdh2 and RpL32) and randomly selected genes (white, cut, CG9568, crc, and CG16926) (Fig. 3). In all but one case, a less-than-twofold change in gene expression was observed between the wild-type and insulator mutant backgrounds. The one exception was white expression in a mod(mdg4)u1 background, where we observed a significant decrease in RNA levels. These data are consistent with previous observations that a mod(mdg4) mutant background represses white expression (11). Taken together, our control data demonstrate that the loss of the Su(Hw) and Mod67.2 insulator proteins does not cause global alterations in gene expression. Thus, any observed change in expression of genes adjacent to pSIs in these mutant backgrounds will support a role for the pSI in establishing an endogenous insulator.
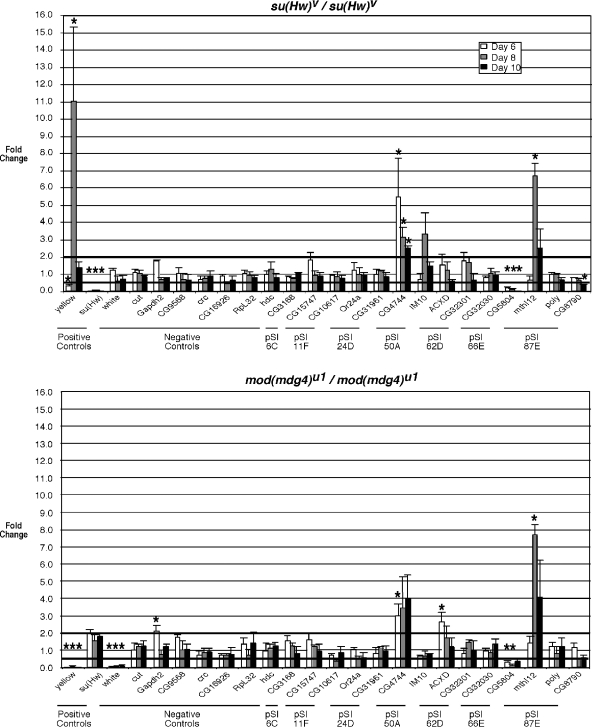
FIG. 3.
pSIs contribute to regulation of endogenous gene expression. (Top) Results from real-time PCR analysis of RNA y2 and y2; su(Hw)v/su(Hw)v flies, isolated from larvae and pupae 6 (white bars), 8 (gray bars), and 10 (black bars) days after egg deposition, are shown, using primers to the indicated genes. These values were normalized to levels of Ras2 RNA accumulation, and the change was determined by dividing the value obtained with RNA isolated from su(Hw) mutants by the value from y2 RNA. A value of 1.0 represents no change in RNA accumulation in the two genetic backgrounds. Solid bars represent the position of a twofold increase or decrease in change of expression. Several genes were analyzed, including two positive controls [genes expected to show different RNA accumulation in the su(Hw) mutant background], eight negative controls [genes expected to show similar levels of RNA accumulation in su(Hw) and y2], two nonbinding Su(Hw) pSIs (6C and 11F) and five binding Su(Hw) pSIs (24D, 50A, 62D, 66E, and 87E). (Bottom) Results from real-time PCR analysis of y2 and y2; mod(mdg4)u1/mod(mdg4)u1 RNA are shown, using the same analysis as described for panel A. Asterisks indicate P values of <0.05.
Seven pSIs were chosen for study, and changes in expression of neighboring genes were determined (Table 3; Fig. 3). First, we studied two in vivo nonbinding pSIs (6C and 11F). As predicted by our control experiments, expression of genes adjacent to these nonbinding pSIs did not change in the su(Hw) or mod(mdg4) mutant backgrounds. Next, we tested the five pSIs that bound the Su(Hw) protein in vivo. For two of these (24D and 62D), loss of the insulator proteins had minor if any effects on gene expression. In contrast, we found that genes near the remaining three pSIs showed a greater-than-twofold change in expression in one or both insulator mutant backgrounds (Fig. 3; Table 3). In two cases, increased expression was observed (CG4744 for 50A; CG32853 for 87E), and in one case, decreased expression was found (CG5804 for 66E). These data provide strong support of the postulate that at least some pSIs represent insulators within their natural location.
TABLE 3.
Changes in expression of genes adjacent to pSIs for developmental day 8a
| pSI | Location | 5′ gene
|
3′ gene
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Distance (kb)b | Change in expression (n-fold)
|
Name | Distance (kb)b | Change in expression (n-fold)
|
||||
| su(Hw)v/su(Hw)v | mod(mdg4)u1/mod(mdg4)u1 | vol/vol | mod | ||||||
| 6C | Intronic | CG3168 | 1.7 | −1.4 | 1.2 | NA | NA | NA | NA |
| 11F | Intergenic | CG15747 | 8.7 | −1.1 | 1.2 | CG10617 | 12.1 | −1.2 | −3.2 |
| 24D | Intergenic | Or24a | 13.4 | 1.0 | −2.1 | CG31961 | 8.0 | 1.2 | 1.2 |
| 50A | Intergenic | CG4744 | 4.4 | 3.2 | 3.4 | IM10 | 3.1 | 3.3 | −1.8 |
| 62D | Intergenic | ACXD | 6.5 | 1.2 | 1.7 | CG32301 | 0.2 | 1.7 | 1.4 |
| 66E | Intronic | CG32030 | 21.0 | 1.0 | 1.2 | CG5804 | 1.1 | −11 | −14 |
| 87E | Intergenic | mthl12 | 6.5 | 6.7 | 7.7 | Poly | 6.6 | 1.0 | −1.3 |
| CG8790 | 4.6 | −1.6 | −2.4 | ||||||
Boldface indicates significant changes in gene expression (P < 0.1), as determined by a Student t test. NA, not applicable, as the pSI resides in an intron.
Distance from pSI to start site of transcription.
DISCUSSION
The Su(Hw) protein associates with hundreds of sites within the Drosophila polytene chromosomes. Using a bioinformatics approach, we identified 37 genomic regions that contained at least two Su(Hw) sites within 250 bp; this number was far lower than expected. DNA sequence analysis of these pSIs showed that the majority of regions (27 of 37; 73%) contained only two Su(Hw) sites (Table 1). Further, in most cases, the spacing between Su(Hw) sites was larger than that found in the gypsy insulator, which ranges from 14 to 23 nucleotides. These data demonstrate that genomic sites of Su(Hw) association differ from the gypsy insulator.
Chromosomal Su(Hw) localization is complex.
ChIP demonstrated that 7 of the 35 pSIs tested bound the Su(Hw) protein in vivo (Table 1). The absence of in vivo binding for the majority of pSIs did not reflect an inability of these sequences to bind the Su(Hw) protein in vitro (Table 1; Fig. 2). In fact, 11F and 50A have identical Su(Hw) consensus sequences, yet only 50A binds the Su(Hw) protein in vivo. These observations suggest that DNA sequence recognition is only one determinant of in vivo association. Other factors may have a major influence on chromosomal occupancy of the Su(Hw) protein, including the nucleosomal organization of the region, which may affect accessibility of the Su(Hw) binding sites, or the presence of binding sites for other factors that may facilitate or inhibit Su(Hw) localization. Further, we predict that computational as well as in vitro DNA SELEX (systematic evolution of ligands by exponential enrichment) approaches may be limited in their ability to identify in vivo binding sites.
The Su(Hw) protein is a twelve-zinc-finger protein and may use different combinations of fingers in DNA recognition, as has been previously demonstrated for the vertebrate CTCF insulator protein (8, 44, 56, 74). As such, the limited number of identified Su(Hw) binding site clusters may indicate that the sequence requirements for binding the endogenous sites of DNA are more relaxed than represented by the gypsy consensus sequence. This supposition is supported by two recent findings. First, the endogenous Su(Hw) insulator, 1A2, contains two Su(Hw) binding sites, of which only one site carries the invariant gypsy core sequence (28, 54). Second, modified SELEX analyses indicate that although the gypsy consensus is representative of the sequence specificity of the Su(Hw) protein, the true consensus sequence is invariant in only three nucleotides of the core (57). Using this predicted Su(Hw) consensus sequence to identify clustered genomic Su(Hw) sites, only ∼20 regions were found that contained immediately adjacent or overlapping Su(Hw) sites, whereas ∼2,500 Su(Hw) single sites were found. Taken together, emerging data implies that the organization of Su(Hw) sites in the genome differs from that of the gypsy insulator.
Two underlying assumptions led to the prediction that bioinformatic screens should identify hundreds of clustered Su(Hw) sites in the genome. First, we hypothesized that only clustered sites establish genomic insulators because such features are required for production of a functional synthetic gypsy insulator (65). However, our observation that pSI 87E, which contains only one binding site in Canton S flies, binds Su(Hw) in vivo and confers insulation (Table 1; Fig. 2 and 3), suggests that single Su(Hw) consensus sites could represent endogenous Su(Hw) insulators; a postulate supported by other recent studies (57). Second, we hypothesized that Su(Hw) association depends only on direct DNA binding. This postulate is predicated on the demonstration that mutations in the zinc finger domain abolish chromosome association (32). However, it remains possible that localization of Su(Hw) to some genomic regions depends upon protein-protein or protein-RNA interactions, as zinc finger motifs are flexible interaction domains (7, 12). For example, the Su(Hw) protein directly interacts with CP190, another zinc finger DNA binding protein, which may direct the Su(Hw) protein to chromosomal regions devoid of Su(Hw) binding sites. A complete understanding of the genomic distribution of endogenous Su(Hw) sites will require analysis of ChIP preparations using genomic tiling arrays.
Endogenous pSIs contribute to neighboring gene expression.
Our goal in identifying pSIs was to determine whether non-gypsy sites of Su(Hw) association represent genomic locations of endogenous insulators. To this end, we tested whether pSIs prevented enhancer-activated transcription. Our data demonstrate that four of the five pSIs that bind the Su(Hw) protein in vivo confer some degree of enhancer blocking. These effects ranged from intermediate to strong blocking of the yellow wing and body enhancers, with pSI 62D, an insulator with four Su(Hw) sites, showing the strongest effects (Table 2). These data imply that genomic clusters with fewer than four Su(Hw) sites do establish insulators, even though synthetic binding regions comprised of a reiteration of site 3 from the gypsy insulator do not. These data can be reconciled if factors in addition to the Su(Hw) protein contribute to the formation of endogenous insulators. For example, pSIs may be composite elements that contain binding sites for other insulator proteins, as has been previously identified for the chicken globin HS4 insulator, which contains binding sites for two insulator proteins, CTCF and USF (1, 60, 76). Alternatively, pSIs may contain MARs that could increase the effectiveness of enhancer blocking by clusters of less than four Su(Hw) sites (57). This prediction is based on the observation that several insulators show MAR activity, including the gypsy insulator (52, 77, 78). Analysis of our pSIs further supports this possibility, as three of the four pSIs that demonstrate enhancer-blocking activity (50A, 62D, and 87E) overlap with predicted MAR sequences (data not shown). Importantly, our data suggest that Su(Hw) association alone is not sufficient for insulator effects. We found that although pSI 24D was associated with Su(Hw) in vivo, these sequences did not demonstrate enhancer-blocking activity. These studies imply that the properties of Su(Hw)-associated regions are complex.
Enhancer-blocking assays are a powerful but synthetic method to test for insulator effects. As such, these assays do not address the important question of whether an element establishes an insulator within its natural location. For this reason, we undertook a second, complementary approach to address the function of the pSIs. In these studies, we determined whether RNA production of genes residing next to pSIs was affected in su(Hw) and/or mod(mdg4) mutants by comparing levels of RNA accumulation in control (y2) flies and flies carrying mutations in the insulator genes. These studies demonstrated that the loss of the Su(Hw) and Mod67.2 insulator proteins produced changes in gene expression specifically in genes adjacent to in vivo associated pSIs, without causing global effects on RNA levels (Fig. 3). Interestingly, we found that the direction of change in expression of genes next to 50A, 66E, and 87E was the same in both insulator mutant backgrounds, implying that the absence of either protein caused the loss of an endogenous insulator. The Su(Hw) binding sites were located up to 6.5 kb away from the promoters of the genes that were affected in su(Hw) and mod(mdg4) mutant backgrounds, suggesting that changes in gene expression depended on long-range effects, consistent with the properties of an insulator. These data provide the first direct evidence that gypsy insulator proteins affect regulation of gene expression by the formation of insulators in their natural location.
We present evidence that the genomic Su(Hw) insulators are composite elements. We postulate that this organization might have evolved to prevent the collapse of transcriptional regulation by loss of any one insulator protein. This proposal is consistent with the observation that the absence of either the Su(Hw) or Mod67.2 protein is not lethal, as might be expected if these proteins were solely responsible for the definition of transcriptional regulatory domains. Identification of genomic Su(Hw) sites provides an important step toward understanding the role of insulator proteins in genome organization and gene expression.
Acknowledgments
We thank Lori Wallrath and members of the Geyer laboratory for critically reading the manuscript. We thank Marie Stelzer, Jeff Miller, and Eric Schultz for their outstanding technical assistance.
This work was supported by National Institutes of Health grant GM42539 to P.K.G. and a Carver College of Medicine collaborative grant to P.K.G. and M.S.W.
REFERENCES
- 1.Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. [DOI] [PubMed] [Google Scholar]
- 2.Berman, B. P., Y. Nibu, B. D. Pfeiffer, P. Tomancak, S. E. Celniker, M. Levine, G. M. Rubin, and M. B. Eisen. 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA 99:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman, B. P., B. D. Pfeiffer, T. R. Laverty, S. L. Salzberg, G. M. Rubin, M. B. Eisen, and S. E. Celniker. 2004. Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 5:R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biessmann, H. 1985. Molecular analysis of the yellow gene (y) region of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 82:7369-7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutanaev, A. M., A. I. Kalmykova, Y. Y. Shevelyov, and D. I. Nurminsky. 2002. Large clusters of co-expressed genes in the Drosophila genome. Nature 420:666-669. [DOI] [PubMed] [Google Scholar]
- 6.Brasset, E., and C. Vaury. 2005. Insulators are fundamental components of the eukaryotic genomes. Heredity 94:571-576. [DOI] [PubMed] [Google Scholar]
- 7.Brown, R. S. 2005. Zinc finger proteins: getting a grip on RNA. Curr. Opin. Struct. Biol. 15:94-98. [DOI] [PubMed] [Google Scholar]
- 8.Burcin, M., R. Arnold, M. Lutz, B. Kaiser, D. Runge, F. Lottspeich, G. N. Filippova, V. V. Lobanenkov, and R. Renkawitz. 1997. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol. Cell Biol. 17:1281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrd, K., and V. G. Corces. 2003. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 162:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai, H., and M. Levine. 1995. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature 376:533-536. [DOI] [PubMed] [Google Scholar]
- 11.Cai, H. N., and M. Levine. 1997. The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 16:1732-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantor, A. B., and S. H. Orkin. 2005. Coregulation of GATA factors by the friend of GATA (FOG) family of multitype zinc finger proteins. Semin. Cell Dev. Biol. 16:117-128. [DOI] [PubMed] [Google Scholar]
- 13.Capelson, M., and V. G. Corces. 2004. Boundary elements and nuclear organization. Biol. Cell 96:617-629. [DOI] [PubMed] [Google Scholar]
- 14.Chen, J. L., K. L. Huisinga, M. M. Viering, S. A. Ou, C. T. Wu, and P. K. Geyer. 2002. Enhancer action in trans is permitted throughout the Drosophila genome. Proc. Natl. Acad. Sci. USA 99:3723-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chia, W., G. Howes, M. Martin, Y. B. Meng, K. Moses, and S. Tsubota. 1986. Molecular analysis of the yellow locus of Drosophila. EMBO J. 5:3597-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuvier, O., C. M. Hart, and U. K. Laemmli. 1998. Identification of a class of chromatin boundary elements. Mol. Cell. Biol. 18:7478-7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engel, N., and M. S. Bartolomei. 2003. Mechanisms of insulator function in gene regulation and genomic imprinting. Int. Rev. Cytol. 232:89-127. [DOI] [PubMed] [Google Scholar]
- 18.Erives, A., and M. Levine. 2004. Coordinate enhancers share common organizational features in the Drosophila genome. Proc. Natl. Acad. Sci. USA 101:3851-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaszner, M., J. Vazquez, and P. Schedl. 1999. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 13:2098-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gause, M., P. Morcillo, and D. Dorsett. 2001. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol. Cell. Biol. 21:4807-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgiev, P., and M. Kozycina. 1996. Interaction between mutations in the suppressor of Hairy wing and modifier of mdg4 genes of Drosophila melanogaster affecting the phenotype of gypsy-induced mutations. Genetics 142:425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgiev, P. G., and T. I. Gerasimova. 1989. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol. Gen. Genet. 220:121-126. [DOI] [PubMed] [Google Scholar]
- 23.Gerasimova, T. I., and V. G. Corces. 2001. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet. 35:193-208. [DOI] [PubMed] [Google Scholar]
- 24.Geyer, P. K., and V. G. Corces. 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6:1865-1873. [DOI] [PubMed] [Google Scholar]
- 25.Geyer, P. K., and V. G. Corces. 1987. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1:996-1004. [DOI] [PubMed] [Google Scholar]
- 26.Geyer, P. K., C. Spana, and V. G. Corces. 1986. On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. EMBO J. 5:2657-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh, D., T. I. Gerasimova, and V. G. Corces. 2001. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 20:2518-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golovnin, A., I. Birukova, O. Romanova, M. Silicheva, A. Parshikov, E. Savitskaya, V. Pirrotta, and P. Georgiev. 2003. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development 130:3249-3258. [DOI] [PubMed] [Google Scholar]
- 29.Golovnin, A., M. Gause, S. Georgieva, E. Gracheva, and P. Georgiev. 1999. The su(Hw) insulator can disrupt enhancer-promoter interactions when located more than 20 kilobases away from the Drosophila achaete-scute complex. Mol. Cell. Biol. 19:3443-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Granok, H., B. A. Leibovitch, C. D. Shaffer, and S. C. Elgin. 1995. Chromatin. Ga-ga over GAGA factor. Curr. Biol. 5:238-241. [DOI] [PubMed] [Google Scholar]
- 31.Hagstrom, K., M. Muller, and P. Schedl. 1996. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 10:3202-3215. [DOI] [PubMed] [Google Scholar]
- 32.Harrison, D. A., D. A. Gdula, R. S. Coyne, and V. G. Corces. 1993. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 7:1966-1978. [DOI] [PubMed] [Google Scholar]
- 33.Harrison, D. A., P. K. Geyer, C. Spana, and V. G. Corces. 1989. The gypsy retrotransposon of Drosophila melanogaster: mechanisms of mutagenesis and interaction with the suppressor of Hairy-wing locus. Dev. Genet. 10:239-248. [DOI] [PubMed] [Google Scholar]
- 34.Harrison, D. A., M. A. Mortin, and V. G. Corces. 1992. The RNA polymerase II 15-kilodalton subunit is essential for viability in Drosophila melanogaster. Mol. Cell. Biol. 12:928-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hart, C. M., K. Zhao, and U. K. Laemmli. 1997. The scs′ boundary element: characterization of boundary element-associated factors. Mol. Cell. Biol. 17:999-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holdridge, C., and D. Dorsett. 1991. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell. Biol. 11:1894-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoover, K. K., T. I. Gerasimova, A. J. Chien, and V. G. Corces. 1992. Dominant effects of suppressor of Hairy-wing mutations on gypsy-induced alleles of forked and cut in Drosophila melanogaster. Genetics 132:691-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurst, L. D., C. Pal, and M. J. Lercher. 2004. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 5:299-310. [DOI] [PubMed] [Google Scholar]
- 39.Kalmykova, A. I., D. I. Nurminsky, D. V. Ryzhov, and Y. Y. Shevelyov. 2005. Regulated chromatin domain comprising cluster of co-expressed genes in Drosophila melanogaster. Nucleic Acids Res. 33:1435-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, C., B. F. Paulus, and M. S. Wold. 1994. Interactions of human replication protein A with oligonucleotides. Biochemistry 33:14197-14206. [DOI] [PubMed] [Google Scholar]
- 41.Kuhn, E. J., and P. K. Geyer. 2003. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 15:259-265. [DOI] [PubMed] [Google Scholar]
- 42.Kuhn, E. J., M. M. Viering, K. M. Rhodes, and P. K. Geyer. 2003. A test of insulator interactions in Drosophila. EMBO J. 22:2463-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lercher, M. J., A. O. Urrutia, and L. D. Hurst. 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 31:180-183. [DOI] [PubMed] [Google Scholar]
- 44.Lobanenkov, V. V., R. H. Nicolas, V. V. Adler, H. Paterson, E. M. Klenova, A. V. Polotskaja, and G. H. Goodwin. 1990. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5:1743-1753. [PubMed] [Google Scholar]
- 45.Mallin, D. R., J. S. Myung, J. S. Patton, and P. K. Geyer. 1998. Polycomb group repression is blocked by the Drosophila suppressor of Hairy-wing [su(Hw)] insulator. Genetics 148:331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Markstein, M., and M. Levine. 2002. Decoding cis-regulatory DNAs in the Drosophila genome. Curr. Opin. Genet. Dev. 12:601-606. [DOI] [PubMed] [Google Scholar]
- 47.Markstein, M., P. Markstein, V. Markstein, and M. S. Levine. 2002. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 99:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Modolell, J., W. Bender, and M. Meselson. 1983. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc. Natl. Acad. Sci. USA 80:1678-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morris, J. R., J. L. Chen, P. K. Geyer, and C. T. Wu. 1998. Two modes of transvection: enhancer action in trans and bypass of a chromatin insulator in cis. Proc. Natl. Acad. Sci. USA 95:10740-10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mozer, B., R. Marlor, S. Parkhurst, and V. Corces. 1985. Characterization and developmental expression of a Drosophila ras oncogene. Mol. Cell. Biol. 5:885-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muravyova, E., A. Golovnin, E. Gracheva, A. Parshikov, T. Belenkaya, V. Pirrotta, and P. Georgiev. 2001. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291:495-498. [DOI] [PubMed] [Google Scholar]
- 52.Nabirochkin, S., M. Ossokina, and T. Heidmann. 1998. A nuclear matrix/scaffold attachment region co-localizes with the gypsy retrotransposon insulator sequence. J. Biol. Chem. 273:2473-2479. [DOI] [PubMed] [Google Scholar]
- 53.Pai, C. Y., E. P. Lei, D. Ghosh, and V. G. Corces. 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 16:737-748. [DOI] [PubMed] [Google Scholar]
- 54.Parnell, T. J., M. M. Viering, A. Skjesol, C. Helou, E. J. Kuhn, and P. K. Geyer. 2003. An endogenous Suppressor of Hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl. Acad. Sci. USA 100:13436-13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quitschke, W. W., M. J. Taheny, L. J. Fochtmann, and A. A. Vostrov. 2000. Differential effect of zinc finger deletions on the binding of CTCF to the promoter of the amyloid precursor protein gene. Nucleic Acids Res. 28:3370-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramos, E., D. Ghosh, E. Baxter, and V. Corces. 2006. Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics 172:2337-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rebeiz, M., and J. W. Posakony. 2004. GenePalette: a universal software tool for genome sequence visualization and analysis. Dev. Biol. 271:431-438. [DOI] [PubMed] [Google Scholar]
- 59.Rebeiz, M., N. L. Reeves, and J. W. Posakony. 2002. SCORE: a computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Site clustering over random expectation. Proc. Natl. Acad. Sci. USA 99:9888-9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Recillas-Targa, F., M. J. Pikaart, B. Burgess-Beusse, A. C. Bell, M. D. Litt, A. G. West, M. Gaszner, and G. Felsenfeld. 2002. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl. Acad. Sci. USA 99:6883-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ren, X. Y., M. W. Fiers, W. J. Stiekema, and J. P. Nap. 2005. Local coexpression domains of two to four genes in the genome of Arabidopsis. Plant Physiol. 138:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roseman, R. R., E. A. Johnson, C. K. Rodesch, M. Bjerke, R. N. Nagoshi, and P. K. Geyer. 1995. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics 141:1061-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Savitskaya, E., L. Melnikova, M. Kostuchenko, E. Kravchenko, E. Pomerantseva, T. Boikova, D. Chetverina, A. Parshikov, P. Zobacheva, E. Gracheva, A. Galkin, and P. Georgiev. 2006. Study of long-distance functional interactions between Su(Hw) insulators that can regulate enhancer-promoter communication in Drosophila melanogaster. Mol. Cell. Biol. 26:754-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schweinsberg, S., K. Hagstrom, D. Gohl, P. Schedl, R. P. Kumar, R. Mishra, and F. Karch. 2004. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168:1371-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scott, K. C., A. D. Taubman, and P. K. Geyer. 1999. Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics 153:787-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scott, K. S., and P. K. Geyer. 1995. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J. 14:6258-6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Senger, K., G. W. Armstrong, W. J. Rowell, J. M. Kwan, M. Markstein, and M. Levine. 2004. Immunity regulatory DNAs share common organizational features in Drosophila. Mol. Cell 13:19-32. [DOI] [PubMed] [Google Scholar]
- 68.Shen, B., J. Kim, and D. Dorsett. 1994. The enhancer-blocking suppressor of Hairy-wing zinc finger protein of Drosophila melanogaster alters DNA structure. Mol. Cell. Biol. 14:5645-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sigrist, C. J., and V. Pirrotta. 1997. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147:209-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh, G. B., J. A. Kramer, and S. A. Krawetz. 1997. Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res. 25:1419-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spana, C., and V. G. Corces. 1990. DNA bending is a determinant of binding specificity for a Drosophila zinc finger protein. Genes Dev. 4:1505-1515. [DOI] [PubMed] [Google Scholar]
- 72.Spana, C., D. A. Harrison, and V. G. Corces. 1988. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2:1414-1423. [DOI] [PubMed] [Google Scholar]
- 73.Spellman, P. T., and G. M. Rubin. 2002. Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vostrov, A. A., and W. W. Quitschke. 1997. The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J. Biol. Chem. 272:33353-33359. [DOI] [PubMed] [Google Scholar]
- 75.West, A. G., M. Gaszner, and G. Felsenfeld. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16:271-288. [DOI] [PubMed] [Google Scholar]
- 76.West, A. G., S. Huang, M. Gaszner, M. D. Litt, and G. Felsenfeld. 2004. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell 16:453-463. [DOI] [PubMed] [Google Scholar]
- 77.Yusufzai, T. M., and G. Felsenfeld. 2004. The 5′-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl. Acad. Sci. USA 101:8620-8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yusufzai, T. M., H. Tagami, Y. Nakatani, and G. Felsenfeld. 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13:291-298. [DOI] [PubMed] [Google Scholar]
- 79.Zhao, H., and A. Dean. 2005. Organizing the genome: enhancers and insulators. Biochem. Cell Biol. 83:516-524. [DOI] [PubMed] [Google Scholar]