Abstract
Hedgehog (HH)/GLI signaling plays a critical role in epidermal development and basal cell carcinoma. Here, we provide evidence that epidermal growth factor receptor (EGFR) signaling modulates the target gene expression profile of GLI transcription factors in epidermal cells. Using expression profiling and quantitative reverse transcriptase PCR, we identified a set of 19 genes whose transcription is synergistically induced by GLI1 and parallel EGF treatment. Promoter studies of a subset of GLI/EGF-regulated genes, including the genes encoding interleukin-1 antagonist IL1R2, Jagged 2, cyclin D1, S100A7, and S100A9, suggest convergence of EGFR and HH/GLI signaling at the level of promoters of selected direct GLI target genes. Inhibition of EGFR and MEK/ERK but not of phosphatidylinositol 3-kinase/AKT abrogated synergistic activation of GLI/EGF target genes, showing that EGFR can signal via RAF/MEK/ERK to cooperate with GLI proteins in selective target gene regulation. Coexpression of the GLI/EGF target IL1R2, EGFR, and activated ERK1/2 in human anagen hair follicles argues for a cooperative role of EGFR and HH/GLI signaling in specifying the fate of outer root sheath (ORS) cells. We also show that EGF treatment neutralizes GLI-mediated induction of epidermal stem cell marker expression and provide evidence that EGFR signaling is essential for GLI-induced cell cycle progression in epidermal cells. The results suggest that EGFR signaling modulates GLI target gene profiles which may play an important regulatory role in ORS specification, hair growth, and possibly cancer.
The behavior of a given cell in a multicellular organism is to a large extent determined by its microenvironment, which provides a rich source of distinct stimuli. The precise sensing of and responding to this variety of extracellular stimuli is a fundamental process in metazoans. The controlled integration of these molecular cues via activation of distinct signaling networks implies intricate regulatory mechanisms that orchestrate the multitude of cellular inputs to accomplish the appropriate transcriptional responses (41). In line with the importance of such networks in development, growth, and tissue homeostasis, perturbation or aberrant activation of signaling cascades, such as Wnt, transforming growth factor beta, epidermal growth factor receptor (EGFR), or the Hedgehog (HH) pathway, is a major causative factor in diseases such as cancer (for reviews, see references 37, 56, 83, and 88).
In recent years, the HH/GLI signaling pathway has gained significant importance, as it has been shown to play a prominent role in a number of different developmental processes and in the growth and maintenance of a variety of common tumors whose growth can be efficiently prevented by selective inhibition of constitutive pathway activity (reviewed in references 7, 39, 43, 71, 82, 83, 86, and 101).
HH signal transduction involves binding of processed HH proteins to the HH receptor Patched (PTCH), a 12-pass transmembrane protein that, in the absence of ligand, represses pathway activity by inhibiting the activity of the seven-transmembrane domain protein Smoothened (SMOH). Binding of HH protein to PTCH triggers the signaling activity of SMOH, which eventually converts the latent GLI zinc finger transcription factors GLI2 and GLI3 into transcriptional activators to control HH target gene expression. Activation of latent GLI proteins is an intricate process that involves modifications and interactions of a number of positive and negative pathway regulators and is not fully understood (5, 6, 12; for reviews see references 32, 39, 43, and 50). Following the activation of latent GLI proteins, transcription of the third member of the vertebrate GLI family, GLI1, is directly induced by the activator forms of GLI2 and GLI3. The concerted activity of the GLI proteins is thought to control the transcriptional program in response to HH signaling in a highly context-dependent manner (3, 12, 20, 38, 61).
In mammalian skin, HH/GLI signaling plays a prominent role in the development of epidermal appendages and skin cancer. Loss of HH signaling in murine epidermis results in an arrest of hair follicle growth at an early stage of follicle development due to reduced proliferation (16, 58, 95), while constitutive pathway activation in epidermal cells induces (with high frequency) tumors with basal cell carcinoma (BCC)-like features (22, 26, 36, 63, 66, 106). The causative role of aberrant HH/GLI signaling in BCC is underlined by the fact that the majority of human BCCs show increased HH/GLI signaling due to inactivating mutations in PTCH or activating mutations in SMOH (24, 27, 40, 47, 106).
The transcriptional programs activated in response to HH signaling in epidermal development and BCC are mainly controlled by GLI1 and GLI2. The skin phenotype of Gli2−/− mice largely resembles that of Shh-deficient mice, suggesting that the Gli2 gene encodes the major transcriptional effector of HH signaling during epidermal development. By contrast, the loss of Gli1 or Gli3 does not affect epidermal development (58, 70).
Although dispensable for normal development, GLI1 is likely to play a key role in HH-driven cancers. GLI1 is expressed at elevated levels in BCC and other types of HH-driven tumors. Its forced expression induces BCC-like tumors in transgenic mice, transforms epithelial cells, enhances the metastatic potential of prostate cancer cells, and induces expression of prosurvival genes, such as the BCL2 gene (9, 18, 42, 63, 65, 76, 77, 84). Moreover, RNA interference-mediated inhibition of GLI1 function has been shown to be sufficient to inhibit proliferation of prostate cancer cells (87).
A remarkable feature of GLI proteins is their context-dependent biological activity (3, 61), which suggests that the cellular microenvironment and the entity of regulatory factors coexpressed with GLI proteins eventually determine the transcriptional programs activated by HH signaling. Besides proteolytic processing of GLI proteins that regulates the precise balance of GLI activator and repressor activity (3, 4, 93, 105), the transcriptional activity of GLI proteins appears to be also controlled by interactions with a number of other factors. The dual-specificity Yak1-related kinase Dyrk1 has been shown to increase Gli1-dependent target gene activation by retaining Gli1 in the nucleus and by enhancing its transcriptional activity (55). The actin-binding protein Missing in Metastasis is encoded by a Shh-responsive gene that enhances the activity of GLI1 and GLI2 in skin development and tumorigenesis (13). Furthermore, several intraflagellar transport proteins act as regulators of Gli protein activity by controlling both Gli activator and repressor activity (33, 34, 48).
While regulatory factors, such as Missing in Metastasis and intraflagellar proteins, are acting within the HH pathway itself to control GLI activity, little is known about the role of parallel signaling cascades in modulating GLI target gene profiles and target gene specificities. Recent reports have provided evidence that the biological effect of HH/GLI signaling can be modulated by parallel activation of additional intracellular signaling routes, such as those triggered by EGF or fibroblast growth factor (10, 44, 51, 68, 69). Inputs from the local microenvironment may thus interact with HH/GLI signaling and account for at least part of the context-dependent properties of GLI proteins. In line with this concept, recent reports suggest a requirement of phosphatidylinositol 3-kinase (PI3K)/AKT and MEK/ERK activation for HH/GLI signaling (78, 79), though the molecular basis and biological consequences of HH/GLI pathway modulation by parallel signaling cascades are still largely elusive.
In this study, we addressed whether parallel activation of EGFR signaling by EGF affects the transcriptional activity of the major epidermal HH effectors GLI1 and GLI2. We focused on EGF signaling, as EGF has already been shown to stimulate the proliferative activity of Shh on neural cells with stem cell characteristics and to enhance the invasive properties of epidermal cells expressing Shh (10, 68, 69). We present evidence that the transcriptional activity and target gene profiles of GLI1 and GLI2 are modulated by concomitant EGFR signaling in human keratinocytes and hair follicles and propose that combined EGFR-HH/GLI signaling is involved in keratinocyte proliferation, outer root sheath specification, and hair follicle growth.
MATERIALS AND METHODS
Cell culture and retroviral transgene expression.
Retroviral expression of GLI1 and GLI2* in primary human keratinocytes and N/TERT-1 keratinocytes was done as described previously (75). Following retroviral transduction, cells were first cultured for 36 h in defined, serum-free keratinocyte medium (SFM) in the presence of growth supplement (GS) (Invitrogen) and then in SFM without GS. Twenty-four hours prior to harvesting, cells were either exposed to 10 ng/ml EGF (Sigma) or kept in SFM without GS. Doxycycline-inducible GLI1 and GLI2* HaCaT keratinocytes were cultured as described previously (77). For analysis of ERK1/2 function, cells were seeded at a density of 30 percent confluence in Dulbecco's modified Eagle's medium-10% fetal bovine serum and treated with 50 ng/ml doxycycline (Sigma) to induce transgene expression. 36 h after transgene induction culture media were changed into defined SFM without GS. Twenty-four hours prior to harvesting, cells were treated with 10 ng/ml EGF, 10 μM UO126, or 20 μM LY294002 (Sigma). For protein stability assays, GLI1 and GLI2 expression was induced in tetracycline-regulated HaCaT keratinocytes by doxycycline in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. The next day, medium was replaced with SFM and cells were further kept in the absence of doxycycline to cease transgene expression. Cells were treated overnight with 10 ng/ml EGF, 10 μM UO126, or 25 μM MG132 (Sigma) or a combination thereof as indicated below.
RNA isolation and qRT-PCR analysis.
Real-time quantitative reverse transcriptase PCR (qRT-PCR) analysis was done as described previously (77). In brief, total RNA was isolated using High Pure RNA isolation kit (Boehringer Mannheim). cDNA was synthesized with SuperScript II (RNase H−) reverse transcriptase (Invitrogen) according to the manufacturer's instructions. qRT-PCR analysis was done on a Rotorgene 3000 (Corbett Research) using SYBR green supermix (Bio-Rad Laboratories). The sequences of the primers used for amplification are listed in Table S2 in the supplemental material. Primers used that are not listed in Table S2 in the supplemental material were as described previously (75, 77).
DNA array analysis.
mRNA expression profiling was done as described in references 1 and 75. Array images were analyzed by using AIDA Metrix Array Suite (Raytest). Data were normalized for total signal intensity and statistically analyzed using Significance Analysis of Microarrays 2.1 software (104).
Promoter constructs and luciferase reporter assays.
For the construction of the IL1R2 reporter plasmid, a 255-bp fragment of the 5′ cis-regulatory and untranslated region of human IL1R2 (position −81 to +174; GenBank accession no. NM_004633) was cloned into KpnI/XhoI-digested pGL3 basic vector (Promega) by PCR using the following primers sequences: forward, 5′CATCCTCGAGTCTCACCTTGTTGCTG CTCA3′; and reverse, 5′GAGGGTACCAACTTATGCGGCGTTTCCTT3′. GLI binding sites were mutated by using the QuikChange multisite-directed mutagenesis kit (Stratagene). S100A7, S100A9, CCND1, and JAG2 reporter constructs were generated by PCR-based cloning of the promoter regions as shown in Fig. 3A into pGL3-basic (JAG2) or pGL3-promoter (S100A7, S100A9, and CCND1) vector (Promega) by using the primers listed in Table S2 in the supplemental material. Luciferase reporter assays in HaCaT keratinocytes (11) cultured in complete medium and 293 cells were done as described previously (76). To measure the effect of EGF treatment on GLI-induced activity, doxycycline-inducible GLI1 or GLI2 HaCaT cells were grown to 80-percent confluence. Gene expression was induced by adding 50 ng/ml doxycycline for 24 h. Cells were then transfected with respective reporter constructs, mutated controls (IL1R2) or PTCH reporter in combination with renilla pRL-SV40 plasmid (Promega) for normalization and treated with 10 ng/ml EGF for 24 h or left untreated in defined SFM with GS. Luciferase assays were done using a dual-luciferase reporter assay system (Promega) and signals measured on a Lucy II luminometer (Anthos).
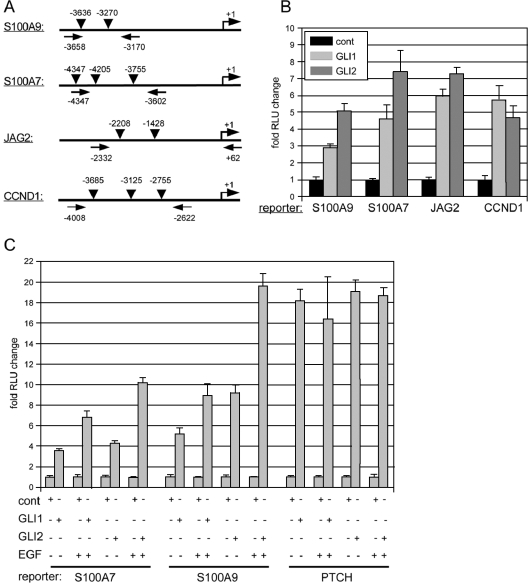
FIG. 3.
Activation of S100A9, S100A7, JAG2, and CCND1 promoters in response to GLI and EGF signaling. RLU, relative light units; cont, control. (A) Illustration of the location of Gli binding sites in the promoter region of S100A9, S100A7, JAG2, and CCND1. Arrows mark the positions of forward and reverse PCR primers used for cloning of the corresponding luciferase reporter constructs. (B) Reporter assays showing activation of luciferase activity in response to GLI1 and GLI2* in 293 cells. (C) Enhanced induction of S100A7 and S100A9 but not of PTCH reporter activity by combinatorial GLI1 (GLI2*)/EGF signaling in HaCaT keratinocytes.
Protein expression and EMSAs.
For the production of recombinant GLI1 protein, a region encompassing part of the N terminus and the entire five-zinc finger DNA binding domain (amino acids 209 to 397) was PCR amplified using GLI1 cDNA as the template (45) and the following primers: forward, 5′GAACTAG TTTCAACTCGATGAC3′; and reverse, 5′TAGCAAGCTTGTGCCGTTTGGTCA3′. The amplicon was subcloned into the Escherichia coli expression vector pHIS-Parallel1 (92). Protein expression, purification, and electrophoretic mobility shift assays (EMSAs) were done as described previously (38).
Western blot analysis, protein stability, and proliferation assays.
Western blot analysis of GLI1 and GLI2 protein expression was done according to standard procedures using polyclonal goat anti-GLI1 antibody (C-18; Santa Cruz Biotechnology), polyclonal goat anti-GLI2 antibody (N-20; Santa Cruz Biotechnology), and a secondary donkey anti-goat horseradish peroxidase (HRP)-conjugated antibody (Santa Cruz Biotechnology). Phosphorylated ERK1/2 (p-ERK1/2) protein was detected using an anti-phospho-p44/42 mitogen-activated protein kinase (MAPK) antibody (Thr202/Tyr204 [E10]; Cell Signaling Technology), phospho-AKT (Ser473) protein with monoclonal mouse antibody (587F11; Cell Signaling Technology). All incubations were done in the presence of 100 μM NaVO4. For total ERK1/2 and for total AKT protein detection, polyclonal rabbit anti-p44/42 MAPK and anti-AKT antibody, respectively (Cell Signaling Technology), and a secondary goat anti-rabbit HRP-conjugated antibody were used (Santa Cruz Biotechnology). Proteins were visualized with ECL detection system (Amersham Biosciences). Densitometric quantification of GLI1 and GLI2 proteins was done using Aida Image Analyzer version 3.10 software (Raytest). S-phase activity of HaCaT keratinocytes was determined by bromodeoxyuridine (BrdU) incorporation as described previously (77), except that Alexa Fluor 594-conjugated antibromodeoxyuridine mouse antibody (Molecular Probes) was used.
Immunohistochemistry.
Specimens from paraffin-embedded skin tissue were stained for IL1R2, EGFR, and phospho-ERK1/2 by using standard streptavidin-biotin-peroxidase (StreptABComplex/HRP Duet kit; DakoCytomation, Glostrup, Denmark) and double-immunofluorescence techniques.
Before antibody application unspecific staining was prevented by incubation with rabbit normal serum or goat normal serum (DakoCytomation) for 10 min at room temperature. Following preincubation for 10 min at room temperature with rabbit (Novacastra) or goat normal serum (DakoCytomation), proteins were detected with polyclonal goat anti-IL1R2 antibody (1:25; RnD Systems), monoclonal mouse anti-EGFR antibody (1:20; DakoCytomation), and monoclonal mouse anti-phospho-p44/42 MAPK (1:50) (Thr202/Tyr204 [E10]; Cell Signaling Technology) antibody. Primary antibodies were incubated at 4°C overnight. Detection was performed using biotinylated goat anti-mouse (1:200) or biotinylated rabbit anti-goat (1:400) antibody and horseradish peroxidase-streptavidin-biotin complex (DakoCytomation), followed by development with diaminobenzidine. Sections were counterstained with hematoxylin. For double-immunofluorescence staining primary antibodies were applied simultaneously followed by incubation with biotinylated rabbit anti-goat antibody (1:200), Alexa Fluor 555-goat anti-goat antibody (1:100; Molecular Probes) and fluorescein isothiocyanate-streptavidin (1:50; Molecular Probes). Microscopic analysis was done on an Olympus IX 70 microscope equipped with a SPOT charge-coupled-device camera (Diagnostic Instruments Inc.).
RESULTS
EGF signaling modulates GLI-regulated gene expression patterns in human keratinocytes.
We first addressed whether EGF signaling modifies the transcriptional response of epidermal cells to GLI1. To analyze this on a more global scale, we performed gene expression profiling of human keratinocytes with either GLI1 or EGF signaling or a combination of both signals activated. For a biological system that allows the controlled activation of distinct signaling pathways under serum-free conditions, we used the human keratinocyte line N/TERT-1, which has been immortalized by stable hTERT expression (53). N/TERT-1 keratinocytes have retained their responsiveness to typical growth control and differentiation signals yet show enhanced survival in the absence of exogenous growth stimuli compared to primary keratinocytes (53) (M. Kasper and F. Aberger, unpublished). N/TERT-1 keratinocytes therefore represent an excellent model system with which to study changes in gene expression in response to well-defined stimuli.
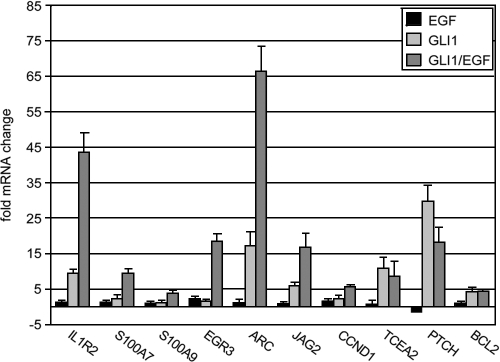
To analyze whether EGF signaling can modulate the biological activity of HH/GLI in keratinocytes, N/TERT cells were retrovirally transduced with human GLI1 (45) or control transduced with inactive GLI1 lacking the nuclear localization signal (GLI1mNLS). Transduced cells were either treated with 10 ng/ml EGF for 24 h or left untreated. Changes in the transcriptional profiles were analyzed by microarrays representing a set of 2,135 human transcripts (75) (see Table S1 in the supplemental material) followed by qRT-PCR validation, the results of which are shown in Table 1. According to the transcriptional response to GLI1 and EGF treatment, we defined two classes of differentially regulated genes: class I genes (n = 19) showed synergistic transcriptional induction by GLI1 and EGF (GLI1/EGF > GLI1 + EGF), e.g., IL1R2 mRNA levels were induced 2.4-fold (ARC, 1.7-fold; S100A9, 1.2-fold; S100A7, 2.3-fold; EGR3, 7.7-fold; JAG2, 1.9-fold) by EGF, 9.9-fold (ARC, 1,097.5-fold; S100A9, −1.6-fold; S100A7, 9.8-fold; EGR3, 1.2-fold; JAG2, 19.0-fold) by GLI1, and 999.5-fold (ARC, 11,585.2-fold; S100A9, 24.3-fold; S100A7, 1,351.1-fold; EGR3, 652.6-fold; JAG2, 90.5-fold) by simultaneous GLI1/EGF treatment.
TABLE 1.
qRT-PCR analysis of GLI1/EGF-regulated target gene expression in human keratinocytes
| HUGO gene designation | Basis for nomenclaturea | NCBI accession no. | Fold mRNA induction change compared to untreated controls (SD)
|
||
|---|---|---|---|---|---|
| EGF | GLI1 | GLI1/EGF | |||
| Class I (GLI1/EGF > GLI1 + EGF) | |||||
| EGR3 | Early growth response 3 | NM_004430 | 7.7 (2.1) | 1.2 (0.3) | 652.6 (192.2) |
| S100A7 | S100 calcium binding protein A7 (psoriasin 1) | NM_002963 | 2.3 (0.6) | 9.8 (2.5) | 1,351.1 (156.3) |
| IL1R2 | Interleukin-1 receptor, type II | NM_004633 | 2.4 (0.3) | 9.9 (0.4) | 999.5 (165.3) |
| S100A9 | S100 calcium binding protein A9 (calgranulin B) | NM_002965 | 1.2 (0.2) | −1.6 (0.3) | 24.3 (3.2) |
| FOXA2 | Forkhead box A2 | NM_021784 | 76.1 (12.9) | 14.4 (2.4) | 494.6 (65.6) |
| ARC | Activity-regulated cytoskeleton-associated protein | NM_015193 | 1.7 (0.4) | 1,097.5 (98.4) | 11,585.2 (967.3) |
| LNK | Lymphocyte specific adapter protein Lnk (signal transduction protein Lnk) (lymphocyte adapter protein) | NM_005475 | 2.5 (0.5) | 1.5 (0.4) | 9.2 (0.9) |
| ID2 | Inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | NM_002166 | 1.7 (0.6) | 6.5 (0.9) | 33.1 (4.9) |
| JAG2 | Jagged 2 | NM_002226 | 1.9 (0.3) | 19.0 (2.4) | 90.5 (11.3) |
| MAPK11 | Mitogen-activated protein kinase 11 | NM_002751 | −1.1 (0.2) | −2 (0.7) | 2.1 (0.2) |
| TUFM | Tu translation elongation factor, mitochondrial | NM_003321 | 1.3 (0.2) | −1.2 (0.3) | 2.9 (0.4) |
| PHLDA2 | Pleckstrin homology-like domain, family A, member 2 | NM_003311 | 2.7 (0.4) | 5.3 (0.4) | 17.1 (1.7) |
| IDS | Iduronate 2-sulfatase (Hunter syndrome) | NM_000202 | 1.9 (0.2) | 2.2 (0.1) | 7.0 (0.4) |
| CDC34 | Cell division cycle 34 | NM_004359 | 1.0 (0.2) | 3.7 (0.2) | 10.6 (1.7) |
| ULK1 | Unc-51-like kinase 1 (C. elegans) | NM_003565 | −1.4 (0.3) | 8.3 (1.6) | 21.9 (2.1) |
| CCND1 | Cyclin D1 | NM_053056 | 1.5 (0.1) | 2.5 (0.3) | 5.1 (0.6) |
| RXRA | Retinoid X receptor, alpha | NM_002957 | −1.1 (0.3) | 6.1 (0.8) | 11.3 (1.9) |
| CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | NM_004360 | 1.0 (0.3) | 1.9 (0.2) | 3.5 (0.4) |
| RPS6KA1 | Ribosomal protein S6 kinase, 90-kDa, polypeptide 1 | NM_002953 | 1.2 (0.4) | 1.1 (0.1) | 1.8 (0.1) |
| Class II (GLI1/ EGF ∼ GLI) | |||||
| PTCH | Patched homolog (Drosophila) | NM_000264 | −2.1 (0.3) | 29.9 (3.1) | 19.7 (2.7) |
| BCL2 | B-cell CLL/lymphoma 2 | NM_000633 | 2.2 (0.4) | 7.2 (0.9) | 7.7 (0.6) |
| CTSL | Cathepsin L | NM_001912 | 1.3 (0.2) | 12.7 (2.0) | 8.8 (1.4) |
| TCEA2 | Transcription elongation factor A (SII) 2 | NM_198723 | −1.6 (0.2) | 123.6 (12.1) | 119.4 (17.6) |
| FYN | FYN oncogene protein related to SRC, FGR, YES | NM_002037 | 1.2 (0.2) | 4.8 (0.3) | 8.3 (0.5) |
| DGCR6 | DiGeorge syndrome critical region gene 6 protein | NM_005675 | −1.2 (0.2) | 2.3 (0.1) | 2.3 (0.2) |
| PITRM1 | Pitrilysin metallopeptidase 1 | NM_014889 | 1.5 (0.1) | 3.7 (0.4) | 2.4 (0.5) |
| MYLK | Myosin, light polypeptide kinase | NM_053025 | −2.2 (0.7) | 5.1 (1.5) | 3.6 (1.4) |
| MNAT1 | Menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | NM_002431 | 1.4 (0.4) | 1.7 (0.2) | 2.1 (0.3) |
| MYC | v-myc myelocytomatosis viral oncogene homolog protein (avian) | NM_002467 | 1 (0.2) | −4.3 (0.6) | −2.5 (0.2) |
| BNC1 | Basonuclin 1 | NM_001717 | 1.8 (0.2) | −2.8 (0.14) | −2.4 (0.3) |
Nomenclature is per the HUGO database (approved gene names).
By contrast, class II genes (n = 11) were regulated by GLI1 but were largely unchanged by parallel treatment with EGF (for induction, GLI1/EGF ∼ GLI1) (e.g., induction for BCL2, 2.2-fold by EGF, 7.2-fold by GLI1, and 7.7-fold by simultaneous GLI1/EGF treatment). Comparable results were obtained with primary human keratinocytes. Here, the effect of EGF treatment was less pronounced than in N/TERT-1 keratinocytes, which may be due to the significantly lower levels of EGFR in primary keratinocytes (Fig. 1) (data not shown).
FIG. 1.
qRT-PCR validation of GLI1/EGF-regulated gene expression in primary human keratinocytes. Note that the lower induction values compared to N/TERT keratinocytes (see Table 1) are likely to be due to lower expression levels of EGFR.
HH/GLI and EGF signaling converge upstream or at the level of direct GLI target genes.
Having shown that treatment of human keratinocytes with EGF has dramatic effects on the mRNA profile of GLI1-expressing keratinocytes, we next addressed the levels at which EGF and HH/GLI signaling may interact. Though the unresponsiveness of the BCL2 and PTCH direct GLI target genes to EGF (Table 1) points to convergence below the level of downstream target genes, signaling induced via EGF may well converge at the level of direct GLI target genes in a promoter-specific manner. To test whether convergence of the two signal transduction pathways can occur at the level of direct GLI target gene promoters, we used the ScanAce algorithm (81) to scan 5 kb of the upstream regulatory region of all 19 GLI1/EGF-regulated genes for the presence of multiple GLI binding sequences. As shown in Table 2, eight GLI1/EGF-regulated genes had at least two GLI binding sites in their promoter region. This suggests that these genes represent putative direct GLI target genes whose activation depends on concomitant EGF signaling.
TABLE 2.
Putative GLI binding sites in GLI/EGF-regulated promoters
| Gene | GLI binding site sequence | Position relative to TSSa | Reference or source |
|---|---|---|---|
| IL1R2 | CACCACCCA | −2998 | 110 |
| GATCACCCA | −45 | This report | |
| TGCCACCCA | 28 | This report | |
| CACCACCCG | 51 | This report | |
| ARC | GCCCACCCA | −1890 | 110 |
| GCCCACCCA | −469 | 110 | |
| TGCCACCCA | −4245 | This report | |
| S100A7 | CACCACCCA | −4205 | 110 |
| CACCACCCA | −3755 | 110 | |
| GAACACCCA | −4347 | 89 | |
| TGCCACCCA | −3915 | This report | |
| S100A9 | GCCCACCCA | −3270 | 110 |
| TGCCACCCA | −3636 | This report | |
| LNK | GATCACCCA | −3054 | This report |
| CACCGCCCA | −522 | 98; C. Schmid and F. Aberger, unpublished | |
| JAG2 | CACCGCCCA | −2208 | 98; C. Schmid and F. Aberger, unpublished |
| CACCGCCCA | −1428 | 98; C. Schmid and F. Aberger, unpublished | |
| ULK1 | CACCACCCG | −1854 | This report |
| GCCCACCCG | −260 | 21 | |
| CCND1 | GACCTCCCA | −4520 | 110 |
| CACCACCCG | −2755 | This report | |
| GCCCACCCG | −3685 | 21 | |
| GCCCACCCG | −3125 | 21 | |
| Consensus | GACCACCCA | 46 |
TSS, transcriptional start site.
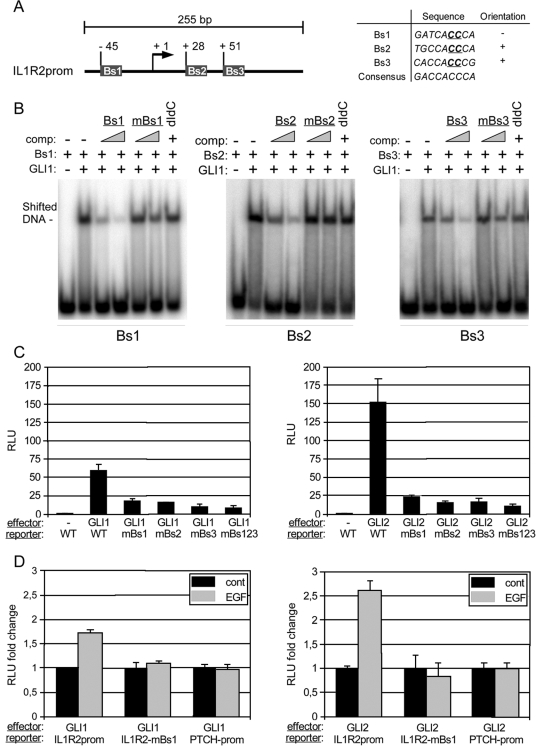
To experimentally verify that EGF signaling can modulate the transcriptional response of a subset of direct GLI target genes, we analyzed in detail a 255-base-pair fragment (position −81 to +184) of the IL1R2 promoter region comprising a cluster of three predicted GLI binding sites in the proximal region of the IL1R2 promoter (Fig. A2). By EMSA, we found that the DNA-binding domain of GLI1 specifically bound to all three binding sites in vitro (Fig. 2B). In luciferase assays, GLI1 activated the IL1R2 promoter more than 20-fold compared to controls. Of note, induction of reporter activity by GLI1 was dramatically reduced upon mutation of any of the three GLI binding sites (Fig. 2C, left graph). We also analyzed activation of the IL1R2 promoter in response to an activator form of GLI2 (GLI2*) (80, 97). Similar to GLI1, GLI2* induced IL1R2 reporter activation, which was abolished by mutation of any of the three GLI binding sites (Fig. 2C, right graph).
FIG. 2.
Direct activation of the IL1R2 promoter by combinatorial GLI1/EGF signaling. Bs, binding site; mBs, mutated binding site; RLU, relative light units. (A) Illustration of the localization of three putative GLI binding sites in the 255-bp promoter (prom) fragment of the IL1R2 upstream regulatory region. The sequence of the three binding sites is shown in the table (right panel). For mutated binding sites used in EMSA (mBs1, mBs2, and mBs3), two essential C nucleotides (underlined) were changed to two G nucleotides. (B) EMSA analysis of the three GLI binding sites showing that all three sites are bound by recombinant DNA binding domain of GLI1. To demonstrate specific binding, competition experiments (comp) with increasing amounts of oligonucleotide corresponding either to wild-type or mutated binding site sequences were done. Furthermore, poly(dI-dC) · poly(dI-dC) (dIdC) was added to the binding reactions to test for unspecific binding. (C) Luciferase reporter assays showing that both GLI1 (left) and active GLI2* (right) efficiently stimulate reporter activity from the 255-bp IL1R2 promoter fragment. Note that mutation of any of the three GLI binding sites is sufficient to abrogate reporter activation. (D) Activation of the IL1R2 promoter by GLI1 and active GLI2* is enhanced by simultaneous activation of EGF signaling. Mutation of Bs1 in the IL1R2 promoter (IL1R2-mBs1) inhibits the stimulatory effect of EGF signaling. cont, control. Promoter (prom) activation of the known direct GLI target PTCH is unaffected by EGF signaling. Reporter assays shown in panels C and D were done in HaCaT keratinocytes.
We next tested whether the treatment of epidermal cells with EGF is able to boost the GLI-mediated induction of the IL1R2 promoter. For this purpose, we transfected HaCaT keratinocytes expressing GLI1 or GLI2* under tetracycline control (75, 77), either with IL1R2 wild-type or mutated promoter constructs and monitored luciferase activation in response to the addition of EGF. For comparison, the same experiment was done with a human PTCH luciferase reporter, containing either wild-type or mutated GLI binding sites. As shown in Fig. 2D, addition of EGF resulted in 60-percent and 250-percent increases of IL1R2 promoter activity in GLI1 and GLI2* expressing cells, respectively. By contrast, activation of the PTCH promoter was not affected by EGF treatment.
To provide further evidence that EGF and HH/GLI signaling synergistically act on the promoters of selected direct GLI target genes, we cloned the upstream-regulatory regions of S100A7, S100A9, JAG2, and CCND1 comprising the Gli binding sites and performed luciferase reporter assays. Figure 3A illustrates the location of Gli binding sites in the respective promoter regions and the position of the primers used for cloning. The reporter constructs were first tested in 293 human embryonic kidney cells for induction by GLI1 and GLI2*. As shown in Fig. 3B, all reporter constructs were activated by GLI1 and GLI2*. In addition, we assayed the S100A7 and S100A9 promoter regions comprising the Gli binding sites for hyperinduction by parallel EGF signaling. GLI1 and GLI2* expressing HaCaT keratinocytes were transfected with S100A7 or S100A9 reporter constructs and subsequently treated with EGF or left untreated. Similar to the case with the IL1R2 promoter fragment, the addition of EGF significantly increased GLI1/GLI2*-induced reporter activity driven by the S100A7 and S100A9 promoter fragments, while GLI1/GLI2*-induced PTCH reporter activity was unaffected by EGF. In the absence of GLI effector, EGF had no effect on reporter activity (Fig. 3C).
Taken together, these data identify IL1R2 (an antagonist of interleukin-1 [IL-1] signaling [17]), S100A7, S100A9, JAG2, and CCND1 genes as direct GLI/EGF-regulated target genes, providing evidence that EGF signaling converges upstream or at the level of GLI target promoters to selectively modulate the target gene profiles for GLI1 and GLI2 in epidermal cells.
EGFR and MEK/ERK signaling are required for synergistic activation of GLI target genes by EGF.
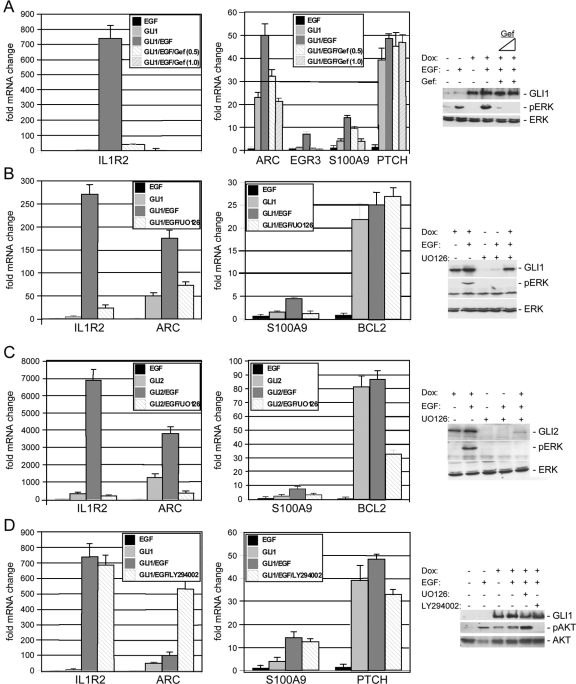
EGF initiates signal transduction by binding to its specific tyrosine kinase receptor, EGFR. Signaling by the EGFR is an intricate process and is relayed towards the nucleus mainly by the RAF/MEK/ERK and PI3K/AKT signaling routes (14, 91, 109). To analyze which signaling pathways are required for the synergistic activation of GLI target genes by EGF, we first addressed whether the effect of EGF on GLI target genes is specifically relayed via EGFR. As shown in Fig. 4A, cooperative activation of GLI/EGF targets (IL1R2, ARC, EGR3, and S100A9 genes) but not of PTCH and BCL2 GLI target genes (data not shown) was efficiently blocked by the addition of the highly specific EGFR inhibitor gefitinib (IRESSA; AstraZeneca), showing that EGFR signaling cooperates with GLI to regulate selected target genes. The efficacy of gefitinib as an EGFR inhibitor was also demonstrated by preventing phosphorylation of ERK1/2 in response to EGF (Fig. 4A, right panel). We next assessed the impact of pharmacologic inhibition of MEK/ERK and PI3K/AKT signaling on the transcriptional response of GLI/EGF target genes. We found that inhibition of MEK/ERK signaling by treatment of GLI1 or GLI2* expressing keratinocytes with the selective MEK1/2 inhibitor UO126 abolished the synergistic effect of EGF on the transcription of IL1R2, ARC, and S100A9 genes (Fig. 4B and C). Inhibition of PI3K/AKT by LY294002 treatment did not affect the EGF-dependent activation of GLI/EGF targets (Fig. 4D). We noticed that LY294002 treatment caused a dramatic upregulation of ARC mRNA levels, suggesting that PI3K/AKT signaling may play a role in repressing ARC transcription. Consistent with their EGF-independent activation, induction of BCL2 and PTCH mRNA expression by GLI1 was affected neither by UO126 nor by LY294002 treatment (Fig. 4B and D). These data suggest that modulation of GLI target gene transcription by EGFR signaling involves activation of the RAF/MEK/ERK pathway.
FIG. 4.
Inhibition of EGFR and MEK/ERK signaling abrogates synergistic activation of GLI/EGF target genes. (A through D) qRT-PCR analysis of GLI/EGF (IL1R2, S100A9, and ARC [and EGR3 in panel A]) and EGF-insensitive GLI target genes (PTCH and BCL2 genes) in HaCaT keratinocytes expressing a doxycycline (Dox)-regulated conditional allele of GLI1 (A, B, and D) or GLI2* (C). (A) Inhibition of EGFR signaling by gefitinib (Gef) (at 0.5 μM and 1.0 μM) abrogates synergistic activation of GLI1/EGF target genes (IL1R2 [left graph] and ARC, EGR3, and S100A9 [right graph] genes) but does not affect the induction of PTCH mRNA. (Right panel) Control Western blot showing efficient inhibition of EGF-induced ERK phosphorylation by gefitinib (0.5 μM and 1.0 μM). (B and C) Synergistic induction of GLI/EGF target gene expression by EGF is abolished by simultaneous treatment with UO126, a selective inhibitor of MEK1/2 function that prevents activation and phosphorylation of ERK1/2 (pERK), respectively. mRNA expression of BCL2 was unaffected by EGF and UO126 treatment in GLI1-expressing cells (B) yet was reduced by UO126 in GLI2-expressing cells (C). Control Western blot analysis of transgene and activated ERK1/2 (pERK) expression in GLI1 (B) and GLI2 (C) HaCaT cells treated with EGF and/or UO126, showing that UO126 treatment efficiently blocked phosphorylation of ERK1/2. Note that the GLI2 protein level is decreased by UO126 treatment. Total ERK protein was monitored as loading control. (D) Inhibition of PI3K/AKT signaling by LY294002 treatment reduces neither synergistic activation of GLI1/EGF target genes nor GLI1-mediated activation of PTCH mRNA expression. (Right panel) Control Western blot showing inhibition of AKT phosphorylation (pAKT) by the PI3K inhibitor LY294002.
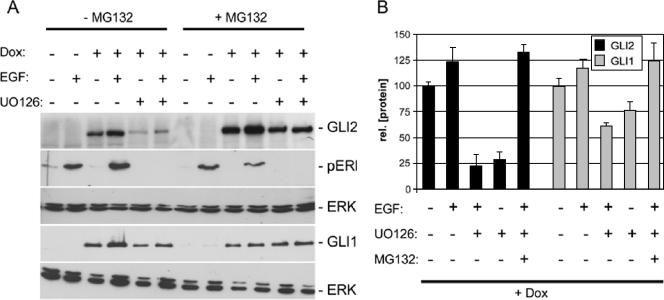
When we analyzed the effect of MEK/ERK inhibition in GLI2*-expressing cells, we obtained comparable results for the EGF/GLI-regulated IL1R2, ARC, and S100A9 genes but noticed a significant decrease in the BCL2 and PTCH mRNA levels upon inhibition of MEK/ERK signaling (Fig. 4C) (data not shown). Control Western blots showed that simultaneous treatment of GLI2* expressing cells with EGF and UO126 results in a dramatic decrease in GLI2* protein levels. To test whether this is due to decreased GLI protein stability in the absence of active ERK1/2, we quantitatively analyzed the effect of ERK1/2 inhibition on the stability of GLI1 and GLI2*. We used a conditional expression system that allows temporal control of GLI1 and GLI2 protein production in a doxycycline-dependent manner (Fig. 5) (77). We induced GLI1 and GLI2 expression by adding doxycycline for 72 h. Following the removal of doxycycline to cease transgene production, cells were treated for 24 h with EGF or a combination of EGF and UO126. Controls were treated with solvent only (dimethyl sulfoxide). To analyze whether the decrease in GLI protein levels in the absence of ERK1/2 function is due to degradation by the proteasome machinery, one sample set was also treated with the proteasome inhibitor MG132. Figure 5 shows that GLI1 and GLI2 are stable after doxycycline withdrawal. The addition of EGF resulted in a slight increase in the amount of GLI1 or GLI2 protein. Inhibition of ERK1/2 function by UO126 treatment reduced the amount of GLI2* protein by about 75 percent. GLI1 protein levels decreased by only 35 to 40 percent. Simultaneous inhibition of the proteasome by MG132 treatment efficiently reversed the destabilizing effect of UO126, suggesting that functional MEK/ERK signaling is required to prevent degradation of GLI2 and also of GLI1 by the proteasome. In this study, we analyzed an activator form of GLI2 lacking part of the N-terminal domain including a conserved destabilizing signal that has recently been identified to contribute to ubiquitin/proteasome-mediated degradation of Gli1 protein (35). The more pronounced decrease in GLI2* compared to GLI1 may therefore in part be due to the use of a possibly stabilized GLI2 form as reference value.
FIG. 5.
ERK function is required for GLI protein stability. Dox, doxycycline. (A) Western blot analysis of GLI1 and GLI2 protein stability (rel. [protein]) in response to EGF signaling and ERK1/2 inactivation. GLI2 protein is destabilized upon inhibition of ERK1/2. The decrease in GLI1 protein levels by UO126 treatment is less pronounced. p-ERK and total ERK were monitored to control for the effect UO126 and for protein loading, respectively. Concomitant inhibition of proteasome function by MG132 treatment prevents UO126-induced degradation of GLI2 and GLI1. (B) Quantitative analysis of GLI1 and GLI2 protein stability in response to EGF treatment, ERK1/2 inhibition and MG132 treatment. Data represent the mean values and standard deviations calculated from five independent experiments.
Taken together, our results reveal a dual role of MEK/ERK signaling in modulating the response of epidermal cells to HH/GLI pathway activation: first, MEK/ERK signal transduction is essential for EGFR-mediated modulation of GLI1 and GLI2 target gene transcription, and second, active MEK/ERK signaling is involved in the stabilization of GLI1 and GLI2 proteins.
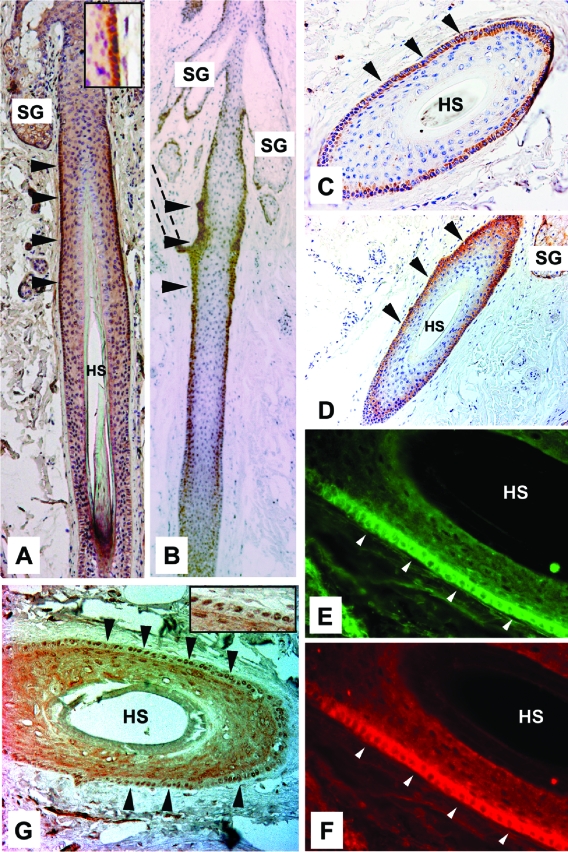
IL1R2, EGFR, and p-ERK are coexpressed in the ORS of human hair follicles.
To assess the in vivo relevance of our data on EGF-dependent modulation of GLI target gene specificity, we analyzed the protein expression patterns of EGFR, active ERK1/2 (p-ERK), and the GLI/EGF target IL1R2 in human skin (Fig. 6). GLI1 and GLI2 have previously been shown to be expressed in the outer root sheath (ORS) of human anagen hair follicles (25, 38). Consistent with our in vitro data, we found strong expression of IL1R2 in the ORS of anagen hair follicles, where staining was restricted to the region below the sebaceous gland (Fig. 6A and C). A similar pattern was observed for EGFR, though staining extended further towards the hair bulb (Fig. 6B and D). Double immunofluorescence analysis clearly revealed coexpression of IL1R2 (Fig. 6E) and EGFR (Fig. 6F) in ORS cells. In line with the activation of IL1R2 expression by synergistic GLI and EGFR/ERK signaling, phosphorylated nuclear p-ERK1/2 protein also localized to the ORS (Fig. 6G). Together with the findings that the JAG2 and CCND1 GLI/EGF target genes are also expressed in the ORS region of anagen hair follicles (74, 107), the results suggest that synergistic signaling of the EGFR and HH/GLI signal transduction pathways shown in vitro may also operate in vivo. However, the data do not rule out the possibility that ERK1/2 may also be activated by signaling pathways other than EGFR.
FIG. 6.
Coexpression of IL1R2, EGFR, and p-ERK in the ORS of human anagen hair follicles. (A) IL1R2 protein is localized to the ORS of human hair follicles below the sebaceous gland. Inset shows intense IL1R2 staining in the ORS at higher magnification. (B) EGFR expression in human hair follicles is primarily detected in the ORS region. The dashed lines outline the arrector pili muscle. (C through G) Cross sections of anagen hair follicles cut at the level of high IL1R2 expression below the sebaceous gland. (C and D) Cross sections of anagen hair follicle stained for IL1R2 (C) and EGFR (D). Staining for IL1R2 and EGFR is restricted to the ORS. (E and F) Double-immunofluorescence analysis showing coexpression of IL1R2 and EGFR in the ORS. (G) p-ERK expression in anagen hair follicles localized in the nucleus of ORS cells. Inset shows nuclear p-ERK1/2 staining at higher magnification. Arrowheads in all panels point at the ORS stained with the corresponding antibody. HS, hair shaft; SG, sebaceous gland.
Modulation of epidermal stem cell marker expression and keratinocyte proliferation by GLI1/EGF signaling.
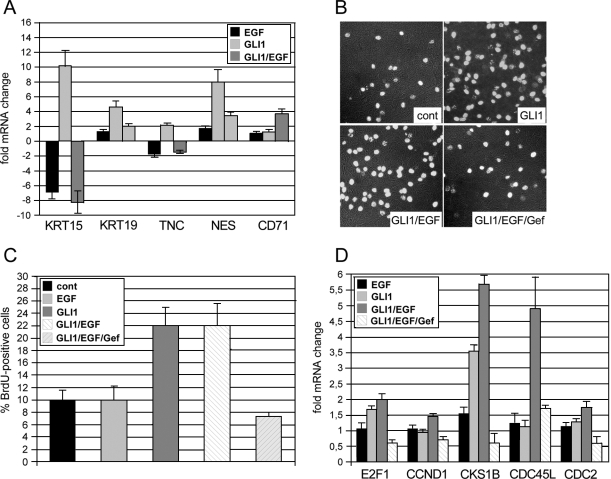
The expression patterns of IL1R2 and EGFR in a region of the ORS that lies in or close to the putative stem cell niche (bulge region) (23) of human hair follicles may point to a role of combined HH/GLI and EGFR signaling in modulating the fate of ORS cells. We addressed this speculation by analyzing the expression of hair follicle stem cell markers (KRT15, KRT19, NES, TNC, and CD71) (52, 60, 96, 103) in human N/TERT-1 keratinocytes in response to GLI1 and EGFR signaling. As shown in Fig. 7A, expression of GLI1 in the absence of extracellular stimuli induced elevated mRNA levels of all stem cell markers tested. Intriguingly, the addition of EGF essentially neutralized the effect of GLI1 such that expression levels of stem cell markers were comparable to control cells transduced with EGFP-expressing retrovirus. Accordingly, mRNA levels of the CD71 gene, which encodes the transferrin receptor that is absent in stem cells (96), increased in response to GLI1/EGF. These results show that GLI1 can elicit a stem cell-like expression signature in epidermal cells and that this signature can be prevented by simultaneous EGF signaling.
FIG. 7.
Regulation of epidermal stemcell marker expression and keratinocyte proliferation by combinatorial GLI/EGFR signaling. cont, control. (A) Expression of GLI1 in the absence of extracellular stimuli resulted in transcriptional activation of the entire panel of epidermal stem cell markers. Simultaneous activation of EGF signaling (GLI1/EGF) abolished GLI1-mediated induction of stem cell marker genes, while the negative stem cell marker CD71 increased in response to combined GLI1/EGF signaling. (B and C) BrdU incorporation analysis showing that GLI1-induced G1/S-phase progression of keratinocytes requires activation of EGFR signaling. Mean values of three independent experiments are shown. (D) qRT-PCR analysis of cell cycle progression genes regulated in response to GLI1 either in the absence or presence of EGF and/or gefitinib. Inhibition of EGFR by gefitinib (0.5 μM) prevents GLI1-mediated activation of cell cycle progression genes.
Loss of stem cell-like expression signatures by treatment of GLI1 keratinocytes with EGF may point to a proliferative role of combinatorial GLI/EGF signaling in human skin. This is supported by the observation that both HH/GLI and EGFR signaling have previously been implicated in proliferative control and growth of human keratinocytes and anagen hair follicles, respectively, and by our own data presented in this study showing enhanced induction of CCND1 expression by concomitant GLI1 and EGF signaling (16, 28, 58, 67, 72, 94, 95). Furthermore, we have previously shown that GLI1 and GLI2 promote G1-phase-to-S-phase progression in serum-containing medium, while they fail to do so under starving conditions, suggesting that additional mitogenic stimuli are required to allow GLI proteins to exert their proliferative activity (75, 77) (G. Regl and F. Aberger, unpublished data). We therefore asked whether the proliferative effect of GLI1 on human keratinocytes depends on parallel EGFR signaling. As shown in Fig. 7B and C, expression of tetracycline-inducible GLI1 in confluent HaCaT keratinocyte cultures induced entry into S phase as monitored by BrdU incorporation. EGF alone or in combination with GLI1 did not further increase the number of cells in S phase. However, presence of the selective EGFR-inhibitor gefitinib (0.5 μM) completely abrogated the S-phase-promoting effect of GLI1, suggesting that activation of EGFR signaling is essential for GLI-induced proliferation. At this concentration (and even up to 1.5 μM), gefitinib affects neither the expression of EGF-independent GLI targets, such as PTCH and BCL2 genes, nor the overall cell viability (also see Fig. 4A) (data not shown); concentrations higher than 1.5 μM were not tested. qRT-PCR analysis of cell cycle progression genes revealed, to a variable extent, enhanced expression in response to combinatorial GLI1/EGF signaling compared to either stimulus alone. Consistent with results obtained with BrdU incorporation assays, the addition of 0.5 μM gefitinib inhibited GLI1- and GLI1/EGF-induced expression of cell cycle progression genes, most notably expression of CKS1B and CDC45L (Fig. 7D). The results uncover an essential role for EGFR signaling in GLI-induced cell cycle progression and suggest that combinatorial HH/GLI and EGFR signaling promote keratinocyte proliferation by cooperative induction of cell cycle progression genes.
DISCUSSION
Modulation of GLI target gene profiles by EGF signaling.
Using a microarray-based approach, we set out to investigate the molecular mechanisms by which EGF signaling may modify the transcriptional response of human epidermal cells to GLI1, a key effector of HH signaling.
Activation of EGF signaling in human keratinocytes expressing GLI1 resulted in a number of distinct transcriptional changes that were observed only if both signals, EGF and GLI1, were active. By screening 2,135 genes, we identified a set of 19 genes whose transcription was synergistically induced by simultaneous activation of GLI1 and EGFR signaling. Assuming there are 25,000 to 30,000 genes in the human genome, the total number of genes regulated by combinatorial GLI1/EGFR signaling in human keratinocytes is likely to be in the range of 200 to 300, suggesting that EGFR signaling has a substantial impact on the transcriptional response of epidermal cells to HH/GLI signaling. Analysis of EGF-dependent target gene expression in keratinocytes expressing an activator form of GLI2 showed that the effect of EGFR signaling on GLI target gene specificity was not GLI1 specific but applied to the major epidermal HH effector GLI2.
By analyzing the promoter regions of GLI/EGF-regulated genes, we identified IL1R2, S100A7, S100A9, CCND1, and JAG2 genes as direct GLI target genes, suggesting that EGFR and HH/GLI signaling can converge at the level of or upstream of the promoters of direct HH/GLI target genes. Of note, the known PTCH (2) and BCL2 (9, 76) direct GLI target genes were not affected by EGF. This clearly shows that only a subset of direct GLI target genes is synergistically regulated by parallel EGFR signaling. Based on these observations, we propose that EGFR signaling alters the target gene profiles for GLI proteins either by directly modifying GLI activator proteins or by regulating other transcription factors or cofactors that may interact with GLI1 and GLI2 to control promoter activation of selected target genes. Alternatively, activation of ERK1/2 via EGF signaling may also enhance the disassembly and degradation of putative transcriptional repressor complexes, such as multimeric YAN/Tel proteins, members of the Ets family of transcriptional regulators (102), which could then allow GLI activators to access the target promoters and stimulate transcription.
The role of EGFR and MEK/ERK signaling in selective modulation of GLI target gene expression and GLI protein stability.
We found that pharmacologic inhibition of EGFR and ERK1/2 but not of PI3K/AKT activation abolished the synergistic activity of EGF signaling on the induction of GLI/EGF target genes, without affecting the activation of the EGF-independent PTCH and BCL2 GLI targets. Two recent reports on the role of MEK/ERK and PI3K/AKT signaling in regulating HH/GLI signaling have suggested a more general involvement of both intracellular pathways in contributing to HH/GLI activity (78, 79). As these data were obtained with cell types other than keratinocytes, this opens up the interesting speculation that the context-dependent behavior of GLI proteins may at least in part result from cell-type-specific cross talk with other signaling pathways.
The data presented in our study clearly show that modulation of selective GLI target gene regulation by EGFR signaling is relayed towards the nucleus via the RAF/MEK/ERK route. Activated ERK1/2 has many substrates and controls gene expression by phosphorylating transcriptional regulators such as MYC, AP1 factors, and members of the Ets, STAT, and Rel family (reviewed in reference 15). Using the Transfac algorithm and binding matrices (Transfac Professional 10.1; Biobase, Germany), we performed in silico analysis of the IL1R2, S100A7, S100A9, CCND1, and JAG2 promoter regions to identify putative binding sequences for ERK-regulated transcriptional regulators. Putative binding sites for several of the transcription factors mentioned above can be localized within the 5-kb upstream cis-regulatory regions, but the analysis did not reveal high-scoring binding motifs in the GLI/EGF-responsive promoter regions tested (data not shown). Although the algorithms available for in silico promoter analysis may miss such motifs, the data support the suggestion that EGFR signaling can modify GLI proteins. Such modifications could enable GLI proteins to interact with cofactors required for the activation of selected target gene promoters or even control interactions among GLI proteins themselves, which have recently been shown to undergo homotypic and heterotypic interactions via their zinc finger domains (61). In this context, it is noteworthy that mutation of any of the three GLI binding sites in the IL1R2 promoter is sufficient to abolish reporter activation in response to GLI1 and GLI2*. As EGFR signaling acts via ERK1/2 activation, a plausible scenario would be direct phosphorylation of GLI proteins by activated ERK1/2. This, however, appears unlikely to be the case, as Riobo et al. recently reported that in vitro kinasing assays with GLI1 as the ERK substrate failed to provide evidence for direct modification of GLI proteins (78). Consistent with this finding, we were unable to identify by phospho-shift gel electrophoresis analysis phosphorylated forms of GLI1 or GLI2 in response to EGF treatment (data not shown). Further studies of additional GLI/EGF target promoters and of potential posttranslational modifications of GLI proteins in response to EGF treatment are needed to address the detailed molecular mechanisms involved in the EGF-dependent regulation of GLI target gene expression.
We provided evidence that ERK1/2 activity, in addition to its requirement for EGFR-modulated GLI target gene expression, stabilizes GLI proteins, particularly GLI2*, by preventing its degradation via the proteasome pathway. Degradation of GLI2* in the absence of ERK1/2 activity did not yield any detectable putative GLI repressor fragments, suggesting that a lack of active ERK1/2 decreases the overall level of GLI2 protein. Compared to GLI2, the stability of GLI1 was less strongly affected by inhibition of ERK1/2 activity. As GLI2 is considered a latent transcription factor that becomes activated upon HH pathway stimulation (12, 58, 90), we propose that ERK1/2 activity augments the stability of latent GLI2 by preventing its degradation by the proteasome, which may contribute to establish sustained competence of a cell to respond to HH ligand stimulation. Recent work on the role of degradation signals in Gli1 has revealed that Gli1 is targeted for ubiquitin/proteasome-mediated degradation by binding to β-TrCP (35). Whether ERK1/2 stabilizes GLI1 and GLI2 proteins by interfering with the formation of β-TrCP-ubiquitin ligase complexes will be an important question to be addressed in future studies.
Putative role of GLI/EGF signaling in skin development and disease.
HH/GLI signaling has been shown to be essential for hair follicle development and morphogenesis (16, 58, 95). Conversely, aberrant expression of HH effectors in the basal layer of the epidermis and the ORS of hair follicles results in hyperproliferation and BCC-like tumors (26, 36, 63, 66, 106). Recent studies suggested that HH/GLI signaling is required in the ORS during the anagen growth phase of the hair follicle and that BCC may preferentially arise from this region of the hair follicle (25, 36, 38, 65), although there is also evidence that these tumors may originate in the interfollicular epidermis (54, 65). We showed that the GLI/EGF target IL1R2 gene is specifically expressed in the ORS of human anagen hair follicles in close proximity to the stem cell region (23). Coexpression of IL1R2 and EGFR in the ORS suggests that in vivo, synergistic HH/GLI and EGFR signaling may specify the fate of ORS cells. Of note, we showed that in the absence of growth stimuli, GLI1 induced expression of a comprehensive panel of epidermal stem cell markers, which was neutralized by the addition of EGF. Furthermore, we provided evidence that activation of EGFR signaling is essential for GLI-induced keratinocyte proliferation, which is likely to involve synergistic activation of key regulators of cell cycle progression, such as CCND1, CDC45L, and CKS1B. It is therefore tempting to speculate that the combined activity of HH/GLI and EGFR signaling provides molecular cues that determine the fate of ORS cells which have left the stem cell niche to promote hair follicle growth. Two additional lines of evidence support this model: (i) IL1r2 expression in murine hair follicles is downregulated in the quiescent population of bulge stem cells compared to nonbulge cells (60) and (ii) both EGFR signaling and HH/GLI signaling have been implicated in the control of human ORS cell proliferation and human hair follicle growth (67, 72). Consistently, Egfr−/− mice display abnormal hair development, most likely due to a deficiency in hair follicle cycling, although the overall EGFR loss-of-function phenotype is highly variable and dependent on the genetic background (28, 57, 94, 100). Experiments addressing the role of HH/GLI signaling in Egfr-deficient mutants are required to better understand the (patho)physiological interaction of these pathways in vivo.
HH/GLI-EGFR signaling in the ORS may also protect anagen hair follicles from IL-1-induced inhibition of growth and hair loss (29, 30, 73) by upregulating the expression of the IL-1-antagonist IL1R2. Another consequence of combined GLI/EGFR signaling may be the activation of Notch signaling by the Notch ligand JAG2. Consistent with the localization of EGFR and GLI proteins in the ORS of anagen hair follicles, Jagged 2 has been shown to be expressed in the ORS of murine hair follicles (74). As Notch signaling appears to play multiple roles in skin and hair follicle development, ranging from control of epidermal proliferation and differentiation to the regulation of hair follicle ORS and stem cell fate (49, 62, 64, 108), additional approaches addressing the relationship of HH/GLI-EGFR and Notch signaling will have to be taken to obtain a better understanding of how, when, and where these pathways possibly interact to control skin development.
Our study also argues for combined inhibition of EGFR and HH/GLI signaling as a putative efficient strategy in the treatment of various malignancies. As both signaling pathways have been implicated in various malignancies, including basal cell carcinoma, pancreatic cancer, prostate cancer, and glioma (8, 10, 19, 26, 31, 37, 42, 63, 66, 85, 87, 99, 106), it is well possible that EGFR and HH/GLI also cooperate in these cancers to promote tumor growth and malignancy. Consistent with this notion, combined administration of the EGFR inhibitor gefitinib and the HH pathway antagonist cyclopamine has recently been shown to be more efficient in inhibiting invasiveness and inducing apoptosis of metastatic prostate cancer cells than either drug alone (59).
Future studies of the detailed molecular events involved in the control of GLI activity by EGFR signaling will help to shed light on the context-dependent activity of these transcription factors during development and will provide a basis for the design of efficient targeted therapies for malignant diseases.
ADDENDUM
While this paper was being revised, two groups have found that Gli2 protein also binds to β-TrCP, resulting in degradation via the ubiquitin-proteasome pathway (8a, 69a). Furthermore, during revision of this paper Eichberger et al. published expression profiling data showing transcriptional regulation of IL1R2, ARC, S100A7, and TCEA2 by GLI1 and GLI2 in human keratinocytes (21a).
Supplementary Material
Acknowledgments
We are grateful to Anna-Maria Frischauf and members of her group for helpful discussions on the project and for critical reading of the manuscript.
This work was supported by the Austrian Science Fund (FWF grant P16518-B14), the Austrian Genome Program GEN-AU, and the priority program “Life Sciences and Health” of the University of Salzburg.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aberger, F., A. P. Costa-Pereira, J. F. Schlaak, T. M. Williams, R. F. O'Shaughnessy, G. Hollaus, I. M. Kerr, and A. M. Frischauf. 2001. Analysis of gene expression using high-density and IFN-gamma-specific low-density cDNA arrays. Genomics 77:50-57. [DOI] [PubMed] [Google Scholar]
- 2.Agren, M., P. Kogerman, M. I. Kleman, M. Wessling, and R. Toftgard. 2004. Expression of the PTCH1 tumor suppressor gene is regulated by alternative promoters and a single functional Gli-binding site. Gene 330:101-114. [DOI] [PubMed] [Google Scholar]
- 3.Altaba, A. R. 1999. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development 126:3205-3216. [DOI] [PubMed] [Google Scholar]
- 4.Aza-Blanc, P., F. A. Ramirez-Weber, M. P. Laget, C. Schwartz, and T. B. Kornberg. 1997. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89:1043-1053. [DOI] [PubMed] [Google Scholar]
- 5.Bai, C. B., W. Auerbach, J. S. Lee, D. Stephen, and A. L. Joyner. 2002. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129:4753-4761. [DOI] [PubMed] [Google Scholar]
- 6.Bai, C. B., D. Stephen, and A. L. Joyner. 2004. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell 6:103-115. [DOI] [PubMed] [Google Scholar]
- 7.Beachy, P. A., S. S. Karhadkar, and D. M. Berman. 2004. Tissue repair and stem cell renewal in carcinogenesis. Nature 432:324-331. [DOI] [PubMed] [Google Scholar]
- 8.Berman, D. M., S. S. Karhadkar, A. Maitra, R. Montes De Oca, M. R. Gerstenblith, K. Briggs, A. R. Parker, Y. Shimada, J. R. Eshleman, D. N. Watkins, and P. A. Beachy. 2003. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature 425:846-851. [DOI] [PubMed] [Google Scholar]
- 8a.Bhatia, N., S. Thiyagarajan, I. Elcheva, M. Saleem, A. Dlugosz, H. Mukhtar, and V. S. Spiegelman. 1 May 2006, posting date. GLi2 is targeted for ubiquitination and degradation by beta-TrCP ubiquitin ligase. J. Biol. Chem. [Online.] doi: 10.1074/jbc.M513203200. [DOI] [PubMed]
- 9.Bigelow, R. L., N. S. Chari, A. B. Unden, K. B. Spurgers, S. Lee, D. R. Roop, R. Toftgard, and T. J. McDonnell. 2004. Transcriptional regulation of bcl-2 mediated by the sonic hedgehog signaling pathway through gli-1. J. Biol. Chem. 279:1197-1205. [DOI] [PubMed] [Google Scholar]
- 10.Bigelow, R. L., E. Y. Jen, M. Delehedde, N. S. Chari, and T. J. McDonnell. 2005. Sonic hedgehog induces epidermal growth factor dependent matrix infiltration in HaCaT keratinocytes. J. Investig. Dermatol. 124:457-465. [DOI] [PubMed] [Google Scholar]
- 11.Boukamp, P., R. T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N. E. Fusenig. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buttitta, L., R. Mo, C. C. Hui, and C. M. Fan. 2003. Interplays of Gli2 and Gli3 and their requirement in mediating Shh-dependent sclerotome induction. Development 130:6233-6243. [DOI] [PubMed] [Google Scholar]
- 13.Callahan, C. A., T. Ofstad, L. Horng, J. K. Wang, H. H. Zhen, P. A. Coulombe, and A. E. Oro. 2004. MIM/BEG4, a Sonic hedgehog-responsive gene that potentiates Gli-dependent transcription. Genes Dev. 18:2724-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carpenter, G. 2000. The EGF receptor: a nexus for trafficking and signaling. Bioessays 22:697-707. [DOI] [PubMed] [Google Scholar]
- 15.Chang, F., L. S. Steelman, J. T. Lee, J. G. Shelton, P. M. Navolanic, W. L. Blalock, R. A. Franklin, and J. A. McCubrey. 2003. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia 17:1263-1293. [DOI] [PubMed] [Google Scholar]
- 16.Chiang, C., R. Z. Swan, M. Grachtchouk, M. Bolinger, Y. Litingtung, E. K. Robertson, M. K. Cooper, W. Gaffield, H. Westphal, P. A. Beachy, and A. A. Dlugosz. 1999. Essential role for Sonic hedgehog during hair follicle morphogenesis. Dev. Biol. 205:1-9. [DOI] [PubMed] [Google Scholar]
- 17.Colotta, F., F. Re, M. Muzio, R. Bertini, N. Polentarutti, M. Sironi, J. G. Giri, S. K. Dower, J. E. Sims, and A. Mantovani. 1993. Interleukin-1 type II receptor: a decoy target for IL-1 that is regulated by IL-4. Science 261:472-475. [DOI] [PubMed] [Google Scholar]
- 18.Dahmane, N., J. Lee, P. Robins, P. Heller, and A. Ruiz i Altaba. 1997. Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature 389:876-881. (Erratum, 390:536.) [DOI] [PubMed] [Google Scholar]
- 19.Dahmane, N., P. Sanchez, Y. Gitton, V. Palma, T. Sun, M. Beyna, H. Weiner, and A. Ruiz i Altaba. 2001. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development 128:5201-5212. [DOI] [PubMed] [Google Scholar]
- 20.Dai, P., H. Akimaru, Y. Tanaka, T. Maekawa, M. Nakafuku, and S. Ishii. 1999. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 274:8143-8152. [DOI] [PubMed] [Google Scholar]
- 21.Eichberger, T., G. Regl, M. S. Ikram, G. W. Neill, M. P. Philpott, F. Aberger, and A. M. Frischauf. 2004. FOXE1, a new transcriptional target of GLI2 is expressed in human epidermis and basal cell carcinoma. J. Investig. Dermatol. 122:1180-1187. [DOI] [PubMed] [Google Scholar]
- 21a.Eichberger, T., V. Sander, H. Schnidar, G. Regl, M. Kasper, C. Schmid, S. Plamberger, A. Kaser, F. Aberger, and A. M. Frischauf. 2006. Overlapping and distinct transcriptional regulator properties of the GLI1 and GLI2 oncogenes. Genomics 87:616-632. [DOI] [PubMed] [Google Scholar]
- 22.Fan, H., A. E. Oro, M. P. Scott, and P. A. Khavari. 1997. Induction of basal cell carcinoma features in transgenic human skin expressing Sonic Hedgehog. Nat. Med. 3:788-792. [DOI] [PubMed] [Google Scholar]
- 23.Fuchs, E., and S. Raghavan. 2002. Getting under the skin of epidermal morphogenesis. Nat. Rev. Genet. 3:199-209. [DOI] [PubMed] [Google Scholar]
- 24.Gailani, M. R., M. Stahle-Backdahl, D. J. Leffell, M. Glynn, P. G. Zaphiropoulos, C. Pressman, A. B. Unden, M. Dean, D. E. Brash, A. E. Bale, and R. Toftgard. 1996. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat. Genet. 14:78-81. [DOI] [PubMed] [Google Scholar]
- 25.Ghali, L., S. T. Wong, J. Green, N. Tidman, and A. G. Quinn. 1999. Gli1 protein is expressed in basal cell carcinomas, outer root sheath keratinocytes and a subpopulation of mesenchymal cells in normal human skin. J. Investig. Dermatol. 113:595-599. [DOI] [PubMed] [Google Scholar]
- 26.Grachtchouk, M., R. Mo, S. Yu, X. Zhang, H. Sasaki, C. C. Hui, and A. A. Dlugosz. 2000. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat. Genet. 24:216-217. [DOI] [PubMed] [Google Scholar]
- 27.Hahn, H., C. Wicking, P. G. Zaphiropoulous, M. R. Gailani, S. Shanley, A. Chidambaram, I. Vorechovsky, E. Holmberg, A. B. Unden, S. Gillies, K. Negus, I. Smyth, C. Pressman, D. J. Leffell, B. Gerrard, A. M. Goldstein, M. Dean, R. Toftgard, G. Chenevix-Trench, B. Wainwright, and A. E. Bale. 1996. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 85:841-851. [DOI] [PubMed] [Google Scholar]
- 28.Hansen, L. A., N. Alexander, M. E. Hogan, J. P. Sundberg, A. Dlugosz, D. W. Threadgill, T. Magnuson, and S. H. Yuspa. 1997. Genetically null mice reveal a central role for epidermal growth factor receptor in the differentiation of the hair follicle and normal hair development. Am. J. Pathol. 150:1959-1975. [PMC free article] [PubMed] [Google Scholar]
- 29.Harmon, C. S., and T. D. Nevins. 1993. IL-1 alpha inhibits human hair follicle growth and hair fiber production in whole-organ cultures. Lymphokine Cytokine Res. 12:197-203. [PubMed] [Google Scholar]
- 30.Hoffmann, R. 1999. The potential role of cytokines and T cells in alopecia areata. J. Investig. Dermatol. Symp. Proc. 4:235-238. [DOI] [PubMed] [Google Scholar]
- 31.Holbro, T., and N. E. Hynes. 2004. ErbB receptors: directing key signaling networks throughout life. Annu. Rev. Pharmacol. Toxicol. 44:195-217. [DOI] [PubMed] [Google Scholar]
- 32.Hooper, J. E., and M. P. Scott. 2005. Developmental cell biology: communicating with hedgehogs. Nat. Rev. Mol. Cell Biol. 6:306-317. [DOI] [PubMed] [Google Scholar]
- 33.Huangfu, D., and K. V. Anderson. 2005. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. USA 102:11325-11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huangfu, D., A. Liu, A. S. Rakeman, N. S. Murcia, L. Niswander, and K. V. Anderson. 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426:83-87. [DOI] [PubMed] [Google Scholar]
- 35.Huntzicker, E. G., I. S. Estay, H. Zhen, L. A. Lokteva, P. K. Jackson, and A. E. Oro. 2006. Dual degradation signals control Gli protein stability and tumor formation. Genes Dev. 20:276-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hutchin, M. E., M. S. Kariapper, M. Grachtchouk, A. Wang, L. Wei, D. Cummings, J. Liu, L. E. Michael, A. Glick, and A. A. Dlugosz. 2005. Sustained Hedgehog signaling is required for basal cell carcinoma proliferation and survival: conditional skin tumorigenesis recapitulates the hair growth cycle. Genes Dev. 19:214-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hynes, N. E., and H. A. Lane. 2005. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer 5:341-354. [DOI] [PubMed] [Google Scholar]
- 38.Ikram, M. S., G. W. Neill, G. Regl, T. Eichberger, A. M. Frischauf, F. Aberger, A. Quinn, and M. Philpott. 2004. GLI2 is expressed in normal human epidermis and BCC and induces GLI1 expression by binding to its promoter. J. Investig. Dermatol. 122:1503-1509. [DOI] [PubMed] [Google Scholar]
- 39.Ingham, P. W., and A. P. McMahon. 2001. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15:3059-3087. [DOI] [PubMed] [Google Scholar]
- 40.Johnson, R. L., A. L. Rothman, J. Xie, L. V. Goodrich, J. W. Bare, J. M. Bonifas, A. G. Quinn, R. M. Myers, D. R. Cox, E. H. Epstein, Jr., and M. P. Scott. 1996. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272:1668-1671. [DOI] [PubMed] [Google Scholar]
- 41.Jordan, J. D., E. M. Landau, and R. Iyengar. 2000. Signaling networks: the origins of cellular multitasking. Cell 103:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karhadkar, S. S., G. S. Bova, N. Abdallah, S. Dhara, D. Gardner, A. Maitra, J. T. Isaacs, D. M. Berman, and P. A. Beachy. 2004. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature 431:707-712. [DOI] [PubMed] [Google Scholar]
- 43.Kasper, M., G. Regl, A. M. Frischauf, and F. Aberger. 2006. GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur. J. Cancer 42:437-445. [DOI] [PubMed] [Google Scholar]
- 44.Kessaris, N., F. Jamen, L. L. Rubin, and W. D. Richardson. 2004. Cooperation between sonic hedgehog and fibroblast growth factor/MAPK signalling pathways in neocortical precursors. Development 131:1289-1298. [DOI] [PubMed] [Google Scholar]
- 45.Kinzler, K. W., J. M. Ruppert, S. H. Bigner, and B. Vogelstein. 1988. The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature 332:371-374. [DOI] [PubMed] [Google Scholar]
- 46.Kinzler, K. W., and B. Vogelstein. 1990. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol. Cell. Biol. 10:634-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lam, C. W., J. Xie, K. F. To, H. K. Ng, K. C. Lee, N. W. Yuen, P. L. Lim, L. Y. Chan, S. F. Tong, and F. McCormick. 1999. A frequent activated smoothened mutation in sporadic basal cell carcinomas. Oncogene 18:833-836. [DOI] [PubMed] [Google Scholar]
- 48.Liu, A., B. Wang, and L. A. Niswander. 2005. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development 132:3103-3111. [DOI] [PubMed] [Google Scholar]
- 49.Lowell, S., P. Jones, I. Le Roux, J. Dunne, and F. M. Watt. 2000. Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr. Biol. 10:491-500. [DOI] [PubMed] [Google Scholar]
- 50.Lum, L., and P. A. Beachy. 2004. The Hedgehog response network: sensors, switches, and routers. Science 304:1755-1759. [DOI] [PubMed] [Google Scholar]
- 51.Lupo, G., Y. Liu, R. Qiu, R. A. Chandraratna, G. Barsacchi, R. Q. He, and W. A. Harris. 2005. Dorsoventral patterning of the Xenopus eye: a collaboration of Retinoid, Hedgehog and FGF receptor signaling. Development 132:1737-1748. [DOI] [PubMed] [Google Scholar]
- 52.Lyle, S., M. Christofidou-Solomidou, Y. Liu, D. E. Elder, S. Albelda, and G. Cotsarelis. 1999. Human hair follicle bulge cells are biochemically distinct and possess an epithelial stem cell phenotype. J. Investig. Dermatol. Symp. Proc. 4:296-301. [DOI] [PubMed] [Google Scholar]
- 53.Mahmood, R., J. Bresnick, A. Hornbruch, C. Mahony, N. Morton, K. Colquhoun, P. Martin, A. Lumsden, C. Dickson, and I. Mason. 1995. A role for FGF-8 in the initiation and maintenance of vertebrate limb bud outgrowth. Curr. Biol. 5:797-806. [DOI] [PubMed] [Google Scholar]
- 54.Mancuso, M., S. Pazzaglia, M. Tanori, H. Hahn, P. Merola, S. Rebessi, M. J. Atkinson, V. Di Majo, V. Covelli, and A. Saran. 2004. Basal cell carcinoma and its development: insights from radiation-induced tumors in Ptch1-deficient mice. Cancer Res. 64:934-941. [DOI] [PubMed] [Google Scholar]
- 55.Mao, J., P. Maye, P. Kogerman, F. J. Tejedor, R. Toftgard, W. Xie, G. Wu, and D. Wu. 2002. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J. Biol. Chem. 22:22. [DOI] [PubMed] [Google Scholar]
- 56.Massague, J., S. W. Blain, and R. S. Lo. 2000. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 103:295-309. [DOI] [PubMed] [Google Scholar]
- 57.Miettinen, P. J., J. E. Berger, J. Meneses, Y. Phung, R. A. Pedersen, Z. Werb, and R. Derynck. 1995. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature 376:337-341. [DOI] [PubMed] [Google Scholar]
- 58.Mill, P., R. Mo, H. Fu, M. Grachtchouk, P. C. Kim, A. A. Dlugosz, and C. C. Hui. 2003. Sonic hedgehog-dependent activation of Gli2 is essential for embryonic hair follicle development. Genes Dev. 17:282-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mimeault, M., E. Moore, N. Moniaux, J. P. Henichart, P. Depreux, M. F. Lin, and S. K. Batra. 2006. Cytotoxic effects induced by a combination of cyclopamine and gefitinib, the selective hedgehog and epidermal growth factor receptor signaling inhibitors, in prostate cancer cells. Int. J. Cancer 118:1022-1031. [DOI] [PubMed] [Google Scholar]
- 60.Morris, R. J., Y. Liu, L. Marles, Z. Yang, C. Trempus, S. Li, J. S. Lin, J. A. Sawicki, and G. Cotsarelis. 2004. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22:411-417. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen, V., A. L. Chokas, B. Stecca, and A. R. Altaba. 2005. Cooperative requirement of the Gli proteins in neurogenesis. Development 132:3267-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nicolas, M., A. Wolfer, K. Raj, J. A. Kummer, P. Mill, M. van Noort, C. C. Hui, H. Clevers, G. P. Dotto, and F. Radtke. 2003. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 33:416-421. [DOI] [PubMed] [Google Scholar]
- 63.Nilsson, M., A. B. Unden, D. Krause, U. Malmqwist, K. Raza, P. G. Zaphiropoulos, and R. Toftgard. 2000. Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing GLI-1. Proc. Natl. Acad. Sci. USA 97:3438-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Okuyama, R., B. C. Nguyen, C. Talora, E. Ogawa, A. Tommasi di Vignano, M. Lioumi, G. Chiorino, H. Tagami, M. Woo, and G. P. Dotto. 2004. High commitment of embryonic keratinocytes to terminal differentiation through a Notch1-caspase 3 regulatory mechanism. Dev. Cell 6:551-562. [DOI] [PubMed] [Google Scholar]
- 65.Oro, A. E., and K. Higgins. 2003. Hair cycle regulation of Hedgehog signal reception. Dev. Biol. 255:238-248. [DOI] [PubMed] [Google Scholar]
- 66.Oro, A. E., K. M. Higgins, Z. Hu, J. M. Bonifas, E. H. Epstein, Jr., and M. P. Scott. 1997. Basal cell carcinomas in mice overexpressing sonic hedgehog. Science 276:817-821. [DOI] [PubMed] [Google Scholar]
- 67.Paladini, R. D., J. Saleh, C. Qian, G. X. Xu, and L. L. Rubin. 2005. Modulation of hair growth with small molecule agonists of the hedgehog signaling pathway. J. Investig. Dermatol. 125:638-646. [DOI] [PubMed] [Google Scholar]
- 68.Palma, V., D. A. Lim, N. Dahmane, P. Sanchez, T. C. Brionne, C. D. Herzberg, Y. Gitton, A. Carleton, A. Alvarez-Buylla, and A. Ruiz i Altaba. 2005. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development 132:335-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Palma, V., and A. Ruiz i Altaba. 2004. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development 131:337-345. [DOI] [PubMed] [Google Scholar]
- 69a.Pan, Y., C. B. Bai, A. L. Joyner, and B. Wang. 2006. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol. Cell. Biol. 26:3365-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park, H. L., C. Bai, K. A. Platt, M. P. Matise, A. Beeghly, C. C. Hui, M. Nakashima, and A. L. Joyner. 2000. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127:1593-1605. [DOI] [PubMed] [Google Scholar]
- 71.Pasca di Magliano, M., and M. Hebrok. 2003. Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer 3:903-911. [DOI] [PubMed] [Google Scholar]
- 72.Philpott, M. P., and T. Kealey. 1994. Effects of EGF on the morphology and patterns of DNA synthesis in isolated human hair follicles. J. Investig. Dermatol. 102:186-191. [DOI] [PubMed] [Google Scholar]
- 73.Philpott, M. P., D. A. Sanders, J. Bowen, and T. Kealey. 1996. Effects of interleukins, colony-stimulating factor and tumour necrosis factor on human hair follicle growth in vitro: a possible role for interleukin-1 and tumour necrosis factor-alpha in alopecia areata. Br. J. Dermatol. 135:942-948. [DOI] [PubMed] [Google Scholar]
- 74.Powell, B. C., E. A. Passmore, A. Nesci, and S. M. Dunn. 1998. The Notch signalling pathway in hair growth. Mech. Dev. 78:189-192. [DOI] [PubMed] [Google Scholar]
- 75.Regl, G., M. Kasper, H. Schnidar, T. Eichberger, G. W. Neill, M. S. Ikram, A. G. Quinn, M. P. Philpott, A. M. Frischauf, and F. Aberger. 2004. The zinc-finger transcription factor GLI2 antagonizes contact inhibition and differentiation of human epidermal cells. Oncogene 23:1263-1274. [DOI] [PubMed] [Google Scholar]
- 76.Regl, G., M. Kasper, H. Schnidar, T. Eichberger, G. W. Neill, M. P. Philpott, H. Esterbauer, C. Hauser-Kronberger, A. M. Frischauf, and F. Aberger. 2004. Activation of the BCL2 promoter in response to Hedgehog/GLI signal transduction is predominantly mediated by GLI2. Cancer Res. 64:7724-7731. [DOI] [PubMed] [Google Scholar]
- 77.Regl, G., G. W. Neill, T. Eichberger, M. Kasper, M. S. Ikram, J. Koller, H. Hintner, A. G. Quinn, A. M. Frischauf, and F. Aberger. 2002. Human GLI2 and GLI1 are part of a positive feedback mechanism in basal cell carcinoma. Oncogene 21:5529-5539. [DOI] [PubMed] [Google Scholar]
- 78.Riobo, N. A., G. M. Haines, and C. P. Emerson, Jr. 2006. Protein kinase C-delta and mitogen-activated protein/extracellular signal-regulated kinase-1 control GLI activation in hedgehog signaling. Cancer Res. 66:839-845. [DOI] [PubMed] [Google Scholar]
- 79.Riobo, N. A., K. Lu, X. Ai, G. M. Haines, and C. P. Emerson, Jr. 2006. Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. USA 103:4505-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roessler, E., A. N. Ermilov, D. K. Grange, A. Wang, M. Grachtchouk, A. A. Dlugosz, and M. Muenke. 2005. A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum. Mol. Genet. 14:2181-2188. [DOI] [PubMed] [Google Scholar]
- 81.Roth, F. P., J. D. Hughes, P. W. Estep, and G. M. Church. 1998. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 16:939-945. [DOI] [PubMed] [Google Scholar]
- 82.Ruiz i Altaba, A., V. Nguyen, and V. Palma. 2003. The emergent design of the neural tube: prepattern, SHH morphogen and GLI code. Curr. Opin. Genet. Dev. 13:513-521. [DOI] [PubMed] [Google Scholar]
- 83.Ruiz i Altaba, A., P. Sanchez, and N. Dahmane. 2002. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat. Rev. Cancer 2:361-372. [DOI] [PubMed] [Google Scholar]
- 84.Ruppert, J. M., B. Vogelstein, and K. W. Kinzler. 1991. The zinc finger protein GLI transforms primary cells in cooperation with adenovirus E1A. Mol. Cell. Biol. 11:1724-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Salomon, D. S., R. Brandt, F. Ciardiello, and N. Normanno. 1995. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit. Rev. Oncol. Hematol. 19:183-232. [DOI] [PubMed] [Google Scholar]
- 86.Sanchez, P., V. Clement, and A. Ruiz i Altaba. 2005. Therapeutic targeting of the Hedgehog-GLI pathway in prostate cancer. Cancer Res. 65:2990-2992. [DOI] [PubMed] [Google Scholar]
- 87.Sanchez, P., A. M. Hernandez, B. Stecca, A. J. Kahler, A. M. DeGueme, A. Barrett, M. Beyna, M. W. Datta, S. Datta, and A. Altaba. 2004. Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. 101:12561-12566. [DOI] [PMC free article] [PubMed]
- 88.Sancho, E., E. Batlle, and H. Clevers. 2004. Signaling pathways in intestinal development and cancer. Annu. Rev. Cell Dev. Biol. 20:695-723. [DOI] [PubMed] [Google Scholar]
- 89.Sasaki, H., C. Hui, M. Nakafuku, and H. Kondoh. 1997. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 124:1313-1322. [DOI] [PubMed] [Google Scholar]
- 90.Sasaki, H., Y. Nishizaki, C. Hui, M. Nakafuku, and H. Kondoh. 1999. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 126:3915-3924. [DOI] [PubMed] [Google Scholar]
- 91.Schlessinger, J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103:211-225. [DOI] [PubMed] [Google Scholar]
- 92.Sheffield, P., S. Garrard, and Z. Derewenda. 1999. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15:34-39. [DOI] [PubMed] [Google Scholar]
- 93.Shin, S. H., P. Kogerman, E. Lindstrom, R. Toftgard, and L. G. Biesecker. 1999. GLI3 mutations in human disorders mimic Drosophila cubitus interruptus protein functions and localization. Proc. Natl. Acad. Sci. USA 96:2880-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sibilia, M., and E. F. Wagner. 1995. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science 269:234-238. [DOI] [PubMed] [Google Scholar]
- 95.St.-Jacques, B., H. R. Dassule, I. Karavanova, V. A. Botchkarev, J. Li, P. S. Danielian, J. A. McMahon, P. M. Lewis, R. Paus, and A. P. McMahon. 1998. Sonic hedgehog signaling is essential for hair development. Curr. Biol. 8:1058-1068. [DOI] [PubMed] [Google Scholar]
- 96.Tani, H., R. J. Morris, and P. Kaur. 2000. Enrichment for murine keratinocyte stem cells based on cell surface phenotype. Proc. Natl. Acad. Sci. USA 97:10960-10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tanimura, A., S. Dan, and M. Yoshida. 1998. Cloning of novel isoforms of the human Gli2 oncogene and their activities to enhance tax-dependent transcription of the human T-cell leukemia virus type 1 genome. J. Virol. 72:3958-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Teh, M. T., S. T. Wong, G. W. Neill, L. R. Ghali, M. P. Philpott, and A. G. Quinn. 2002. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 62:4773-4780. [PubMed] [Google Scholar]
- 99.Thayer, S. P., M. P. di Magliano, P. W. Heiser, C. M. Nielsen, D. J. Roberts, G. Y. Lauwers, Y. P. Qi, S. Gysin, C. Fernandez-del Castillo, V. Yajnik, B. Antoniu, M. McMahon, A. L. Warshaw, and M. Hebrok. 2003. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature 425:851-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Threadgill, D. W., A. A. Dlugosz, L. A. Hansen, T. Tennenbaum, U. Lichti, D. Yee, C. LaMantia, T. Mourton, K. Herrup, R. C. Harris, et al. 1995. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269:230-234. [DOI] [PubMed] [Google Scholar]
- 101.Toftgard, R. 2000. Hedgehog signalling in cancer. Cell Mol. Life Sci. 57:1720-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tootle, T. L., and I. Rebay. 2005. Posttranslational modifications influence transcription factor activity: a view from the ETS superfamily. Bioessays 27:285-298. [DOI] [PubMed] [Google Scholar]
- 103.Tumbar, T., G. Guasch, V. Greco, C. Blanpain, W. E. Lowry, M. Rendl, and E. Fuchs. 2004. Defining the epithelial stem cell niche in skin. Science 303:359-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang, B., J. F. Fallon, and P. A. Beachy. 2000. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100:423-434. [DOI] [PubMed] [Google Scholar]
- 106.Xie, J., M. Murone, S. M. Luoh, A. Ryan, Q. Gu, C. Zhang, J. M. Bonifas, C. W. Lam, M. Hynes, A. Goddard, A. Rosenthal, E. H. Epstein, Jr., and F. J. de Sauvage. 1998. Activating smoothened mutations in sporadic basal-cell carcinoma. Nature 391:90-92. [DOI] [PubMed] [Google Scholar]
- 107.Xu, X., S. Lyle, Y. Liu, B. Solky, and G. Cotsarelis. 2003. Differential expression of cyclin D1 in the human hair follicle. Am. J. Pathol. 163:969-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yamamoto, N., K. Tanigaki, H. Han, H. Hiai, and T. Honjo. 2003. Notch/RBP-J signaling regulates epidermis/hair fate determination of hair follicular stem cells. Curr. Biol. 13:333-338. [DOI] [PubMed] [Google Scholar]
- 109.Yarden, Y., and M. X. Sliwkowski. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2:127-137. [DOI] [PubMed] [Google Scholar]
- 110.Yoon, J. W., Y. Kita, D. J. Frank, R. R. Majewski, B. A. Konicek, M. A. Nobrega, H. Jacob, D. Walterhouse, and P. Iannaccone. 2002. Gene expression profiling leads to identification of GLI1-binding elements in target genes and a role for multiple downstream pathways in GLI1-induced cell transformation. J. Biol. Chem. 277:5548-5555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.