Abstract
In Saccharomyces cerevisiae, zinc cluster protein Pdr1 can form homodimers as well as heterodimers with Pdr3 and Stb5, suggesting that different combinations of these proteins may regulate the expression of different genes. To gain insight into the interplay among these regulators, we performed genome-wide location analysis (chromatin immunoprecipitation with hybridization to DNA microarrays) and gene expression profiling. Unexpectedly, we observed that Stb5 shares only a few target genes with Pdr1 or Pdr3 in rich medium. Interestingly, upon oxidative stress, Stb5 binds and regulates the expression of most genes of the pentose phosphate pathway as well as of genes involved in the production of NADPH, a metabolite required for oxidative stress resistance. Importantly, deletion of STB5 results in sensitivity to diamide and hydrogen peroxide. Our data suggest that Stb5 acts both as an activator and as a repressor in the presence of oxidative stress. Furthermore, we show that Stb5 activation is not mediated by known regulators of the oxidative stress response. Integrity of the pentose phosphate pathway is required for the activation of Stb5 target genes but is not necessary for the increased DNA binding of Stb5 in the presence of diamide. These data suggest that Stb5 is a key player in the control of NADPH production for resistance to oxidative stress.
In the yeast Saccharomyces cerevisiae, a number of transcription factors that are members of the binuclear zinc cluster family regulate transcription of genes involved in a wide variety of cellular processes. These transcriptional regulators contain six well-conserved cysteines that bind two zinc atoms (CysX2CysX6CysX5-12CysX2CysX6-8Cys), coordinating folding of the domain involved in DNA binding (18, 49). The cysteine-rich region is usually followed by a short linker sequence that bridges the zinc finger to a dimerization domain. In most cases, the DNA binding domain is located at the N terminus, while an acidic activation domain is found at the C terminus tail (54). A region of low homology of approximately 80 amino acids, called the middle homology region, is found in many zinc cluster proteins, is located between the cysteine-rich region and the acidic portion, and may be involved in controlling their transcriptional activities (54). Members of the binuclear zinc cluster family are unique to fungi and are involved in a wide range of processes, including primary and secondary metabolism, drug resistance, and meiotic development (58). A well-characterized member is Gal4, a positive regulator of the expression of genes involved in galactose catabolism (38). Gal4 binds as a homodimer to inverted CGG triplets with a spacing of 11 bp, as revealed by the crystal structure of its DNA binding domain (44). The yeast genome contains 55 genes encoding putative zinc cluster proteins, and many of them have unknown functions (4, 5).
Two highly homologous zinc cluster proteins, Pdr1 and Pdr3, positively control the expression of genes involved in the regulation of multidrug resistance (47). Targets of Pdr1 and Pdr3 include the ATP-binding cassette transporter genes PDR5 and SNQ2, as well as HXT9 and HXT11, which are members of the major facilitator superfamily of transporters (8, 12, 31, 40). Deletion of PDR1 or PDR3 results in increased sensitivity to drugs, while a double knockout results in hypersensitivity to drugs (11, 31). We recently characterized Stb5 as a regulator of drug resistance. Stb5, like Pdr1 and Pdr3, is a transcriptional activator of SNQ2, which encodes an ABC transporter required to translocate a wide variety of compounds outside the cell (3). Deletion of this activator leads to a variety of phenotypes, such as sensitivity to cold, caffeine, and the translation inhibitor cycloheximide (3, 4).
Many zinc cluster proteins bind DNA as homodimers or heterodimers to recognition sites that usually fall within three types: inverted, direct, and everted repeats (22, 41, 44). Both Pdr1 and Pdr3 recognize CGG triplets oriented in opposite directions (CCGCGG) to form an everted repeat (9, 19, 22, 31, 32, 40). This motif (called PDRE [for pleiotropic drug response element]) is found in the target genes of Pdr1 and Pdr3, such as PDR5 and SNQ2 (31, 32). Pdr1 and Pdr3 have been shown to be able to form homo- and heterodimers (42). In addition, the zinc cluster protein Stb5 interacts in vivo with Pdr1 (but not Pdr3), suggesting that different combinations of homo- and heterodimers may differentially regulate gene expression (2).
To better understand the interplay among Pdr1, Pdr3, and Stb5, we used a combination of chromatin immunoprecipitation (ChIP) coupled with microarrays containing intergenic and coding regions, a method called genome-wide location analysis or ChIP-chip (53). Our data show that while Pdr1 and Pdr3 largely overlap, Stb5 colocalizes with Pdr1 and/or Pdr3 at only a few genes. Interestingly, we observed that Stb5 is bound to promoters of genes of the pentose phosphate pathway, which is important for oxidative stress resistance. Importantly, an STB5 deletion strain shows sensitivity to the oxidative agents hydrogen peroxide and diamide. We also show that Stb5 binding at some promoters is increased following diamide addition. Combined global Stb5 binding data and transcriptome profiling in the presence of diamide show that Stb5 acts as both an activator and a repressor in the presence of oxidative stress. Intriguingly, Stb5 activation does not appear to be mediated by a mitogen-activated protein (MAP) kinase pathway or by a mechanism similar to the transcription factor Yap1, two well-known signaling modules for oxidative stress response. However, loss of some pentose phosphate pathway genes impaired the induction of various Stb5 target genes. Our data suggest that the dual activator/repressor function of Stb5 is important for the control of the pentose phosphate pathway and NADPH production.
MATERIALS AND METHODS
Yeast strains and media.
Yeast strains used in this study are isogenic to BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) (61) except for the Δtrx1/Δtrx2 and Δgrx1/Δgrx2 strains, which are isogenic to YPH250 (MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1) (a gift from Yoshiharu Inoue, Kyoto University) (27). Other deletion strains were obtained from Research Genetics, and were all in the BY4741 background (61). Hemagglutinin (HA)-PDR1, PDR3-Myc, and Myc-STB5 open reading frames were N-terminally tagged (for PDR1 and STB5) or C-terminally tagged (for PDR3) at their natural locations with triple Myc or HA epitopes as previously described (2). Tagged Stb5 protein is functional; its expression was confirmed by Western blotting, and the Myc-STB5 strain grows as a wild-type (WT) strain on diamide and hydrogen peroxide plates (data not shown). The Myc-STB5 Δgnd1 strain was created by replacing the wild-type GND1 open reading frame with the KanMX4 cassette in the Myc-STB5 strain. Cells were routinely grown in YPD (1% yeast extract, 2% peptone, and 2% destrose).
Phenotypic assays.
For hydrogen peroxide and diamide sensitivity growth assays, yeast cells were grown overnight in YPD. Cells were washed, serially diluted, and spotted on rich medium (YPD) with or without 5 mM H2O2 or 1.8 mM diamide (highly fresh plates). Plates were incubated at 30°C as indicated.
Gene induction conditions and primer extension assays.
Cells were grown in YPD medium to an optical density at 600 nm of 0.6, and diamide was added to a final concentration of 1.5 mM for 30 or 60 min. For primer extension, 25 or 40 μg of RNA was used, and the assays were carried out as previously described (39). The sequences of the oligonucleotide primers used were as follows: ZWF1, CGCACCAAAGACAGATATGACGGTA; GND1, GACCCATGACGGCCAAACCAAT; GND2, GACCCATCACGGCTAAACCAAC; TAL1, GTTCTAGAGAGTTGTTAGCAACCTT; TKL1, GCCAAAATTCTTATGGTGGAGACGG; YEL041W, GCATCTCCCTTGGTACATGTCTC; ADH6, GTGTGATTGAATAGCGATACCTTC; ALD4, GTCTCCCAATGGAGGATGCAGA; ALD6, CATTTGGAAGTGTGATCTTGACTGG; YNL274C, GGTCACCAAAGGCATCCTTTCC; PGI1, GCTGGCAATTCAGTGGCCAGTT; TRX2, GCCAGATGCTAAAGCACTGTCG; GPX2, GATTCGCCTTTCTTGTCCTTGCA; and CTT1, GAACACGTTCATTTGTGAAGCTGA. For RNA loading controls, 15 μg of total RNA was loaded onto agarose-formaldehyde gels.
Protein expression and EMSA.
Expression, purification, and electrophoretic mobility shift assays (EMSA) with the Stb5 DNA binding domain (amino acids 1 to 163) were performed as described previously (3, 22). The probes for EMSA were obtained by annealing oligonucleotides and filling in with Klenow fragment and dGTP, dTTP, dATP, and [32P]dCTP. The following oligonucleotides were used as probes: ALD6-AB, TCGATGCGATATAGCACCGACCAT and TCGAATGGTCGGTGCTATATCGCA (positions −278 to −297 bp relative to the ATG); GND1-AB, TCGAACTTAATTAAGGCCGAAAAAT and TCGAATTTTTCGGCCTTAATTAAGT (positions −892 to −912 bp relative to the ATG); GND1-CD, TCGATGGCTGTCGGTGTTACATTACGG and TCGACCGTAATGTAACACCGACAGCCA (positions −267 to −289 bp relative to the ATG); GND1-EF, TCGATGCCTCGGTGTTACGCGGTG and TCGACACCGCGTAACACCGAGGCA (positions −247 to −266 bp relative to the ATG); GND1-EF1, TCGATGCCTCTGTGTTACGCGGTG and TCGACACCGCGTAACACAGAGGCA; GND1-EF2, TCGATGCCTCAGTGTTACGCGGTG and TCGACACCGCGTAACACTGAGGCA; and GND1-EF3, TCGATGCCTCGGTGTGACGCGGTG and TCGACACCGCGTCACACCGAGGCA (mutations are underlined).
Chromatin immunoprecipitations.
ChIP experiments were performed as described previously with some modifications (1). Briefly, cells were grown to an optical density at 600 nm of 0.6 in YPD. This was followed by the addition (or not) of diamide (final concentration, 1.5 mM) or hydrogen peroxide (final concentration, 0.4 mM) for 30 min or 20 min, respectively. Since diamide was dissolved in dimethyl sulfoxide, we added this solvent as a control. Five hundred microliters of whole-cell extract was incubated with either anti-HA (Roche) or anti-Myc (9E10; Upstate Technologies) antibody coupled to magnetic beads (Dynal). Immunoprecipitated DNA was used for either genome-wide location analysis or gene-specific PCR analysis. Three microliters of DNA was used for PCR. The sequences of the oligonucleotides used for ChIP were as follows: ZFW1, TTTGCACCCGTGTACATAAGC and GAAGAATTCGCCACCCTTAGA; SOL3, CATTTCTGGGCTCCTACTTTG and GCGTTTATCTGCCCGTAGTAA; GND1, AGAGAGACCTAAACGTAAGAG and AGCTCAGGAACAATACTGCAG; RKI1, TACTCAATCATTATCTGCCGG and CGCTGGTTGATATACCACATT; RPE1, TAACAACAGCCAACGACCGAT and GTTCCAGCTTACGGATCAAGA; TKL1, GGAATGAAGCATCCGAAAACG and GAATCATGCCTACGATCGTAC; TAL1, GAAAACAAGAACCGAAACGCG and ATACGATTCGTTGGAAAGGGG; PGI1, ATGTCAATGAGGCAAGAACCG and GCATTGATTATGAAGAAAGGACC; and GAL1-10, TACGCTTAACTGCTCATTGCT and GCCAATTTTTCCTCTTCATAACC.
Labeling of ChIP samples.
Labeling for ChIP-chip was performed as described previously (13). Briefly, immunoprecipitated DNA from tagged and isogenic nontagged (control) cells was blunted, ligated to a unidirectional linker, and amplified by PCR using linker-specific primers. A modified base [5-(3-aminoallyl)-dUTP] was included in the PCR at a ratio of two 5-(3-aminoallyl)-dUTP molecules to three dTTP molecules. The PCR products were purified, and the DNA was labeled with either Cy3 (control) or Cy5 monoreactive NHS esters. The Cy3- and Cy5-labeled DNAs were mixed and hybridized to a DNA microarray containing approximately 13,000 spots representing each intergenic and coding region of the S. cerevisiae genome. Detailed protocols and complete information about the microarray content can be found at http://www.ircm.qc.ca/microsites/francoisrobert/en.
Gene expression profiling.
Expression analysis was done as described previously (21) (using the QIAGEN cleanup method), starting from total RNA extracted by the hot acid phenol method, except that an 18-dT primer was used in the reverse transcription reaction instead of random hexamers and that RNA degradation was done by using two units of RNase H and 0.5 μg RNase A per reaction. The labeled cDNA products were hybridized to a DNA microarray containing approximately 7,000 spots (70-mer oligonucleotides) representing all yeast open reading frames (oligonucleotides purchased from QIAGEN). Detailed protocols and complete information about the microarray content can be found at http://www.ircm.qc.ca/microsites/francoisrobert/en.
Microarray hybridization and data analysis.
The labeled DNA was resuspended in 20 μl water and 430 μl of a solution of digoxigenin Easy Hyb buffer (Roche) containing 50 ng/μl salmon sperm DNA, and 40 ng/μl yeast tRNA was added. The hybridizations were done at 42°C for 16 to 20 h using Agilent hybridization chambers. Slides were washed first for 15 min at 37°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate and then (at room temperature) twice in 0.1× SSC, 0.1% sodium dodecyl sulfate and twice in 0.1× SSC. Then, slides were dried by centrifugation. Slides were scanned using an Axon 4000b scanner and analyzed with Genepix Pro 4.1.
The analysis of the ChIP-chip results was done as described in reference 53. The P value cutoff value used was ≤0.005. The expression data were preprocessed with ArrayPipe 1.7 (23). The background was subtracted using the “by-subgrid background correction” method. The data were normalized using the “linear model for microarray analysis (limma) loess (subgrid)” method. Replicate slides were merged and the data exported to the Significance Analysis of Microarrays (SAM) 2.20 Microsoft Excel add-on package (59). The “two-class unpaired” response was used, and a cutoff false discovery rate (FDR) of 0.45% was used to determine significance in the wild-type-versus-self-on-self experiment (1,825/6,748 genes differentially expressed), an FDR of 0.46% was used for the Δstb5-versus-self-on-self experiment (2,372/6,748 genes differentially expressed), and an FDR of 4.94% was used to determine which genes were differentially expressed between the wild-type and the Δstb5 experiment results (1,257/6,748 genes differentially expressed). Genesis 1.50 (55) was used for microarray data visualization and to generate some of the figures. All data for ChIP-chip and expression are available (see the supplemental material).
Overlaps with gene ontology processes were performed using the SGD Gene Ontology Term Finder tool (http://db.yeastgenome.org/cgi-bin/GO/goTermFinder).
Promoter sequence analysis.
To find a motif highly represented in the sequences we discovered by genome-wide location, we used MDscan, an algorithm for protein-DNA binding sites (37). Sequences were ranked according to the P value of Stb5 binding before the program was run. The logo picture was generated using WEBLOGO (http://weblogo.Berkeley.edu/logo.cgi). Following the motif identification, we used Regulatory Sequence Analysis Tools (http://rsat.ulb.ac.be/rsat/) to search for the CGGNSNTA motif in all Stb5 target promoters (between −100 and −800 bp relative to the ATG).
RESULTS
Stb5 targets are distinct from those of Pdr1 and Pdr3.
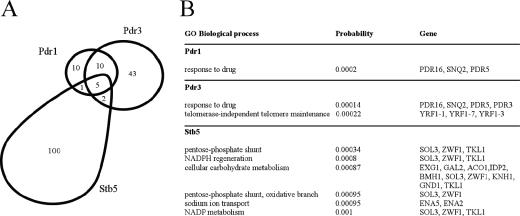
Zinc cluster protein Pdr1 is able to form homodimers or heterodimers with Pdr3 and Stb5, suggesting that different protein combinations may regulate the expression of different genes (2, 42). To gain insight into the interplay among the Pdr1, Pdr3, and Stb5 activators, we used genome-wide location analysis (ChIP-chip), a procedure that combines ChIP with hybridization to DNA microarrays (chip) (26, 53). For this purpose, Pdr1, Pdr3, and Stb5 were tagged at their natural locations with a triple HA or Myc epitope, and ChIP-chip experiments were performed with DNA microarrays covering the entire yeast genome (see Materials and Methods). As expected, all three transcription factors predominantly bind intergenic regions that contain promoters, while virtually no enrichment is observed in coding regions (data not shown). In rich medium, we have identified 26, 60, and 108 target promoters for Pdr1, Pdr3, and Stb5, respectively (P ≤ 0.005) (Fig. 1) (see the supplemental material for complete data sets). As expected, we identified known targets for these factors, such as PDR5, SNQ2, HXT9, HXT11, and PDR3 for Pdr1 and/or Pdr3 (8, 9, 12, 31, 32, 40) and SNQ2 for Stb5 (3). As shown in Fig. 1, the overlap between the promoters bound by Pdr1 and Pdr3 is quite large (15/26 for Pdr1 and 15/60 for Pdr3). This is consistent with the fact that these two factors can recognize the same DNA element (9, 19, 22, 31, 32, 40) and can heterodimerize (42). However, a number of promoters are bound exclusively by Pdr1 or Pdr3, suggesting that these factors may also act as homodimers or have additional dimerization partners. More importantly, Stb5 binds almost exclusively without Pdr1 or Prd3 (100/108 targets), suggesting that it functions mostly by itself or as a heterodimer with as-yet-unidentified partners.
FIG. 1.
Stb5 targets are distinct from those of Pdr1 and Pdr3. (A) The overlap between the promoter regions bound by Pdr1, Pdr3, and Stb5 in rich medium (P ≤ 0.005) is shown. (B) Gene ontology (GO) processes that overlap significantly (P ≤ 0.001) with the Pdr1, Pdr3, and Stb5 target genes are shown.
In order to gain insight into the physiological role of these factors, we looked at the overlap between the genes bound by Pdr1, Pdr3, or Stb5 and the gene ontology processes. The overlaps were scored statistically, and the most significant gene categories are shown in Fig. 1B. In agreement with previous observations, both Pdr1 and Pdr3 targets are enriched for genes involved in response to drugs. Also, Pdr3, but not Pdr1, binds to genes involved in telomere maintenance. More interestingly, and consistent with the lack of overlap with Pdr1/Pdr3, the target genes for Stb5 are enriched for different gene categories, namely the pentose phosphate pathway, NADPH regeneration and metabolism, carbohydrate metabolism, and sodium transport. Taken together, these data suggest a role for Stb5 in oxidative stress response, a function which is further explored below.
Stb5 is required for resistance to oxidative agents.
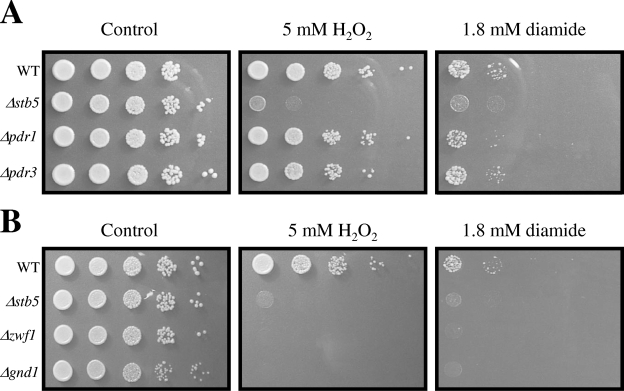
Our genome-wide location analysis performed in rich medium revealed binding of Stb5 to the promoters of genes of the pentose phosphate pathway, which generates NADPH, a molecule required for oxidative stress resistance (Fig. 1). This result suggests that Stb5 may be important for an appropriate response to oxidative stress. We thus decided to investigate the behavior of an STB5 deletion strain by growth assays in the presence of oxidative agents. As expected, Stb5 (but not Pdr1 or Pdr3) is required for growth on plates containing hydrogen peroxide or diamide (Fig. 2A). Moreover, two genes of the pentose phosphate pathway, ZWF1 (encoding a glucose-6-phosphate dehydrogenase) and GND1 (encoding a phosphogluconate dehydrogenase) are required for proper growth on oxidative plates (Fig. 2B) (29, 57). These results confirm an important role for Stb5 in oxidative stress resistance and a link with the pentose phosphate pathway.
FIG. 2.
Deletion of STB5 or genes ZWF1 and GND1 of the pentose phosphate pathway confers sensitivity to oxidative reagents. (A) Growth of cells in the absence or the presence of diamide or hydrogen peroxide. Serial dilutions (10-fold) of yeast cell suspensions were spotted on YPD plates supplemented or not with 1.8 mM diamide or 5 mM hydrogen peroxide. Plates were incubated at 30°C for 2 to 5 days. (B) Genes of the pentose phosphate pathway are required for proper growth in the presence of oxidative agents. The growth assay was performed as described for panel A.
Stb5 acts as an activator and a repressor in the presence of diamide.
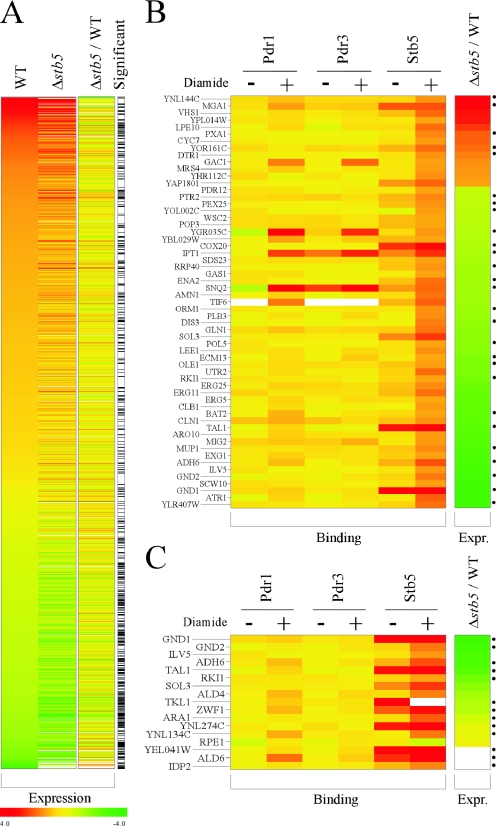
Genome-wide location analysis of Stb5 in rich medium led to the identification of genes involved in the production of NADPH through the pentose phosphate pathway (ZWF1, SOL3, GND1, and TKL1) or via other pathways (ALD6, YEL041W, YNL274C, and IDP2). Given the growth defect of a Δstb5 strain in the presence of diamide, we investigated the influence of diamide on the binding and activity of Stb5. Yeast cells were grown in rich medium, and diamide was added to a final concentration of 1.5 mM for 30 min. The binding of Stb5, Pdr1, and Pdr3 was analyzed by use of ChIP-chip, and gene expression changes were profiled in both wild-type and Δstb5 cells (see the supplemental material for complete data).
Treatment with diamide for 30 min induced a massive remodeling of gene expression, with more than 1,700 genes being significantly induced or repressed (FDR = 0.45% using SAM; see Materials and Methods) (Fig. 3A, WT). Among these genes, expression of about 500 depends largely on Stb5, since their response to diamide is significantly attenuated in a Δstb5 strain (FDR using SAM was 4.5%) (Fig. 3A, Δstb5). These genes may be either directly or indirectly regulated by Stb5 upon diamide stress. In order to identify the direct targets, we performed ChIP-chip experiments for Stb5 in diamide-treated cells. Figure 3B shows 59 genes that (i) are affected by diamide, (ii) depend on Stb5, and (iii) are directly bound by Stb5 (FDR = 0.45%) when cells are treated with diamide. As shown in Fig. 3B, Pdr1 and Pdr3 do not bind to a significant number of these genes even in the presence of diamide, again arguing that Stb5 plays a role distinct from that of these two zinc cluster proteins. Importantly, the binding of Stb5 is enhanced at most of these genes when cells are treated with diamide compared to vehicle alone (Fig. 3B, compare diamide-treated Stb5 and vehicle-treated Stb5). Most of these genes (46/59) are poorly induced by diamide in the absence of Stb5 compared to what is seen for the wild type (Fig. 3B, Δstb5/WT, green), in agreement with Stb5 being a transcriptional activator. For example, Stb5 is required for proper induction of many pentose phosphate pathway genes, such as SOL3, GND1, GND2, RKI1, and TAL1, as well as other NADPH-producing genes, such as ADH6 and ILV5. Stb5 binding sites (Fig. 3, right) can be detected upstream of approximately half of these genes, further arguing that Stb5 directly regulates these genes. Surprisingly, a nonnegligible fraction of the direct target genes of Stb5 (13/59) is hyperinduced in Δstb5 cells compared to wild-type cells (Fig. 3B, Δstb5/WT, red), strongly suggesting that Stb5 is a direct repressor of these genes. MGA1, encoding a protein similar to a heat shock factor, is a good example of a gene in that category. Taken together, these data show that Stb5 has both activator and repressor functions, coordinating a relatively important remodeling of gene expression induced by diamide.
FIG. 3.
Stb5 acts as an activator and a repressor upon diamide induction. (A) Effect of diamide on gene expression and contribution of Stb5. The effect of diamide on gene expression is shown (log2 changes [n-fold]) for all genes having significant changes in expression (relative to self-on-self controls using SAM analysis; FDR = 0.45%). Data (log2 changes [n-fold]) are shown for experiments performed with WT (first column) and STB5 deletion (Δstb5, second column) strains. The contribution of Stb5 to gene expression upon diamide treatment is displayed in the third column (Δstb5/WT), which shows the difference between WT (first column) and Δstb5 (second column) strains. Genes showing significant expression changes between the Δstb5 strain and the wild-type strain using the same SAM analysis parameters are shown as black bars (FDR = 4.9%) (significant). (B) Direct targets of Stb5 in diamide-treated cells. The binding of Pdr1, Pdr3, and Stb5 is shown for cells grown in the presence of diamide (+) or in the presence of vehicle alone (−) for all genes that show both Stb5 binding to their promoter (P ≤ 0.005) and a significant difference in expression between WT and Δstb5 strains (as per panel A [significant]). The difference between the expression level for the Δstb5 strain and that for the wild-type strain is shown (Δstb5/WT) as for panel A. Stb5 binding sites are indicated with black dots (see Fig. 5). (C) Binding of Pdr1, Pdr3, and Stb5 to the promoters of the pentose phosphate pathway and other NADPH-producing genes. The binding of Pdr1, Pdr3, and Stb5 is shown for cells grown in the presence of diamide (+) or in the presence of vehicle alone (−) for all genes of the pentose pathway and other NADPH-producing genes. The difference between the expression level for the Δstb5 mutant strain and that for the wild-type strain is shown (Δstb5/WT) as for panels A and B. Stb5 binding sites are indicated with black dots (see Fig. 5).
We next wanted to determine if Stb5 plays a role in oxidative stress induced by agents other than diamide. We therefore investigated the binding of Stb5 upon addition of hydrogen peroxide by ChIP-chip analysis. Cells were grown to mid-log phase, and hydrogen peroxide was added at a concentration of 0.4 mM for 20 min, a condition known to activate the oxidative stress transcriptional activator Yap1 (20) and genes associated with hydrogen peroxide stress, such as CTT1, GPX2, and TRX2 (data not shown). As expected, Stb5 binds to many genes under these conditions. Interestingly, most of these genes are the same as those bound by Stb5 in diamide-induced stress (not shown). These results suggest that Stb5 responds to either stress by regulating a common set of genes. More experiments are required to determine whether Stb5 also regulates genes that are specific to one particular stress.
Stb5 plays a pivotal role in the production of NADPH during oxidative stress.
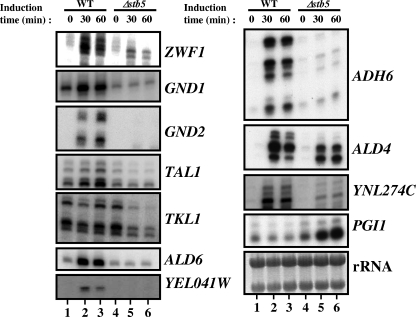
Many direct Stb5 target genes in diamide-treated cells (Fig. 3B) are genes of the NADPH pathway. Figure 3C—which shows the binding of Pdr1, Pdr3, and Stb5, as well as the effect of diamide on the expression of all genes of the pentose pathway and of other NADPH-encoding genes—suggests that Stb5 is a key regulator of that pathway. We confirmed these observations by standard ChIPs (see Fig. S1 in the supplemental material) and by primer extension analysis for some genes involved in NADPH production (Fig. 4). Upon addition of diamide, the RNA levels of ZWF1, GND1, GND2, TAL1, YNL274C, ALD6, YEL041W, ADH6, and ALD4 are increased in wild-type cells (Fig. 4, lanes 1 to 3), whereas deletion of STB5 prevented or reduced this induction (lanes 4 to 6). In the presence of diamide, TKL1 is not induced in wild-type cells, but Stb5 is required to maintain its expression level (Fig. 4, TKL1 panel, compare lanes 2 and 3 to lanes 5 and 6). Stb5 is also required for basal expression of GND1 (GND1 panel, compare lanes 1 and 4). Taken together with our ChIP-chip data, these results show that Stb5 is required for the regulation of all genes of the pentose phosphate pathway but one (RPE1) as well as for the regulation of many other genes involved in the production of NADPH through alternate pathways.
FIG. 4.
Confirmation of microarray results by gene-specific analysis. Primer extension analysis of pentose phosphate pathway genes, as well as of other genes involved in NADPH production, was performed in the presence of diamide as described in Materials and Methods. Induction time in the presence of 1.5 mM diamide is shown in minutes. The rRNAs are shown at the bottom of the figure as a loading control.
We also wished to determine if the transcriptional repressor function of Stb5 could play a role in the production of NADPH. We focused on the PGI1 gene, which encodes a glucose-6-phosphate isomerase at the interplay between the pentose phosphate and the glycolysis pathways. PGI1 drives glucose-6-phosphate towards the glycolysis pathway, while its inhibition would favor the utilization of glucose-6-phosphate for the production of NADPH via the pentose phosphate pathway. The PGI1 promoter is bound by Stb5 in diamide-treated cells (see Fig. S1 in the supplemental material), but its expression, despite its hyperactivation in Δstb5 cells, had missed our statistical cutoff and was therefore not included in Fig. 3B. However, primer extension analysis confirmed that the PGI1 gene is hyperactivated in Δstb5 cells after treatment with diamide (Fig. 4, PGI1 panel). Taken together, these data clearly demonstrate that Stb5 is required to actively repress the expression of PGI1 and to activate many other genes in order to coordinate the production of NADPH.
Identification of Stb5 binding motif.
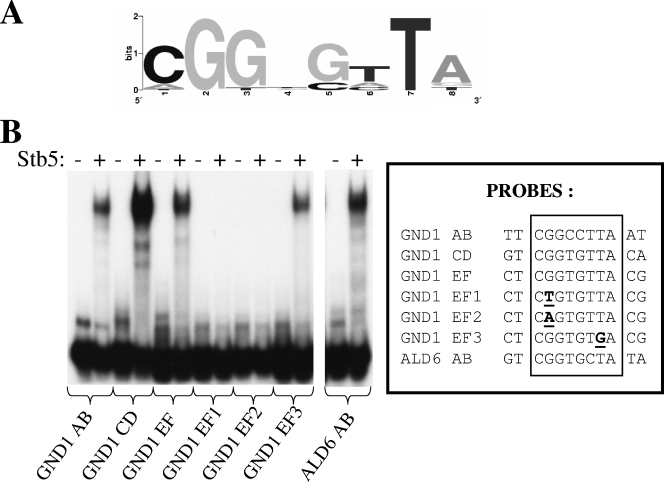
It has been suggested that Stb5 could recognize a CGGNStTAta motif (20) (lowercase letters indicate less conserved nucleotides). We observed a similar motif by promoter analysis of our ChIP-chip data (Fig. 5A). Because no experiments were done to demonstrate direct binding of Stb5 to this motif, we performed an electrophoretic mobility shift assay with the purified Stb5 DNA binding domain (Fig. 5B). The GND1 promoter contains three potential binding sites for Stb5. Stb5 can bind to any of these sites in vitro (GND1 AB, CD, and EF probes). Mutations in the middle of the CGG triplet completely abolished Stb5 binding (compare GND1 EF with GND1 EF1 and EF2), while mutation of the highly conserved T residue at position 7 had virtually no effect (compare GND1 EF with GND1 EF3). We also confirmed Stb5 binding at a motif found in the ALD6 promoter (ALD6 AB probe). These results suggest that the motif identified by promoter analysis is important for Stb5 binding in vitro.
FIG. 5.
Binding of the DNA binding domain of Stb5 in vitro to the sequences identified by ChIP-chip analysis. (A) Motif discovered by genome-wide location and computer analysis. (B) Binding of Stb5 (amino acids 1 to 163) to the motifs found in ALD6 and GND1 promoters and mutants. EMSA was performed with various probes (as indicated at the bottom of the figure) in the presence (+) or absence (−) of the purified DNA binding domain of Stb5. Mutations in probes (boxed) are underlined and in bold.
Stb5 activation is mediated neither by the Mpk1, Hog1, and Kss1 MAP kinase pathways nor by a Yap1-like mechanism.
Two major mechanisms are known to regulate transcriptional changes following oxidative stress: the MAP kinase signaling pathways and the Yap1-like transcription factor-sensing oxidants (24, 36). We thus decided to investigate the importance of these two mechanisms in the Stb5 activation process following diamide stress.
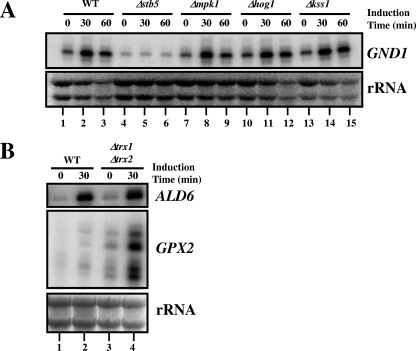
We first investigated the importance of various MAP kinase pathways on Stb5 activation. If a MAP kinase pathway is required for that process, eliminating a component of the pathway should phenocopy the expression defect observed for Δstb5 cells. Figure 6A shows that Hog1, Mpk1, and Kss1, three MAP kinases known to transduce reactive oxygen species (ROS) signals, are not required for activation of GND1, a gene regulated by Stb5 in the presence of diamide (compare lanes 1 to 3 with lanes 4 to 6, 7 to 9, 10 to 12, and 13 to 15). Moreover, Lcb4 and Mek1, two kinases that interact with Stb5 in a two-hybrid system (25), are both dispensable for GND1 induction (data not shown). We also tested others kinases (Pfk26, Fab1, Dun1, Dbf2, and Mrk1) required for diamide resistance according to an S. cerevisiae deletion mutant screening for cellular viability to ROS (57), but none of the kinases tested were required to induce GND1 (data not shown). These results strongly suggest that Stb5 activation is not mediated by these kinases.
FIG. 6.
Stb5 activation is mediated neither by the Mpk1, Hog1, and Kss1 MAP kinase pathways nor by a Yap1-like mechanism. (A) Effect of various deletions of genes encoding MAP kinases on GND1 expression. Primer extension analyses were done as described for Fig. 4. The rRNAs are shown at the bottom of the figure as a loading control. (B) Thioredoxin deficiency of a Δtrx1/Δtrx2 strain does not cause a constitutive activation of Stb5 like that observed for Yap1. Primer extension analysis of the ALD6 and GPX2 genes was performed with RNA isolated from WT or thioredoxin-deficient cells (Δtrx1/Δtrx2) treated or not with diamide for 30 min. The rRNAs are shown at the bottom of the figure as a loading control.
Thioredoxin deficiency of a Δtrx1/Δtrx2 strain leads to an oxidized form of the regulator Yap1. This form causes a constitutive activation of the regulator, resulting in nuclear localization and activation of its target genes in nonstressed cells, suggesting that the thioredoxin system negatively regulates the Yap1 activity (10, 27, 35). We thus decided to investigate if the activity of Stb5 could be regulated by the thioredoxin and/or glutaredoxin systems. We used primer extension experiments to compare the expression levels of ALD6 in wild-type, Δtrx1/Δtrx2, and Δgrx1/Δgrx2 cells in the absence and the presence of diamide (Fig. 6B and data not shown). As a positive control, we looked for the expression of GPX2, a Yap1 target gene. Zinc cluster protein Stb5, in contrast to Yap1, is not constitutively active in the absence of stress in a Δtrx1/Δtrx2 strain or in a Δgrx1/Δgrx2 strain, since we did not observe constitutive expression of ALD6 (Fig. 6B, compare lanes 1 and 3). In contrast, we observed increased GPX2 expression in the absence of stress in the Δtrx1/Δtrx2 strain (compare lanes 1 and 3) coupled to a hyperactivation in the presence of diamide (compare lanes 2 and 4). These results suggest that Stb5 and Yap1 are not activated by a similar mechanism.
Pentose phosphate pathway integrity is required to activate Stb5 target genes.
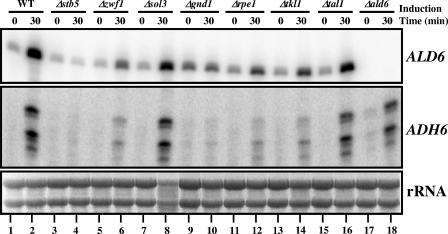
In an effort to gain some understanding of the mechanism by which Stb5 is activated by diamide, we examined the importance of the pentose phosphate pathway in that process. Leu3 and Lys14, two zinc cluster protein regulators involved in leucine and lysine biosynthesis, respectively, are activated by pathway intermediates (14, 15, 33, 56). We thus decided to analyze the requirement of many pentose phosphate pathway genes in the induction of different Stb5 target genes (Fig. 7). Interestingly, we found that the induction of ALD6 and ADH6, two NADPH production genes, via alternative pathways is negatively affected in Δzwf1, Δgnd1, Δrpe1, and Δtkl1 strains (compare lanes 1 and 2 to lanes 5 and 6, 9 and 10, 11 and 12, and 13 and 14). The ALD6 and ADH6 genes can still be efficiently induced even in the absence of SOL3 and TAL1 (compare lanes 1 and 2 to lanes 7 and 8 and 15 and 16). Deletion of ALD6 had no effect on ADH6 expression, suggesting that a deletion of an alternative NADPH production gene (outside the pentose phosphate pathway) is dispensable for the activation of Stb5 target genes (ADH6 panel, compare lanes 1 and 2 to lanes 17 and 18). Given the importance of some pentose phosphate pathway genes for activation of Stb5 target genes, we examined the binding of Stb5 in vivo in the absence of GND1. To achieve that condition, ChIP-chip assays of Stb5 in a Δgnd1 strain were performed in the absence and in the presence of diamide. The Stb5 binding profiles in the presence of diamide are very similar in WT and in Δgnd1 cells (data not shown), suggesting that the pentose phosphate pathway is not involved in Stb5 binding, despite being required for activation of Stb5 target genes.
FIG. 7.
Requirement of the pentose phosphate pathway to activate Stb5 target genes. The effect of some pentose phosphate pathway gene deletions on ALD6 and ADH6 expression is shown. Primer extension analysis was done as described for Fig. 4. The rRNAs are shown at the bottom of the figure as a loading control.
DISCUSSION
In this work, we used the ChIP-chip technology to better understand the interplay among three zinc cluster proteins: Pdr1, Pdr3, and Stb5. We have shown that the overlap between Pdr1 and Pdr3, two important regulators involved in drug resistance, is large, in agreement with the facts that (i) a double-knockout strain is hypersensitive to drugs compared to single-deletion strains (11, 31), (ii) both regulators have the same targets reported in the literature (8, 12, 32, 40), and (iii) these factors can heterodimerize (42). Pdr1 and Stb5 are able to form heterodimers in vivo (2). In agreement with this observation, Pdr1 and Stb5 are both bound to the SNQ2 promoter. However, Stb5 binds almost exclusively without Pdr1 or Pdr3 in rich medium, suggesting that Stb5 may be involved in functions different from those of Pdr1 and Pdr3. Indeed, we observed that Stb5 targets are enriched for genes involved in the pentose phosphate pathway and production of NADPH, an important cofactor required for oxidative stress resistance (Fig. 1B). However, we cannot exclude the possibility that Pdr1 and Stb5 share more common target genes in other conditions.
Cells growing aerobically are exposed to ROS such as the superoxide anion radical (O2−), the hydroxyl radical (OH · ), and hydrogen peroxide, leading to lipid, protein, and DNA damage and eventually to cell death. To overcome oxidative stress, cells possess a variety of defenses (7, 28). These include both nonenzymatic defenses, such as the small protein thioredoxin and the tripeptide glutathione (reductant molecules), and enzymatic defense systems, such as catalase and superoxide dismutase, to maintain the cellular redox state. One other enzymatic defense relies on the pentose phosphate pathway, which is generally accepted to be the major cellular source of NADPH. Connection between the pentose phosphate pathway and oxidative stress resistance is well established, since mutations in some genes of this pathway render cells hypersensitive to oxidants (Fig. 2B) (29, 57). ChIP-chip and expression profiling experiments in the presence of the oxidative agent diamide have demonstrated that Stb5 regulates genes encoding enzymes of the pentose phosphate pathway and other genes involved in NADPH production (Fig. 3). Importantly, an STB5 deletion strain is sensitive to both hydrogen peroxide and diamide (Fig. 2A). Phenotypic analyses of single and double knockouts of the ZWF1, ALD6, and IDP2 genes regulated by Stb5 and encoding NADPH-producing enzymes have shown their importance for hydrogen peroxide resistance with different carbon sources (45). Another NADPH gene regulated by Stb5, GND1, not only is important for tolerance in the presence of diamide and peroxide (Fig. 2B) but also is required for resistance to menadione and linoleic acid 13-hydroperoxide, two other oxidative agents (57). Collectively, these results demonstrate an important role for reduced NADPH in oxidative stress resistance. Our data point to Stb5 as a key player in the maintenance of a significant cytosolic pool of the cofactor NADPH following diamide stress.
Surprisingly, combined global Stb5 binding data and the mRNA expression profile in the presence of diamide suggest that it also acts as a repressor (Fig. 3 and 4). A number of studies have provided models to explain how a transcriptional activator can become a repressor. For example, ZRT2 gene regulation by Zap1 is mediated by the antagonistic effect of Zap1 binding to a low-affinity repressor site downstream of the TATA box and high-affinity sites required for activation upstream of the same element (6). The Ume6 transcriptional regulator acts as a repressor by interacting with the Sin3/Rpd3 complex but acts as an activator if the interaction occurs with Ime1 (60). Another mechanism to convert a repressor into an activator is posttranslational modification. For example, Rgt1 phosphorylation in response to glucose is required to abolish the Rgt1-mediated repression of HXT genes and to convert Rgt1 from a transcriptional repressor to an activator (46, 48). The Hog1 kinase is able to convert the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits chromatin-remodeling complexes in response to osmotic stress (51).
How Stb5 acts as a repressor/activator is unknown, but it has been suggested that the middle homology region found in many binuclear zinc cluster proteins could control their transcriptional activities. Stb5 contains such a region between amino acids 397 and 475 (54). One example of this phenomenon is that deletion of that region in the binuclear zinc cluster Hap1 renders the protein active even in the absence of the inducer heme (50). Similar results were also obtained with another binuclear zinc cluster, Leu3 (17, 62). The middle homology region could serve as a platform for posttranslational modification and/or protein-protein interaction. It is interesting that Stb5 is able to interact in a two-hybrid system with Sin3, a component of the large protein complex containing the histone deacetylase Rpd3 (30).
The data presented here strongly suggest that the transcriptional regulator Stb5 is activated upon exposure to diamide (Fig. 3). The stress-responsive MAP kinase pathways and the Yap1-like mechanism are two important signaling modules for oxidative stress response (24, 36). We have shown here that MAP kinases involved in ROS signaling as well as many other kinases important for diamide resistance are not required for Stb5 activation, suggesting that the activation process is not mediated by these kinases (Fig. 6A and data not shown). In agreement with this point, none of the kinases tested are able to phosphorylate Stb5, according to a recent global analysis of protein phosphorylation in yeast (52). In S. cerevisiae, the transcription factor Yap1 has been implicated in the control of the oxidative stress response. Yap1 is able to sense hydrogen peroxide and diamide through reversible disulfide linkage between cysteine residues (10, 35). Yap1 is oxidized in mutants of the thioredoxin system and becomes constitutively active in the absence of stress (10, 27, 35). Thioredoxin and glutathione deficiencies do not cause constitutive activation of Stb5, suggesting that the mechanisms of activation of these two important oxidative stress regulators differ (Fig. 6B and data not shown). Intriguingly, loss of many pentose phosphate pathway genes affected the induction of different Stb5 target genes (Fig. 7). However, Stb5 binding is independent of that pathway (data not shown), a result suggesting that increased Stb5 binding is not sufficient for the activation of target genes. Interestingly, activation of Lys14 target genes is dependent on α-aminoadipate semialdehyde, an intermediate of the pathway controlled by this regulator (14, 15). Similar results were also obtained with another zinc cluster protein, Leu3, whose activation depends on the presence of the precursor α-isopropylmalate (33, 56). Our results suggest that Stb5 activation mechanism could be regulated in a similar fashion.
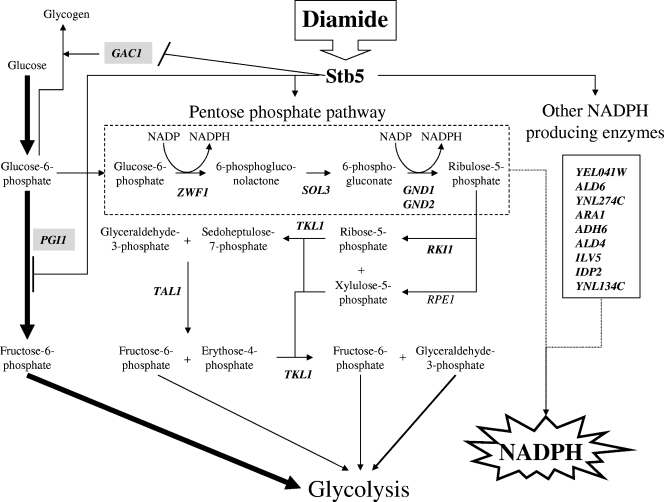
Figure 8 shows a model of cooperation between the activator and the repressor roles of Stb5 in the control of genes involved in NADPH production for oxidative stress resistance. Diamide can rapidly cross biological membranes (34) and activate Stb5, leading to repression of GAC1, which encodes a positive regulator of glycogen accumulation (16) (Fig. 3B). Stb5 also acts as a repressor of PGI1, which encodes a glucose 6-phosphate isomerase required to catalyze the interconversion of glucose-6-phosphate and fructose-6-phosphate (Fig. 4) (see Fig. S1 in the supplemental material). This repression would partially block the normal glycolysis pathway and drive glucose-6-phosphate into the pentose phosphate pathway. Diamide induces the activation of the pentose phosphate pathway in an Stb5-dependent manner (Fig. 3C and 4). Stb5 binding increases at some promoters following the stress. In the oxidative part of the pentose phosphate pathway, the reactions catalyzed by glucose-6-phosphate dehydrogenase (ZWF1) and 6-phosphogluconate dehydrogenase (GND1 and GND2) generate NADPH. The nonoxidative part of the pentose phosphate pathway produces fructose-6-phosphate and glyceraldehyde-3-phosphate, which can then be utilized in the pathway of glycolysis. Stb5 is also important for the activation of nine genes involved in the production of NADPH via other pathways (Fig. 3 and 4). RPE1 is the only gene of the pentose phosphate pathway not bound by Stb5. Most of these genes listed in this figure contain Stb5 binding sites (Fig. 3C).
FIG. 8.
Schematic overview of Stb5-bound genes and pathway function for NADPH production following diamide stress. In the presence of diamide, Stb5 binds to all these genes with the exception of RPE1. Stb5 acts as a repressor of the GAC1 and PGI1 genes (gray boxes) and as an activator of all other genes (in bold). The glycolysis pathway is represented by the thick arrows. The dashed box represents the oxidative part of the pentose phosphate pathway that generates NADPH. See the text for more details.
Glutathione is possibly the most abundant redox scavenging molecule in cells (43) and is known to react rapidly with diamide to produce glutathione disulfide, which is formed upon its oxidation (34). Glutathione disulfide is then reduced by glutathione reductase at the expense of NADPH. NADPH is an essential cofactor for the glutathione- and thioredoxin-dependent enzymes that constitute major cellular defenses against oxidative stress (7, 28). From our data, it is clear that the transcriptional regulator Stb5 is also a major player in the oxidative stress response, since it regulates genes involved in NADPH production.
Supplementary Material
Acknowledgments
We thank Karen Hellauer for critical reading of the manuscript. We are grateful to Yoshiharu Inoue (Kyoto University) for providing yeast strains.
This work was supported by grants from the Canadian Institute of Health Research (CIHR) to B.T. (grant no. 62702) and F.R. (grant no. 64230). M.L. is a recipient of a postdoctoral fellowship from the Research Institute of the McGill University Health Center. S.D. holds a doctoral studentship from the Institut de recherches cliniques de Montréal/CIHR cancer research program. F.R. holds a CIHR New Investigator Award.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adam, M., F. Robert, M. Larochelle, and L. Gaudreau. 2001. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 21:6270-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akache, B., S. MacPherson, M. A. Sylvain, and B. Turcotte. 2004. Complex interplay among regulators of drug resistance genes in Saccharomyces cerevisiae. J. Biol. Chem. 279:27855-27860. [DOI] [PubMed] [Google Scholar]
- 3.Akache, B., and B. Turcotte. 2002. New regulators of drug sensitivity in the family of yeast zinc cluster proteins. J. Biol. Chem. 277:21254-21260. [DOI] [PubMed] [Google Scholar]
- 4.Akache, B., K. Wu, and B. Turcotte. 2001. Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res. 29:2181-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. [DOI] [PubMed] [Google Scholar]
- 6.Bird, A. J., E. Blankman, D. J. Stillman, D. J. Eide, and D. R. Winge. 2004. The Zap1 transcriptional activator also acts as a repressor by binding downstream of the TATA box in ZRT2. EMBO J. 23:1123-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmel-Harel, O., and G. Storz. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54:439-461. [DOI] [PubMed] [Google Scholar]
- 8.Decottignies, A., L. Lambert, P. Catty, H. Degand, E. A. Epping, W. S. Moye-Rowley, E. Balzi, and A. Goffeau. 1995. Identification and characterization of SNQ2, a new multidrug ATP binding cassette transporter of the yeast plasma membrane. J. Biol. Chem. 270:18150-18157. [DOI] [PubMed] [Google Scholar]
- 9.Delahodde, A., T. Delaveau, and C. Jacq. 1995. Positive autoregulation of the yeast transcription factor Pdr3p, which is involved in control of drug resistance. Mol. Cell. Biol. 15:4043-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delaunay, A., A. D. Isnard, and M. B. Toledano. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19:5157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delaveau, T., A. Delahodde, E. Carvajal, J. Subik, and C. Jacq. 1994. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol. Gen. Genet. 244:501-511. [DOI] [PubMed] [Google Scholar]
- 12.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470:156-160. [DOI] [PubMed] [Google Scholar]
- 13.Drouin, S., and F. Robert. Genome-wide location analysis of chromatin-associated proteins by ChIP on chip: controls matter. Methods, in press.
- 14.Feller, A., E. Dubois, F. Ramos, and A. Pierard. 1994. Repression of the genes for lysine biosynthesis in Saccharomyces cerevisiae is caused by limitation of Lys14-dependent transcriptional activation. Mol. Cell. Biol. 14:6411-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feller, A., F. Ramos, A. Pierard, and E. Dubois. 1999. In Saccharomyces cerevisae [sic], feedback inhibition of homocitrate synthase isoenzymes by lysine modulates the activation of LYS gene expression by Lys14p. Eur. J. Biochem. 261:163-170. [DOI] [PubMed] [Google Scholar]
- 16.Francois, J. M., S. Thompson-Jaeger, J. Skroch, U. Zellenka, W. Spevak, and K. Tatchell. 1992. GAC1 may encode a regulatory subunit for protein phosphatase type 1 in Saccharomyces cerevisiae. EMBO J. 11:87-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friden, P., C. Reynolds, and P. Schimmel. 1989. A large internal deletion converts yeast LEU3 to a constitutive transcriptional activator. Mol. Cell. Biol. 9:4056-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner, K. H., T. Pan, S. Narula, E. Rivera, and J. E. Coleman. 1991. Structure of the binuclear metal-binding site in the GAL4 transcription factor. Biochemistry 30:11292-11302. [DOI] [PubMed] [Google Scholar]
- 19.Hallstrom, T. C., and W. S. Moye-Rowley. 1998. Divergent transcriptional control of multidrug resistance genes in Saccharomyces cerevisiae. J. Biol. Chem. 273:2098-2104. [DOI] [PubMed] [Google Scholar]
- 20.Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac, T. W. Danford, N. M. Hannett, J. B. Tagne, D. B. Reynolds, J. Yoo, E. G. Jennings, J. Zeitlinger, D. K. Pokholok, M. Kellis, P. A. Rolfe, K. T. Takusagawa, E. S. Lander, D. K. Gifford, E. Fraenkel, and R. A. Young. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasseman, J. 2002. Aminoallyl labeling of RNA for microarrays. SOP #M0004, The Institute for Genomic Research. [Online.] http://pga.tigr.org/sop/M004_1a.pdf.
- 22.Hellauer, K., M. H. Rochon, and B. Turcotte. 1996. A novel DNA binding motif for yeast zinc cluster proteins: the Leu3p and Pdr3p transcriptional activators recognize everted repeats. Mol. Cell. Biol. 16:6096-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hokamp, K., F. M. Roche, M. Acab, M. E. Rousseau, B. Kuo, D. Goode, D. Aeschliman, J. Bryan, L. A. Babiuk, R. E. Hancock, and F. S. Brinkman. 2004. ArrayPipe: a flexible processing pipeline for microarray data. Nucleic Acids Res. 32:457-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikner, A., and K. Shiozaki. 2005. Yeast signaling pathways in the oxidative stress response. Mutat. Res. 569:13-27. [DOI] [PubMed] [Google Scholar]
- 25.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 27.Izawa, S., K. Maeda, K. Sugiyama, J. Mano, Y. Inoue, and A. Kimura. 1999. Thioredoxin deficiency causes the constitutive activation of Yap1, an AP-1-like transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 274:28459-28465. [DOI] [PubMed] [Google Scholar]
- 28.Jamieson, D. J. 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14:1511-1527. [DOI] [PubMed] [Google Scholar]
- 29.Juhnke, H., B. Krems, P. Kotter, and K. D. Entian. 1996. Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress. Mol. Gen. Genet. 252:456-464. [DOI] [PubMed] [Google Scholar]
- 30.Kasten, M. M., and D. J. Stillman. 1997. Identification of the Saccharomyces cerevisiae genes STB1-STB5 encoding Sin3p binding proteins. Mol. Gen. Genet. 256:376-386. [DOI] [PubMed] [Google Scholar]
- 31.Katzmann, D. J., P. E. Burnett, J. Golin, Y. Mahe, and W. S. Moye-Rowley. 1994. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol. Cell. Biol. 14:4653-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katzmann, D. J., T. C. Hallstrom, Y. Mahe, and W. S. Moye-Rowley. 1996. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J. Biol. Chem. 271:23049-23054. [DOI] [PubMed] [Google Scholar]
- 33.Kirkpatrick, C. R., and P. Schimmel. 1995. Detection of leucine-independent DNA site occupancy of the yeast Leu3p transcriptional activator in vivo. Mol. Cell. Biol. 15:4021-4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosower, N. S., and E. M. Kosower. 1995. Diamide: an oxidant probe for thiols. Methods Enzymol. 251:123-133. [DOI] [PubMed] [Google Scholar]
- 35.Kuge, S., M. Arita, A. Murayama, K. Maeta, S. Izawa, Y. Inoue, and A. Nomoto. 2001. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 21:6139-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, H., R. Colavitti, I. I. Rovira, and T. Finkel. 2005. Redox-dependent transcriptional regulation. Circ. Res. 97:967-974. [DOI] [PubMed] [Google Scholar]
- 37.Liu, X. S., D. L. Brutlag, and J. S. Liu. 2002. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 20:835-839. [DOI] [PubMed] [Google Scholar]
- 38.Lohr, D., P. Venkov, and J. Zlatanova. 1995. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 9:777-787. [DOI] [PubMed] [Google Scholar]
- 39.Ma, J., and M. Ptashne. 1987. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48:847-853. [DOI] [PubMed] [Google Scholar]
- 40.Mahe, Y., A. Parle-McDermott, A. Nourani, A. Delahodde, A. Lamprecht, and K. Kuchler. 1996. The ATP-binding cassette multidrug transporter Snq2 of Saccharomyces cerevisiae: a novel target for the transcription factors Pdr1 and Pdr3. Mol. Microbiol. 20:109-117. [DOI] [PubMed] [Google Scholar]
- 41.Mamane, Y., K. Hellauer, M. H. Rochon, and B. Turcotte. 1998. A linker region of the yeast zinc cluster protein Leu3p specifies binding to everted repeat DNA. J. Biol. Chem. 273:18556-18561. [DOI] [PubMed] [Google Scholar]
- 42.Mamnun, Y. M., R. Pandjaitan, Y. Mahe, A. Delahodde, and K. Kuchler. 2002. The yeast zinc finger regulators Pdr1p and Pdr3p control pleiotropic drug resistance (PDR) as homo- and heterodimers in vivo. Mol. Microbiol. 46:1429-1440. [DOI] [PubMed] [Google Scholar]
- 43.Marchler, G., C. Schuller, G. Adam, and H. Ruis. 1993. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 12:1997-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marmorstein, R., M. Carey, M. Ptashne, and S. C. Harrison. 1992. DNA recognition by GAL4: structure of a protein-DNA complex. Nature 356:408-414. [DOI] [PubMed] [Google Scholar]
- 45.Minard, K. I., and L. McAlister-Henn. 2005. Sources of NADPH in yeast vary with carbon source. J. Biol. Chem. 280:39890-39896. [DOI] [PubMed] [Google Scholar]
- 46.Mosley, A. L., J. Lakshmanan, B. K. Aryal, and S. Ozcan. 2003. Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J. Biol. Chem. 278:10322-10327. [DOI] [PubMed] [Google Scholar]
- 47.Moye-Rowley, W. S. 2003. Transcriptional control of multidrug resistance in the yeast Saccharomyces. Prog. Nucleic Acid Res. Mol. Biol. 73:251-279. [DOI] [PubMed] [Google Scholar]
- 48.Ozcan, S., T. Leong, and M. Johnston. 1996. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol. Cell. Biol. 16:6419-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan, T., and J. E. Coleman. 1990. GAL4 transcription factor is not a “zinc finger” but forms a Zn(II)2Cys6 binuclear cluster. Proc. Natl. Acad. Sci. USA 87:2077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfeifer, K., K. S. Kim, S. Kogan, and L. Guarente. 1989. Functional dissection and sequence of yeast HAP1 activator. Cell 56:291-301. [DOI] [PubMed] [Google Scholar]
- 51.Proft, M., and K. Struhl. 2002. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9:1307-1317. [DOI] [PubMed] [Google Scholar]
- 52.Ptacek, J., G. Devgan, G. Michaud, H. Zhu, X. Zhu, J. Fasolo, H. Guo, G. Jona, A. Breitkreutz, R. Sopko, R. R. McCartney, M. C. Schmidt, N. Rachidi, S. J. Lee, A. S. Mah, L. Meng, M. J. Stark, D. F. Stern, C. De Virgilio, M. Tyers, B. Andrews, M. Gerstein, B. Schweitzer, P. F. Predki, and M. Snyder. 2005. Global analysis of protein phosphorylation in yeast. Nature 438:679-684. [DOI] [PubMed] [Google Scholar]
- 53.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 54.Schjerling, P., and S. Holmberg. 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24:4599-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sturn, A., J. Quackenbush, and Z. Trajanoski. 2002. Genesis: cluster analysis of microarray data. Bioinformatics 18:207-208. [DOI] [PubMed] [Google Scholar]
- 56.Sze, J. Y., M. Woontner, J. A. Jaehning, and G. B. Kohlhaw. 1992. In vitro transcriptional activation by a metabolic intermediate: activation by Leu3 depends on alpha-isopropylmalate. Science 258:1143-1145. [DOI] [PubMed] [Google Scholar]
- 57.Thorpe, G. W., C. S. Fong, N. Alic, V. J. Higgins, and I. W. Dawes. 2004. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc. Natl. Acad. Sci. USA 101:6564-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Todd, R. B., and A. Andrianopoulos. 1997. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21:388-405. [DOI] [PubMed] [Google Scholar]
- 59.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Washburn, B. K., and R. E. Esposito. 2001. Identification of the Sin3-binding site in Ume6 defines a two-step process for conversion of Ume6 from a transcriptional repressor to an activator in yeast. Mol. Cell. Biol. 21:2057-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 62.Zhou, K. M., and G. B. Kohlhaw. 1990. Transcriptional activator LEU3 of yeast. Mapping of the transcriptional activation function and significance of activation domain tryptophans. J. Biol. Chem. 265:17409-17412. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.