Abstract
Drosophila N-cadherin (CadN) is an evolutionarily conserved classic cadherin which has a large, complex extracellular domain and a catenin-binding cytoplasmic domain. The CadN locus contains three modules of alternative exons (7a/b, 13a/b, and 18a/b) and undergoes alternative splicing to generate multiple isoforms. Using quantitative transcript analyses and green fluorescent protein-based cell sorting, we found that during development CadN alternative splicing is regulated in a temporal but not cell-type-specific fashion. In particular, exon 18b is predominantly expressed during early developmental stages, while exon 18a is prevalent at the late developmental and adult stages. All CadN isoforms share the same molecular architecture but have different sequences in their extracellular and transmembrane domains, suggesting functional diversity. In vitro quantitative cell aggregation assays revealed that all CadN isoforms mediate homophilic interactions, but the isoforms encoded by exon 18b have a higher adhesive activity than those by its alternative, 18a. Domain-swapping experiments further revealed that the different sequences in the transmembrane domains of isoforms are responsible for their differential adhesive activities. CadN alternative splicing might provide a novel mechanism to fine-tune its adhesive activity at different developmental stages or to restrict the use of high-affinity 18b-type isoforms at the adult stage.
Cadherins are calcium-dependent cell adhesion receptors. By mediating homophilic or heterophilic interactions, cadherins play key roles in a wide spectrum of developmental processes, including morphogenesis, axonal guidance, and synaptogenesis (7, 31, 34). The Drosophila N-cadherin (CadN) belongs to a unique type of classic cadherin that is found in worms, insects, and vertebrates (35) and is likely the most ancient form of classic cadherin (11, 12). This type of cadherin has large, complex extracellular domains and cytoplasmic domains that bind to catenins. In Drosophila and Caenorhabditis elegans, CadN has been shown to regulate axonal patterning and fasciculation in embryonic motor neurons (2, 12). Furthermore, Drosophila CadN is required for photoreceptor and olfactory axons to select correct targets during development (10, 14, 48).
The Drosophila CadN gene contains three modules of alternative exons and undergoes alternative splicing to generate 12 isoforms. Although complex alternative splicing is not unusual for transcription factors and ion channels, it has been reported for only a small number of genes encoding surface receptors (30, 39). Among these are the vertebrate neurexins and insect Dscam (Down syndrome cell adhesion molecule), which can generate more than 1,000 and 38,000 isoforms, respectively (16, 29). In addition, the vertebrate cadherin-related neuronal receptor (CNR) genes utilize different promoters to generate approximately 50 isoforms (8, 45, 46). Though these findings suggest that alternative splicing in these receptor genes could conceivably generate extensive molecular complexity on the cell surfaces, the functions of receptor diversity are just being elucidated (30). Single-cell transcript analyses showed that individual Purkinje cells expressed different sets of CNR isoforms in mice (6), and individual photoreceptor neurons express different subsets of Dscam isoforms in flies (20). Furthermore, Dscam isoforms mediate strong homophilic but not heterophilic interactions (44). By expressing a distinct subset of Dscam or CNR isoforms, individual neurons could carry a unique combination of molecular tags or a unique identity, which would facilitate neuron-target or neuron-self recognition. Interestingly, vertebrate Dscam and CadN and insect neurexin do not appear to undergo extensive alternative splicing (32). It is likely that arthropods and vertebrates each selected different surface receptors to expand their repertoires during evolution.
The modular organization, and likely alternative splicing, of the CadN gene are conserved in Arthropoda, including mosquitoes and beetles (21, 37). Their conservation over 380 million years of evolutionary time is not restricted to the genomic structure but is extended to the amino acid sequence level, suggesting functional significance. All CadN isoforms generated by alternative splicing share the same molecular architecture but have different sequences in their extracellular and transmembrane domains. Although the differences in sequence in CadN isoforms could conceivably result in distinct properties, previous studies have not revealed any isoform specificity. Using the Drosophila visual and olfactory systems as models, previous studies showed that expression of a single CadN isoform is sufficient to rescue the CadN axonal-mistargeting phenotypes (19, 25, 37, 48). Furthermore, an in vitro cell aggregation study revealed that CadN isoforms mediate promiscuous heterophilic interactions (37). Thus, the functions of CadN molecular diversity remain to be determined.
In this study, we investigate the function of CadN molecular diversity. We find that the alternative splicing of CadN is regulated in a developmental, but not a cell-type-specific, fashion. In addition, CadN isoforms mediate graded homophilic interactions. We propose that CadN isoforms are differentially expressed during different developmental stages to provide adhesive interactions of different strengths.
MATERIALS AND METHODS
Cell sorting, RNA isolation, and reverse transcription.
Eye discs were hand dissected from the flies at various developmental stages. Populations of R3/R4 or R7 cells at the third instar stage were isolated by cell sorting as previously described (20). Total RNA was isolated using TRIZOL (Invitrogen) and reverse-transcribed using the Thermoscript III RT system (Invitrogen). For synthesizing cDNA from whole-animal RNA samples, random hexamers were used. For eye disc and specific R-cell-type RNA samples, CadN-specific primers were used to increase the yield. Control experiments using embryonic RNA samples showed that CadN profiles assessed using CadN-specific primers or random hexamers are indistinguishable. The CadN-specific primers were as follows: for exon 7b/7a, 5′-GCATTATGTACACGGTCG-3′; for exon 13a/13b, 5′-GTCCTGGGCGAATACAGTGA-3′; and for exon 18a/18b, 5′-GAGGGAGCAGCAGTATCAA-3′. We used 100 ng of reverse transcribed product for each TaqMan real-time PCR analysis.
Real-time PCR.
Primers and TaqMan probes were designed using PrimerExpress 1.0 software (PerkinElmer Life Sciences). Each assay was designed to detect one specific alternative exon and contained one general primer and one exon-specific primer to amplify 100 to 150 bp of the CadN transcript. To increase specificity, the designed TaqMan probes encompassed the junction sequence between the alternative and common exons (see Table 1 for primer sequences). TaqMan probes, primers, and universal TaqMan master mix were obtained from Applied Biosystems and used according to the manufacturer's instructions. An ABI Prism 7000 sequence detection system was used for real-time PCR analyses. Controlled amounts (10 to 3000 fg) of CadN isoform cDNAs were used as templates to derive standard curve and PCR efficiency and to test cross-reactivity for each assay. To construct the standard curve for the internal control, 18S rRNA, we used 10 to 1000 fg of the cDNAs from the 12-h and 24-h embryonic stages. The following thermocycling program was used for PCR amplification: one cycle at 50°C for 2 min and 95°C for 10 min and 50 cycles at 95°C for 15 s and 60°C for 1 min. Experiments were performed in triplicate. The data were analyzed using ABI Prism 7000 sequence detection system software. The standard errors of the ratio {SE[a/(a + b)]} were calculated using the δ method (26) with the following formulation: SE[a/(a + b)] = [ûb2 × σa2/(ûa + ûb)4 + μa2 × σb2/(ûa + ûb)4]1/2, where û and σ are the sample mean and standard error, respectively. The primers and TaqMan probes used for real-time PCR are listed in Table 1.
TABLE 1.
Primers and TaqMan probes used for real-time PCR
| Alternative exon | Forward primer (5′-3′) | Reverse primer (5′-3′) | TaqMan probe (5′-FAM, 3′-TAMRA) |
|---|---|---|---|
| 7a | TGTCACTGTGGGTACGGTTGTAA | TCGATAAAAAACTGTGGGATTGC | ATCGGTTAAGGCAAGTTCGGGTATCGAA |
| 7b | CCCGTTGGCGGCAAA | ACTGTGGGATTGCCTTCGAT | TCGTCTCGATCAAAGCCAGTTCGGG |
| 13a | CCAGATTACCGAAGAGGACGAT | CGATCCTTGTCGCCATCTGT | TGCCCAAACGCGTCTTGCAGGTTA |
| 13b | TCGACGAGGGAATGACCAA | CGATCCTTGTCGCCATCTGT | ACGCCCTTTACCATTATGCAGGTTACGG |
| 18a | CGTAGACCTGTGGAATGTCTACGA | CCTTGGAATCGGGTGACATAA | TGCACTTGCGGCGAAGGACG |
| 18b | CGTAGACCTGTGGAATGTCTACGA | TGCTGCCCGAGCTTTTGT | TGCACCTGTCCGGCGGGC |
Construction of CadN expression vectors.
The process for isolation of the CadN isoform cDNAs has been described previously. Standard PCR amplification and subcloning procedures were used to construct the expression vectors for various CadN mutants and CadN-green fluorescent protein (GFP). The chimera EGF-CA(18a)-TM(18b) is the CadN 7b-13a-18a isoform, except that the N-terminal half of the transmembrane region (residues 2,914 to 2,929) is encoded by the exon 18b (see Fig. 1). The chimera EGF-CA(18b)-TM(18a) is CadN 7b-13a-18a except that one and one-half of the EGF-CA repeats (residues 2850 to 2913) are encoded by the exon 18b. Constructs and detailed cloning procedures are available upon request. These cDNAs were inserted into the S2 expression vector, pRmHa3.
FIG. 1.
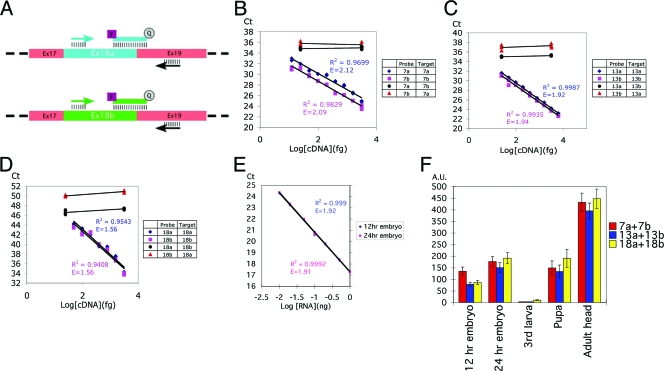
Exon organization of the Drosophila CadN gene. The CadN gene contains three exon modules corresponding to exons 7 (7a, 7a′,7b), 13 (13a, 13b), and 18 (18a, 18b) (37). Mutually exclusive alternative splicing results in variable transcripts that encode 12 different isoforms, each sharing the same molecular architecture but having different sequences in their cadherin repeats (exons 7 and 13) or EGF-CA repeats and transmembrane domains (exons 18a and 18b). Amino acid sequences encoded by the alternative exons 18a and 18b are shown with the conserved residues colored red and the divergent residues blue or brown. The putative “AXXXG” motif in the transmembrane domain encoded by the exon 18b is marked with yellow plus signs. Constant exons are shown as orange boxes and alternative exons as green or blue boxes. SP, signal peptide; CA, cadherin repeat; NC, nonchordate domain; EGF, EGF-like calcium-binding repeat; LamG, laminin-G-like domain; TM, transmembrane domain; JX, juxtamembrane region; βcat, β-catenin binding region; cleavage, proteolytic cleavage site.
Cell aggregation assay.
Suspension S2 cells used for cell aggregation assays were a generous gift from James Clemens. Cell culture and transfection were performed according to the Invitrogen DES and QIAGEN Effectene manuals. For expressing different CadN isoforms in S2 suspension cells, different CadN isoform expression vectors (pRmHa3/CadN isoforms) were cotransfected with a GFP vector (pRmHa3/GFP). CuSO4 (0.7 μM) was added 24 h after transfection to induce the expression of CadN isoforms and the GFP marker. The S2 cells were induced for 48 h and then subjected to cell aggregation assays as described previously by Oda (22) except for the following modifications. The S2 cells at a concentration of 1.2 × 106 cell/ml were incubated in 2 ml of BBS buffer (10 mM HEPES, 55 mM NaCl, 40 mM KCl, 15 mM MgSO4, 20 mM glucose, 50 mM sucrose, and 5 or 10 mM of CaCl2), an adult Drosophila hemolymph-like solution (13, 40). The cell suspensions were placed in a 35-mm polystyrene dish and agitated, using a gyratory shaker at 100 rpm for 1 h. The formation of cell aggregates was visualized and analyzed under a Zeiss M2bio fluorescence microscope. The cells in each aggregate were counted using the M2bio microscope in micro mode with a 10× lens objective. The image of these cell aggregates was captured using a Zeiss AxioCam digital camera and processed using Openlab software. The cell aggregates were divided into five categories based on the number of cells in each aggregate (25 to 50, 50 to 100, 100 to 150, 150 to 200, and >200 cells/per aggregate). Experiments were performed in triplicate. CadN mutants were compared with the control (full-length CadN 7b-13a-18a), and the confidence interval was calculated for each category.
FACS analysis.
Cells were fixed for 10 min with 1% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature. For surface fluorescence-activated-cell-sorter (FACS) analysis, cells were fixed, labeled using a Rat monoclonal antibody against the CadN extracellular domain [anti-CadN(EX), a gift from Tadashi Uemura] in a nondetergent buffer (5% goat serum in PBS), and then washed with this buffer before being labeling with Alexa 647-conjugated anti-rat immunoglobulin G secondary antibody and propidium iodide (PI). For GFP FACS analysis, cells were directly stained with a fluorescein isothiocyanate-conjugated anti-GFP antibody in a wash buffer (5% goat serum, 0.1% Triton X-100 in PBS) and then washed with the washing buffer to remove unbound antibodies. Cells were treated with RNase in PBS for 30 min at 37°C and then stained with PI for 20 min at room temperature. CadN expression, GFP, and DNA content (PI signal) were measured using a FACSCalibur flow cytometer (Becton Dickinson) with the appropriate filter sets. For each sample, 10,000 cells were analyzed. Data acquisition and analysis were performed using CELLQuest software (Becton Dickinson).
Immunoprecipitation and Western blotting.
Cell lysis, immunoprecipitation, and immunoblotting were performed as previously described (3), except that we used a different lysis buffer (50 mM Tris-Cl [pH 7.4], 150 mM NaCl, 1 mM dithiothreitol (DTT), and 1% NP-40 with or without 5 mM CaCl2). For each lane, a protein sample equivalent to 200,000 S2 cells was loaded. Rat monoclonal antibody against the cytoplasmic domain of CadN proteins (a gift from Tadashi Uemura) and rabbit polyclonal anti-GFP antibody (Molecular Probe) were used to detect the CadN and CadN-GFP fusion proteins, respectively.
RESULTS
CadN isoform expression can be accurately profiled using custom-tailored quantitative transcript assays.
The genomic structure of the CadN locus (Fig. 1) reveals three modules of alternatively spliced exons (7a/b, 13a/b, and 18a/b), which are evolutionarily conserved in Arthropoda, and a small alternative exon, 7a′, which is found only in Drosophila. By alternative use of these exons, the CadN locus is capable of generating 12 isoforms. In this study, we investigated the CadN isoforms generated by alternative splicing in the three conserved exon pairs. To address whether CadN alternative splicing is regulated, we developed a quantitative transcript assay based on TaqMan real-time PCR analysis (9). For each alternative exon, one common primer (complementary to the neighboring constant exon) and one alternative exon-specific primer were used to amplify 100 to 150 bp of the CadN transcript (Fig. 2A). To increase specificity and to avoid cross-reaction to genomic DNA, the TaqMan probes were designed to encompass the splicing junction sequence, thus including parts of both constant and variable exons (Table 1). In these assays, the total mRNA was reverse transcribed and the resulting cDNA was then subjected to real-time PCR analysis. The cycle-by-cycle detection of the increase in the amount of CadN PCR product was quantified (as a function of the FAM [6-carboxyfluorescein] fluorescence signal) in real time as the Taqman probes were cleaved to separate the reporter dye FAM from the quencher, TAMRA (6-carboxytetramethylrhodamine), during PCR extension.
FIG. 2.
TaqMan-based real-time PCR analysis provides an accurate measurement of the expression levels of the CadN alternative exons. (A) A schematic for primer and probe design is shown for exons 18a and 18b. Each assay used one alternative-exon-specific probe (blue or green line), one alternative-exon-specific primer (blue or green arrow), and one common primer (black arrows). In order to ensure target specificity, the TaqMan probes were designed to match the exon-exon boundary. Common exons 17 and 19 are shown in pink boxes, and alternative exons 18a and 18b in a blue or green box, respectively. (B to D). Standard curves and cross-reactivity tests for exons 7a/7b, 13a/13b, and 18a/18b, respectively. The accuracy and specificity of the TaqMan probes were tested using the cDNAs of the CadN isoform 7b-13a-18a or 7a-13b-18b in control amounts (as indicated). The x axis represents the logarithm of the input cDNA; the y axis represents the number of PCR cycles required to reach a given fluorescent signal level (Ct). When the probes were tested against their cognate target cDNAs (blue rhomboids and red squares), an inverse linear relationship between Ct and the logarithm of the input cDNA was observed over a range of 10 to 3,000 fg of CadN cDNA. Cross-reactivity, tested using TaqMan probes with their noncognate targets (blue discs and red triangles), was found to be minimal. Standard linear regression analysis was performed to calculate the standard curve, the coefficient of determination (R2), and PCR efficiency (E) (as indicated). The standard curves were used to calculate the experimental data (Fig. 3) by interpolation. (E) The standard curve for the internal control, 18S rRNA. Experiments were performed as for panels A to C except that embryo cDNAs were used as the target. (F) The CadN expression levels at different developmental stages were normalized using 18S rRNA as an internal reference (E). The sums of the expression levels of the CadN alternative exons 7a/7b, 13a/13b, and 18a/18b are shown as red, blue, and yellow bars, respectively. The x axis represents the developmental stages; the y axis represents CadN expression levels in an arbitrary unit (A.U.) with respect to the 18S rRNA level in the same sample.
The accuracy and specificity of these assays were tested with different CadN isoform cDNAs in controlled amounts. The apparent linearity in the derived standard curves indicated that the assays were accurate over a large range (10 to 3,000 fg) of CadN cDNA, and cross-reactivity was found to be minimal (Fig. 2B to D). The levels of CadN cDNA relative to those of an internal control (18S rRNA) in whole animals at various developmental stages are presented in Fig. 2F. The total CadN cDNA levels, calculated by adding the two alternatives (i.e., 7a + 7b; 13a + 13b, etc.), were found to be similar, regardless of which exon pair was used. This indicated that there was no significant bias in the reverse transcriptase reactions and that the assays provided a good estimate for the levels of CadN isoform mRNA. In the following, we describe the expression levels of alternative exons as a percentage of the total CadN cDNA in the given sample.
CadN alternative splicing is regulated in a temporal fashion during development.
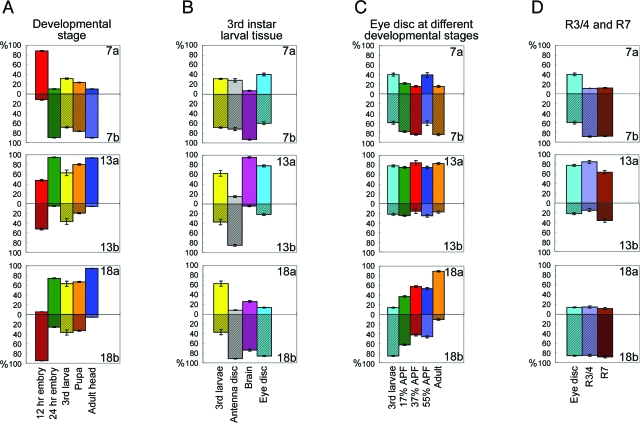
We first used this assay to determine the CadN isoform expression profiles in whole animals at different developmental stages. We found that all six alternative exons are expressed at some level throughout development (Fig. 3A). However, there were several significant differences in the CadN expression profiles at different developmental stages, indicating that CadN alternative splicing is developmentally regulated. At the early embryonic stage (Fig. 3A), exons 7a and 18b were predominantly used (88.3% and 94.8%, respectively) while exons 13a and 13b were expressed at approximately equal levels. In sharp contrast, at the late embryonic stage, exons 7b, 13a, and 18a (Fig. 3A) are used predominantly (89.7%, 94.7%, and 74.1%, respectively). Moreover, from the larval to pupal stages (Fig. 3A), the relative levels of 7b, 13a, and 18a gradually increased, and in the adult stage they constituted more than 90% of the total CadN transcripts (90.0%, 93.6%, and 94.7%, respectively). These data indicate that the isoform 7b-13a-18a is prevalent in the adult stage, while the other isoforms encoded by exons 7a, 13b, and 18b are expressed primarily during development.
FIG. 3.
CadN alternative splicing is developmentally regulated. The expression profiles of CadN alternative exons in wild-type animals were examined with TaqMan real-time PCR. The expression levels of alternative exons are described as percentages of the total CadN transcript levels. (A) CadN expression profiles of whole animals were assessed at the following five developmental stages: early embryo (embryos collected 0 to 12 h after egg laying), late embryo (embryos collected 12 to 24 h after egg laying), third larvae (wandering third instar larvae), pupae (20 to 80 h after pupal formation), and adult (heads of 1- to 3-day-old adult flies). (B) CadN expression profiles of different tissues of third instar larvae. The third larvae is as described for panel A; the antenna disc, brain, and eye disc are as indicated. (C) CadN expression profiles of the eye discs at the five different developmental stages, as indicated. (D) CadN expression profiles were assessed in two R-cell populations, R3/4 and R7 neurons, and in eye discs as described for panel B. All data present means and standard errors (error bars) of triplicate samples.
Next, we examined the use of CadN alternative exons in different tissues during development. Eye imaginal discs, antenna imaginal discs, and brains were isolated from the late third instar larvae. Quantitative analyses on RNA samples extracted from these tissues revealed that the expression of exons 13a and 13b, but not 7a/b or 18a/b, varies significantly in different tissues: exon 13b is the predominant form in antenna discs, while 13a dominates in the eye discs and brains (Fig. 3B).
We next examined CadN isoform profiles in the retina at different developmental stages. As in embryos, similar changes in the CadN expression profiles of the developing eye discs were observed for exons 18a and 18b: from the third instar larval, to the pupal, to the adult stages, the exon 18a level gradually increased from 15% to 92% (Fig. 3C). The 7b-13a-18b and 7b-13a-18a isoforms were the predominant forms in the larval eye discs and in the adult eyes, respectively; however, these two forms were expressed at similar levels during the pupal stages. Unlike that in the embryos, levels of exons 7a/b and 13a/b remain largely unchanged in developing retina.
We next examined whether different subtypes of photoreceptor neurons express different isoforms. Using a previously established cell-sorting method (20), we isolated R3/4 and R7 neurons from the third instar larval eye discs (see Fig. S1 in the supplemental material) and subjected the extracted RNA to quantitative PCR analyses. We found little difference between the expression profiles of the two R-cell types: R3/4 and R7 neurons exhibited virtually identical expression profiles on exons 7a/7b and 18a/18b and a similar preference for exon 13a over 13b (Fig. 3D). In summary, the alternative splicing of exons 18a/18b, but not of the other alternative exons, is dynamically regulated in the developing eye discs. However, the R3/4 and R7 types of photoreceptor neurons have similar expression profiles.
CadN isoforms mediate graded homophilic interactions.
The developmental regulation of CadN alternative splicing suggests that the isoforms might be differentially expressed to serve different functions if they have different biological activities. The CadN 7b-13a-18a isoform has been reported to mediate homophilic interaction in an S2 cell-based cell aggregation assay (12). The CadN isoforms have different amino acid sequences in their extracellular and transmembrane domains (Fig. 1), which could potentially alter their binding activities. Therefore, we set up a quantitative cell aggregation assay to measure the adhesive activity of different CadN isoforms.
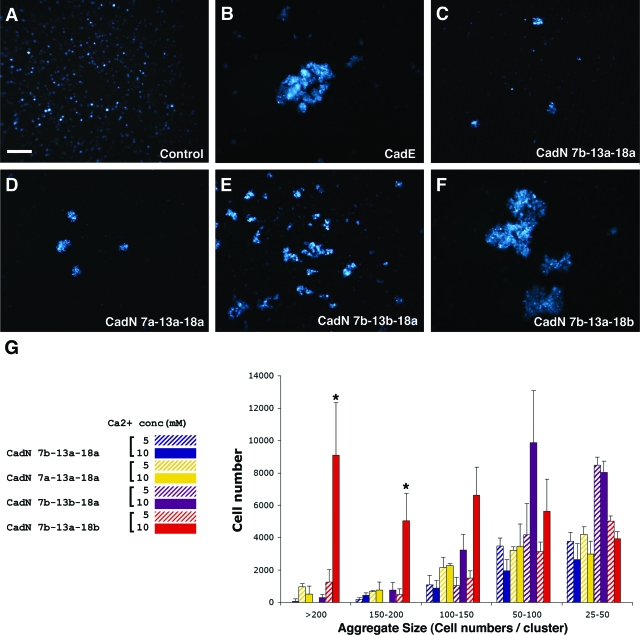
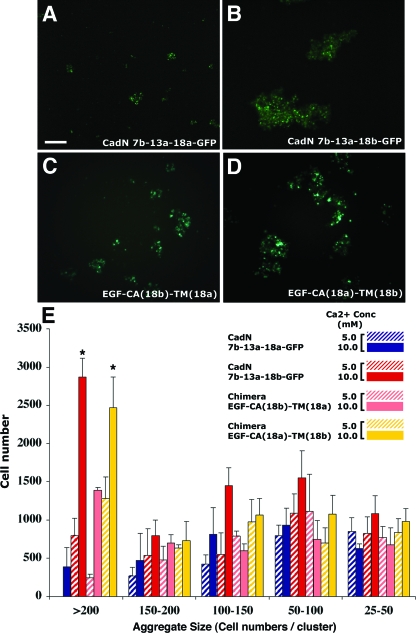
We found that S2 cells expressing the CadN 7b-13a-18a isoform or the alternative exon-substituted isoforms 7a-13a-18a, 7b-13b-18a, and 7b-13a-18b formed aggregates in the presence of calcium, indicating that they mediate homophilic interactions (Fig. 4C to F). The sizes of the cell aggregates varied significantly, depending on the specific CadN isoform the S2 cells expressed. The protein expression levels of different CadN isoforms, assayed by Western blotting, were comparable (Fig. 5A). Furthermore, FACS analysis using an antibody against the CadN extracellular domain revealed that the surface expression profiles of the CadN 7b-13a-18a and 7b-13a-18b isoforms were indistinguishable (Fig. 5B to E). The data indicated that the observed differences in the aggregate sizes were due to the intrinsic properties of different CadN isoforms rather than differences in their expression levels or surface presentations. We quantified the sizes of the cell aggregates by counting the number of S2 cells in each aggregate (Fig. 4G; see Materials and Methods for details). We observed that the CadN 7b-13a-18b isoform was capable of inducing very large aggregates, while the 7b-13a-18a isoform induced mostly small ones (P < 0.005). It is likely that the 18b-type CadN′s ability to induce larger cell aggregates reflects its stronger adhesive activity (or avidity) or faster forward-binding kinetics than that of the 18a type.
FIG. 4.
CadN isoforms mediate graded homophilic interactions. S2 cells coexpressing a GFP marker and CadE (E-cadherin) or different CadN isoforms were assessed for their abilities to induce cell aggregation (see Materials and Methods for details). (A to F). Representative images of cell aggregates induced by the expression of CadN isoforms or CadE and the addition of 10 mM of calcium. (A) GFP as a negative control; (B) CadE as a positive control; (C to F) CadN isoforms as indicated. Scale bar: 100 μm. (G) A bar chart of the size of cell aggregates formed by S2 cells expressing different CadN isoforms in the presence of 5 (striped bars) or 10 (solid bars) mM of calcium. Cell aggregates are divided into five categories according to the number of cells in each aggregate. The x axis represents the cell aggregate size (25 to 50, 50 to 100, 100 to 150, 150 to 200, and greater than 200 cells per cell cluster). The y axis represents the total number of cells that form a given range of cell cluster size. The bars indicate the means of three independent results. The error bars indicate the standard error. *, P < 0.005.
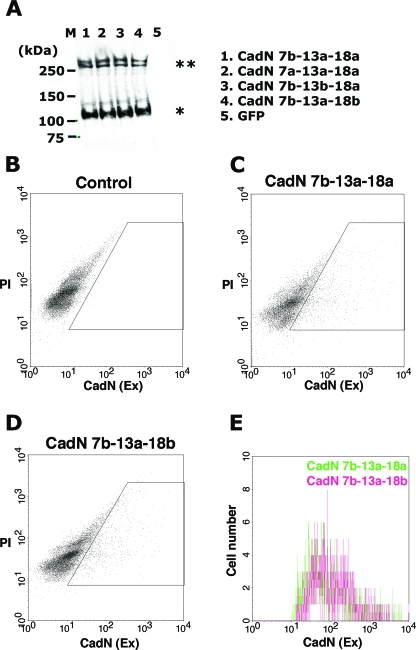
FIG. 5.
CadN isoforms exhibit similar surface representations. (A) Western blot analysis of the S2 cells expressing CadN 7b-13a-18a (lane 1), CadN 7a-13a-18a (lane 2), CadN 7b-13b-18a (lane 3), CadN 7b-13a-18b (lane 4), and GFP (lane 5). Rat monoclonal antibodies against the intracellular domain of the CadN protein were used to quantify the CadN expression. Mature CadN protein is proteolytically cleaved in the extracellular domain (Fig. 1) (12), resulting in two fragments that are associated noncovalently. Double and single asterisks indicate the unprocessed form and the processed C-terminal fragment of CadN proteins, respectively. M, molecular mass markers. (B to E) The surface expression of CadN isoforms was assessed using surface FACS (S-FACS) analysis. Anti-CadN (EX) antibody, which recognizes the extracellular domain of CadN, was used to label CadN protein on cell surfaces under non-cell-permeation conditions (see Materials and Methods for details). (B to D) Scatter plots of S-FACS analyses of the control S2 cells (B), the S2 cells expressing CadN 7b-13a-18a (C), and the CadN 7b-13a-18b isoform (D). The x axis represents the CadN surface expression level; the y axis represents the PI signal for the DNA content. S2 cells are heterogeneous, and large polyploid cells exhibit a low level of autofluorescence. The gate (boxed area) was established to exclude these cells in the analyses. For each sample, 10,000 cells were analyzed. The CadN 7b-13a-18a (C) and CadN 7b-13a-18b isoforms (D) were expressed by 13.2% and 12.8%, respectively, of S2 cells. (E) A histogram showing the CadN-positive cells in the gated area of panels C and D. The x axis represents CadN signal intensity; the y axis represents the cell number.
To determine the kinetics of cell aggregation induced by the 18a- and 18b-type CadN isoforms, we conducted time-course experiments. To facilitate the quantification of CadN expression, we fused a GFP moiety to the CadN carboxyl termini, allowing direct visualization of CadN expression. We found that cells expressing CadN 7b-13a-18a-GFP or its 18b counterpart aggregated in a time-dependent fashion (see Fig. S2 in the supplemental material). The 18b-expressing cells appeared to aggregate more rapidly than the 18a-expressing cells, as judged by the total number of aggregated cells (see Fig. S2A in the supplemental material) in the initial adhesion phase (the first 10 and 20 min). After 30 to 45 min, the apparent steady state was reached, when CadN-expressing cells continued to aggregate and dissociate but the total number of aggregated cells (or number of cells in each size category) remained constant (see Fig. S2A and B in the supplemental material). The sizes of the cell aggregates induced by CadN 7b-13a-18a-GFP CadN-7b-13a-18b-GFP recapitulated the differences observed in their untagged counterparts (P < 0.005) at all time points. We reasoned that, in order to form aggregates under constant rotating speed, the S2 cells must counter the shearing force that varies in proportion to the size of the aggregates. Thus, the aggregate size at the steady state should provide an estimate for the adhesive activity (see Discussion for the limitations of the cell aggregation assay). Therefore, we concluded that CadN isoforms mediate homophilic interaction, and the region encoded by exons 18a and 18b regulates their adhesive activity.
The transmembrane domain of CadN regulates its adhesive activity.
Exons 18a and 18b each encode one and one-half EGF-like repeats and one half of the transmembrane domain (Fig. 1). To determine which of these regions is responsible for the differential adhesive activity between the exon 18a- and 18b-encoding isoforms, we conducted a domain-swapping experiment. Interestingly, the CadN chimera 7b-13a-EGF-CA(18a)-TM(18b) (which contains the transmembrane domain encoded by exon 18b but which is otherwise CadN 7b-13a-18a) induced large cell aggregates essentially indistinguishable from those induced by CadN 7b-13a-18b (Fig. 6D and E). Conversely, the chimera 7b-13a-EGF-CA(18b)-TM(18a) primarily induced small and intermediate-sized cell clusters (Fig. 6C and E). The overall expression levels (assayed by Western blotting) and the expression profiles at the individual cell level (assayed by FACS analysis) were indistinguishable in the four cell populations (see Fig. S3A to H in the supplemental material). These data support the role of transmembrane domains in regulating CadN-mediated adhesion.
FIG. 6.
The transmembrane domains of CadN isoforms determine the strength of the homophilic adhesive activity. Representative images of cell aggregates formed by the S2 cells expressing CadN 7b-13a-18a-GFP (A), CadN 7b-13a-18b-GFP (B), or the domain-swapped CadN, EGF-CA(18b)-TM(18a) (C) or EGF-CA(18a)-TM(18b) (D), in the presence of 10 mM of calcium. Scale bar in panel A, 100 μm. (E) Bar chart comparing the sizes of cell aggregates formed by S2 cells expressing different CadN isoforms in the presence of 5 (striped bars) or 10 (solid bars) mM of calcium. The cell aggregates were divided into five categories according to the number of cells in each aggregate. The x axis represents cell aggregate sizes 25 to 50, 50 to 100, 100 to 150, 150 to 200, and greater than 200 cells per cell cluster. The y axis represents the total number of cells that form a given range of cell cluster size. All data present means and standard errors (error bars) of triplicate samples. *, P < 0.005.
CadN isoforms mediate cis interaction.
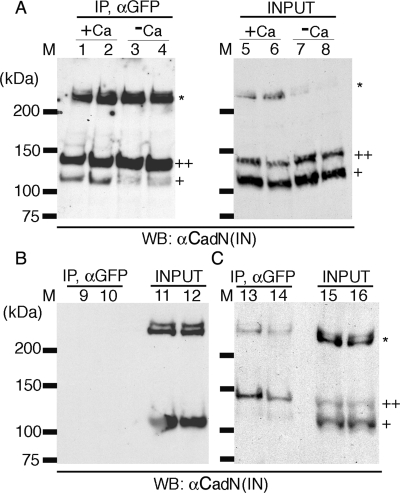
Because the transmembrane domains regulate CadN′s adhesive activity, we reasoned that the transmembrane domains might exert their effect by regulating cis interactions (on the same cell surface). Previous studies have demonstrated that vertebrate cadherins, which are distinct from CadN, form cis dimers before forming trans tetramers (between different cell surfaces) (1, 33). We therefore examined whether Drosophila CadN mediates cis interactions. The CadN isoform 7b-13a-18a and its GFP-tagged version (7b-13a-18a-GFP) were coexpressed in S2 cells, and the GFP-tagged CadN was immunoprecipitated using anti-GFP antibody in the presence of calcium. We found that the native form of CadN (corresponding to ∼25% of GFP-CadN) coprecipitated with the GFP-tagged CadN, suggesting that they form cis homodimers or oligomers (Fig. 7, lane 1). The precipitation of native CadN depended on the presence of CadN-GFP, confirming the specificity of the immunoprecipitation (Fig. 7, lane 9). Furthermore, the formation of CadN cis homodimers (or oligomers) was largely dependent on calcium because only small amounts of CadN could be coimmunoprecipitated without calcium in the incubation and lysis buffers (Fig. 7, lane 3). To exclude the possibility that coimmunoprecipitation (co-IP) of CadN and CadN-GFP reflects trans interaction rather than cis interaction, we mixed two populations of S2 cells, one expressing the CadN native form and the other CadN-GFP, and performed immunoprecipitation in the presence of calcium as before. We found very little CadN (corresponding to ∼1% of CadN-GFP) coprecipitated with CadN-GFP (Fig. 7, lane 13), indicating that cis but not trans interaction is responsible for the observed co-IP. These data are consistent with a previous report that most of the trans interactions of cadherins are broken during immunoprecipitation processes (1).
FIG. 7.
CadN mediates calcium-dependent cis interactions. (A) CadN 7b-13a-18a and CadN 7b-13a-18a-GFP (lanes 1, 3, 5, and 7) or CadN 7b-13a-18b (lanes 2, 4, 6, and 8) and its GFP-tagged version were coexpressed in S2 cells. The association of CadN and CadN-GFP, in the presence (lanes 1, 2, 5, and 6) or absence (lanes 3, 4, 7, and 8) of 5 mM of calcium, was assessed by immunoprecipitation. Extracts (lanes 5 to 8) or anti-GFP immunoprecipitates (lanes 1 to 4) were analyzed by Western blotting and probed with an antibody recognizing the CadN cytoplasmic domain [αCadN (IN)]. CadN 7b-13a-18a (or CadN 7b-13a-18b) coimmunoprecipitated with CadN 7b-13a-18a-GFP (or CadN 7b-13a-18b-GFP) in the presence of calcium. In the absence of calcium, only a small fraction of CadN coimmunoprecipitated with CadN-GFP. Mature CadN protein is proteolytically cleaved to form N-terminal and C-terminal fragments (12). The C-terminal fragments of CadN (∼110 kDa) and CadN-GFP (∼140 kDa) are indicated by single and double plus signs, respectively. Some CadN protein remained unprocessed (asterisk). (B) In the absence of CadN-GFP, CadN 7b-13a-18a (lane 9) and CadN 7b-13a-18b (lane 10) were not found in the anti-GFP immunoprecipitates, indicating that the anti-GFP antibody is specific. Extracts of the S2 cells expressing CadN 7b-13a-18a (lane 11) or CadN 7b-13a-18b (lane 12) used for the immunoprecipitate experiments were analyzed with anti-CadN (IN) Western blotting. (C) trans interactions between CadN and CadN-GFP were assessed using anti-GFP immunoprecipitation (Ip). Two separate populations of S2 cells, one expressing CadN and the other CadN-GFP (7b-13a-18a, lanes 13 and 15; 7b-13a-18b, lanes 14 and 16), were mixed and subjected to anti-GFP Ip and Western blot analysis as described before. Only a very small amount of CadN coimmunoprecipitated with CadN-GFP, indicating that the trans association between CadN and CadN-GFP was largely disrupted by the experimental procedures.
To examine the possibility that the exon 18b-encoding transmembrane domains induce strong cis interactions, which translate into high cell-aggregating activity, we examined the co-IP between CadN 7b-13a-18b and its GFP-tagged version. We found that CadN 7b-13a-18b behaved similarly to CadN 7b-13a-18a in the co-IP experiments (Fig. 6, lanes 2, 4, 6, 8, 10, 12, 14, and 16). This suggests that, within the limitations of our experiments, there were no observed differences in the cis-interaction activity between the 18a- and 18b-encoding CadN isoforms. In summary, both CadN 7b-13a-18a and 7b-13a-18b isoforms form calcium-dependent cis-dimers (or -oligomers) with similar strengths.
DISCUSSION
CadN alternative splicing might provide isoforms of different adhesive activities at different developmental stages.
In insects, CadN and Dscam are the only two known receptor genes that have modular exon organizations and undergo alternative splicing to generate multiple isoforms (29, 37). However, they appear to use alternative splicing to alter the isoform functions in very different ways. The Dscam isoforms exhibit isoform-specific homophilic activity, specified by their extracellular domains (44). In contrast, CadN isoforms mediate graded homophilic interactions as well as promiscuous heterophilic interactions (37). Furthermore, two different Dscam transmembrane/juxtamembrane domains, encoded by the alternative exon 17.1/17.2, determine the subcellular localization of the receptors (42). On the other hand, the CadN transmembrane domains, encoded by the exon 18a/18b, regulate the adhesive activity. It has been proposed that the isoform-specific homophilic activity of Dscam mediates self-versus-nonself recognition in neurites (47). How the differential adhesive activities of CadN isoforms relate to their in vivo functions remains to be elucidated.
Previous studies demonstrated that expression of a single CadN isoform is sufficient to rescue targeting defects in the CadN mutant photoreceptor and olfactory neurons (19, 25, 37, 48). While it is possible that in these transgene rescue experiments quantitative differences among the adhesive activities of CadN isoforms are masked by the artificially high expression levels mediated by transgenes, these results clearly establish that the CadN isoforms do not form synaptic codes to direct target selection in the visual and olfactory systems. This is further supported by our quantitative transcript analyses showing that different R-cell subtypes have similar CadN isoform expression profiles.
Interestingly, our quantitative transcript analyses revealed that CadN alternative splicing is developmentally regulated. Overall, the exon 18a-encoding CadN isoforms are primarily expressed during the late developmental and adult stages, while the exon 18b-encoding isoforms are expressed during the early developmental stage. In the retina, CadN 7b-13a-18b dominates during the third instar larval stage when R-cells project axons, and it gradually switches to 7b-13a-18a from the early pupal to the adult stages. This developmental switch from exon 18b to 18a is of particular interest in light of the observed differences in the adhesive activities of the isoforms encoded by these two exons. We hypothesize that 7b-13a-18b provides strong homophilic interactions among the axons during axon extension, while 7b-13a-18a mediates weak, and perhaps tunable, interactions between growth cones and their targets. It has been revealed that, in vertebrates, synaptic adhesion mediated by N-cadherin is modulated by synaptic activity (36). Therefore, the alternative hypothesis is that the high-affinity 18b-type isoforms are replaced with the low-affinity 18a type at the late developmental stage to ensure synaptic plasticity in the adults.
The transmembrane domain of CadN regulates adhesive activity.
Using an in vitro cell aggregation assay to study CadN-mediated adhesion, we demonstrated that the S2 cells expressing the 18b-type CadN aggregate faster and form larger clusters than those expressing the 18a type do. Although it is tempting to directly translate these measurements into affinity and kinetic parameters, note that this widely used aggregation assay has two limitations. First, because the cells have only limited time to form aggregates, this and other similar in vitro adhesion assays are intrinsically biased toward fast-association events (5, 17). The in vitro conditions clearly do not reflect slow in vivo events such as cell sorting but perhaps mimic those of fast events, such as the interactions between nerve growth cones and their targets. During the establishment of neuronal connections, nerve growth cones project highly motile filopodia to interact with dynamic filopodium-like structures of the target cells (18, 27, 41). Second, the cell aggregation assay described in this study and another (24) does not depend on the cytoplasmic domain of cadherins and thus their interactions with catenins and the actin cytoskeleton (S. Yonekura and C.-H. Lee, unpublished observation). While many in vivo functions of cadherins depend on their abilities to bind catenin (24), some might not. In the Drosophila visual system, the cytoplasmic domain of CadN is not required for R7 growth cones to reach the target layer (a CadN-dependent process) (Yonekura and Lee, unpublished). Taken together, these findings indicate that the in vitro cell aggregation assay described here and elsewhere likely assesses cadherin's ability to initiate adhesion, an important attribute of cadherin function.
Using domain swap experiments, we demonstrate that the transmembrane domains of CadN isoforms regulate their adhesive activities. To our knowledge, this is the first report documenting that transmembrane domains regulate adhesion. It has been suggested that interactions between transmembrane domains stabilize homodimer formation of the epidermal growth factor receptor (ErbB1) and the erythropoietin receptor (4, 43). Several transmembrane domains, including that of ErbB1, form dimers in the absence of their extracellular domains (15), and the dimer formation involves a G(S/A)XXXG motif in which transmembrane helix interactions are mediated (28). Interestingly, the Drosophilia CadN exon 18b- but not 18a-encoding transmembrane region contains an AXXXG motif (Fig. 1), and CadN protein forms cis dimers in culture cells. In classic vertebrate cadherins, cis dimer formation is a prerequisite for generating the adhesive tetramers in trans (23, 38). It is tempting to speculate that Drosophila CadN functions in a similar fashion. However, our co-IP experiment failed to reveal any differences in the abilities of exon 18a- and 18b-encoding CadN isoforms to form cis dimers. While it is possible that our co-IP assay was not sensitive enough to reveal the differences, the alternative explanation is that exon 18b-encoding transmembrane domains align the CadN in an orientation that facilitates trans-tetramer formation. The current study suggests that the cis interaction between the transmembrane domains could potentially modulate CadN avidity. Further experiments will be needed to determine the structural basis of the cis-interaction by transmembrane sequences.
Supplementary Material
Acknowledgments
We thank Tadashi Uemura and James Clemens for providing reagents, without which this project could not be completed. We give special thanks to Aiyi Liu for advice on statistics. We thank Larry Zipursky and Thomas Clandinin for communicating results prior to publication and Benjamin White, Alan Hinnebusch, Mark Lemmon, and Henry Levin for helpful discussions. We thank Margaret Dieringer for manuscript handling and editing.
This work is supported by the Intramural Research Program of the NIH, National Institute of Child Health and Human Development (grant HD008748-03 to C.-H.L.) S.Y. is a fellow of the Japan Society for the Promotion of Science.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Brieher, W. M., A. S. Yap, and B. M. Gumbiner. 1996. Lateral dimerization is required for the homophilic binding activity of C-cadherin. J. Cell Biol. 135:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broadbent, I. D., and J. Pettitt. 2002. The C. elegans hmr-1 gene can encode a neuronal classic cadherin involved in the regulation of axon fasciculation. Curr. Biol. 12:59-63. [DOI] [PubMed] [Google Scholar]
- 3.Chitaev, N. A., and S. M. Troyanovsky. 1998. Adhesive but not lateral E-cadherin complexes require calcium and catenins for their formation. J. Cell Biol. 142:837-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curran, A. R., and D. M. Engelman. 2003. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr. Opin. Struct. Biol. 13:412-417. [DOI] [PubMed] [Google Scholar]
- 5.Duguay, D., R. A. Foty, and M. S. Steinberg. 2003. Cadherin-mediated cell adhesion and tissue segregation: qualitative and quantitative determinants. Dev. Biol. 253:309-323. [DOI] [PubMed] [Google Scholar]
- 6.Esumi, S., N. Kakazu, Y. Taguchi, T. Hirayama, A. Sasaki, T. Hirabayashi, T. Koide, T. Kitsukawa, S. Hamada, and T. Yagi. 2005. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat. Genet. 37:171-176. [DOI] [PubMed] [Google Scholar]
- 7.Gumbiner, B. M. 1996. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84:345-357. [DOI] [PubMed] [Google Scholar]
- 8.Hamada, S., and T. Yagi. 2001. The cadherin-related neuronal receptor family: a novel diversified cadherin family at the synapse. Neurosci. Res. 41:207-215. [DOI] [PubMed] [Google Scholar]
- 9.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 10.Hummel, T., and S. L. Zipursky. 2004. Afferent induction of olfactory glomeruli requires N-cadherin. Neuron 42:77-88. [DOI] [PubMed] [Google Scholar]
- 11.Iwai, Y., Y. Hirota, K. Ozaki, H. Okano, M. Takeichi, and T. Uemura. 2002. DN-cadherin is required for spatial arrangement ultrastructural organization of synapses. Mol. Cell Neurosci. 19:375-388. [DOI] [PubMed] [Google Scholar]
- 12.Iwai, Y., T. Usui, S. Hirano, R. Steward, M. Takeichi, and T. Uemura. 1997. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron 19:77-89. [DOI] [PubMed] [Google Scholar]
- 13.Larrivee, D. C. 1979. A biochemical analysis of the Drosophila rhabdomere and its extracellular environment. Ph.D. dissertation. Purdue University, West Lafayette, Ind.
- 14.Lee, C. H., T. Herman, T. R. Clandinin, R. Lee, and S. L. Zipursky. 2001. N-cadherin regulates target specificity in the Drosophila visual system. Neuron 30:437-450. [DOI] [PubMed] [Google Scholar]
- 15.Mendrola, J. M., M. B. Berger, M. C. King, and M. A. Lemmon. 2002. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J. Biol. Chem. 277:4704-4712. [DOI] [PubMed] [Google Scholar]
- 16.Missler, M., and T. C. Sudhof. 1998. Neurexins: three genes and 1001 products. Trends Genet. 14:20-26. [DOI] [PubMed] [Google Scholar]
- 17.Moyer, W. A., and M. S. Steinberg. 1976. Do rates of intercellular adhesion measure the cell affinities reflected in cell-sorting and tissue-spreading configurations? Dev. Biol. 52:246-262. [DOI] [PubMed] [Google Scholar]
- 18.Murray, M. J., D. J. Merritt, A. H. Brand, and P. M. Whitington. 1998. In vivo dynamics of axon pathfinding in the Drosophilia CNS: a time-lapse study of an identified motorneuron. J. Neurobiol. 37:607-621. [PubMed] [Google Scholar]
- 19.Nern, A., L. V. Nguyen, T. Herman, S. Prakash, T. R. Clandinin, and S. L. Zipursky. 2005. An isoform-specific allele of Drosophila N-cadherin disrupts a late step of R7 targeting. Proc. Natl. Acad. Sci. USA 102:12944-12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neves, G., J. Zucker, M. Daly, and A. Chess. 2004. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat. Genet. 36:240-246. [DOI] [PubMed] [Google Scholar]
- 21.Oda, H., K. Tagawa, and Y. Akiyama-Oda. 2005. Diversification of epithelial adherens junctions with independent reductive changes in cadherin form: identification of potential molecular synapomorphies among bilaterians. Evol. Dev. 7:376-389. [DOI] [PubMed] [Google Scholar]
- 22.Oda, H., T. Uemura, Y. Harada, Y. Iwai, and M. Takeichi. 1994. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev. Biol. 165:716-726. [DOI] [PubMed] [Google Scholar]
- 23.Ozawa, M. 26. March 2002. Lateral dimerization of the E-cadherin extracellular domain is necessary but not sufficient for adhesive activity. J. Biol. Chem. 277:19600-19608. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 24.Pacquelet, A., L. Lin, and P. Rorth. 2003. Binding site for p120/delta-catenin is not required for Drosophila E-cadherin function in vivo. J. Cell Biol. 160:313-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prakash, S., J. C. Caldwell, D. F. Eberl, and T. R. Clandinin. 2005. Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nat. Neurosci. 8:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radhakrishna, R. C. 1965. Linear statistical inference and its applications. John Wiley & Sons Inc, New York, N.Y.
- 27.Ritzenthaler, S., E. Suzuki, and A. Chiba. 2000. Postsynaptic filopodia in muscle cells interact with innervating motoneuron axons. Nat. Neurosci. 3:1012-1017. [DOI] [PubMed] [Google Scholar]
- 28.Russ, W. P., and D. M. Engelman. 2000. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 296:911-919. [DOI] [PubMed] [Google Scholar]
- 29.Schmucker, D., J. C. Clemens, H. Shu, C. A. Worby, J. Xiao, M. Muda, J. E. Dixon, and S. L. Zipursky. 2000. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101:671-684. [DOI] [PubMed] [Google Scholar]
- 30.Schmucker, D., and J. G. Flanagan. 2004. Generation of recognition diversity in the nervous system. Neuron 44:219-222. [DOI] [PubMed] [Google Scholar]
- 31.Shapiro, L., and D. R. Colman. 1999. The diversity of cadherins and implications for a synaptic adhesive code in the CNS. Neuron 23:427-430. [DOI] [PubMed] [Google Scholar]
- 32.Tabuchi, K., and T. C. Sudhof. 2002. Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics 79:849-859. [DOI] [PubMed] [Google Scholar]
- 33.Takeda, H., Y. Shimoyama, A. Nagafuchi, and S. Hirohashi. 1999. E-cadherin functions as a cis-dimer at the cell-cell adhesive interface in vivo. Nat. Struct. Biol. 6:310-312. [DOI] [PubMed] [Google Scholar]
- 34.Takeichi, M. 1991. Cadherin cell adhesion receptors as a morphogenetic regulator. Science 251:1451-1455. [DOI] [PubMed] [Google Scholar]
- 35.Tanabe, K., M. Takeichi, and S. Nakagawa. 2004. Identification of a nonchordate-type classic cadherin in vertebrates: chicken Hz-cadherin is expressed in horizontal cells of the neural retina and contains a nonchordate-specific domain complex. Dev. Dyn. 229:899-906. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka, H., W. Shan, G. R. Phillips, K. Arndt, O. Bozdagi, L. Shapiro, G. W. Huntley, D. L. Benson, and D. R. Colman. 2000. Molecular modification of N-cadherin in response to synaptic activity. Neuron 25:93-107. [DOI] [PubMed] [Google Scholar]
- 37.Ting, C. Y., S. Yonekura, P. Chung, S. N. Hsu, H. M. Robertson, A. Chiba, and C. H. Lee. 2005. Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development 132:953-963. [DOI] [PubMed] [Google Scholar]
- 38.Troyanovsky, R. B., E. Sokolov, and S. M. Troyanovsky. 2003. Adhesive and lateral E-cadherin dimers are mediated by the same interface. Mol. Cell. Biol. 23:7965-7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ullrich, B., Y. A. Ushkaryov, and T. C. Sudhof. 1995. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron 14:497-507. [DOI] [PubMed] [Google Scholar]
- 40.van der Meer, J. M., and L. F. Jaffe. 1983. Elemental composition of the perivitelline fluid in early Drosophila embryos. Dev. Biol. 95:249-252. [DOI] [PubMed] [Google Scholar]
- 41.Vasenkova, I., D. Luginbuhl, and A. Chiba. 2006. Gliopodia extend the range of direct glia-neuron communication during the CNS development in Drosophila. Mol. Cell. Neurosci. 31:123-130. [DOI] [PubMed] [Google Scholar]
- 42.Wang, J., X. Ma, J. S. Yang, X. Zheng, C. T. Zugates, C. H. Lee, and T. Lee. 2004. Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron 43:663-672. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, I. A., and L. K. Jolliffe. 1999. The structure, organization, activation and plasticity of the erythropoietin receptor. Curr. Opin. Struct. Biol. 9:696-704. [DOI] [PubMed] [Google Scholar]
- 44.Wojtowicz, W. M., J. J. Flanagan, S. S. Millard, S. L. Zipursky, and J. C. Clemens. 2004. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell 118:619-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, Q., and T. Maniatis. 1999. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 97:779-790. [DOI] [PubMed] [Google Scholar]
- 46.Yagi, T., and M. Takeichi. 2000. Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev. 14:1169-1180. [PubMed] [Google Scholar]
- 47.Zhan, X. L., J. C. Clemens, G. Neves, D. Hattori, J. J. Flanagan, T. Hummel, M. L. Vasconcelos, A. Chess, and S. L. Zipursky. 2004. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron 43:673-686. [DOI] [PubMed] [Google Scholar]
- 48.Zhu, H., and L. Luo. 2004. Diverse functions of N-cadherin in dendritic and axonal terminal arborization of olfactory projection neurons. Neuron 42:63-75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.