Abstract
Although vertebrate POT1 is thought to play a role in both telomere capping and length regulation, its function has proved difficult to analyze. We therefore generated a conditional cell line that lacks wild-type POT1 but expresses an estrogen receptor-POT1 fusion. The cells grow normally in tamoxifen, but drug removal causes loss of POT1 from the telomere, rapid cell cycle arrest, and eventual cell death. The arrested cells have a 4N DNA content, and addition of caffeine causes immediate entry into mitosis, suggesting a G2 arrest due to an ATM- and/or ATR-mediated checkpoint. γH2AX accumulates at telomeres, indicating a telomeric DNA damage response, the likely cause of the checkpoint. However, POT1 loss does not cause degradation of the G-strand overhang. Instead, the amount of G overhang increases two- to threefold. Some cells eventually escape the cell cycle arrest and enter mitosis. They rarely exhibit telomere fusions but show severe chromosome segregation defects due to centrosome amplification. Our data indicate that vertebrate POT1 is required for telomere capping but that it functions quite differently from TRF2. Instead of being required for G-overhang protection, POT1 is required to suppress a telomeric DNA damage response. Our results also indicate significant functional similarities between POT1 and Cdc13 from budding yeast (Saccharomyces cerevisiae).
Telomeres exist as complex chromatin structures that form a protective cap for the chromosome end (43). This cap contains a number of specialized telomere proteins that regulate access to the enzymes required for telomere maintenance and prevent the chromosome terminus from being subject to DNA repair reactions such as nonhomologous end joining (NHEJ). The telomeric DNA consists of a duplex region of simple repeat sequence that terminates in a single-strand overhang on the 3′ G-rich strand. In vertebrates and other organisms with long telomeres, the telomeric DNA is folded back into a lariat-like structure in which the G overhang is tucked into the duplex region of the telomeric tract (17, 37). During DNA replication, passage of the replication fork is expected to open the T-loop structure so as to expose the overhang to telomerase and other proteins required for telomere replication (56). While the telomere is in this open configuration, the G overhang is thought to be protected by POT1 (protection of telomeres), a protein that specifically recognizes telomeric G-strand DNA (2, 56).
In vertebrate cells, the double-stranded region of the telomeric DNA is bound by a core complex of six proteins: TRF1, TRF2, RAP1, TIN2, TPP1, and POT1 (10, 32, 61). TRF1 and TRF2 bind directly to the duplex T2AG3 repeats (4), while POT1 is anchored to the complex via TPP1 (21, 33, 62). Loss of TRF2 leads to a catastrophic telomere deprotection phenotype that involves removal of G overhangs by the nucleotide excision repair nuclease ERCC1/XPF followed by extensive telomere fusion via the NHEJ pathway (5, 63). Thus, TRF2 plays a key role in telomere capping. In contrast, TRF1 is more important for telomere length regulation, with overexpression leading to telomere shortening and removal leading to lengthening (50). The interplay between the components of the core TRF/RAP/TIN/TPP/POT complex in telomere length regulation appears complicated, as the length set point can be changed in unexpected ways by overexpressing wild-type and mutant versions of the various proteins (7, 21, 31, 34).
In addition to the core telomere protein complex, a number of double-strand break repair proteins associate with telomeres (9). During replication of a functional telomere, the open telomere structure is transiently recognized as DNA damage and hence able to recruit proteins such as ATM and/or ATR and the MRX complex (47, 54). However, subsequent loading of specific telomere proteins somehow limits the DNA damage response and prevents activation of downstream transducers such as p53, Chk1, and Chk2. In contrast, when telomeres become dysfunctional due to critical DNA shortening or loss of TRF2, the full DNA damage response is activated, resulting in accumulation of γH2AX and other damage response proteins at the uncapped telomere (8, 18, 45) and hence formation of telomere dysfunction-induced foci (45).
Telomeric G-strand binding proteins have been identified for a wide range of organisms including vertebrates, plants, fission yeast (Schizosaccharomyces pombe) (the POT1 proteins), ciliates (the telomere end binding proteins), and budding yeast (Saccharomyces cerevisiae) (Cdc13) (42, 56). All of these proteins bind telomeric G-strand DNA with high affinity and show specificity for their cognate telomeric sequence. Although they share relatively low levels of sequence identity, their DNA binding domains consist of structurally conserved oligonucleotide/oligosaccharide binding (OB) folds (20, 29, 30, 36, 49).
POT1 proteins are essential for telomere protection in both fission yeast and plants. Deletion of S. pombe POT1 leads to rapid loss of telomeric and subtelomeric sequences and widespread cell death (2). Likewise, expression of a dominant negative allele of one of the Arabidopsis thaliana POT1 orthologs, Pot2, leads to extreme telomere shortening, telomere fusions, and massive genome instability (42). These telomere uncapping phenotypes, together with the highly conserved DNA binding specificity of the telomere end binding proteins and POT1 proteins, suggested that POT1 family members promote telomere protection by binding to the G-strand overhang. However, studies with human POT1 (hPOT1) did not confirm this hypothesis and revealed alternative functions. Deletion of the N-terminal OB fold resulted in an hPOT1 mutant that could not bind G-strand DNA; however, it could still associate with telomeres in vivo (34). It also caused telomere lengthening, indicating an unexpected role for hPOT1 in telomere length regulation. It is now apparent that hPOT1 binds to TPP1 through its C-terminal domain and is part of the core protein complex that coats the duplex telomeric DNA (33, 62). This finding explains why the ΔOB mutant binds to telomeres and may also explain the effects of hPOT1 on telomere length.
A variety of phenotypes have been observed when hPOT1 levels are decreased by use of short-hairpin RNA (shRNA) or small interfering RNA (19, 52, 60). In general, the knockdown leads to telomere lengthening, a decrease in proliferation, and a stable 40 to 60% decrease in the amount of G-strand overhang. Surprisingly, multiple studies report only a modest (∼2-fold) increase in telomere fusions, indicating that POT1 depletion has much less effect on telomere capping than TRF2 depletion. However, one caveat of the RNA interference (RNAi) experiments is that only 70 to 95% of the POT1 was depleted; thus, the protein could have been lost preferentially from the double-stranded telomeric DNA, leaving sufficient residual protein to bind and protect the G overhangs.
In light of this uncertainty, we have taken a different approach to remove POT1 from telomeres, namely, disruption of the POT1 gene in chicken DT40 cells. We used chicken DT40 cells because this avian leukosis virus-transformed B-cell line exhibits high levels of homologous recombination and hence allows efficient gene targeting (59). Contrary to early reports, DT40 cells are not p53 deficient but have functional p53 that can activate downstream targets (44). The chicken genome is similar to the human genome in that it appears to contain only one POT1 gene. Moreover, previous studies have shown that the chicken and human POT1 proteins are similar in structure, telomeric distribution, and biochemical properties (3, 35, 55). As we found that chicken POT1 is essential, we generated a conditional cell line that expresses an estrogen receptor (ER)-POT1 fusion protein (ER-POT1). This fusion protein is removed from the nucleus during growth in the absence of tamoxifen. The conditional cell line has allowed us to examine the role of POT1 in telomere protection. Our results indicate that POT1 is essential for telomere capping but that, in contrast to TRF2, POT1 is not needed to prevent G-overhang loss or telomere fusions. Rather, POT1 prevents G-overhang elongation, activation of a cell cycle checkpoint, centrosome amplification, and subsequent defects in chromosome segregation.
MATERIALS AND METHODS
Generation of the POT1 gene-targeting constructs and the ER-POT1 cell line.
To make the ER-POT1 gene replacement construct, the mouse ER ligand binding domain was cloned into pcDNA3 (Invitrogen) in frame with Flag-tagged chicken POT1 cDNA. The ER-Flag-POT1 gene plus adjacent polyadenylation sequence and neomycin marker cassette were removed from pcDNA3 and subcloned into pBluescript (Stratagene). A 3.2-kb 5′ segment of POT1 genomic DNA that terminated at the start codon within exon 4 was cloned immediately upstream of ER-Flag-POT1, while a 2.8-kb segment of POT1 genomic DNA starting immediately after exon 6 was cloned 3′ of the neomycin expression construct. To make the gene disruption construct, the ER-Flag-POT1 and neomycin marker cassettes were replaced with a blasticydin (Bsr) marker cassette.
The ER-POT1 gene replacement construct was electroporated into wild-type chicken DT40 cells, and transformants were selected on G418. Genomic DNA from G418-resistant clones was screened for targeted integration by Southern hybridization. Clones that had the ER-POT1 expression cassette integrated into the native POT1 gene locus were screened for expression of the ER-Flag-POT1 fusion protein and tamoxifen-dependent telomere localization by indirect immunofluorescence with ER or Flag (M2) antibody. One clone was then electroporated with the POT1 gene-targeting constructs, and transformants were selected on Bsr in the presence of 100 nM tamoxifen. Positive clones were again screened by Southern hybridization.
Cell culture and FACS analysis.
Cells were cultured in RPMI plus 10% fetal calf serum, 1% chicken serum, and 50 μM β-mercaptoethanol as previously described (55). Where applicable, tamoxifen was added to 100 nM, caffeine to 2 mM, nocodazole to 0.5 μg/ml, and aphidicolin to 1 μg/ml. To obtain cultures in G1, S, and G2, cells were blocked in M with nocodazole for 8 to 10 h, the nocodazole was removed, and aphidicolin was added 1 h later. The aphidicolin was removed after 4 h, and samples were harvested periodically for fluorescence-activated cell sorter (FACS) analysis and DNA isolation. For FACS analysis, cells were fixed in 90% ethanol, treated with RNase, stained with propidium iodide, and processed with a FACSCalibur apparatus. Data were plotted using CellQuest software.
Immunofluorescence, fluorescent in situ hybridization (FISH), and chromosome spreads.
Cells were fixed in 2% formaldehyde, spun onto Alcian blue-coated coverslips, and permeabilized with 0.5% Triton X-100 or methanol at −20°C as described previously (55). POT1, TRF1, and RAP1 were detected with rabbit polyclonal antibodies to the chicken telomere proteins (11, 48, 55). ER-Flag-POT1 was detected with a rabbit polyclonal antibody to the ER (Santa Cruz Biotechnology) or a mouse monoclonal (M2) antibody to the Flag tag (Sigma). Phosphorylated histone H3 (phospho-H3) and γH2AX antibodies were from Upstate Cell Signaling, γ-tubulin antibody was a gift from Kenji Fukasawa, and α-tubulin antibody was from the University of Iowa DSHB. Secondary antibodies were Cy2-conjugated anti-rabbit and Cy2- or Rhodamine Red-X (RRX)-conjugated anti-mouse. Controls for the γH2AX staining were irradiated with 1 to 3 Gy and fixed 30, 60, and 120 min later. Colocalization of γH2AX and telomere staining was performed essentially as described previously (18, 54). Following γH2AX staining, the slides were refixed in 4% paraformaldehyde for 20 min, dehydrated in 70%, 90%, and 100% ethanol, air dried, and rehydrated in hybridization buffer containing a fluorescein isothiocyanate-conjugated peptide nucleic acid (PNA) telomere probe (Applied Biosystems). The slides were denatured for 3 min at 80°C, incubated overnight at room temperature, and washed with 70% formamide-1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
For metaphase spreads, ER-POT1 cells were grown with or without tamoxifen for 24 or 48 h and then blocked with 0.1 μg/ml Colcemid for 1.5 h. Cells were fixed in methanol-acetic acid and dropped onto glass slides. Slides were aged overnight at 65°C, treated with 0.05% trypsin for 20 s, and stained with Giemsa stain for 1.25 min.
G-overhang assay.
G overhangs were detected by in-gel hybridization with a (TA2C3)4 G-strand probe and the amount of signal quantified with a phosphorimager and ImageQuant software as previously described (57). To compare the relative amounts of G overhang in different lanes, the gel was denatured and rehybridized with the same (TA2C3)4 probe. A block of bands corresponding to interstitial telomeric DNA was quantified and used to normalize for differences in loading. Digestion with exonuclease 1 (Exo1) and mung bean nuclease was performed prior to restriction digestion. DNA was digested with 10 U Exo1 per μg DNA for 24 h at 37°C and with 0.5 U mung bean nuclease per μg DNA for 15 to 60 min at 30°C.
RESULTS
Generation of a conditional POT1 cell line.
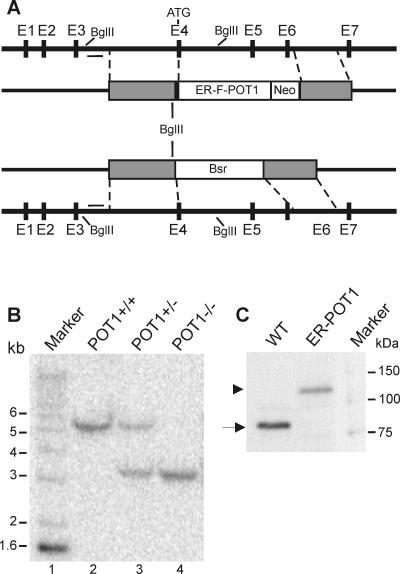
To investigate the in vivo function of vertebrate POT1, we attempted to generate a simple knockout cell line by disrupting both POT1 alleles. Despite high levels of gene targeting for the first allele, we were not able to disrupt the second allele, suggesting that the POT1 gene is essential. To circumvent this problem, we generated a conditional cell line that expresses an estrogen receptor-chicken POT1 fusion protein (ER-POT1). The fusion protein is active in the presence of 4-hydroxy tamoxifen (see below), thus making it possible to disrupt both wild-type POT1 alleles. To make the conditional cell line, a cDNA encoding the ER-POT1 fusion protein was integrated into one allele of the wild-type POT1 gene locus immediately adjacent to the initiating methionine in exon 4 (Fig. 1A). This allowed expression of the ER-POT1 cDNA from the native POT1 promoter. The second POT1 allele was disrupted by replacing exons 4 to 6 with a drug resistance cassette.
FIG. 1.
POT1 gene-targeting strategy. (A) Organization of the chicken POT1 gene locus (black line), the gene replacement, and the gene disruption construct. Black boxes in the gene locus represent exons (E1 to E7), the initiation codon in E4 is marked, and the bar represents the intron 3 probe. The white box represents the ER-POT1 and neomycin (Neo) expression cassettes in the gene replacement construct and the Bsr resistance gene in the disruption construct. Gray boxes represent segments of POT1 genomic DNA. Regions of homology with the wild-type gene locus are marked by dashed lines. (B) Southern blot of BglII-digested genomic DNA hybridized with probe to intron 3. Lane 1, 1-kb marker; lane 2, wild-type (WT) DT40 cells; lane 3, ER-POT1 replacement cell line; lane 4, conditional cell line with POT1 disruption and ER-POT1 replacement. (C) Western blot of total nuclear proteins from WT and ER-POT1 cells probed with POT1 antibody. Nuclei (5 × 106) were loaded in each lane. The arrow marks endogenous POT1 (∼87 kDa), and the arrowhead marks ER-POT1 (∼127 kDa).
The ER-POT1 gene replacement construct contained the ligand binding domain of the mouse estrogen receptor fused in frame to a Flag tag and the POT1 cDNA (Fig. 1A). A neomycin marker cassette was placed downstream, and the two genes were flanked by 5′ and 3′ segments of genomic sequence. Wild-type DT40 cells were transfected first with the gene replacement construct. Clones were selected on neomycin, screened for targeted integration by Southern hybridization (Fig. 1B), and tested for ER-POT1 expression (see below). One clone was then transfected with the gene disruption construct, and transformants were selected with Bsr and screened by Southern hybridization. Targeted integration of both the gene replacement and gene disruption constructs caused a change in the BglII digestion pattern of the POT1 locus such that a wild-type band of 5.5 kb was reduced in size to 3.2 kb. Western blot analysis using antibody to amino acids 1 to 253 of POT1 confirmed that the ER-POT1 cells expressed the ER-POT1 fusion protein but not the endogenous protein (Fig. 1C).
ER-POT1 can substitute for wild-type POT1.
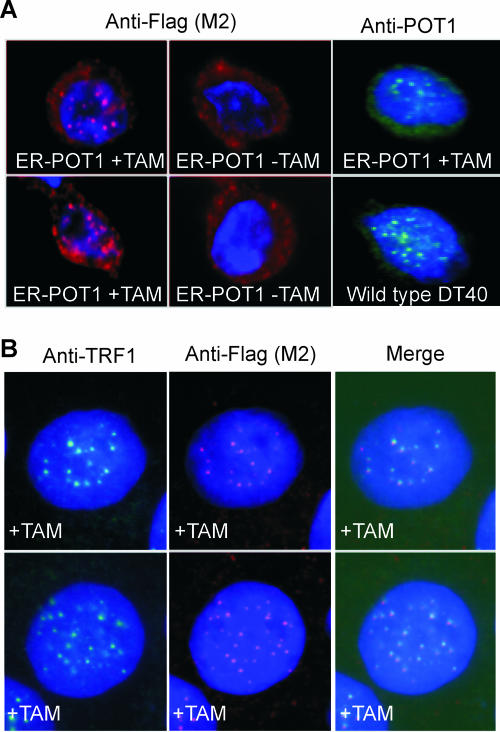
To determine whether the ER-POT1 fusion protein was expressed and showed the expected regulation by tamoxifen, we used indirect immunofluorescence to examine the protein distribution in the presence and absence of drug. In the presence of estrogen or the estrogen analog tamoxifen, the estrogen receptor and many estrogen receptor fusion proteins are free to diffuse into the nucleus, but in the absence of drug they are sequestered in the cytoplasm in a complex with heat shock proteins (15, 23). As shown in Fig. 2, this was the case for the ER-POT1 protein. ER-POT1 cells grown in tamoxifen and subsequently stained with antibody to Flag, POT1 (Fig. 2A), or ER (data not shown) gave rise to a punctuate nuclear pattern, but this nuclear staining was lost when tamoxifen was removed from the growth medium and all staining became cytoplasmic (Fig. 2A). The punctuate nuclear staining observed in the presence of drug was indistinguishable from the telomeric POT1 staining in wild-type cells, and the individual speckles colocalized with TRF1, confirming that ER-POT1 was present at telomeres (Fig. 2A and B). Thus, when bound by tamoxifen, ER-POT1 can both enter the nucleus and bind to telomeres, but in the absence of tamoxifen it redistributes to the cytoplasm.
FIG. 2.
Localization of the ER-POT1 fusion protein. (A) POT1 staining patterns in ER-POT1 or wild-type DT40 cells. Cells grown with or without tamoxifen (+TAM or −TAM, respectively) were fixed, incubated with antibody to POT1 or the Flag tag (M2) on ER-POT1, and counterstained with DAPI. (B) Colocalization of ER-POT1 and TRF1 in cells stained with antibody to TRF1 or Flag (M2).
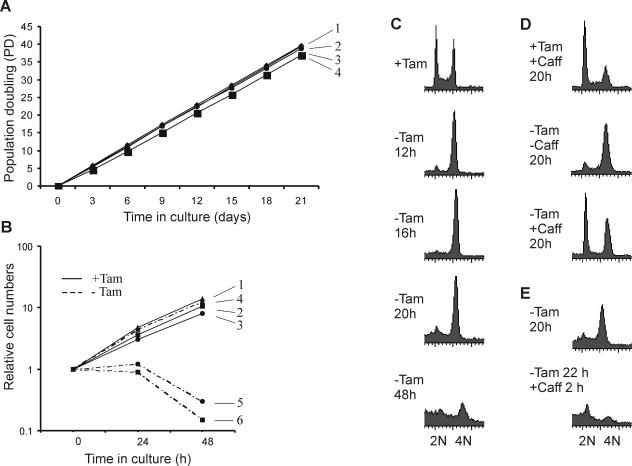
We next analyzed the growth rate of the ER-POT1 cells to determine whether the fusion protein was sufficient to allow normal growth. As shown in Fig. 3A, wild-type DT40 and ER-POT1 cells exhibited essentially the same growth rate when cultured in tamoxifen for a 3-week period. Moreover, staining with vital dyes (trypan blue or 7-amino-actinomycin) revealed a similar level of viability (>95% [data not shown]). Thus, the ER-POT1 fusion protein seems to substitute quite effectively for wild-type POT1.
FIG. 3.
Growth phenotype of ER-POT1 cells. (A) Growth in the presence of tamoxifen. 1, wild-type DT40; symbols 2 to 4, ER-POT1 clones 37, 16, and 25, respectively. (B) Growth after tamoxifen removal. Symbols 1 and 4, wild-type cells with or without tamoxifen (+Tam or −Tam, respectively); symbols 2 and 3, ER-POT1 clones 37 and 16, respectively, with drug; symbols 5 and 6, clones 37 and 16, respectively, without drug. (C to E) FACS analysis showing DNA content of ER-POT1 cells. (C) Cells were grown for 12, 16, 20, or 48 h without tamoxifen. (D) Caffeine (Caff) was added at the time of tamoxifen removal and the cells were harvested 20 h later. (E) Cells were grown for 20 h without tamoxifen and then caffeine was added for 2 h.
POT1 loss leads to a G2 checkpoint and cell death.
Although growth of ER-POT1 and wild-type cells was almost indistinguishable in the presence of tamoxifen, we observed a dramatic decrease in the growth rate of the ER-POT1 cells after drug removal (Fig. 3B). During the first 24 h, the number of ER-POT1 cells remained stationary whereas the number of wild-type cells continued to increase at a normal rate. Continued maintenance in the absence of drug led to a decline in ER-POT1 cell number over the next several days until by day 3 almost all of the cells were dead. Readdition of drug after 48 h failed to rescue the cells, but if the drug was added back after 24 h some cells survived (data not shown). If the cultures were maintained without drug for 3 to 4 weeks, survivors eventually appeared. Staining with ER and POT1 antibodies revealed that these cells had lost all or part of the ER segment of the ER-POT1 fusion protein and that POT1 was no longer sequestered in the cytoplasm in the absence of drug (data not shown). These findings indicate that POT1 is essential for cell survival and there is strong selection to maintain POT1 in an active form.
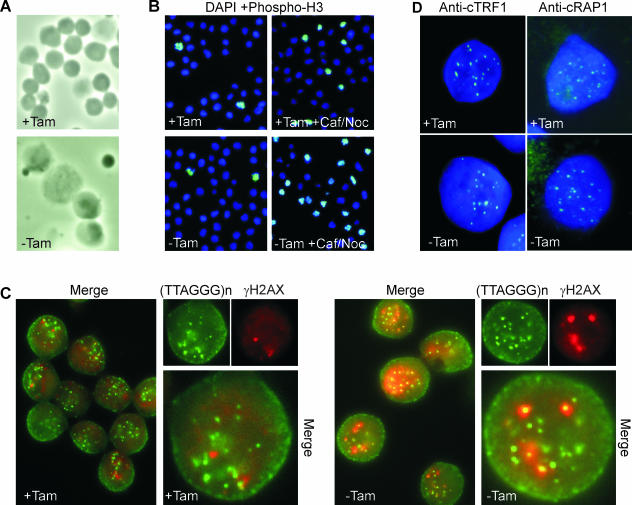
We next investigated whether the failure of the cells to proliferate after POT1 loss might be caused by a block in the cell cycle. Examination of the DNA content by propidium iodide staining and FACS analysis revealed that this was the case, as most cells arrested with a 4N DNA content within 12 h of drug removal (Fig. 3C). The time taken to reach maximum arrest (∼12 h) was similar to the time DT40 cells take to complete one cell cycle (10 to 12 h). The arrest was maintained for about 24 h but seemed to then break down, as the 4N peak declined at later time points. Phase-contrast microscopy of the arrested cultures revealed that many of the cells became abnormally large (Fig. 4A), raising the possibility that they might have arrested in M rather than G2. However, staining with antibody to phosphorylated histone H3 (a marker for mitotic chromosomes) (Fig. 4B) showed that this was not the case, as the mitotic index was low (<8% at 24 h). Moreover, if Colcemid was added to the cells when the arrest was beginning to break down (24 h after tamoxifen removal), we observed an immediate increase in the mitotic index (see below). This suggested that removal of POT1 from the telomere causes an initial G2 arrest but subsequent breakdown of the arrest allows some cells to progress into mitosis.
FIG. 4.
Cell cycle status and telomere integrity in ER-POT1 cells. (A and B) Size and mitotic index of ER-POT1 cells. (A) Phase-contrast images of ER-POT1 cells grown with or without tamoxifen (+Tam or −Tam, respectively) for 24 h. (B) Staining of metaphase cells with phospho-H3 antibody and DAPI. Cells were grown with or without tamoxifen for 20 h (left panels); caffeine (Caf) and nocodazole (Noc) were then added and cells harvested 1 h later (right panels). (C) Telomeric dysfunction-induced foci induced by POT1 removal. Cells were grown with or without tamoxifen for 16 h, fixed, and stained with γH2AX primary and RRX-conjugated secondary antibody. Cells were refixed, and telomeric DNA was visualized by FISH using fluorescein isothiocyanate-conjugated PNA probe. Note that only a fraction of the FISH signals correspond to actual telomeres, because chicken cells contain large blocks of interstitial telomeric DNA (53). (D) Localization of TRF1 and RAP1 after POT1 removal. Cells were grown with or without tamoxifen for 48 h, fixed, and stained with DAPI and antibody to TRF1 or RAP1.
As ATM and/or ATR is present at yeast and mammalian telomeres and acts as a transducer of the response to defective telomeres (18, 39, 46, 54), we next examined whether the cell cycle arrest could be prevented by addition of caffeine. Although caffeine has pleiotropic effects on cells, it is an effective phosphatidylinositol 3-kinase inhibitor and is frequently used to abrogate DNA damage checkpoints (26, 27, 54). FACS analysis revealed that if caffeine was added to the ER-POT1 cells at the time of tamoxifen removal, the arrest was largely prevented (Fig. 3D). Moreover, caffeine could release the cells from a preexisting arrest, as addition of the drug to arrested cultures caused almost complete loss of the 4N peak within 2 h (Fig. 3E), while addition of both caffeine and nocodazole caused the cells to accumulate in M. The increase in mitotic index after caffeine and nocodazole addition was obvious within 1 h (Fig. 4B), and by 4 h ∼60% of the cells were in M (data not shown). The ability of caffeine to release the cell cycle arrest suggests that loss of POT1 from the telomere elicits a checkpoint that is mediated by phosphatidylinositol 3-kinases. Moreover, because a large fraction of the cells enter mitosis within a few hours of checkpoint release, we conclude that loss of POT1 causes the majority of cells to arrest in G2. At present, we cannot tell whether the remaining cells also arrest in G2 but are unable to enter M because they are dying (25 to 30% of the cells are dead after 24 h without tamoxifen) or whether they enter G1 after mitotic catastrophe (failure of chromosome segregation [1]). We observed a very small but reproducible population of the cells with an 8N DNA content (data not shown), suggesting that some cells manage to complete a round of replication without undergoing chromosome segregation.
POT1 loss causes a telomeric DNA damage response but does not disrupt the core telomere protein complex.
The strong cell cycle arrest observed after tamoxifen removal suggested that loss of POT1 from the telomere might result in activation of a telomeric DNA damage response. As accumulation of γH2AX is commonly observed at dysfunctional telomeres (5, 8, 18, 45), we tested for a DNA damage response by looking for colocalization of histone γH2AX with telomeric DNA. γH2AX staining of wild-type DT40 and ER-POT1 cells grown with tamoxifen revealed a low basal level of DNA damage foci that increased dramatically after gamma irradiation (IR) (Fig. 4C and data not shown). A similar increase in focus formation was observed when tamoxifen was removed from the ER-POT1 cells. Multiple small foci became apparent within 6 h of drug removal; these then increased in size and intensity so that by 12 h all cells had multiple foci that were comparable in size to the foci present 1 h after IR. However, unlike the foci generated in response to IR, they did not then gradually disappear but instead persisted and increased in size. When the γH2AX-stained cells were subjected to telomere FISH using a PNA probe, it became apparent that in cells lacking active POT1 most of the γH2AX foci colocalized with telomeres (Fig. 4C). This was not the case for the sporadic foci observed in cells with active POT1. We therefore conclude that loss of POT1 from the telomeres causes a strong telomeric DNA damage response and that this damage response is most likely the cause of the cell cycle arrest.
While POT1 binds specifically to G-overhang DNA, it also associates with the duplex region of the telomeric tract via interactions with the core TRF/RAP1/TIN2/TPP1 complex (21, 32, 33, 62). Since sequestration of ER-POT1 in the cytoplasm appears to cause loss of POT1 from all regions of the telomere, it was possible that removal of POT1 from the core telomere protein complex would affect the binding of other core proteins. However, when we examined TRF1 and RAP1 localization, we saw the same punctuate telomeric staining pattern in ER-POT1 cells grown with and without tamoxifen (Fig. 4D). Since RAP1 binds to telomeres via TRF2, this observation indicates that loss of POT1 from the telomere does not also cause loss of TRF1 or TRF2. Thus, the phenotype of the ER-POT1 cells cannot be attributed to uncapping of the telomere by removal of the core duplex telomere binding proteins.
G-strand overhang signal is increased after POT1 removal.
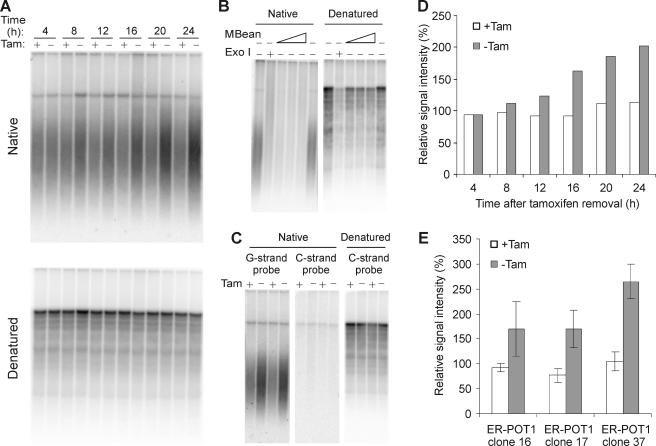
Given that POT1 proteins bind and protect telomeric G-strand DNA and loss of S. pombe POT1 leads to extensive degradation of the telomeric tract (56), it seemed likely that removal of chicken POT1 would result in deprotection and degradation of the G-strand overhang. To test for changes in G-overhang integrity, we isolated DNA from ER-POT1 cells harvested at sequential time points after tamoxifen removal and used nondenaturing in-gel hybridization to monitor the relative amounts of overhang at the telomere. Samples of the same cultures were subjected to FACS analysis to ensure that growth without tamoxifen resulted in the expected cell cycle arrest (data not shown). The DNA was restriction digested and separated in agarose gels, and the gels were then hybridized with a (TA2C3)4 G-strand probe under nondenaturing conditions. Surprisingly, a robust hybridization signal was observed regardless of whether tamoxifen had been added to the culture media (Fig. 5A). Moreover, the strength of the signal actually appeared to become greater with increased time of culture without drug. To ensure that the probe had hybridized only to the G-strand overhang, samples were treated with Exo1 or mung bean nuclease prior to restriction digestion. As anticipated, this abolished the signal (Fig. 5B). Gels were also hybridized with a C-strand probe, and again no telomeric signal was observed (Fig. 5C).
FIG. 5.
G-overhang maintenance in ER-POT1 cells. (A to C) In-gel hybridization of (TA2C3)4 G-strand probe to MspI-, HinfI-, and HaeIII-digested DNA from ER-POT1 cells. (A) ER-POT1 clone 37 grown with (+) or without (−) tamoxifen (Tam) for 4, 8, 12, 16, 20, or 24 h. (B) Samples were treated with Exo1 or mung bean nuclease (MBean) prior to restriction digestion. (C) Gels were hybridized with telomeric G-strand or C-strand probe. (D) Histogram showing relative G-overhang signals for the experiment shown in panel A. The signal in each lane was quantified with a phosphorimager and normalized for loading. The average normalized signal for the +Tam lanes was set to 100%. (E) Histogram showing the G-overhang signals 24 h after tamoxifen removal for ER-POT1 clones 16, 17, and 37. Values are means from three or more independent experiments, and error bars show standard deviations.
To quantify the relative amount of G-overhang signal for each sample, the DNA in each gel was then denatured and reprobed with the G-strand probe, and the signal from a block of bands corresponding to interstitial telomeric DNA was used to normalize for loading. This analysis confirmed that the relative amounts of G-overhang signal increased with time after tamoxifen removal (Fig. 5D). A similar increase in signal was observed when we examined the G overhangs from two other ER-POT1 cell lines. In each case, the relative amount of signal had increased two- to threefold 24 h after drug removal (Fig. 5E). Thus, despite causing a strong DNA damage checkpoint, loss of POT1 from the telomere does not result in widespread loss of the G-strand overhang.
It is not known whether G-overhang length is subject to cell cycle regulation in chicken cells; however, in budding yeast overhang length increases during G2/M (28, 58). If a similar change occurs in chicken cells, the increase in overhang observed with the ER-POT1 cells might simply reflect a natural change caused by synchronization of the culture in G2. To examine whether DT40 cells exhibit cell cycle-related changes in overhang length, we used nocodazole and aphidicolin blocks to obtain cells synchronized in G1, S, and G2. Subsequent G-overhang analysis revealed a small increase in overhang signal in some experiments; however, this was not reproducible and was much smaller than the increase observed after POT1 removal (data not shown). We therefore conclude that in the absence of POT1 the telomere becomes more susceptible to C-strand resection and/or G-strand extension.
Removal of POT1 from the telomere did not cause any obvious change in telomere length (Fig. 5A and data not shown). Three of the ER-POT1 clones had telomeres of length similar to those of the parental POTwt/ER-POT1 cells, while one clone had longer telomeres (data not shown). While the longer telomeres in the latter clone could reflect POT1 haploinsufficiency, this is not necessarily the case, as wild-type DT40 cells can show significant variation in telomere length upon prolonged culture.
Infrequent telomere-telomere association and aneuploidy after POT1 removal.
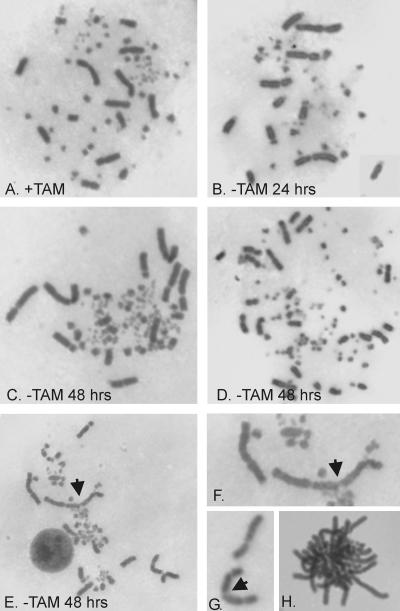
One caveat of the in-gel hybridization technique is that it measures changes in total overhang signal rather than changes in overhang length at individual telomeres. Thus, it was possible that the increased overhang signal reflected a large gain in overhang length at some telomeres that was balanced by overhang loss at other telomeres. As overhang loss leads to chromosome fusions, we prepared metaphase spreads from Colcemid-treated ER-POT1 cells and examined their karyotypes. The control ER-POT1 cells cultured with tamoxifen gave well-spread mitotic figures in which the five largest pairs of macrochromosomes, one Z chromosome, and many microchromosomes were clearly visible (Fig. 6A). Cells cultured without tamoxifen for 24 h also gave mostly normal spreads with few if any obvious telomere associations (Fig. 6B). However, when cells were grown without drug for 48 h we did observe occasional telomere associations between macrochromosomes where two or more chromosomes were clearly aligned to form the linear structures characteristic of a telomere fusion (Fig. 6E to G). We also saw occasional ring chromosomes (data not shown). However, these telomere associations were quite rare and associations between pairs of macrochromosomes were obvious in <1% of metaphases. We could not determine the exact frequency of association because it was impossible to detect associations involving the small but abundant microchromosomes. Nonetheless, the frequency was clearly dramatically lower than the fusion frequency observed after TRF2 removal when long chains of chromosomes are observed (5). Thus, despite its specificity for G-strand DNA, POT1 does not appear to be required to protect against overhang removal and telomere fusions.
FIG. 6.
Giemsa-stained metaphase spreads of ER-POT1 cells. Cells were grown with tamoxifen (+TAM) (A) or without tamoxifen (−TAM) (B to H) for 24 or 48 h. (A to C) Normal karyotypes. (D) Karyotype of an aneuploid cell with an elevated number of macrochromosomes. (E) Cell with telomere-telomere association (marked with arrowhead). (F) Enlargement of telomere association shown in panel E. (G) Association between long arms of two acrocentric chromosomes. (H) Aggregated/entangled chromosomes.
In addition to occasional telomere associations, cells grown without drug for 48 h showed two other interesting phenotypes. First, many of the cells were obviously aneuploid, with an elevated number of macrochromosomes (Fig. 6D). Second, a number of the mitotic figures appeared to have some or all of the chromosomes tightly aggregated so that they could not spread apart (Fig. 6H). The cells with aggregated chromosomes were frequently found beside cells with well-spread chromosomes, so they did not appear to be the result of poor spreading technique. For reasons discussed later, we think they reflect cells which had entered mitosis prior to Colcemid addition but which then experienced problems with chromosome segregation.
The slight increase in telomere associations after 48 h without drug correlated with the gradual breakdown of the G2 arrest and the opportunity for some cells to traverse the cell cycle. This observation, together with the strong G1 bias for NHEJ-mediated DNA repair and telomere fusions (14, 24), led us to ask whether cells lacking functional POT1 might become more susceptible to G-overhang loss and chromosome fusions when they entered G1 or S phase. To test this possibility, we used caffeine to prevent the G2 arrest and allow the ER-POT1 cells to cycle (Fig. 3D) before isolating cells to make metaphase spreads. However, the caffeine treatment did not increase the frequency of telomere associations, and spreads made from cells grown with and without tamoxifen for 24 h looked normal (data not shown). Thus, POT1 does not appear to be required to prevent chromosome fusions in any stage of the cell cycle.
Removal of POT1 causes defects in chromosome segregation and centrosome duplication.
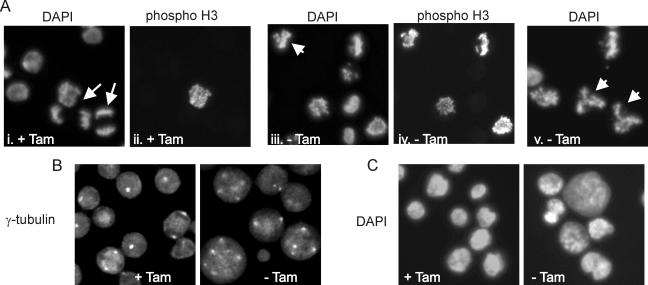
To better understand why loss of POT1 ultimately caused all the cells to die even though many were able to escape the cell cycle arrest, we examined what happened when previously arrested cells entered mitosis. Cells were grown with or without tamoxifen for 20 h, and then caffeine and nocodazole were added to release the arrested cells from G2 and accumulate them in M. The nocodazole was removed 4 h later, and samples were fixed at sequential time points to monitor progression through mitosis. Staining with DAPI (4′,6′-diamidino-2-phenylindole) and antibody to phospho-H3 revealed that many of the cells grown with tamoxifen started anaphase within 60 min of nocodazole removal (Fig. 7A) and mitosis was essentially complete by 90 min (data not shown). However, cells grown without tamoxifen progressed through mitosis more slowly, as there were fewer anaphase figures at 60 min and more cells retained phospho-H3 staining at both 60 and 90 min. Moreover, a large number of cells had obviously abnormal mitotic figures (Fig. 7A). Some cells had chromosome clusters with three or four arms, suggesting they might have tri- or tetrapolar spindles, while many others had entangled chromosomes that appeared unable to segregate. Thus, loss of POT1 from the telomere causes severe defects in chromosome segregation.
FIG. 7.
Defects in chromosome segregation and centrosome amplification. Caffeine and nocodazole were added to ER-POT1 cells grown with or without tamoxifen (+Tam or −Tam, respectively) for 20 h. The nocodazole was removed after 4 h, and cells were fixed 60 min (A and B) or 120 min (C) later. (A) DAPI and phospho-H3 staining showing delayed anaphase and abnormal mitotic figures. Panels i and ii and iii and iv are matched pairs. Arrows indicate anaphase cells, and arrowheads indicate abnormal mitotic figures. (B) γ-Tubulin staining showing centrosome amplification. (C) DAPI staining showing abnormal nuclei following missegregation or mitotic catastrophe.
As a prolonged G2 phase can induce centrosome amplification (12), we suspected that the defects in chromosome segregation might be caused by supernumerary centrosomes. We therefore visualized centrosomes in the caffeine- and nocodazole-treated cells with antibody to γ-tubulin (Fig. 7B). While metaphase and anaphase cells with functional POT1 had the expected two centrosomes, the majority of cells lacking POT1 exhibited striking centrosome amplification. The cells with three- or four-arm chromosome clusters had three or four centrosomes, while the large cells with entangled chromosomes had many more. Staining of G2-arrested cells confirmed that amplification occurred during the arrest, as many cells had supernumerary centrosomes after a 20-h culture without tamoxifen (data not shown). To confirm that the amplified centrosomes gave rise to abnormal mitotic spindles, we stained the caffeine- and nocodazole-treated cells with antibody to α-tubulin. As anticipated, cells grown with tamoxifen formed normal spindles, but cells grown without tamoxifen formed highly aberrant multipolar structures (data not shown). These results indicate that the defects in chromosome segregation observed after POT1 loss are likely to be an indirect effect caused by centrosome amplification during the G2 arrest rather than a direct effect of POT1 removal on telomere capping.
Defects in chromosome segregation commonly lead to aneuploidy and mitotic catastrophe. Examination of cells that had exited mitosis indicated that a lack of active POT1 caused both problems to occur. Phospho-H3 staining indicated that most cells had exited mitosis 120 min after release of the nocodazole block (data not shown). However, DAPI staining revealed that the nuclear morphology was very uneven. Some nuclei were unusually small, some were extremely large, others were lobate, and some cells had several nuclei (Fig. 7C). This morphology indicated that some cells with multipolar spindles had managed to complete an abnormal chromosome segregation whereas others had failed and undergone mitotic catastrophe (1, 22). FACS analysis also indicated uneven chromosome segregation, as cultures that eventually escaped the G2 block either naturally (Fig. 3C) or through caffeine treatment (Fig. 3E) showed an extremely broad G1 peak. These results explain both the increase in aneuploid cells in the metaphase spreads (Fig. 6) and the eventual death of ER-POT1 cells that manage to escape the initial G2 arrest.
DISCUSSION
It has proved difficult to analyze the role of POT1 in higher eukaryotes because attempts to remove the protein from the telomere by use of RNAi or dominant negative approaches have resulted in only partial POT1 depletion (19, 34, 52, 60). Here we have used a different approach to remove POT1, and we now show that POT1 is essential both for telomere capping and cellular survival. However, the role of POT1 in telomere capping is quite different from that of TRF2, which protects against G-overhang loss and telomere fusions (5, 51). Although POT1 is a telomeric G-strand DNA binding protein, loss of POT1 from the telomere does not result in G-overhang degradation or frequent telomere fusions. Instead, POT1 loss causes an increase in G-overhang length and activation of a telomeric DNA damage response that leads to a cell cycle arrest. Thus, POT1 is required to regulate G-overhang length and appears to play a key role in preventing checkpoint activation by G-strand DNA. These functions could not have been predicted from the phenotype of the S. pombe Pot1 gene disruption, as this leads to complete loss of the telomeric tract and chromosome circularization (2). However, our results indicate that vertebrate POT1 resembles Cdc13 from S. cerevisiae, because inactivation of Cdc13 also results in abnormally long overhangs and a cell cycle arrest rather than telomere fusions (16, 38).
G-strand protection and checkpoint activation.
The unexpected finding that POT1 removal does not lead to detectable G-overhang loss strongly suggests that other single-stranded DNA (ssDNA) binding proteins must bind and protect the overhang from degradation by nucleases such as ERCC1, the nuclease that removes the overhang after TRF2 depletion (63). Candidate proteins that might protect the overhang include RPA and several abundant hnRNPs that have a preference for G-rich sequence (13, 25, 40). Indeed, recent studies defining the terminal DNA structure of human telomeres have raised the possibility that POT1 might act in conjunction with these proteins in wild-type cells. Analysis of the DNA at the termini of the G and C strands revealed that the sequence at the end of the G strand is quite variable whereas that of the C strand is precisely defined (41). Since the majority of G strands terminate in a sequence that is not the preferred binding site for POT1 (35), it is possible that other proteins protect the 3′ end of the overhang when it is not buried in a T-loop structure.
Given that a specialized telomeric G-strand binding protein is not required for overhang protection, why should this class of protein be ubiquitous in widely diverse organisms? The answer may be that other ssDNA binding proteins are not capable of preventing single-stranded telomeric DNA from activating a DNA damage response. For example, RPA is known to recruit ATR and cause checkpoint activation in response to ssDNA at double-strand breaks or stalled replication forks (64). Since T loops are almost certainly disrupted during telomere DNA replication, if POT1 is unavailable, the G overhangs might become coated by RPA at this time. The resulting long stretches of RPA-DNA complex would then recruit ATR and activate a checkpoint in late S or G2. Whether or not T loops can form in the absence of POT1 is unclear; however, if they do form, RPA might then bind the displaced segment of G-strand DNA and continue the DNA damage signal.
Chromatin immunoprecipitation studies of synchronized cells have revealed that ATM and/or ATR is recruited to telomeres in late S or G2 phase, where it initiates a DNA damage response (46, 47, 54). This response is transient and appears to be quenched when telomere proteins are loaded back onto the telomere. Thus, the G2 arrest seen after POT1 removal may be an extension of this natural damage response. This implies that a key function of POT1 proteins is to quench the transient DNA damage response that is activated as a result of telomere replication. This could be achieved either by preventing checkpoint-inducing proteins from binding the telomeric G-strand DNA or by displacing them during reassembly of the fully capped telomere.
C-strand protection and overhang length regulation.
Although POT1 is not needed for overhang protection, it does play a role in regulating overhang length since in the absence of POT1 the total amount of G-overhang DNA increases by two- to threefold. This increase can be detected within 8 h of tamoxifen removal, and the G-overhang DNA continues to grow even after all of the cells have arrested. At present, it is unclear whether the increase in G-overhang DNA is a result of G-strand extension by telomerase or C-strand resection by a nuclease. However, it seems likely that C-strand resection is responsible for at least part of the increase because depletion of human POT1 by RNAi leads to randomization of the normal 5′-terminal CCCAATC sequence (19). This result implies that POT1 is responsible for defining the 5′ end of the telomere by somehow modulating the activity of a processing nuclease. While we have not yet examined the C-strand terminus after POT1 removal, we would anticipate a similar randomization of the 5′ nucleotide, as chicken and human POT1 are quite conserved (3, 35, 55). Given the importance of POT1 for 5′-end definition, it seems likely that POT1 binding may determine both overhang length and the 5′ terminus by directly blocking C-strand processing. Under normal conditions, the 5′ terminus may be generated by POT1 binding and blocking the processing nuclease once the POT1 ′(GGT)TAGGGTTAG 3′ binding site becomes exposed. However in the absence of POT1, processing would continue and hence generate random 5′ termini and longer overhangs.
It is interesting that depletion of human POT1 by shRNA leads to a stable decrease in G-strand DNA (19, 60), whereas we observed an immediate increase after POT1 removal. Perhaps the decrease in G-strand DNA in the human cells reflects an adaptation to very low levels of POT1 during prolonged growth, whereas we observed an acute response to POT1 removal. An adaptive response leading to changes in overhang length was observed when shRNA was used to deplete MRN (6). In this case, the overhangs initially became much shorter but then returned to the normal size. While the increase in G-strand DNA caused by POT1 loss is striking, it is actually quite modest (<1 kb) compared to the many kilobases of C-strand DNA that are lost after Cdc13 inactivation (16). It is also very modest compared to the complete loss of telomeric DNA observed after disruption of S. pombe Pot1 (2). It is possible that the lower level of C-strand degradation in vertebrate cells reflects the robust protection offered by the TRF/TIN2/RAP1/TPP1 core complex. In budding and fission yeasts, the corresponding Rap1 or Taz1 complexes may be more easily displaced.
In conclusion, our work has uncovered an unexpected role for vertebrate POT1 in suppressing a DNA damage response and subsequent G2 cell cycle arrest. G2 appears to be when telomeres undergo a major restructuring following DNA replication, and a natural but transient telomeric DNA damage response has been detected to occur at this time in both yeast and human cells (46, 47, 54). At present, it is unclear how this transient response is terminated and data concerning the contribution of ATM versus ATR to the response differ for yeast and human cells. One interesting possibility is that ATM and ATR are both activated as the telomere becomes restructured. The ATM response may then be inhibited by TRF2 reloading onto the duplex region of the telomeric tract (26), while the ATR response may be blocked by POT1 binding to the G-strand overhang.
Acknowledgments
We thank William Brown for the mouse ER cDNA, Kenji Fukasawa for the γ-tubulin antibody, and Dorothy Shippen and Yolanda Sanchez for helpful comments.
This work was supported by NIH grants AG17212 and GM041803 to C.M.P.
REFERENCES
- 1.Andreassen, P. R., F. B. Lacroix, O. D. Lohez, and R. L. Margolis. 2001. Neither p21WAF1 nor 14-3-3sigma prevents G2 progression to mitotic catastrophe in human colon carcinoma cells after DNA damage, but p21WAF1 induces stable G1 arrest in resulting tetraploid cells. Cancer Res. 61:7660-7668. [PubMed] [Google Scholar]
- 2.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, P., E. Podell, and T. R. Cech. 2002. Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol. Cell. Biol. 22:8079-8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broccoli, D., A. Smogorzewska, L. Chong, and T. de Lange. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17:231-235. [DOI] [PubMed] [Google Scholar]
- 5.Celli, G. B., and T. de Lange. 2005. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7:712-718. [DOI] [PubMed] [Google Scholar]
- 6.Chai, W., A. J. Sfeir, H. Hoshiyama, J. W. Shay, and W. E. Wright. 2006. The involvement of the Mre11/Rad50/Nbs1 complex in the generation of G-overhangs at human telomeres. EMBO Rep. 7:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colgin, L. M., C. Wilkinson, A. Englezou, A. Kilian, M. O. Robinson, and R. R. Reddel. 2000. The hTERTalpha splice variant is a dominant negative inhibitor of telomerase activity. Neoplasia 2:426-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.d'Adda di Fagagna, F., P. M. Reaper, L. Clay-Farrace, H. Fiegler, P. Carr, T. Von Zglinicki, G. Saretzki, N. P. Carter, and S. P. Jackson. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426:194-198. [DOI] [PubMed] [Google Scholar]
- 9.d'Adda di Fagagna, F., S. H. Teo, and S. P. Jackson. 2004. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 18:1781-1799. [DOI] [PubMed] [Google Scholar]
- 10.de Lange, T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19:2100-2110. [DOI] [PubMed] [Google Scholar]
- 11.De Rycker, M., R. N. Venkatesan, C. Wei, and C. M. Price. 2003. Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization. Biochem. J. 372:87-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dodson, H., E. Bourke, L. J. Jeffers, P. Vagnarelli, E. Sonoda, S. Takeda, W. C. Earnshaw, A. Merdes, and C. Morrison. 2004. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 23:3864-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enokizono, Y., Y. Konishi, K. Nagata, K. Ouhashi, S. Uesugi, F. Ishikawa, and M. Katahira. 2005. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J. Biol. Chem. 280:18862-18870. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira, M. G., and J. P. Cooper. 2004. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 18:2249-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukagawa, T., and W. R. Brown. 1997. Efficient conditional mutation of the vertebrate CENP-C gene. Hum. Mol. Genet. 6:2301-2308. [DOI] [PubMed] [Google Scholar]
- 16.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. (Erratum, 16:457, 1996.) [DOI] [PMC free article] [PubMed]
- 17.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 18.Herbig, U., W. A. Jobling, B. P. Chen, D. J. Chen, and J. M. Sedivy. 2004. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol. Cell 14:501-513. [DOI] [PubMed] [Google Scholar]
- 19.Hockemeyer, D., A. J. Sfeir, J. W. Shay, W. E. Wright, and T. de Lange. 2005. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 24:2667-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horvath, M. P., V. L. Schweiker, J. M. Bevilacqua, J. A. Ruggles, and S. C. Schultz. 1998. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell 95:963-974. [DOI] [PubMed] [Google Scholar]
- 21.Houghtaling, B. R., L. Cuttonaro, W. Chang, and S. Smith. 2004. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 14:1621-1631. [DOI] [PubMed] [Google Scholar]
- 22.Huang, X., T. Tran, L. Zhang, R. Hatcher, and P. Zhang. 2005. DNA damage-induced mitotic catastrophe is mediated by the Chk1-dependent mitotic exit DNA damage checkpoint. Proc. Natl. Acad. Sci. USA 102:1065-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Im, H., J. A. Grass, K. D. Johnson, S. I. Kim, M. E. Boyer, A. N. Imbalzano, J. J. Bieker, and E. H. Bresnick. 2005. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc. Natl. Acad. Sci. USA 102:17065-17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ira, G., A. Pellicioli, A. Balijja, X. Wang, S. Fiorani, W. Carotenuto, G. Liberi, D. Bressan, L. Wan, N. M. Hollingsworth, J. E. Haber, and M. Foiani. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431:1011-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa, F., M. J. Matunis, G. Dreyfuss, and T. R. Cech. 1993. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell. Biol. 13:4301-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karlseder, J., K. Hoke, O. K. Mirzoeva, C. Bakkenist, M. B. Kastan, J. H. Petrini, and T. de Lange. 2004. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2:E240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaufmann, W. K., T. P. Heffernan, L. M. Beaulieu, S. Doherty, A. R. Frank, Y. Zhou, M. F. Bryant, T. Zhou, D. D. Luche, N. Nikolaishvili-Feinberg, D. A. Simpson, and M. Cordeiro-Stone. 2003. Caffeine and human DNA metabolism: the magic and the mystery. Mutat. Res. 532:85-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larrivee, M., C. LeBel, and R. J. Wellinger. 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 18:1391-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei, M., E. R. Podell, P. Baumann, and T. R. Cech. 2003. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature 426:198-203. [DOI] [PubMed] [Google Scholar]
- 30.Lei, M., E. R. Podell, and T. R. Cech. 2004. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 11:1223-1229. [DOI] [PubMed] [Google Scholar]
- 31.Li, B., and T. De Lange. 2003. Rap1 affects the length and heterogeneity of human telomeres. Mol. Biol. Cell 14:5060-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, D., M. S. O'Connor, J. Qin, and Z. Songyang. 2004. Telosome, a mammalian telomere associated complex formed by multiple telomeric proteins. J. Biol. Chem. 279:51338-51342. [DOI] [PubMed] [Google Scholar]
- 33.Liu, D., A. Safari, M. S. O'Connor, D. W. Chan, A. Laegeler, J. Qin, and Z. Songyang. 2004. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6:673-680. [DOI] [PubMed] [Google Scholar]
- 34.Loayza, D., and T. De Lange. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 423:1013-1018. [DOI] [PubMed] [Google Scholar]
- 35.Loayza, D., H. Parsons, J. Donigian, K. Hoke, and T. de Lange. 2004. DNA binding features of human POT1: a nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 279:13241-13248. [DOI] [PubMed] [Google Scholar]
- 36.Mitton-Fry, R. M., E. M. Anderson, T. R. Hughes, V. Lundblad, and D. S. Wuttke. 2002. Conserved structure for single-stranded telomeric DNA recognition. Science 296:145-147. [DOI] [PubMed] [Google Scholar]
- 37.Nikitina, T., and C. L. Woodcock. 2004. Closed chromatin loops at the ends of chromosomes. J. Cell Biol. 166:161-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 39.Qi, L., M. A. Strong, B. O. Karim, M. Armanios, D. L. Huso, and C. W. Greider. 2003. Short telomeres and ataxia-telangiectasia mutated deficiency cooperatively increase telomere dysfunction and suppress tumorigenesis. Cancer Res. 63:8188-8196. [PubMed] [Google Scholar]
- 40.Schramke, V., P. Luciano, V. Brevet, S. Guillot, Y. Corda, M. P. Longhese, E. Gilson, and V. Geli. 2004. RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat. Genet. 36:46-54. [DOI] [PubMed] [Google Scholar]
- 41.Sfeir, A. J., W. Chai, J. W. Shay, and W. E. Wright. 2005. Telomere-end processing the terminal nucleotides of human chromosomes. Mol. Cell 18:131-138. [DOI] [PubMed] [Google Scholar]
- 42.Shakirov, E. V., Y. V. Surovtseva, N. Osbun, and D. E. Shippen. 2005. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol. Cell. Biol. 25:7725-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177-208. [DOI] [PubMed] [Google Scholar]
- 44.Sui, G., E. B. Affar, Y. Shi, C. Brignone, N. R. Wall, P. Yin, M. Donohoe, M. P. Luke, D. Calvo, and S. R. Grossman. 2004. Yin Yang 1 is a negative regulator of p53. Cell 117:859-872. [DOI] [PubMed] [Google Scholar]
- 45.Takai, H., A. Smogorzewska, and T. de Lange. 2003. DNA damage foci at dysfunctional telomeres. Curr. Biol. 13:1549-1556. [DOI] [PubMed] [Google Scholar]
- 46.Takata, H., Y. Kanoh, N. Gunge, K. Shirahige, and A. Matsuura. 2004. Reciprocal association of the budding yeast ATM-related proteins Tel1 and Mec1 with telomeres in vivo. Mol. Cell 14:515-522. [DOI] [PubMed] [Google Scholar]
- 47.Takata, H., Y. Tanaka, and A. Matsuura. 2005. Late S phase-specific recruitment of Mre11 complex triggers hierarchical assembly of telomere replication proteins in Saccharomyces cerevisiae. Mol. Cell 17:573-583. [DOI] [PubMed] [Google Scholar]
- 48.Tan, M., C. Wei, and C. M. Price. 2003. The telomeric protein Rap1 is conserved in vertebrates and is expressed from a bidirectional promoter positioned between the Rap1 and KARS genes. Gene 323:1-10. [DOI] [PubMed] [Google Scholar]
- 49.Trujillo, K. M., J. T. Bunch, and P. Baumann. 2005. Extended DNA binding site in Pot1 broadens sequence specificity to allow recognition of heterogeneous fission yeast telomeres. J. Biol. Chem. 280:9119-9128. [DOI] [PubMed] [Google Scholar]
- 50.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 51.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 52.Veldman, T., K. T. Etheridge, and C. M. Counter. 2004. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr. Biol. 14:2264-2270. [DOI] [PubMed] [Google Scholar]
- 53.Venkatesan, R. N., and C. Price. 1998. Telomerase expression in chickens: constitutive activity in somatic tissues and down-regulation in culture. Proc. Natl. Acad. Sci. USA 95:14763-14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verdun, R. E., L. Crabbe, C. Haggblom, and J. Karlseder. 2005. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol. Cell 20:551-561. [DOI] [PubMed] [Google Scholar]
- 55.Wei, C., and C. M. Price. 2004. Cell cycle localization, dimerization, and binding domain architecture of the telomere protein cPot1. Mol. Cell. Biol. 24:2091-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei, C., and M. Price. 2003. Protecting the terminus: t-loops and telomere end-binding proteins. Cell. Mol. Life Sci. 60:2283-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei, C., R. Skopp, M. Takata, S. Takeda, and C. M. Price. 2002. Effects of double-strand break repair proteins on vertebrate telomere structure. Nucleic Acids Res. 30:2862-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wellinger, R. J., A. J. Wolf, and V. A. Zakian. 1993. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72:51-60. [DOI] [PubMed] [Google Scholar]
- 59.Winding, P., and M. W. Berchtold. 2001. The chicken B cell line DT40: a novel tool for gene disruption experiments. J. Immunol. Methods 249:1-16. [DOI] [PubMed] [Google Scholar]
- 60.Yang, Q., Y. L. Zheng, and C. C. Harris. 2005. POT1 and TRF2 cooperate to maintain telomeric integrity. Mol. Cell. Biol. 25:1070-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye, J. Z., J. R. Donigian, M. van Overbeek, D. Loayza, Y. Luo, A. N. Krutchinsky, B. T. Chait, and T. de Lange. 2004. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 279:47264-47271. [DOI] [PubMed] [Google Scholar]
- 62.Ye, J. Z., D. Hockemeyer, A. N. Krutchinsky, D. Loayza, S. M. Hooper, B. T. Chait, and T. de Lange. 2004. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, X. D., L. Niedernhofer, B. Kuster, M. Mann, J. H. Hoeijmakers, and T. de Lange. 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12:1489-1498. [DOI] [PubMed] [Google Scholar]
- 64.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage throught ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]