Abstract
The secretory-specific poly(A) signal (μs) of the immunoglobulin μ gene plays a central role in regulating alternative RNA processing to produce RNAs that encode membrane-associated and secreted immunoglobulins. This poly(A) signal is in direct competition with a splice reaction, and regulation requires that these two reaction efficiencies be balanced. The μs poly(A) signal has several unique sequence features that may contribute to its strength and regulation. Site-directed mutations and small internal deletions made in the intact μ gene show that an extensive AU/A-rich sequence surrounding AAUAAA enhances signal use and that, of the two potential downstream GU-rich elements, both of which appear suboptimally located, only the proximal GU-rich sequence contributes substantially to use of this signal. A GU-rich sequence placed at a more standard location did not improve μs poly(A) signal use. All μ genes tested that contained modified μs poly(A) signals were developmentally regulated, indicating that the GU-rich sequences, the sequences between them previously identified as suboptimal U1A binding sites, and an upstream suboptimal U1A site do not contribute to μ mRNA processing regulation. Expression of wild-type and modified μ genes in HeLa cells overexpressing U1A also failed to demonstrate that U1A contributes to μs poly(A) signal regulation.
Cleavage-polyadenylation signals, conserved sequences at the ends of eukaryotic genes, are transcribed and then recognized in the nascent RNA by components of the cleavage-polyadenylation machinery. These components recognize and cleave the nascent RNA and then add a poly(A) tail, thus creating the 3′ end of the mRNA molecule. The cleavage-polyadenylation signals of mammalian genes consist of several conserved sequences, including an AAUAAA or closely related sequence located 11 to 23 nucleotides (nt) upstream from the cleavage site and GU-rich and/or U-rich sequences downstream from the cleavage site. The AAUAAA core upstream element is recognized by the four-subunit cleavage-polyadenylation specificity factor, and the downstream sequence is recognized by the three-subunit cleavage stimulatory factor (CstF). These complexes costabilize each other on the nascent RNA and, in combination with cleavage factor I, cleavage factor II, and poly(A) polymerase, carry out the cleavage-polyadenylation reaction (reviewed in references 5, 47, and 49). This process is coupled to transcription, since components of the cleavage-polyadenylation machinery have been shown to associate with elongating RNA polymerase II (Pol II) (6), the C-terminal domain of RNA Pol II is required for in vitro cleavage-polyadenylation reactions (15), and a functional cleavage-polyadenylation signal is required for RNA Pol II termination (reviewed in reference 37).
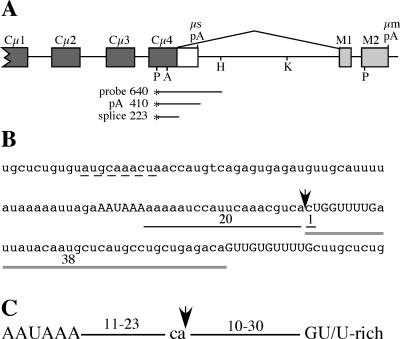
The sequences required for cleavage-polyadenylation have been characterized extensively using several model viral and cellular cleavage-polyadenylation signals. These studies have shown that the AAUAAA core upstream element is relatively invariant; for example, few positions in the simian virus 40 (SV40) late poly(A) signal could be modified without dramatically decreasing its use (41). The GU-rich and/or U-rich sequences downstream from the cleavage site are not as highly conserved. Although one study showed that a sequence element containing 4 out of 5 nt of U residues was sufficient to restore full function to the SV40 late poly(A) signal with a deleted downstream sequence (4), GU sequences are also frequently found in the downstream regions of poly(A) signals and have been shown in several cases to be required for full function (10, 23). The site of pre-mRNA cleavage occurs most often following a CA dinucleotide, but this does not seem to be an absolute requirement, since other nucleotides can substitute for CA (2, 19). The spacing between these sequence elements appears to be critical for poly(A) signal function; while there is some flexibility, optimal distances and distance limits beyond which the sequences no longer function in specific poly(A) signals have been identified (2, 4, 10, 14, 22, 23). Compilations of known poly(A) signal sequences confirm that the locations of the individual sequence elements match the experimentally deduced optimal distances and also reveal the potential diversity of sequences within these regions (11, 16, 44). The spatial arrangement of the required poly(A) signal sequence elements is summarized in Fig. 1C.
FIG. 1.
Structure of the immunoglobulin μ gene 3′ end, the μs poly(A) signal sequence, and poly(A) signal spacing. (A) The 3′ end of the Ig μ gene contains two cleavage-polyadenylation signals (μs pA and μm pA); the upstream μs poly(A) signal is in competition with the splice reaction between the Cμ4 and M1 exons (shown above the diagram). Filled boxes, constant-region exons; open box, μs-specific exon; light gray boxes, μm-specific exons. Restriction sites used in the cloning procedures are shown (P, PstI; A, ApaI; H, HindIII; K, KpnI). The S1 nuclease probe and the fragments protected by μs mRNA cleaved and polyadenylated at the μs poly(A) signal (pA) and μm mRNA that is spliced between Cμ4 and M1 exons (splice) are shown. (B) Sequence surrounding the μs poly(A) signal. The AAUAAA and two downstream GU/U-rich elements are shown in uppercase letters; the arrow indicates the cleavage site. The distances between AAUAAA and the cleavage site and the cleavage site and downstream elements are shown. The sequences suggested to match the consensus Oct1/2 binding site (9) and to contain a suboptimal U1A binding site (33) are underlined with a dashed line. (C) Standard spacing of the essential poly(A) signal sequence elements, as determined by comparison of poly(A) signal sequences and functional analyses as summarized in the text.
The secretory-specific poly(A) signal from the immunoglobulin M (IgM) gene [μs poly(A) signal] has a central role in the regulation of alternative RNA processing to produce RNAs encoding the membrane-associated (μm mRNA) and secreted (μs mRNA) Ig proteins from a single pre-mRNA (Fig. 1A) (reviewed in references 8, 28, and 49). During B-lymphocyte maturation, the ratio of the two RNAs changes from being roughly equal in B cells to heavily favoring μs mRNA in plasma cells. Cleavage-polyadenylation at the μs poly(A) site is in direct competition with splicing between the Cμ4 and M1 exons, and the efficiencies of these two reactions must be balanced for proper regulation (26, 31). Processing regulation does not require μ gene-specific sequences, since a non-Ig gene modified to contain competing cleavage-polyadenylation and splicing reactions was regulated in lymphoid cell lines and in transgenic mice (27, 40). Changes in both general cleavage-polyadenylation and RNA splicing activity have been implicated in controlling this complex regulatory event as B cells mature to plasma cells (1, 7, 43, 46). Therefore, the μs poly(A) signal must have features that allow it to be balanced with the splice reaction in B cells but enable it to be activated and efficiently used in plasma cells. Compared to other poly(A) signals, the μs signal appears to have several unique features that may contribute to its specialized role in μs/μm mRNA regulation (Fig. 1B). First, the AAUAAA core upstream element is embedded in an A-rich sequence. It also has two downstream GU/U-rich sequences that appear to be suboptimally located compared to most other poly(A) signals (Fig. 1C); the proximal and distal GU-rich sequences begin 1 nt and 38 nt, respectively, from the cleavage site (Fig. 1B). Finally, an RNA Pol II pause site that enhances use of the μs poly(A) signal in B-cell, plasma cell, and nonlymphoid cell lines (29) has been identified 50 to 200 nt downstream from the μs cleavage site.
While the μs poly(A) signal is the best-studied regulated poly(A) signal, the features required for its regulation are not well understood. A recent compilation of poly(A) signals has estimated that as many as 54% of human genes and 32% of mouse genes contain multiple poly(A) signals and thus are potentially subject to regulation through the use of alternative poly(A) signals (44). Therefore, it is important to understand features of poly(A) signals that make them responsive to regulation. Also, a better understanding of the features that contribute to the use of the μs poly(A) signal may illuminate details of its regulation during B-lymphocyte development. To investigate the potential role that the unusual features of the μs poly(A) signal may play in the overall use of this signal, we made a series of μs poly(A) signal modifications in the context of the intact μ gene. Importantly, this allows us to evaluate these modifications in competition with the Cμ4-M1 splice reaction and in the local sequence context of the poly(A) signal. By making both small deletions and site-specific mutations, we demonstrate that the AU/A-rich sequences surrounding the AAUAAA core upstream element contribute to μs poly(A) signal use, both in the presence and in the absence of AAUAAA. By specifically mutating the GU-rich downstream sequences individually and together, we find that the proximal sequence is important for μs poly(A) signal usage while the distal GU-rich sequence contributes minimally. Creating a GU-rich sequence at a more consensus distance from the cleavage site did not substantially improve use of the μs poly(A) signal, suggesting that the location of the proximal downstream element is not suboptimal. Importantly, none of the GU-rich sequence mutations disrupted the μs/μm expression switch between B cells and plasma cells, indicating these elements are not specifically required for regulation. We found that the pause site also contributes to the overall use of the modified μs poly(A) signals. Since several of our mutations altered what have been suggested to be suboptimal U1A binding sites (33, 35), we used HeLa cells that stably overexpress U1A as an additional test of the model that U1A represses the μs poly(A) signal through these specific sequences. However, we found little difference in expression of mutant and wild-type μs poly(A) signals in the presence or absence of excess U1A. Overall, our results demonstrate that the μs poly(A) signal is a complex element and that the efficiency with which it is recognized is influenced by multiple sequence elements within and adjacent to the canonical conserved poly(A) signal sequences.
MATERIALS AND METHODS
Plasmid construction.
The μs poly(A) signal deletion pA11 (an 11-nt deletion of the μs core upstream element) was made using a gapped-heteroduplex mutagenesis protocol and the oligonucleotide 5′ GCATTTTATAAAAATTAGATCCATTCAAACGTC 3′ (17). pA21 was made using a megaprimer mutagenesis protocol (38) and the oligonucleotides 5′ GAGATGTTGCATTTTAGATCCATTCAAACGTC 3′ and 5′ ATATGTGCCTGAATGCTGCC 3′. Both of these deletions were constructed in a subclone that contained the 2,318-bp PstI fragment from the Cμ gene subcloned into pUC9. The 1,500-bp ApaI-KpnI fragments containing the mutations were used to replace this same fragment in the wild-type pSV2Cμ gene (32) to generate the constructs that were then transfected into HepG2 cells. The 6-kb BglII fragment was cloned into pSV5neo as described previously (32) for transient transfections into B-cell and plasma cell lines.
The μs poly(A) signal AU/A-rich mutations were made by cloning double-stranded oligonucleotides that had BglII-compatible ends into the pA21 μs poly(A) signal deletion that was modified at one nucleotide position to contain a BglII site. For pA1, the oligonucleotide 5′ GATCAATTAGAAATAAAAAA 3′ and its complement were used, and for pA2, the oligonucleotide 5′ GATCCTCCTAGAATAAACTC 3′ and its complement were used. The sequence surrounding AAUAAA in this oligonucleotide matches that of the secretory-specific signal from the mouse IgA gene. These mutations were cloned back into the pSV2Cμ gene as described above.
The μs poly(A) signal downstream mutations were made using a megaprimer mutagenesis protocol (38), starting with the pUC9 2,318-bp PstI Cμ subclone that contained either the wild type or the ΔU mutation (29). The upstream primer that mutated the proximal GU-rich sequence to make ΔU1 and ΔU2 was 5′ AAACGTCACTGGCATGCATTATACAATGC 3′; the primer that added a new GU-rich sequence to make GU3, ΔUGU, ΔU1GU, and ΔU2GU was 5′ TTTGATTATACCATGTTTTTGGCTGCTGAGACAG 3′. The downstream primer for each of these was 5′ ATATGTGCCTGAATGCTGCC 3′. Because the mutagenic oligonucleotides overlapped, the new GU-rich elements in ΔU1GU and ΔU2ptGU have only four U's instead of five U's. The resulting megaprimer was used in combination with the upstream primer 5′ TATGTGACCAGTGCCCCGAT 3′. The final PCR product was digested with ApaI and HindIII and cloned into the pUC9 2,318-bp PstI subclone that had been digested with the same enzymes. These mutations were cloned back into the pSV2Cμ gene as described above. The DNA substrate used to make the ΔU2GU construct contained a PCR-generated point mutation (see Fig. 4) and is thus named ΔU2ptGU. We found another ΔU2 clone that did not have this point mutation to compare its expression to that of ΔU2pt. This set of mutations was cloned into the intact μ gene of the pR-SP6 plasmid as described previously (31) to stably transfect B-cell and plasma cell lines.
FIG. 4.
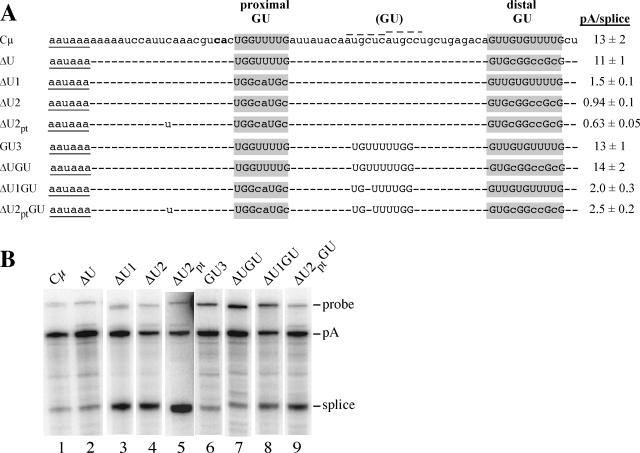
Sequence and expression of the μs poly(A) signals with modified downstream elements. (A) The sequences of the proximal and distal GU-rich downstream elements are labeled and highlighted in gray; the G's and U's in these regions are shown in uppercase letters. The newly inserted GU-rich sequence is labeled (GU), and the G's and U's are also shown in uppercase letters. A dash indicates identity with the Cμ sequence. The AAUAAA sequence is underlined, and the CA cleavage site is shown in bold. During construction of these mutations, a point mutation was introduced between AAUAAA and the cleavage site in the ΔU2 construct, shown as the “u” in ΔU2pt and ΔU2ptGU. The two dotted lines above the Cμ sequence identify the two suboptimal U1A binding sites identified previously; the U1A consensus sequence is AUUGCAC (35). The pA/splice expression ratios are compiled from at least two S1 nuclease analyses of at least three independent transfections. (B) S1 nuclease analysis of μ genes containing the modified downstream elements. RNA from HepG2 cells transiently transfected with the construct shown above each lane was analyzed using the probe diagrammed in Fig. 1A; the probe and protected bands are labeled on the right. This figure was assembled from nonadjacent lanes of three separate gels.
The RNA polymerase pause site that is present just downstream from the μs poly(A) signal was deleted from ΔU2, ΔUGU, and ΔU2ptGU by digesting each with NotI, which was introduced by the ΔU mutation, and HindIII as described previously (29) to create ΔU2ΔNH, ΔUGUΔNH, and ΔU2ptGUΔNH, respectively.
The OCTA mutation was made using the megaprimer mutagenesis protocol as described above, with the same upstream and downstream primers; the mutagenic oligonucleotide was 5′ ATGGTGACCGGTGATACACAGAGCAACTGGACACC 3′.
All of the constructs were confirmed by sequence analysis. Table 1 summarizes the modified μ genes used in this study.
TABLE 1.
List of constructs used in this study
| Construct name | Description | Reference or source |
|---|---|---|
| Cμ | Wild-type gene | 32 |
| pA11 | 11 nt surrounding AAUAAA are deleted | This work |
| pA21 | 21 nt surrounding AAUAAA are deleted | This work |
| pA1 | 5 nt surrounding AAUAAA are mutated | This work |
| pA2 | 19 nt surrounding AAUAAA are mutated away from AU richness | This work |
| ΔU | Distal GU-rich element is mutated | 29 |
| ΔU1 | Proximal GU-rich element is mutated | This work |
| ΔU2 | Both GU-rich elements are mutated | This work |
| ΔU2pt | Same as ΔU2, but with a spurious point mutation | This work |
| GU3 | New GU-rich element inserted between the two natural GU-rich elements | This work |
| ΔUGU | New GU-rich element inserted into ΔU | This work |
| ΔU1GU | New GU-rich element inserted into ΔU1 | This work |
| ΔU2ptGU | New GU-rich element inserted into ΔU2pt | This work |
| ΔNH | NotI-HindIII fragment containing the pause site deleted | 29 |
| ΔU2ΔNH | Pause site deleted from ΔU2 | This work |
| ΔUGUΔNH | Pause site deleted from ΔUGU | This work |
| ΔU2ptGUΔNH | pause site deleted from ΔU2ptGU | This work |
| OCTA | 7 nt located 40 nt upstream from AAUAAA are mutated | This work |
| αs-m | μs poly(A) signal replaced with αs poly(A) signal | 31 |
Cell culture and transfection.
HepG2 human hepatoma cells were maintained in Dulbecco's modified Eagle's medium-F12 medium (1:1) supplemented with 10% heat-inactivated fetal bovine serum and 10 μg/ml insulin. The mouse plasmacytoma line S194 was maintained in Dulbecco's modified Eagle's medium supplemented with 10% horse serum. The mouse B-cell line M12 was maintained in RPMI medium supplemented with 10% fetal bovine serum and 50 μM 2-mercaptoethanol. HepG2 cells were transfected using a calcium phosphate procedure (42), and S194 and M12 cells were transiently transfected using a DEAE-dextran procedure (12). S194 and M12 cells were stably transfected with linear plasmid by electroporation followed by selection in G418 at 500 μg/ml or 300 μg/ml, respectively. RNA was prepared from the HepG2 cells 48 h after transfection by use of a hot phenol procedure (42). Cytoplasmic RNA was prepared from S194 and M12 cells 40 h after transfection (39). Trizol (Invitrogen) was used to prepare RNA from the stable cell lines. HeLa cells stably transfected with tandem affinity purification (TAP)-tagged U1A or the empty TAP-tagged vector were obtained from Carol Lutz (20) and were maintained in RPMI supplemented with 10% fetal bovine serum and 10 μg/ml puromycin. They were transiently transfected with Lipofectamine (Invitrogen) following the manufacturer's instructions; RNA was prepared using Trizol.
RNA analysis.
RNA was analyzed by S1 nuclease protection assays using an end-labeled probe (Fig. 1A) that distinguishes spliced μm mRNA from RNA cleaved and polyadenylated at the μs poly(A) signal, following a protocol described previously (31). The protected fragments were visualized and quantitated by phosphorimager analysis. To map the precise 3′ ends of the pA11 and pA21 RNAs that were cleaved at the mutated poly(A) signal, gene-specific S1 probes were used in the assay and separated on a longer gel with multiple size markers.
RESULTS
Poly(A) signal activity has been studied previously by placing a poly(A) signal in a chimeric context driving a reporter gene as well as by in vitro analyses (34, 36, 41, 48). However, we have chosen to analyze the efficiency of modified μs poly(A) signals in vivo in the context of the intact μs/μm regulatory region. In this context, the μs poly(A) signal remains in competition with the Cμ4-M1 splice (Fig. 1A); a cis-competing reaction provides a sensitive measure of changes in poly(A) signal activity in mutated constructs as a change in the μs/μm (pA/splice) mRNA ratio. Furthermore, the native local sequence context is maintained. HepG2 human hepatoma cells were used for many of these experiments because they have a high transfection efficiency and express transfected μ genes with a pattern similar to that of plasma cells (27, 29). To test whether any of the mutations influence the developmental changes in μs poly(A) signal use, many of the constructs were also analyzed in B cells and plasma cells.
The μs poly(A) signal is difficult to inactivate.
U-to-G changes in the AAUAAA core upstream element are found rarely in natural poly(A) signals and have been shown to inactivate use of the SV40 late poly(A) signal (41, 44, 48). Thus, it was surprising that a U-to-G mutation in the μs poly(A) signal did not inactivate its use in plasma cells (45). One possible explanation is that the μs AAUAAA core upstream element is embedded in an AU/A-rich sequence (97% A or U over 29 nt [Fig. 2A ]) that may have compensated for loss of the original AAUAAA. This AU richness is not a common feature of poly(A) signals, as a compilation of mRNA 3′-end sequences from more than 10,000 mouse and human cDNAs, aligned for the AAUAAA core upstream element, did not show extensive AU-rich sequences adjacent to the hexanucleotide (11). Another bioinformatic analysis comparing strong and weak poly(A) signals proposed a U-rich sequence just downstream of AAUAAA to be a novel cis-acting element enriched in strong poly(A) signals, but a general AU richness was not evident (16). However, this AU richness is an evolutionarily conserved feature among mammalian μs poly(A) signals; the sequences of mouse, human, hamster, cow, and sheep mRNAs all are >91% A or U over 22 nt surrounding AAUAAA (data not shown).
FIG. 2.
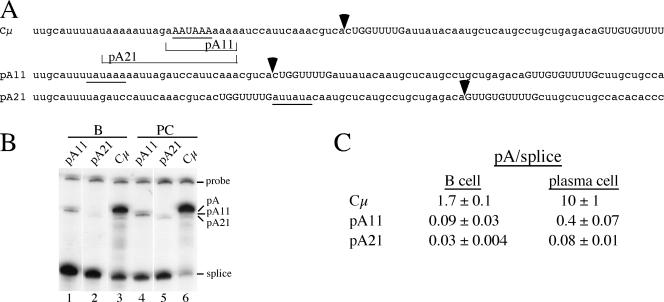
Deletions within the μs poly(A) signal reduce but do not eliminate its use. (A) Sequence of the μs poly(A) signal (Cμ) and the 11-nt deletion (pA11) and 21-nt deletion (pA21) encompassing the AAUAAA core upstream element; the sequences deleted from pA11 and pA21 are bracketed below the Cμ sequence. The cleavage sites for Cμ, pA11, and pA21 were identified using construct-specific S1 nuclease probes and are shown by the arrowheads above each sequence. The core upstream elements or sequences that may fulfill the role of this element in each construct are underlined; the downstream GU-rich sequences are shown in uppercase letters. (B) S1 nuclease analysis of RNA from M12 B cells (B) and S194 plasmacytoma cells (PC) transiently transfected with the constructs shown above each lane. The probe used was from the wild-type sequence, and the bands protected by the pA11 and pA21 RNA are smaller because the sequence of the RNA diverges from the probe at the site of the deletion. (C) Summary of the μs mRNA-to-μm mRNA expression ratios (pA/splice) for these constructs, compiled from at least two S1 nuclease analyses of two independent transfections.
Previously, three U's in the 11 nt upstream from AAUAAA in the μs poly(A) signal were changed to G's (36). This modification had no effect on poly(A) signal use when the AAUAAA core upstream element was intact but further decreased use of the signal when these core sequences were mutated or deleted. Therefore, when we wanted to inactivate the μs poly(A) signal with as minor a modification as possible, as a control for nuclear run-on experiments (29), we deleted 11 or 21 nt that removed the core upstream element and various amounts of the AU/A-rich sequence (Fig. 2A). To determine if either of these deletions would inactivate the μs poly(A) signal, constructs containing them were transiently expressed in B cells and plasma cells. Deleting 11 nt decreased the pA/splice mRNA ratio 20- to 25-fold, but RNA cleaved and polyadenylated at the μs poly(A) site was still detectable (Fig. 2B, lanes 1 and 4, and C). When 21 nt were deleted, which removes most of the AU-rich sequences, the pA/splice ratio decreased another three- to fivefold, but transcripts cleaved and polyadenylated in this region were still detected (Fig. 2B, lanes 2 and 5, and C). A construct-specific S1 nuclease probe showed that pA11 is cleaved at the normal site, even in the absence of a clear core upstream element. A potential element, UAUAAA, remains in pA11 (Fig. 2A); this sequence is found in about 4% of mouse and human poly(A) signals (44). The cleavage site for pA21 is shifted into downstream sequences and occurs just upstream of the more distal GU-rich region (Fig. 2A). There is an AU-rich sequence, AUUAUA, within 30 nt upstream from the cleavage site that may fulfill the core upstream element role. These results suggest that in the absence of the authentic core upstream element, the surrounding AU/A-rich sequences can, at a lower efficiency, fulfill the role of this required element. Thus, the μs poly(A) signal is difficult to totally inactivate; even pA21, with a poor core upstream element, is still able to direct a low level of cleavage-polyadenylation. This suggests that additional positive-acting elements, such as the RNA polymerase pause site (29), contribute to use of the μs poly(A) signal. Also, since the pA/splice ratio is higher in plasma cells than in B cells, the core upstream element and surrounding AU-rich sequences are not specifically required for the developmental changes in RNA processing patterns.
The AU/A-rich sequence surrounding AAUAAA contributes to μs poly(A) signal use.
While the AU/A-rich sequences surrounding the μs poly(A) signal core upstream element could partially compensate for the loss of the conserved hexanucleotide in pA11, it was not clear if these sequences contributed to the μs poly(A) signal use in the presence of the intact AAUAAA core upstream element. To test this, we altered the sequences surrounding AAUAAA by first making a single point mutation in the pA21 μs poly(A) signal deletion to create a BglII site and then inserting double-stranded oligonucleotides to replace the μs poly(A) signal. The pA1 construct, which was slightly less AU rich than the wild-type μs poly(A) signal, had a slightly reduced pA/splice ratio in HepG2 cells (Fig. 3). However, when we inserted an AAUAAA that was not embedded in an AU-rich sequence (pA2), the pA/splice ratio was reduced threefold relative to that of the wild-type gene (Fig. 3). Thus, the AU/A-rich sequences surrounding AAUAAA do enhance the use of the μs poly(A) signal.
FIG. 3.
AU/A-rich sequences surrounding the core upstream element contribute to the strength of the μs poly(A) signal. The AAUAAA core upstream element and proximal downstream GU/U-rich element are shown in uppercase letters, and the CA cleavage site is shown in bold. The nucleotides that are changed from the wild-type sequence are underlined. In pA1, the changes are due to the restriction site used for cloning. In pA2, the entire sequence surrounding AAUAAA has been changed. The pA/splice expression ratios are compiled from at least two S1 nuclease analyses of two independent transfections in HepG2 cells.
The proximal GU-rich element is required for full poly(A) signal activity and is not suboptimally located.
The μs poly(A) signal has two GU-rich sequences that each have four sequential U's (Fig. 1B and 4). However, they are located 1 nt and 38 nt from the cleavage site and thus fall outside the previously defined optimal spacing range of 10 to 30 nt (Fig. 1C). Because of this, they have been called suboptimal (35). Dual suboptimally spaced GU-rich elements are not a conserved feature of mammalian μs poly(A) signals (data not shown). To determine whether either of these sequences contributed to μs poly(A) signal use and whether they are in fact suboptimal, these two elements were mutated by site-directed mutagenesis, individually and in combination, and a more optimally placed GU-rich element was inserted (Fig. 4A). Sequence changes were minimized to better detect the effect of the targeted sequences and to not change the spacing of other elements within the poly(A) signal. Each of the GU-inactivating mutations removes most of the U's and all of the UU dinucleotides, which have been suggested to be important for tight CstF binding (25). These constructs were transiently transfected into HepG2 cells, and the RNA was analyzed by S1 nuclease protection. As we found previously (29), mutating the distal GU-rich sequence to a GC-rich sequence (ΔU) had no effect on use of the poly(A) signal (Fig. 4A and B, lane 2 compared to lane 1). In contrast, when the proximal GU-rich sequence was mutated (ΔU1), the pA/splice mRNA ratio decreased roughly ninefold (Fig. 4A and B, lane 3). When the two mutations were combined in ΔU2, which has no obvious GU-rich sequence, the pA/splice mRNA ratio decreased only slightly compared to that of ΔU1 (Fig. 4A and B, lane 4). This suggests that the distal GU, even when it is the only GU element, does not contribute much to μs poly(A) signal recognition. In constructing additional mutations, we found a PCR-generated mutation in ΔU2 9 nt upstream from the cleavage site, which we named ΔU2pt; this single nucleotide change has a very minor effect on expression (Fig. 4). Overall, these results indicate that the proximal GU-rich element contributes substantially to use of the μs poly(A) signal, whereas we find little evidence for activity contributed by the distal GU-rich element.
To determine whether use of the μs poly(A) signal could be improved by a more optimally placed downstream element, we mutated the sequence 18 nt downstream of the μs cleavage site to a GU/U-rich sequence that contained four to five U's, flanked by GU and GG sequences. This mutation was introduced into the wild-type μs poly(A) signal (GU3) and the μs poly(A) signals that lacked one or both of the natural GU-rich sequences (ΔUGU, ΔU1GU, and ΔU2ptGU) (Fig. 4A). We found that the added, optimally spaced GU/U-rich sequence had a modest effect at best on poly(A) signal usage. When the proximal GU-rich element was intact, the new GU-rich sequences did not improve use of the signal (Fig. 4A and B, compare Cμ to GU3 and ΔU to ΔUGU, lanes 1 and 6 and lanes 2 and 7). In the absence of the proximal GU-rich element, the new GU-rich sequence only slightly improved use of the poly(A) signal (Fig. 4A and B, compare ΔU1 to ΔU1GU, lanes 3 and 8). The added GU-rich sequence was able to improve the poly(A) signal most in ΔU2ptGU, where the new sequence was the only GU-rich sequence in the downstream region; the pA/splice ratio increased about fourfold compared to that of ΔU2pt (Fig. 4A and B, lanes 5 and 9). Thus, the inserted GU-rich sequence could function as a weak downstream element but it did not function nearly as well as the natural proximal GU-rich element.
RNA polymerase pause site deletions further decrease mutated μs poly(A) signal use.
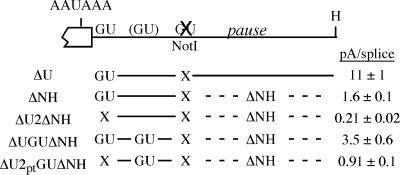
We were surprised that the optimally placed GU-rich sequences did not improve poly(A) signal usage very much. One reason for this may be that some of the μs poly(A) signals were already being used at their maximum potential. We showed previously that an RNA polymerase pause site, located ∼50 to 200 nt downstream of the μs cleavage site, contributes greatly to use of the μs poly(A) signal (29); its removal decreased the pA/splice ratio about sevenfold (Fig. 5, compare ΔU to ΔNH). To decrease use of the μs poly(A) signal, which would potentially allow us to detect a larger effect of the added, optimally placed GU-rich element, we deleted the downstream pause site from ΔUGU, ΔU2, and ΔU2ptGU (Fig. 5). In HepG2 cells, the pA/splice expression ratio of each was reduced by loss of the pause site (compare expression data in Fig. 4 and 5). However, the effect of the new GU-rich element was still rather modest. The new GU sequence improved use of the μs poly(A) signal about twofold in the presence of the proximal GU element (Fig. 5, compare ΔNH to ΔUGUΔNH). When both of the natural GU-rich sequences were mutated, as described before, the inserted GU-rich sequence increased the pA/splice ratio about fourfold (Fig. 5, compare ΔU2ΔNH and ΔU2ptGUΔNH). Nevertheless, the poly(A) signal containing only the native proximal GU-rich element (ΔNH) is still used about twofold more efficiently than the μs poly(A) signal that contained only the newly inserted GU-rich element (ΔU2ptGUΔNH). Therefore, the optimally placed GU-rich element is not able to replace the activity of the natural proximal GU-rich element and the proximal GU-rich element should not be considered to be suboptimal because of its location.
FIG. 5.
Deleting the RNA polymerase pause site further reduces recognition of the modified μs poly(A) signals. The ΔU mutation introduces a NotI restriction site in place of the distal GU-rich element; when the NotI-HindIII fragment containing an RNA Pol II pause site is deleted (ΔNH), the pA/splice ratio decreases (29). The pause site deletion was combined with several of the downstream element mutations that had the NotI restriction site, as shown. “X” indicates that the GU-rich element is mutated; the added GU sequence is indicated in those constructs where present. These constructs were transfected into HepG2 cells, and RNA levels were quantitated by S1 nuclease analysis.
Modified μs poly(A) signals are still regulated in B cells and plasma cells.
Several recent reports have implicated the downstream sequences of the μs poly(A) signal in the regulated use of this signal during B-lymphocyte maturation. These sequences were suggested to be important for hnRNP H, H′, and F competition with binding of the 64-kDa subunit of CstF (CstF-64); B cells were shown to express higher levels of hnRNP F than plasma cells, and this was suggested to contribute to the lower use of the μs poly(A) signal in the B cells (46). Also, two suboptimal U1A binding sites were found between the two downstream GU-rich sequences and were suggested to down-regulate this signal in B cells where U1A expression is higher (21, 35). The constructs we have generated provide an in vivo system to test the roles these downstream sequence elements may play in the developmentally regulated μs/μm mRNA processing switch. If the downstream GU-rich sequences were bound by hnRNP F so as to block CstF-64 binding, then one would predict that at least some of the mutations in these GU-rich sequences should disrupt the B-cell-plasma cell regulation. Furthermore, the mutation we created to add a GU-rich sequence at a more optimally placed distance from the cleavage site also disrupted both U1A-like sites between the GU-rich sequences (Fig. 4A). If, in fact, U1A bound these sites to decrease use of the μs poly(A) signal in B cells but not plasma cells, then all of the constructs containing the added GU-rich element should be impaired for regulation and have a higher pA/splice ratio in B cells than the corresponding constructs without the added GU-rich sequence.
To test these predictions, we cloned the downstream poly(A) signal mutations into a μ gene that includes normal μ promoter and enhancer elements and these constructs were stably transfected into the M12 B-cell and S194 plasma cell lines. RNA was isolated from pools of stable clones, and the pA/splice RNA was measured by S1 nuclease analysis (Table 2). Comparing the different constructs within the same cell line, we found that the pA/splice expression trends among the poly(A) signal mutations were similar to those seen when they were transiently expressed in HepG2 cells. The distal GU-rich sequence contributes minimally to use of this signal (Table 2, compare Cμ to ΔU), while the proximal GU-rich sequences are most important (Table 2, ΔU1 and ΔU2). Also, the newly inserted GU-rich sequences do not improve the use of the μs poly(A) signal (Table 2, compare Cμ to GU3, ΔU1 to ΔU1GU, and ΔU2 to ΔU2ptGU). In fact, in plasma cells, the new GU-rich sequence in ΔU1GU caused a slight decrease in use of the modified signal. Importantly, when the expression of each construct was compared between B cells and plasma cells, we found that all of the constructs were regulated; the pA/splice ratio was always higher in plasma cells than in B cells. Therefore, the normal GU-rich sequences are not specifically required for μs/μm developmental regulation; even ΔU2, which has no obvious GU-rich element, is still regulated between B cells and plasma cells. Thus, while the hnRNP F/H ratio changes during B-cell development (46), the μs poly(A) signal GU-rich elements must not be the cis-acting targets of these factors. Also, since the GU-rich mutation that both added the GU-rich sequence and eliminated the potential inhibitory U1A binding sites had little effect on expression or regulation, it is not likely that these elements play a significant role in μs/μm processing regulation in the intact μ gene.
TABLE 2.
Expression of Cμ poly(A) signal mutations in stable B-cell and plasma cell lines
| Construct | pA/splice ratio (avg ± SD)a
|
Ratio of PC/B regulation | |
|---|---|---|---|
| B cell | PC | ||
| Cμ | 3.9 ± 0.2 | 44 ± 3 | 11 |
| ΔU | 3.7 ± 0.5 | 39 ± 5 | 11 |
| ΔU1 | 0.51 ± 0.04 | 6.7 ± 0.3 | 13 |
| ΔU2 | 0.34 ± 0.02 | 5.1 ± 0.5 | 15 |
| ΔU1GU | 0.61 ± 0.04 | 2.7 ± 0.2 | 4 |
| ΔU2ptGU | 0.50 ± 0.07 | 4.6 ± 0.3 | 9 |
| GU3 | 3.7 ± 0.2 | 43 ± 4 | 12 |
The expression ratios were compiled from at least three independent S1 analyses of RNA from stably transfected cell lines.
No evidence for μs poly(A) signal regulation by U1A.
Over the past several years, the U1A protein has been proposed to play a significant role in the regulation of μs/μm mRNA abundance during lymphocyte development. This conclusion is based mainly on in vitro experiments and in vivo assays of a μs poly(A) signal fragment located downstream of the luciferase gene (33, 35). Suboptimal U1A sites were identified both upstream and downstream of the μs poly(A) signal and proposed to specifically repress use of this signal in B cells but not plasma cells. However, our results with the inserted GU-rich sequence, which also removes the two downstream U1A sites, do not support a role for these sequences in μs/μm regulation.
To further test the model that U1A affects use of the μs poly(A) signal, we obtained (from Carol Lutz) HeLa cells that stably overexpressed TAP-tagged U1A at a level similar to that of the endogenous protein and HeLa cells containing the empty TAP-tagged vector. These cells were used previously to purify U1A-interacting proteins (20). Also, when a construct containing tandem poly(A) signals was expressed in these two cell lines, there was a change in the relative usages of the two sites, indicating that these HeLa cell lines contain biologically significant differences in U1A protein levels (C. Lutz, personal communication). If U1A represses the μs poly(A) signal through the previously identified upstream or downstream elements, less μs RNA should be detected when the μ gene is transfected into HeLa cells overproducing U1A (HeLa+U1A cells) than when it is transfected into control HeLa cells. Furthermore, this U1A-mediated repression should be relieved when the sites are mutated. To test this, we cotransfected the wild-type or mutated μ genes, along with the major histocompatibility complex class I Dd gene as a control for variations in transfection efficiency, into HeLa or HeLa+U1A cells. We quantitated both μ and Dd mRNA levels by S1 nuclease mapping, corrected μ expression for Dd expression, and then determined the corrected μs ratio between the HeLa and HeLa+U1A cells. If the μs poly(A) signal is repressed by U1A, the HeLa/HeLa+U1A cell ratio should be >1, and if it is unaffected by U1A, this ratio should be ∼1. The wild-type μs poly(A) signal was present in both the Cμ and ΔNH pause site deletion constructs, and the suboptimal U1A sites were removed from the GU3 construct. As a control, we transfected the μ gene containing the αs poly(A) signal substituted for the μs poly(A) signal (αs-m [31]) and measured both the pA (αs-m sec) and spliced (αs-m mem) RNAs; we expected that this construct should not respond to U1A levels. The data from four to eight independent transfections are compiled in Table 3. The HeLa/HeLa+U1A cell ratios of αs-m, Cμ, and ΔNH were all very close to 1 and were not significantly different from each other as judged by Student's t test (all pairwise comparisons showed P values of >0.05). When the GU3 mutation was tested, it also was not significantly different from these wild-type and control constructs. This analysis indicates that doubling U1A protein levels had no effect on processing of the μs poly(A) signal in an intact μ gene. Also, by this measure, we detected no effect of the mutation that disrupts potential U1A binding sites downstream of the μs poly(A) signal. Overall, these data fail to support a model in which U1A regulates μs poly(A) signal usage.
TABLE 3.
Expression levels of wild-type and modified Cμ genes in HeLa cells stably overexpressing TAP-tagged U1A or the empty vector
| Construct | Ratio of HeLa/HeLa+U1A expression (avg ± SD)a |
|---|---|
| Cμ | 1.16 ± 0.22 |
| ΔNH | 1.08 ± 0.11 |
| αs-m sec | 0.98 ± 0.08 |
| αs-m mem | 0.98 ± 0.23 |
| GU3 | 1.12 ± 0.30 |
Cμ and Dd constructs were coexpressed in the two cell lines, Cμ mRNA levels were corrected for Dd mRNA levels, and then the corrected Cμ levels in the two cell lines were compared. The results are compiled from four to eight independent experiments.
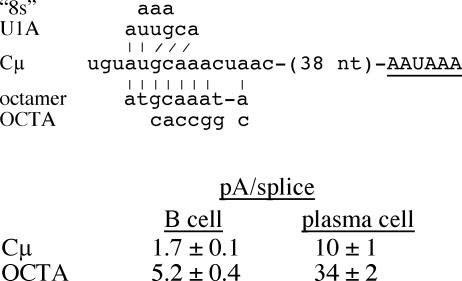
Another mutation we made previously, for other reasons, also addresses the potential regulation of the μs poly(A) signal by U1A. We made the OCTA mutation to test whether the sequence resembling an Oct1/2 binding site upstream of the μs poly(A) signal affected μs/μm expression (9); this mutation also disrupts one of the suboptimal U1A sites upstream from the μs poly(A) signal, the “8s” site (33). We found that the pA/splice ratio was increased about threefold by this mutation compared to that of the wild type in both B cells and plasma cells (Fig. 6). This suggests that a negative-acting element had been disrupted, which is consistent with the effect of the “8s” mutation (33). However, in contrast to the interpretations of Phillips et al. (33), we conclude that this element does not contribute directly to μs/μm regulation because the OCTA mutation affected the pA/splice expression ratio similarly in B cells and plasma cells (Fig. 6).
FIG. 6.
The OCTA mutation upstream of the μs poly(A) signal increases poly(A) signal use but does not affect B-cell-plasma cell regulation. The AAUAAA core upstream element is shown in uppercase letters and is underlined. The sequence 38 nt upstream from this core element is shown (see also Fig. 1B). The upstream sequence that matches the consensus Oct1/2 protein binding site is shown below (octamer) (9), and the nucleotide changes made in the OCTA mutation are shown. Above the Cμ sequence are the consensus U1A binding site (U1A) and the nucleotide changes in the “8s” mutation (33). The matches between the Cμ sequence and that of the consensus octamer and U1A sequences are indicated by vertical lines. The pA/splice expression ratios for the Cμ and OCTA constructs transiently expressed in the M12 B-cell and S194 plasma cell lines were obtained by S1 nuclease analysis of at least three transfections analyzed multiple times.
DISCUSSION
The μs poly(A) signal is a central element in the μ pre-mRNA alternative processing regulation because it is in direct competition with the Cμ4-M1 splice reaction. The efficiencies of these two reactions are balanced, and this is critical for proper regulation (26, 31). The μs poly(A) signal has been shown in vivo to be weaker than both the μm and SV40 late poly(A) signals but stronger than the IgA secretory-specific poly(A) signal (26, 30, 31). Also, the μs poly(A) signal has a lower affinity for CstF-64K and is processed less efficiently than the μm poly(A) signal in vitro (43). In this study, we have identified multiple sequence features that contribute to the overall strength of the μs poly(A) signal. We have shown that the proximal, but not the distal, GU/U-rich element contributes substantially to use of the μs poly(A) signal. If the proximal GU/U-rich element were truly suboptimally located relative to the cleavage site in the μs poly(A) signal, as has been suggested previously (35), we would have expected to improve use of the signal by introducing a GU/U-rich sequence in a more consensus location. However, this was not the case, as introducing a GU/U-rich element 18 nt beyond the cleavage site did not improve use of the μs poly(A) signals that still contained the proximal GU/U-rich element. Also, when the newly inserted GU-rich element was the only GU-rich sequence, the poly(A) signal was not as strong as when the proximal GU-rich sequence was the only GU-rich sequence. Thus, the GU-rich sequences of the μs poly(A) signal do not appear to be suboptimal. For the μs poly(A) signal, perhaps the distance between the AAUAAA core upstream element and the GU/U-rich element (10, 14) is of greater importance than that between the cleavage site and the GU-rich sequences.
It was shown previously that the AU-rich sequence upstream from the μs poly(A) signal could partially compensate for a mutation in the AAUAAA core upstream element (36). Here we show that an evolutionarily conserved AU-rich sequence surrounding AAUAAA also affects the μs poly(A) signal activity in the presence of intact AAUAAA; an extensive mutation of the AU/A-rich sequence (pA2) decreased the pA/splice ratio threefold. This indicates that, even in the presence of the core upstream element, the additional AU/A-rich sequences enhance use of the μs poly(A) signal. While the sequence alteration in pA2 may have changed the poly(A) signal so that AAUAAA was no longer unstructured (3, 13), we believe that this is unlikely since both the μs and pA2 sequences are predicted by an RNA-folding program (http://www.bioinfo.rpi.edu/∼zukerm/) to have AAUAAA and cleavage site in unpaired regions rather than stems of the folded RNA.
While our results have identified a number of sequence features that affect the overall strength of the μs poly(A) signal, none of the mutations we made differentially affected the use of the μs poly(A) signal in B cells and plasma cells. Thus, none of these sequences are significant targets for the developmentally regulated RNA processing switch. During the past several years, it has been proposed that the U1A protein plays a significant role in the regulation of μs/μm mRNA abundance during lymphocyte development (21, 33, 35). Evidence in support of a role for U1A in μs/μm regulation includes the findings (i) of multiple “suboptimal” U1A binding sites upstream and downstream of the μs poly(A) signal that bound purified recombinant U1A, (ii) that recombinant U1A inhibited in vitro polyadenylation of a μs substrate, and (iii) that when the three postulated upstream sites were mutated, use of the μs poly(A) signal increased up to twofold in vivo, as measured by luciferase activity in transfected cells (33, 35). This model proposes that in B cells U1A levels are higher and interfere with polyadenylation of the μs poly(A) signal, whereas U1A levels and its inhibitory activity decline as B cells differentiate to plasma cells. Although we did not make mutations to directly test the role of U1A in μs/μm regulation, several of our modified μs poly(A) signals fortuitously mutated sites identified to be suboptimal U1A binding sites. In contrast to earlier studies, we found no evidence that these potential U1A sites had an effect on μs/μm regulation in our in vivo assays using the intact μ regulatory region. We did find that sequences upstream of the μs poly(A) signal affected use of the μs poly(A) signal, but they had the same effect in both B cells and plasma cells, indicating they are not involved in μs/μm regulation. When the downstream suboptimal U1A sites were mutated, there was little effect on either expression or B-cell-plasma cell regulation. While one previous experiment in which an intact μ gene was transiently cotransfected with increasing levels of a U1A expression vector into M12 B cells claimed to show that U1A repressed the μs poly(A) signal (33), these data were difficult to evaluate because there were no controls for transfection efficiency. The significance of these data is also questioned by the fact that both the μs and μm poly(A) signals were repressed, although μs was repressed to a greater extent.
We also addressed the hypothesis that the μs poly(A) signal is affected by U1A by comparing the activities of μ genes containing mutant and wild-type μs poly(A) signals in HeLa cells that do or do not overexpress U1A. In the overexpressing cell line, U1A protein levels are approximately double that seen in control cells, which is within the range of changes in U1A measured between B cells and plasma cells (21, 24). However, we found little expression difference between the two HeLa cell lines. This again suggests that, in the context of the intact μs/μm regulatory region, U1A protein levels have little effect on use of the μs poly(A) signal. This brings into question the biological relevance of the previous studies, which relied heavily on in vitro assays to measure interactions between μs RNA segments and recombinant U1A protein.
The most striking and perhaps unexpected finding of our work is that the μs poly(A) signal, while not being particularly robust, is very difficult to inactivate. For example, the pA11 and ΔU2 poly(A) signals each lack one of the critical components of standard poly(A) signals, the AAUAAA core upstream element and the downstream GU-rich sequences, respectively. Yet, the pA/splice ratio for pA11 was 0.4 and that for ΔU2 was 0.94, which means that nearly 30% and 50% of the transcripts, respectively, are cleaved and polyadenylated at the mutant poly(A) signal. Use of the ΔU2 poly(A) signal decreased an additional four- to fivefold when the pause site also was deleted (ΔU2ΔNH). These results demonstrate that the μs poly(A) signal is a complex element, containing multiple sequences in addition to the standard poly(A) signal core sequences that contribute to its use, such as the AU-rich sequences around the AAUAAA core upstream element and the downstream RNA polymerase II pause site. There are also likely to be upstream elements that contribute to the recognition of the μs poly(A) signal, since scanning mutagenesis of the region upstream of the μs poly(A) signal revealed the presence of both positive- and negative-acting sequences (33). Large-scale comparisons of sequences surrounding poly(A) signals have identified numerous motifs that may contribute to poly(A) signal recognition (11, 16, 18, 44). While U-rich sequences have been found near AAUAAA (16), AU richness in this area has not been identified as a general element. However, in light of our results, it will be interesting to see how many poly(A) signals have such sequences and whether they have any shared characteristics, such as being regulated poly(A) signals. As so few poly(A) signals have been mutationally dissected, it will be important to combine bioinformatics and biochemical approaches to better understand the complexities of poly(A) signal sequences.
Acknowledgments
We thank Shannon Bertolino and Frankie Davis for making and analyzing, respectively, the pA1 and pA2 mutations, Amanda Ribble for making and analyzing the stable lymphoid cell lines, Songchun Liang and Carol Lutz for the U1A-expressing HeLa cells, and Brett Spear, Rachel Burnside, and Carol Lutz for their helpful comments on the manuscript.
This work was supported by grants MCB-9808637 and MCB-0318047 from the National Science Foundation. G.L.B. was supported in part by a Howard Hughes undergraduate summer research fellowship.
REFERENCES
- 1.Bruce, S. R., R. W. C. Dingle, and M. L. Peterson. 2003. B-cell and plasma-cell splicing differences: a potential role in regulated immunoglobulin RNA processing. RNA 9:1264-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, F., C. C. MacDonald, and J. Wilusz. 1995. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 23:2614-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, F., and J. Wilusz. 1998. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res. 26:2891-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou, Z.-F., F. Chen, and J. Wilusz. 1994. Sequence and position requirements for uridylate-rich downstream elements of polyadenylation signals. Nucleic Acids Res. 22:2525-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755-2766. [DOI] [PubMed] [Google Scholar]
- 6.Dantonel, J. C., K. G. K. Murthy, J. L. Manley, and L. Tora. 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389:399-402. [DOI] [PubMed] [Google Scholar]
- 7.Edwalds-Gilbert, G., and C. Milcarek. 1995. Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol. Cell. Biol. 15:6420-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwalds-Gilbert, G., K. L. Veraldi, and C. Milcarek. 1997. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 25:2547-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falkner, F. G., and H. G. Zachau. 1984. Correct transcription of an immunoglobulin kappa gene requires an upstream fragment containing conserved sequence elements. Nature 310:71-74. [DOI] [PubMed] [Google Scholar]
- 10.Gil, A., and N. J. Proudfoot. 1987. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit β-globin mRNA 3′ end formation. Cell 49:399-406. [DOI] [PubMed] [Google Scholar]
- 11.Graber, J. H., C. R. Cantor, S. C. Mohr, and T. F. Smith. 1999. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. USA 96:14055-14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grosschedl, R., and D. Baltimore. 1985. Cell-type specificity of immunoglobulin gene expression is regulated by at least three DNA sequence elements. Cell 41:885-897. [DOI] [PubMed] [Google Scholar]
- 13.Hans, H., and J. C. Alwine. 2000. Functionally significant structure of the simian virus 40 late polyadenylation signal. Mol. Cell. Biol. 20:2926-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heath, C. V., R. M. Denome, and C. N. Cole. 1990. Spatial constraints on polyadenylation signal function. J. Biol. Chem. 265:9098-9104. [PubMed] [Google Scholar]
- 15.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395:93-96. [DOI] [PubMed] [Google Scholar]
- 16.Hu, J., C. S. Lutz, J. Wilusz, and B. Tian. 2005. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11:1485-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inouye, S., and M. Inouye. 1991. Site-directed mutagenesis using gapped-heteroduplex plasmid DNA, p. 71-82. In M. J. McPherson (ed.), Directed mutagenesis. IRL Press, Oxford, United Kingdom.
- 18.Legendre, M., and D. Gautheret. 2003. Sequence determinants in human polyadenylation site selection. BMC Genomics 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levitt, N., D. Briggs, A. Gil, and N. J. Proudfoot. 1989. Definition of an efficient synthetic poly(A) site. Genes Dev. 3:1019-1025. [DOI] [PubMed] [Google Scholar]
- 20.Liang, S., and C. S. Lutz. 2006. p54nrb is a component of the snRNP-free U1A (SF-A) complex that promotes pre-mRNA cleavage during polyadenylation. RNA 12:111-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma, J., S. I. Gunderson, and C. Phillips. 2006. Non-snRNP U1A levels decrease during mammalian B-cell differentiation and release the IgM secretory poly(A) site from repression. RNA 12:122-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacDonald, C. C., J. Wilusz, and T. Shenk. 1994. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol. 14:6647-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDevitt, M. A., R. P. Hart, W. W. Wong, and J. R. Nevins. 1986. Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J. 5:2907-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milcarek, C., K. Martincic, L.-H. Chung-Ganster, and C. S. Lutz. 2003. The snRNP-associated U1A levels change following IL-6 stimulation of human B cells. Mol. Immunol. 39:809-814. [DOI] [PubMed] [Google Scholar]
- 25.Perez Canadillas, J. M., and G. Varani. 2003. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 22:2821-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson, M. L. 1992. Balanced efficiencies of splicing and cleavage-polyadenylation are required for μs and μm mRNA regulation. Gene Expr. 2:319-327. [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson, M. L. 1994. Regulated immunoglobulin (Ig) RNA processing does not require specific cis-acting sequences: non-Ig genes can be alternatively processed in B cells and plasma cells. Mol. Cell. Biol. 14:7891-7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson, M. L. 1994. RNA processing and the expression of immunoglobulin genes, p. 321-342. In E. C. Snow (ed.), Handbook of B and T lymphocytes. Academic Press, San Diego, Calif.
- 29.Peterson, M. L., S. Bertolino, and F. Davis. 2002. An RNA polymerase pause site is associated with the immunoglobulin μs poly(A) site. Mol. Cell. Biol. 22:5606-5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson, M. L., M. B. Bryman, M. Peiter, and C. Cowan. 1994. Exon size affects competition between splicing and cleavage-polyadenylation in the immunoglobulin μ gene. Mol. Cell. Biol. 14:77-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson, M. L., and R. P. Perry. 1989. The regulated production of μm and μs mRNA is dependent on the relative efficiencies of μs poly(A) site usage and the Cμ4-to-M1 splice. Mol. Cell. Biol. 9:726-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson, M. L., and R. P. Perry. 1986. Regulated production of μm and μs mRNA requires linkage of the poly(A) addition sites and is dependent on the length of the μs-μm intron. Proc. Natl. Acad. Sci. USA 83:8883-8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips, C., S. Jung, and S. I. Gunderson. 2001. Regulation of nuclear poly(A) addition controls the expression of immunoglobulin M secretory mRNA. EMBO J. 20:6443-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips, C., C. B. Kyriakopoulou, and A. Virtanen. 1999. Identification of a stem-loop structure important for polyadenylation at the murine IgM secretory poly(A) site. Nucleic Acids Res. 27:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips, C., N. Pachikara, and S. I. Gunderson. 2004. U1A inhibits cleavage at the immunoglobulin M heavy-chain secretory poly(A) site by binding between the two downstream GU-rich regions. Mol. Cell. Biol. 24:6162-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips, C., and A. Virtanen. 1997. The murine secretory poly(A) site contains dual upstream and downstream elements which affect polyadenylation. Nucleic Acids Res. 25:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proudfoot, N. J. 2000. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 25:290-293. [DOI] [PubMed] [Google Scholar]
- 38.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 39.Schibler, U., K. B. Marcu, and R. P. Perry. 1978. The synthesis and processing of the messenger RNAs specifying heavy and light chain immunoglobulins in MPC-11 cells. Cell 15:1495-1509. [DOI] [PubMed] [Google Scholar]
- 40.Seipelt, R. L., B. T. Spear, E. C. Snow, and M. L. Peterson. 1998. A nonimmunoglobulin transgene and the endogenous immunoglobulin μ gene are coordinately regulated by alternative RNA processing during B-cell maturation. Mol. Cell. Biol. 18:1042-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheets, M. D., S. C. Ogg, and M. P. Wickens. 1990. Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 18:5779-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spear, B. T., and S. M. Tilghman. 1990. Role of α-fetoprotein regulatory elements in transcriptional activation in transient heterokaryons. Mol. Cell. Biol. 10:5047-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takagaki, Y., R. L. Seipelt, M. L. Peterson, and J. L. Manley. 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87:941-952. [DOI] [PubMed] [Google Scholar]
- 44.Tian, B., J. Hu, H. Zhang, and C. S. Lutz. 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33:201-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsurushita, N., N. M. Avdalovic, and L. J. Korn. 1987. Regulation of differential processing of mouse immunoglobulin μ heavy-chain mRNA. Nucleic Acids Res. 15:4603-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veraldi, K. L., G. Arhin, K. Martincic, L.-H. Chung-Ganster, J. Wilusz, and C. Milcarek. 2001. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 21:1228-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wahle, E., and U. Kuhn. 1997. The mechanism of 3′ end cleavage and polyadenylation of eukaryotic pre-mRNA. Prog. Nucleic Acid Res. Mol. Biol. 57:41-70. [DOI] [PubMed] [Google Scholar]
- 48.Wilusz, J., S. M. Pettine, and T. Shenk. 1989. Functional analysis of point mutations in the AAUAAA motif of the SV40 late polyadenylation signal. Nucleic Acids Res. 17:3899-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]