Abstract
Class IIa histone deacetylases (HDACs) are found both in the cytoplasm and in the nucleus where they repress genes involved in several major developmental programs. In response to specific signals, the repressive activity of class IIa HDACs is neutralized through their phosphorylation on multiple N-terminal serine residues and 14-3-3-mediated nuclear exclusion. Here, we demonstrate that class IIa HDACs are subjected to signal-independent nuclear export that relies on their constitutive phosphorylation. We identify EMK and C-TAK1, two members of the microtubule affinity-regulating kinase (MARK)/Par-1 family, as regulators of this process. We further show that EMK and C-TAK1 phosphorylate class IIa HDACs on one of their multiple 14-3-3 binding sites and alter their subcellular localization and repressive function. Using HDAC7 as a paradigm, we extend these findings by demonstrating that signal-independent phosphorylation of the most N-terminal serine residue by the MARK/Par-1 kinases, i.e., Ser155, is a prerequisite for the phosphorylation of the nearby 14-3-3 site, Ser181. We propose that this multisite hierarchical phosphorylation by a variety of kinases allows for sophisticated regulation of class IIa HDACs function.
Deacetylation of histones by histone deacetylases (HDACs) results in a compact chromatin structure that occludes the transcriptional machinery from accessing DNA and, as such, HDACs mostly function as transcriptional repressors (31, 40). The 18 human HDACs identified to date are grouped into four distinct classes, with members of class II further subdivided into two subclasses, IIa and IIb (49). The class IIa HDACs (HDAC4, HDAC5, HDAC7, and HDAC9) have a modular structure, comprising a conserved catalytic region at their C terminus and an “adapter” N-terminal domain that plays a central role in their regulation. First, this region contains conserved amino acid motifs that are specialized for binding an array of transcription factors. As an example, a short motif is implicated in interactions with members of the MEF2 family, and repression of MEF2-targeted promoters via recruitment of class IIa-associated HDAC activity has been extensively documented (8, 17, 22, 25, 28, 45, 50).
The adapter domain of class IIa HDACs is also subject to various posttranslational modifications such as proteolytic cleavage (2, 20, 33), ubiquitination (18), sumoylation (16, 36), and, most importantly, phosphorylation. Phosphorylation has recently emerged as the primary mechanism in the regulation of class IIa HDACs-mediated repression (49). In response to various stimuli, a number of serine residues in the adapter domain of class IIa HDACs are phosphorylated and become docking sites for 14-3-3 proteins. Association with 14-3-3 induces CMR1-dependent nuclear export and cytoplasmic accumulation of class IIa HDACs with concomitant derepression of their target promoters (13, 15, 22, 46). This nuclear export mechanism allows for signal-dependent activation of class IIa HDACs target genes and has proven to be crucial for various developmental programs such as muscle differentiation (24) and activity (27), cardiac hypertrophy (5, 50), T-cell apoptosis (8), bone development (43), and neuron survival (3, 19).
Different signaling pathways converge on the signal-responsive serine residues of class IIa HDACs. It is now well established that members of Ca2+/calmodulin-dependent protein kinases (CaMKs) promote nuclear export of class IIa HDACs (6, 7, 15, 19, 24, 25). Recently, we along with others have reported that specific stimuli can induce nuclear export of class IIa HDACs through a Ca2+-independent mechanism involving protein kinase C (PKC). Protein kinase D (PKD; also known as PKCμ), a downstream effector in PKC signaling, was, indeed, shown to directly phosphorylate HDAC5 and HDAC7 on the serine residues that control their nucleocytoplasmic trafficking (9, 34, 42). In addition, several reports suggest that other protein kinases might also be involved in signal-dependent phosphorylation and subcellular localization of class IIa HDACs (50, 52). More interestingly, recent observations indicate that subcellular localization of class IIa HDACs might also be constitutively regulated in a signal-independent manner (3, 21).
In this study, we identified the hPar-1/MARK (microtubule affinity-regulating kinase) kinases, EMK and C-TAK1, as constitutively active kinases regulating subcellular trafficking of class IIa HDACs. Both kinases directly phosphorylate class IIa HDACs on their N-terminal adapter domain, promoting their nuclear export and leading to derepression of MEF2-dependent transcription. Unexpectedly, we found that, among the multiple, conserved residues previously involved in nucleocytoplasmic shuttling of class IIa HDACs, MARK/Par-1 kinases specifically target a unique site. More importantly, phosphorylation of this site is a prerequisite for subsequent phosphorylation at other serine residues. These results support a model of hierarchical class IIa HDAC phosphorylation and establish a new role for MARK/Par-1 kinases in the control of gene expression.
MATERIALS AND METHODS
Plasmids, antibodies, and chemicals.
C-terminal green fluorescent protein (GFP) fusion proteins of human HDACs have been described elsewhere (8, 11, 12). Glutathione transferase (GST) fusion proteins of the N terminus (amino acids [aa] 1 to 490) and C terminus (aa 490 to 915) of HDAC7 and the N terminus (aa 1 to 661) of HDAC4 have been previously described (9, 12). GST-S155, GST-S181, GST-S321, and GST-S449 constructs contain, respectively, amino acids 130 to 180, 156 to 216, 267 to 345, and 396 to 490 of HDAC7, cloned into pGEX4T1 (Pharmacia) (9). GST-HDAC4 and GST-HDAC5 correspond to amino acids 221 to 272 and 234 to 285 of HDAC4 and HDAC5, respectively. Serine-to-alanine and leucine-to-alanine substitutions were introduced by PCR, and mutations were verified by direct DNA sequencing according to standard methods. GST-Cdc25C and GST-Cdc25C(S216A) fusion proteins have been described previously (1, 35). Expression vectors for active EMK and C-TAK1 have been described previously (32, 35). The Nur77 Luciferase construct has been described elsewhere (8).
Anti-FLAG, anti-panPKC, anti-pan14-3-3, anti-c-jun, anti-HDAC7, anti-HDAC1, anti-actin, and anti-tubulin antibodies were purchased from Santa Cruz Biotechonology (Santa Cruz, CA). Anti-C-TAK1, anti-ribosomal S6 kinase 1 (RSK1) and anti-RSK2, and anti-mitogen- and stress-activated protein kinase 1 (MSK1) and anti-MSK2 were obtained from Upstate Biotech (Lake Placid, NY). Monoclonal antibody against EMK has been described elsewhere (14). Polyclonal antibodies against phosphorylated Ser155 and phosphorylated Ser181 were generated by 21st Century Biochemicals (Marlboro, MA). Rabbits were immunized with the KLH-linked peptides HFPLRKTVpS155EPNLKLRYKP (where pS155 is phosphorylated Ser155) and KNPLLRKEpS181APPSLRRRP, respectively, and sera were collected and purified according to the company's procedures.
Staurosporine (Alexis Biochemical Corp., Lausen, Switzerland) and leptomycin B (Merck Biosciences Inc, Darmstadt, Germany) were used at concentrations of 1 μM and 10 ng/ml, respectively.
Cell culture.
The following cell lines were used in these experiments: Cos7, African green monkey, simian virus 40-transformed kidney cells (ATCC CRL-1651); HeLa, human cervical epitheloid carcinoma (ATCC CCL-2.1); HEK293, transformed human kidney (ATCC CRL-1573); and Do11.10 T-cell hybridomas (47). Cell lines were grown in recommended medium (Dulbecco's modified Eagle's medium [DMEM] for HeLa, HEK293, and Cos7 and RPMI 1640 medium for Do11.10 cells) supplemented with 10% fetal bovine serum, 2 mM glutamine, and 50 U/ml of streptomycin-penicillin at 37°C in a humidified incubator.
RNAi.
Functionally validated Stealth RNA interference (RNAi) Duopacks, each containing two nonoverlapping Stealth RNAi molecules (duplex 1 and duplex 2) directed against C-TAK1 or EMK, and corresponding Stealth RNAi Negative Control were purchased from Invitrogen (Carlsbad, Calif.). HeLa cells were plated to achieve 60 to 80% confluence at the time of transfection. GFP-HDAC7 (for immunofluorescence analysis) or FLAG-HDAC7 (for Western blotting analysis) proteins were transfected along with either pooled small interfering RNA (siRNA) duplexes 1 for EMK and C-TAK1 (50 nM of each duplex) or control siRNA using Lipofectamine 2000 according to the manufacturer's instructions. Twenty-four hours after this first transfection, cells were transfected with pooled siRNA duplexes 2 for each kinase or with control siRNA. Cells were then left to recover in complete medium for an additional 36 to 48 h before being processed for immunofluorescence or Western blotting analysis as described below.
Confocal microscopy.
For steady-state immunofluorescence experiments, GFP constructs were transiently transfected by the standard calcium phosphate method (for Cos7 and 293 HEKcells) or with Lipofectamine 2000 (HeLa cells) according to the manufacturer's instructions (Invitrogen, California). Localization of the fluorescent proteins was assessed on fixed cells (for Cos7 and HeLa) or live cells (for HEK293) by confocal microscopy (Axivert 200 with and LSM 510 laser scanning microscope; Carl Zeiss Microscopy). When indicated, the average percentage of cells showing nuclear exclusion of the GFP-tagged protein was assessed by examining at least three independent fields, each containing more than 50 cells. Imaging of living Cos7 cells was performed after transfection with FUGENE 6 (Roche Diagnostics, Basel, Switzerland) on a confocal laser scanning microscope (LSM 510 META; Carl Zeiss, Jena, Germany) equipped with a 488-nm argon laser. Cells were grown and transfected in MatTek glass-bottomed dishes (MatTek, Ashland, MA). Forty-eight hours after transfection, cells were maintained in DMEM supplemented with 30 mM HEPES, pH 7.0. During the measurements, the medium was kept at 37°C in an atmosphere containing 5% CO2 using an LSM 510 Incubator S (Carl Zeiss). Quantitative analysis of the nuclear and cytoplasmic fluorescence intensity levels was performed on images of the midsection of living cells. The midsection was first determined by a Z-stack. The nuclear/cytoplasmic ratio of fluorescence intensity was quantified using the Image J public domain Java image processing program (http://rsb.info.nih.gov/ij/download.htm). For quantification, the fluorescence intensity in the cytoplasmic or nuclear compartment was determined in a 0.5- by 0.5-μm square that was centered in the nucleus or cytoplasm, respectively (two values per cell). Results are the means of the fluorescence intensity determined in 20 cells expressing the GFP fusion HDAC7 proteins. The relative nuclear fluorescence intensity (FI) was calculated using the following equation: relative FInuc = Σ(FInuc/n)/Σ(FIcyt/n), where FInuc is the fluorescence intensity in the nucleus, FIcyt is the relative fluorescence intensity in the cytoplasm, and n is the number of cells examined.
For fluorescence loss in photobleaching (FLIP) experiments, a time series of images recorded live was collected as described above. The bleach rate for each series of images was calculated and used to correct the fluorescence intensities of the images. For qualitative FLIP analysis, cells were imaged using the 488-nm line of an argon laser of a Zeiss LSM510 META confocal microscope operating at 40% laser power and 5% transmission (imaging intensity). Four imaging scans of a single cell were performed. Then, the cytoplasm was chosen as region of interest by the LSM 510 META software and selectively bleached four times with 100% transmission (bleaching intensity), 30 s each, followed by imaging scans at the times indicated in the figures. At least 10 data sets were analyzed for each time point. The background corrected nuclear fluorescence at the time before the initial photobleaching was set as 100%.
GST fusion proteins: expression, purification, and pull-downs.
HDACs portions and human EMK and C-TAK1 were purified as GST fusion proteins in BL21 RIP (receptor-interacting protein) (Stratagene) according to protocols described elsewhere (9). For in vitro kinase (IVK) assays, purified GST-hEMK and GST-hC-TAK1 were first eluted from the beads by incubation in 25 mM reduced glutathione and 50 mM Tris-HCl, pH 8. Recombinant, active GST-PKD1 was produced and purified from mammalian cells as described previously (44).
Pull-down reactions and immunoaffinity purifications from cell extracts were performed exactly as previously described (8, 9).
SDS-PAGE and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis were performed according to standard procedures (37). Western blots were developed with an ECL detection kit (Amersham Pharmacia Biotech).
IVK assay and in-gel kinase (IGK) assay.
Purified GST fusion proteins were incubated with a dilution of recombinant active protein kinase (0.5 μg/ml) in 30 μl of phosphorylation mix containing 10 μM ATP with 10 μCi of [γ-32P]ATP, 25 mM Tris, pH 7.5, and 10 mM MgCl2. After 30 min at 30°C, reactions were terminated by adding an equal amount of 2× SDS-PAGE sample buffer and resolved by SDS-PAGE analysis, and phosphorylated proteins were visualized by autoradiography.
The IGK assay protocol was a slightly modified version of (38). Briefly, pull-down reactions were resolved by standard SDS-PAGE analysis with no exogenous substrate incorporation in the gel matrix. After electrophoresis, the gel was soaked in 20% 2-propanol in 50 mM Tris-HCl, pH 8.0, and then in buffer A (50 mM Tris-HCl with 5 mM dithiothreitol) to remove SDS. Resolved proteins were denatured by incubating the gel for 1 h in a solution of 6 M guanidine-HCl in buffer A and then renatured in buffer A containing 0.04% Tween 20 overnight at 4°C. The gel was preincubated in the phosphorylation buffer (see above) omitting the [γ-32P]ATP. A 10 μM concentration of ATP with 100 μCi of [γ-32P]ATP was then added, and the phosphorylation was allowed to proceed for 1 h at 30°C. Finally, the gel was washed in 5% (wt/vol) trichloroacetic acid plus 1% (wt/vol) sodium pyrophosphate, dried, and analyzed by autoradiography to detect autophosphorylated proteins.
Reporter assays.
Transient transfections of Do11.10 cells using the DEAE-dextran method and dual luciferase (Promega) reporter assays have been described before (8, 9).
Metabolic labeling and phosphorylation site analysis.
For in vivo phosphorylation site mapping, Do11.10 cells or transfected HEK293 cells were incubated for 3 h at 37°C in phosphate-free DMEM containing 500 μCi of [32P]orthophosphate per ml. Cells were then washed twice in ice-cold phosphate-buffered saline and lysed in IPLS buffer (12).Labeled proteins were purified by immunoaffinity. In vitro phosphorylation reactions of GST-HDAC7Nter by purified GST-hC-TAK1, GST-EMK, or GST-PKD were performed following the IGK protocol described above.
Purified proteins were resolved by SDS-PAGE analysis and Coomassie staining. Phosphorylated proteins were in-gel digested with trypsin. The phosphorylated peptides were separated by high-pressure liquid chromatography (HPLC) on a Thermo Hypercarb column (2.1 mm by 15 cm) in an acetonitrile gradient in 0.1% (vol/vol) trifluoroacetic acid (solvent A). Elution was performed with the following gradient program: 5 to 100% solvent B (70% [vol/vol] acetonitrile in solvent A) over 100 min at a flow rate of 200 μl/min generated by a model 1100 Agilent HPLC system. Radioactive peaks were detected by Cerenkov counting, dried under vacuum, redissolved in 5 μl of 50% (vol/vol) acetonitrile-0.3% (vol/vol) acetic acid, and analyzed by nano-electrospray ionization tandem mass spectrometry (MS/MS) in an LCQ Deca XP Plus ion-trap mass spectrometer (Thermo Finnigan, San Jose, CA). Spectra were taken in full MS and zoom scan mode to determine parent ion monoisotopic masses and their charge states. Phosphopeptides were identified in MS/MS mode as by the loss of H3PO4 (98 Da) under low-collision-induced dissociation energy, and the phosphorylated residue was pinpointed in MS3 mode.
RESULTS
Class IIa HDACs are constitutively phosphorylated and bound to 14-3-3 proteins.
Phosphorylation-dependent nuclear exclusion of class IIa HDACs is thought to be mediated by at least two signal-responsive families of kinases, the CaMKs and PKDs. However, accumulating evidence suggests that class IIa HDACs could also be phosphorylated in the absence of external stimulus and implies the existence of additional HDACs kinases distinct from CaMKs or PKD (3, 15, 21).
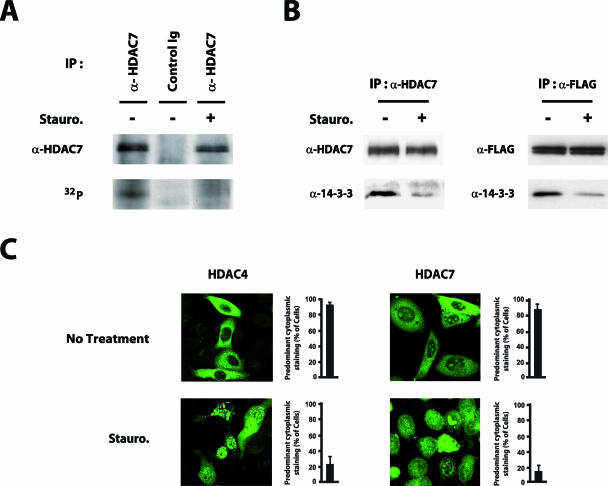
As a step toward the identification of these new class IIa HDACs kinases, we first investigated whether endogenous HDAC7 could be phosphorylated under normal growing conditions. In vivo, HDAC7 is predominantly expressed in double-positive thymocytes (8). Logically, we found that among numerous cell lines tested, T-cell hybridomas express high levels of the HDAC7 protein (data not shown). Do11.10 T-cell hybridoma cells were thus metabolically labeled with inorganic 32P, and endogenous HDAC7 was recovered by immunoprecipitation. As shown in Fig. 1A, HDAC7 appeared as a phosphorylated protein after SDS-PAGE analysis and autoradiography. Interestingly, phosphorylation of endogenous HDAC7 was almost completely abolished by treatment with staurosporine, a general serine/threonine kinase inhibitor (Fig. 1A).
FIG. 1.
Class IIa HDACs are constitutively phosphorylated, bound to 14-3-3 proteins, and subjected to phosphorylation-dependent nucleo-cytoplasmic shuttling. (A) Do11.10 cells were labeled with [32P]orthophosphate and subsequently treated with staurosporine (+) or left untreated (−). Endogenous HDAC7 was immunoprecipitated and analyzed by SDS-PAGE followed by Western blotting (α-HDAC7) or autoradiography (32P). (B) Do11.10 cells, transduced with FLAG-tagged HDAC7 (right) or not (left), were left untreated or treated with staurosporine for 1 h. Endogenous (left) or ectopically expressed HDAC7 (right) was immunoprecipitated from cell lysates using the indicated antibodies. Immunoprecipitated material was then subjected to Western blot analysis with antibodies directed against endogenous HDAC7 (α-HDAC7), the FLAG epitope (α-FLAG), or 14-3-3 proteins (α-14-3-3). (C) HeLa cells expressing GFP-HDAC4 or GFP-HDAC7 were either left untreated (No Treatment) or treated with staurosporine. After 1 h of treatment, the subcellular distribution of GFP-HDACs was determined using confocal microscopy. Bar histograms represent the mean percentages of cells showing predominant cytoplasmic localization of GFP-HDACs in each condition. Stauro, staurosporine; IP, immunoprecipitation.
According to the current model, phosphorylation of specific serine residues in class IIa HDACs creates docking sites for 14-3-3 proteins. To confirm the above observations, we thus examined the interaction between HDAC7 and 14-3-3 proteins in Do11.10 cells. Endogenous HDAC7 (Fig. 1B, left) or ectopically expressed Flag-tagged HDAC7 (Fig. 1B, right) was immunoprecipitated and analyzed for association with endogenous 14-3-3 proteins. Western blot analysis revealed a constitutive association between endogenous 14-3-3 proteins and endogenous or FLAG-HDAC7 (Fig. 1B). Interestingly, treatment with staurosporine resulted in a drastic reduction in the interaction between HDAC7 and 14-3-3 proteins. Similar observations were made with HDAC4 and HDAC5 in HEK293 cells (data available on request). These results thus suggest that class IIa HDACs exists as phosphoproteins in unstimulated cells and are constitutively associated with 14-3-3 proteins in a phosphorylation-dependent manner.
HDAC7 constitutively shuttles from the nucleus to the cytoplasm.
Phosphorylation of class IIa HDACs and association with 14-3-3 proteins controls their distribution between the nucleus and the cytoplasm. We thus examined the steady-state subcellular localization of GFP-HDAC4 and -HDAC7 in HeLa cells (Fig. 1C). Under normal growth conditions, the vast majority of cells showed predominantly cytoplasmic localization of both HDAC4 and HDAC7 (Fig. 1C, No Treatment). In contrast, under hypophosphorylating conditions induced by staurosporine, both HDACs accumulated in the nucleus (Fig. 1C).
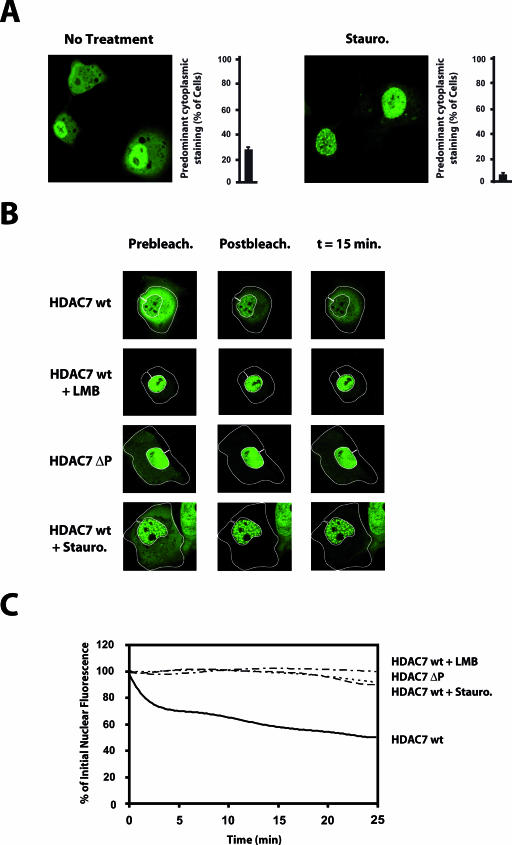
It is now well known that class IIa HDACs show differential subcellular localization depending on the cell line examined (11, 15, 46). This suggests that the machinery controlling nuclear export of class IIa HDACs is differently effective in different cell types. To generalize the above observations, we examined the effect of staurosporine in Cos7 cells, where ectopically expressed class IIa HDACs have been shown to localize primarily in the nucleus (11, 25, 26). In the majority of the transfected cells, GFP-HDAC7 was, for the most part, found in the nucleus, with only a small fraction of the protein present in the cytoplasm. However, in about 25% of the cells, HDAC7 was predominantly detected in the cytoplasm (Fig. 2A, No treatment). Similarly to HeLa cells, treatment with staurosporine was associated with significant nuclear retention of HDAC7. Indeed, after 1 h of treatment, the protein was almost exclusively localized in the nucleus, and the proportion of cells with predominant cytoplasmic staining dropped to less than 5% (Fig. 2A). Of note, staurosporine also increased nuclear retention of GFP-HDAC7 in HEK293 cells, where it distributes equally between the nucleus and the cytoplasm in untreated cells (data available on request). Our observations so far clearly unraveled basal phosphorylation and 14-3-3 binding of class IIa HDACs and point toward a constitutive, phosphorylation-dependent nuclear efflux of these enzymes in various normally growing cell lines. To challenge this model, we examined the intracellular mobility of HDAC7 in Cos7 cells, which appear to be the least potent in exporting class IIa HDACs from the nucleus to the cytoplasm. GFP-HDAC7 was thus expressed in Cos7 cells, and its nuclear efflux was examined on live cells using the FLIP technology. For this purpose, the cytoplasm of transfected cells was selectively and repeatedly bleached, and loss of fluorescence was monitored in the nucleus. FLIP experiments revealed a substantial loss of nuclear fluorescence of GFP-HDAC7 in living Cos7 cells after bleaching of the cytoplasm (Fig. 2B). This loss of nuclear fluorescence was completely blocked by leptomycin B, an inhibitor of CRM1-dependent nuclear export (Fig. 2B). Quantification of the data confirmed these observations and showed a 50% decrease in nuclear fluorescence of GFP-HDAC7 wild type (wt) 25 min after cytoplasmic bleaching (Fig. 2C, HDAC7wt). In accordance with the steady-state localization data (Fig. 2A), staurosporine treatment totally abolished constitutive nuclear export of HDAC7 (Fig. 2B and C). A similar effect was observed when the four 14-3-3 binding sites of HDAC7 (Ser155, Ser181, Ser321, and Ser449 [9]) were mutated to alanines (Fig. 2B and C, HDAC7ΔP). These results clearly demonstrate a constitutive efflux of HDAC7 from the nucleus to the cytoplasm under normal growing conditions. In addition, they are consistent with a model in which the dynamic nuclear export of class IIa HDACs is dependent on the constitutive phosphorylation of their conserved 14-3-3 binding sites. This thus implies the existence of additional class IIa HDACs kinases.
FIG.2.
Class IIa HDACs are subjected to phosphorylation-dependent nucleocytoplasmic efflux. (A) Recombinant GFP-HDAC7 was transfected into Cos7 cells. Forty-eight hours posttransfection, cells were left untreated (No Treatment) or treated with staurosporine (Stauro) for 1 h before the localization of HDAC7 was examined by confocal microscopy. (B) Cos7 cells were transfected with constructs expressing GFP fusion proteins corresponding to HDAC7 (HDAC7wt) or a mutant of HDAC7 in which the residues Ser155, Ser181, Ser321, and Ser449 were mutated to alanines (HDAC7ΔP). Forty-eight hours after transfection, cells were left untreated or treated with staurosporine (Stauro) or leptomycin B (LMB) as indicated. The cytoplasm of transfected cells was repeatedly bleached, and the loss of fluorescence in the nuclear region was assessed by confocal microscopy. (C) Relative loss of fluorescence in the nuclear region was determined as described in Materials and Methods. FLIP data are represented as nonlinear fit curves according to the legend at right of the graph. HDACwt +LMB, HDAC7 wt with leptomycin B treatment; HDAC7wt+Stauro, HDAC7 wt with staurosporine treatment.
An 85-kDa autophosphorylating kinase associates with the N terminus of class IIa HDACs.
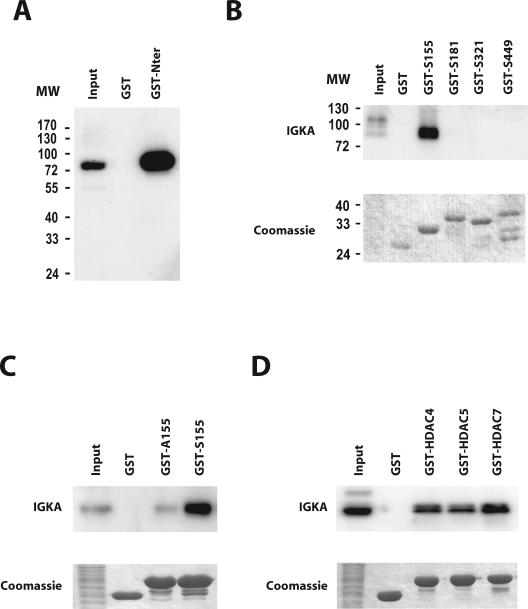
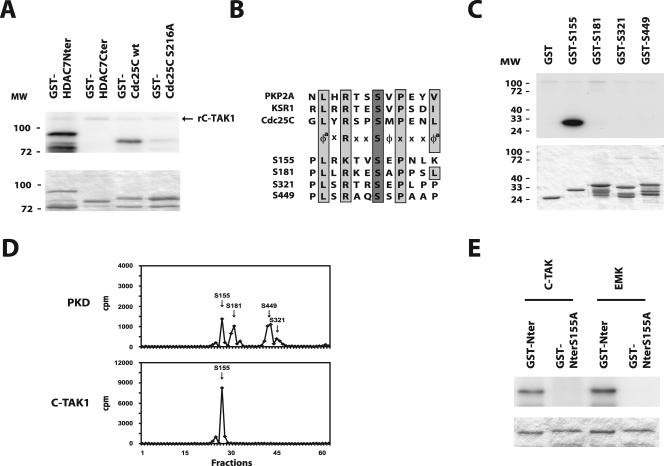
In an attempt to identify the kinases responsible for the constitutive nuclear export of class IIa HDACs, we used the N terminus of HDAC7, which contains the four previously identified phosphorylatable serines (9), in a GST pull-down assay. The material pulled down from unstimulated 293 cell extracts was analyzed by an IGK assay to determine the molecular weight(s) of associated cellular kinase(s). Bands of variable intensity were observed in the input lane, revealing the presence of several constitutively active kinases in the lysate. One band, with an apparent molecular mass of approximately 85 kDa was specifically detected in association with GST-HDAC7Nter (Fig. 3A). Identical results were obtained with extracts from Cos7, NIH 3T3, HeLa, Do11.10, and DPK cells (data not shown).
FIG. 3.
The N terminus of class IIa HDACs associates with an 85-kDa autophosphorylating kinase. (A) The N terminus of HDAC7 was expressed as a GST fusion protein (GST-Nter) and incubated with total cellular extracts from unstimulated, normally growing HEK293 cells. A control reaction was performed in parallel with the GST protein alone. Associated autophosphorylating kinase activities were revealed by IGK assay and autoradiography. The input lane corresponds to 10% of the lysate engaged in the pull-down. (B) GST fusion proteins corresponding to the sequences surrounding the four phosphorylatable serine residues in HDAC7 (GST-S155, GST-S181, GST-S321, and GST-S449) were used in independent pull-down reactions with lysates from HEK293 cells. Autophosphorylating protein kinase activities in the associated material were detected by IGK assay. The gel was stained with Coomassie, dried, and analyzed by autoradiography. The lane marked “input” equals 10% of the cell lysate used in the reaction. (C) A GST fusion protein corresponding to the sequences around Ser155 of HDAC7 (GST-S155) was incubated with HEK293 cell extracts in a pull-down assay. A mutant fusion protein harboring a serine to alanine substitution (GST-A155) was used as control. Pull-down reactions were resolved by SDS-PAGE and analyzed by IGK assay. The gel was stained with Coomassie and dried, and autophosphorylating protein kinase activity was visualized by autoradiography. Input amounts to 10% of the cell lysate were used in each reaction. (D) GST fusion proteins corresponding to Ser246 of HDAC4, Ser259 of HDAC5, and Ser155 of HDAC7 (GST-HDAC4, GST-HDAC5, and GST-HDAC7, respectively) were incubated with extracts from HEK293 cells. A control reaction was performed with the GST protein alone. Pull-down reactions were analyzed by IGK and autoradiography to visualize autophosphorylating kinase activities. The input lane corresponds to 10% of the material used in each reaction. IGKA, IGK assay.
We then expressed the HDAC7 sequences surrounding each phosphorylatable serine residue as GST fusion proteins (respectively, GST-S155, GST-S181, GST-S321, and GST-S449) and examined their ability to recruit the 85-kDa kinase in a pull-down assay. Surprisingly, only the fusion protein corresponding to Ser155 specifically associated with the 85-kDa kinase (Fig. 3B). In addition, when Ser155 was mutated into alanine, association with the constitutively active kinase was greatly impaired, thus indicating that the interaction specifically involves Ser155 (Fig. 3C). To generalize our findings, we tested other class IIa HDACs for their ability to recruit the same 85-kDa kinase activity. Pull-down assays were carried out with GST fusion proteins corresponding to regions of HDAC4 and HDAC5 centered on Ser246 and Ser259, respectively, the residues corresponding to Ser155 of HDAC7. IGK assays revealed that an autophosphorylating kinase with a similar molecular mass of approximately 85 kDa was highly enriched in the material associated with GST-HDAC4, GST-HDAC5, and GST-HDAC7 but not with GST alone (Fig. 3D). These results show that members of class IIa HDACs can associate with a similar, if not identical, 85-kDa autophosphorylating kinase that is constitutively active in numerous mammalian cell lines.
The 85-kDa autophosphorylating kinase activity includes hPar-1 kinases, C-TAK1 and EMK.
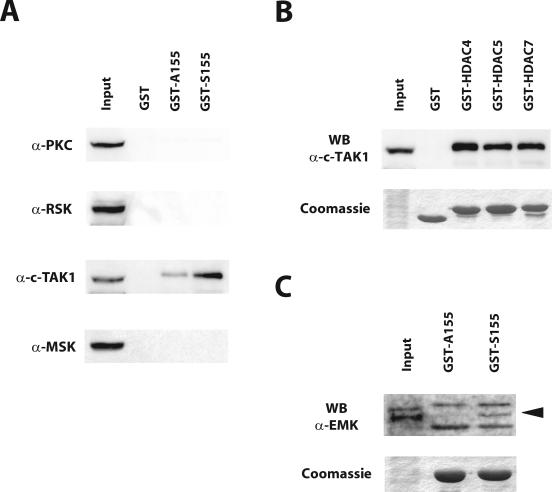
The apparent molecular weight observed in IGK assays precludes the autophosphorylating kinase(s) from being a member of the CaMK or PKD families. To identify this new class IIa HDACs-associated kinase, we screened the human kinome for constitutively active serine/threonine protein kinases with apparent molecular sizes between 80 and 100 kDa (23) and showing autophosphorylation in an IGK assay. This search identified members of the PKC, RSK, MARK/Par-1, and MSK families. We then tested if any of these kinases could interact with Ser155 of HDAC7. Pull-down assays were carried out with GST-S155 or GST-A155 and GST as a control and analyzed by sequential Western blotting with antibodies directed against PKC family members, RSK1/2, the MARK/Par-1 kinase C-TAK1, and MSK1/2. Among these, GST-S155 specifically associated with endogenous C-TAK1 (Fig. 4A). More importantly, mutation of Ser155 into alanine greatly reduced the amount of bound C-TAK1. Because these results strongly suggested that C-TAK1 could be the 85-kDa autophosphorylating kinase described above, we then tested the ability of C-TAK1 to associate with other class IIa members. As expected, GST fusion proteins corresponding to Ser246 and Ser259 of HDAC4 and HDAC5, respectively, were also able to specifically recruit endogenous C-TAK1 in pull-down assays (Fig. 4B).
FIG. 4.
MARK/Par-1 kinases, C-TAK1 and EMK, associate with the N terminus of HDAC7. (A) GST fusion proteins corresponding to Ser155 of HDAC7 (GST-S155), to the serine-to-alanine mutant (GST-A155), or to GST alone were incubated with HEK293 cell extracts in a pull-down assay. Reactions were resolved by SDS-PAGE and analyzed for the presence of endogenous PKC family members (α-PKC), RSK1 and RSK2 (α-RSK), C-TAK1 (α-C-TAK1), and MSK1 and MSK2 (α-MSK) by Western blotting. (B) Binding of endogenous C-TAK1 to GST fusion proteins corresponding to Ser246 of HDAC4, Ser259 of HDAC5, and Ser155 of HDAC7 (GST-HDAC4, GST-HDAC5, and GST-HDAC7, respectively) after a pull-down assay was analyzed by Western blotting or Coomassie staining for loading control. (C) Pull-down reactions described in panel A were analyzed by SDS-PAGE, followed by Western blotting for detection of associated endogenous EMK or by Coomassie staining for control loading. Input equals 10% of the cell extract used in each reaction. The arrow indicates the band corresponding to EMK in the pull-down reactions. Other bands result from cross-reactivity of the anti-EMK antiserum with bacterial proteins copurifying with the GST-fusion proteins. WB, Western blotting.
Other MARK/Par-1 family members include the two closely related hPar-1c and hPar-1b/EMK that phosphorylate the microtubule-associated proteins (MAPs) MAP2, MAP4, and Tau (10). We next investigated if EMK could participate in the 85-kDa kinase activity described above. Pull-down reactions prepared with GST-S155 and GST-A155 were then tested for the presence of EMK. Western blot analysis revealed that while endogenous EMK associated with GST-S155, the interaction was greatly impaired by mutation of Ser155 into alanine (Fig. 4C).
C-TAK1 phosphorylates serine 155 of HDAC7 in vitro.
We next asked whether C-TAK1 could directly phosphorylate the N terminus of HDAC7, which contains the four phosphorylatable serines involved in nucleocytoplasmic shuttling of HDAC7 (i.e., Ser155, Ser181, Ser321, and Ser449). For this purpose, the N- or C-terminal domains of HDAC7 were expressed as GST fusion proteins and incubated with purified recombinant C-TAK1 in an IVK assay. Since C-TAK1 has been shown to phosphorylate Cdc25C on serine 216 (32, 35), we used GST-Cdc25C wt and GST-Cdc25C(S216A) as controls. By comparison with GST-Cdc25C wt, the N terminus of HDAC7 was very efficiently phosphorylated by recombinant C-TAK1 (Fig. 5A). In contrast, C-TAK1 was unable to phosphorylate the C terminus of HDAC7 or the Cdc25C(S216A) mutant (Fig. 5A).
FIG. 5.
C-TAK1 and EMK specifically phosphorylates Ser155 of HDAC7 in vitro. (A) The C- and N-terminal domains of HDAC7 and wild-type or S216A mutant Cdc25C were produced as GST fusion proteins [GST-HDAC7Cter, GST-HDAC7Nter, GST-Cdc25Cwt, and GST-Cdc25C(S216A), respectively]. Equal amounts of purified recombinant proteins were used in IVK assays with recombinant active C-TAK1. IVK reactions were analyzed by SDS-PAGE and Coomassie staining (bottom) prior to autoradiography (top). The arrow indicates the signal resulting from autophosphorylated C-TAK1 (rC-TAK1). (B) Sequences around the four phosphorylatable serine residues in HDAC7 match with the canonical C-TAK1 phosphorylation motif. (C) GST-fusion proteins corresponding to the sequences surrounding Ser155, Ser181, Ser321, and Ser449 of HDAC7 were used as substrates in independent IVK assays with active recombinant C-TAK1. Reactions were resolved by SDS-PAGE, and the gel was stained with Coomassie (bottom), dried, and analyzed by autoradiography (top). (D) The N terminus of HDAC7 was expressed as a GST fusion protein and used as a substrate for recombinant PKD or C-TAK1 in IVK assays. Labeled proteins were resolved by SDS-PAGE and digested with trypsin. The resulting peptides were then separated by HPLC. Positions of the peptides containing each phosphorylatable serine are indicated on the radioactivity profile (as confirmed by mass spectrometry). (E) GST fusion proteins corresponding to the N-terminal domain of HDAC7 (GST-Nter) or to the same region with a serine to alanine substitution at position Ser155 (GST-NterS155A) were phosphorylated in vitro by EMK or C-TAK1. IVK reactions were analyzed by SDS-PAGE and Coomassie staining (bottom) prior to autoradiography (top). MW, molecular weight.
Hydrophobic residues at −5, +1, and + 5, as well as an arginine at the −3 position relative to the phosphorylated serine seem to be crucial for optimal phosphorylation by C-TAK1 (30). Except for the presence of a hydrophobic residue at the +5 and +1 positions, the four phosphorylation sites previously identified in the N terminus of HDAC7 match this consensus phosphorylation motif (Fig. 5B). These observations suggest that C-TAK1 could directly phosphorylate HDAC7 and identify Ser155, Ser181, Ser321, and Ser449 as putative target sites for C-TAK1. To test this hypothesis, GST fusion proteins corresponding to each serine residue (respectively, GST-S155, GST-S181, GST-S321, and GST-S449) were evaluated as potential substrate for C-TAK1 in an IVK assay. Surprisingly, only Ser155 was efficiently phosphorylated by recombinant C-TAK1 (Fig. 5C).
EMK and C-TAK1 display site preference among the four serine residues of HDAC7.
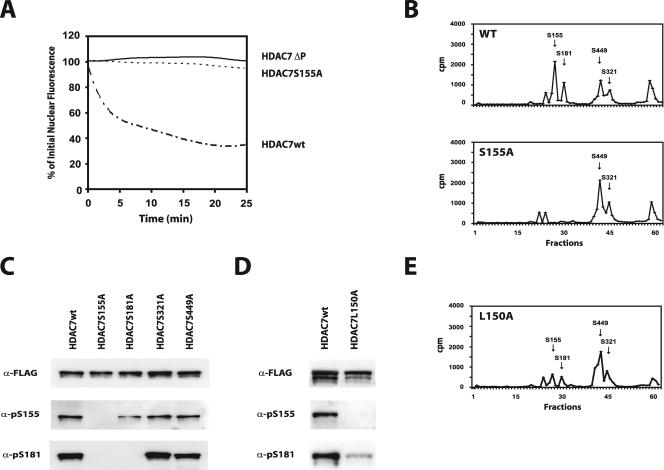
To confirm and extend these observations, we performed an exhaustive analysis of C-TAK1 target sites in the N terminus of HDAC7. This region of HDAC7 was incubated with [γ-32P]ATP and recombinant C-TAK1 in vitro. A control reaction was performed in parallel with PKD, which phosphorylates Ser155, Ser181, Ser321, and Ser449 in vitro (9). Labeled proteins were digested with trypsin, and the resulting peptides were separated by HPLC. Radioactive fractions were then analyzed by mass spectrometry to identify the phosphorylated residue(s). Labeling with PKD led to four major radioactive peaks (Fig. 5D), which corresponded to the formerly identified Ser155, Ser181, Ser321, and Ser449. In contrast, after phosphorylation with C-TAK1, the HPLC profile exhibited a single major phosphorylation peak (Fig. 5D). Mass spectrometry analysis showed that this peak corresponded to phosphorylated Ser155. In contrast to PKD, which targets all four serine residues implicated in the nuclear-cytoplasmic shuttling of HDAC7, these results demonstrate that C-TAK1 specifically phosphorylates Ser155 (Fig. 5C and D).
Our results showed that EMK can associate with the N-terminal Ser155 of HDAC7 (Fig. 4C). In addition, sequences around this serine residue, which are conserved in all class IIa HDACs (Ser246, Ser259, Ser155, and Ser220 of HDAC4, HDAC5, HDAC7, and HDAC9, respectively), match with the KXGS motif phosphorylated by EMK in Tau (10). We therefore investigated whether EMK, similarly to C-TAK1, could phosphorylate Ser155 of HDAC7. As expected, the entire N-terminal domain of HDAC7, which contains all four 14-3-3 binding sites, was very efficiently phosphorylated by purified C-TAK1 or EMK in IVK assays (Fig. 5E, GST-Nter). More importantly, when the sole Ser155 was mutated to alanine, phosphorylation by both kinases was totally abolished (Fig. 5E, GST-NterS155A). Taken together, these results demonstrate the specificity of C-TAK1 and EMK for Ser155 over the three other serine residues previously involved in the nuclear cytoplasmic shuttling of HDAC7.
HDAC7 is phosphorylated on Ser155 in vivo.
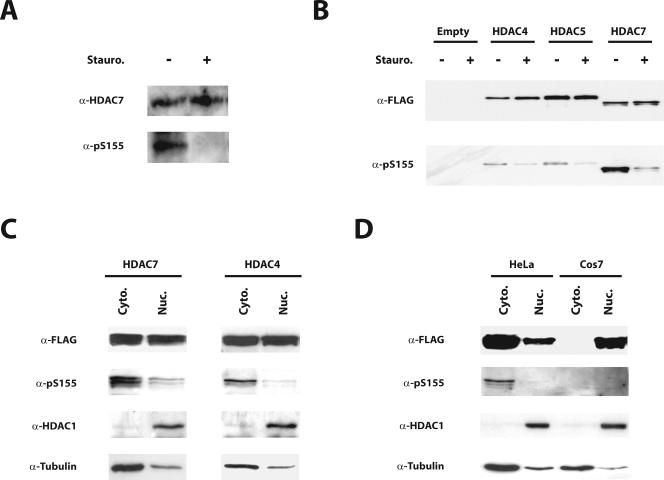
We next examined the phosphorylation status of Ser155 on endogenous HDAC7 in vivo. For this purpose, we first developed an antibody that specifically recognizes the phosphorylated form of HDAC7 Ser155 (data of antibody characterization available on request). Total extracts from Do.11.10 T-cell hybridomas, which express high levels of endogenous HDAC7, were examined by Western blotting with the phospho-specific antibody for Ser155. As shown in Fig. 6A, strong basal phosphorylation of HDAC7 was consistently observed at Ser155 in normally growing cells (Fig. 6A, α-pS155). More importantly, Ser155 phosphorylation was totally lost when cells were treated with staurosporine.
FIG. 6.
HDAC7 is phosphorylated on Ser155 in vivo. (A) Total cell lysates were prepared from Do11.10 cells which were first treated with staurosporine (+) for 1 h or left untreated (−). Phosphorylation of endogenous HDAC7 was detected by Western blotting with the antibody specific for phosphorylated Ser155 (α-pS155). As a loading control, the same membrane was stripped and immunoblotted with an antibody against endogenous HDAC7 (α-HDAC7). (B) HEK293 cells were transiently transfected with expression vectors encoding FLAG-tagged versions of wild-type HDAC4, HDAC5, and HDAC7 or the empty vector (Empty). Total cellular extracts were analyzed by SDS-PAGE and subjected to immunoblotting using antibodies directed either against the FLAG epitope (α-FLAG) or phosphorylated Ser155 (α-pS155). (C) Nuclear (Nuc) and cytoplasmic (Cyto) extracts were prepared from HEK293 cells transiently expressing FLAG-tagged HDAC7 or HDAC4. Both extracts were analyzed by Western blotting with antibodies against the FLAG epitope (α-FLAG) and phosphorylated Ser155 (α-pHDACs). As a control, extracts were also analyzed by Western blotting with antisera against the cytoplasmic protein tubulin (α-tubulin) and the nuclear protein HDAC1 (α-HDAC1). (D) Nuclear (Nuc) and cytoplasmic (Cyto) extracts were prepared from HeLa or Cos7 cells transiently expressing FLAG-tagged HDAC7. Equal protein amounts from each extract were analyzed by Western blotting with antisera against the FLAG epitope (α-FLAG) and phosphorylated Ser155 (α-pS155). As a control, extracts were also analyzed by Western blotting with antibody against the cytoplasmic protein tubulin (α-tubulin) and the nuclear protein HDAC1 (α-HDAC1). Stauro, staurosporine.
Because sequences around Ser155 of HDAC7 are highly conserved in other members of the class IIa, the phospho-specific antibody against Ser155 also recognizes the corresponding phosphorylated serines in HDAC4 and HDAC5 (Ser246 and Ser259, respectively). To generalize our observations to other members of the class IIa, FLAG-tagged versions of HDAC4, HDAC5, and HDAC7 were transiently expressed in HEK293 cells and examined by Western blotting using the phospho-specific antibody (Fig. 6B, α-pS155). Confirming our observations on endogenous HDAC7, all three class IIa HDACs showed basal phosphorylation of their respective serine residue, which was significantly reduced upon treatment with staurosporine.
The above findings suggest that phosphorylation of Ser155 in HDAC7 (or Ser246 and Ser259 in HDAC4 and HDAC5, respectively) might be important for constitutive nuclear export. To test this hypothesis, we fractionated extracts from HEK293 cells transiently transfected with FLAG-tagged HDAC4 or HDAC7 into nuclear and cytoplasmic fractions. Confirming our immunofluorescence data (data available on request), comparable amounts of HDAC4- or HDAC7-FLAG were found in both fractions (Fig. 6C). However, Western blot analysis with the phospho-specific antibody revealed that HDAC7-phosphorylated at Ser155 and HDAC4-phosphorylated at Ser246 were highly enriched in the cytoplasm (Fig. 6C). To confirm and extend these observations, we performed a similar experiment in HeLa and Cos7 cells expressing FLAG-tagged HDAC7. In accordance with the immunofluorescence data (Fig. 1 and 2), HDAC7 localized primarily in the cytoplasm of HeLa cells, where it is phosphorylated on Ser155 (Fig. 6D). In contrast, HDAC7 was found almost exclusively in the nucleus of Cos7 cells, and no phosphorylation of Ser155 could be detected (Fig. 6D). Taken together, these results establish a strong correlation between phosphorylation of Ser155 and cytoplasmic localization of HDAC7.
EMK and C-TAK1 alter nuclear export of class IIa HDACs and regulate their repressive activity.
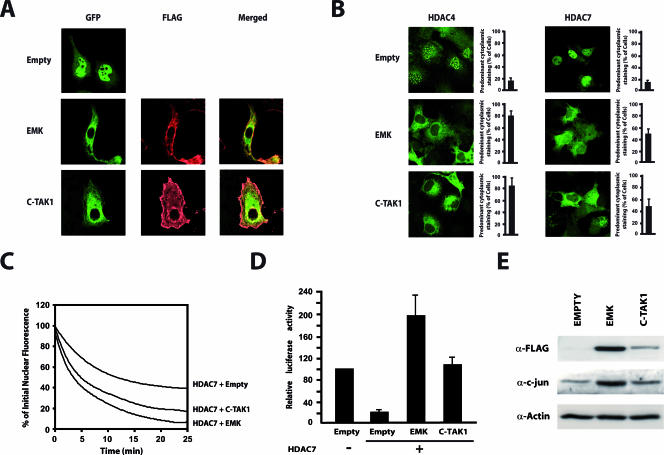
Our results so far strongly suggest that in the absence of extracellular stimuli, EMK and C-TAK1 control localization of class IIa HDACs by phosphorylating their most N-terminal 14-3-3 binding sites (i.e., Ser246, Ser259, and Ser155 in HDAC4, HDAC5, and HDAC7, respectively). In Cos7 cells, class IIa HDACs are mainly found in the nucleus, probably because the mechanisms controlling their nuclear export are poorly efficient in these cells. To test our model, we examined the steady-state localization of HDAC7 in the presence of overexpressed EMK or C-TAK1 in Cos7 cells. As observed before, HDAC7 was mainly found in the nucleus of Cos7 cells when expressed alone (Fig. 7A). In contrast, coexpression of EMK and C-TAK1 induced dramatic cytoplasmic accumulation of HDAC7. However, we did not observe convincing colocalization of HDAC7 with either MARK members in the cytosol (Fig. 7A, merged). To generalize these observations, we tested the effect of EMK and C-TAK1 on the subcellular localization of HDAC4. By comparison with HDAC7, HDAC4 was even more sensitive to cytoplasmic retention by MARK kinases, with approximately 80% of cells showing predominant cytoplasmic staining (Fig. 7B). We next tested whether cytoplasmic accumulation of class IIa HDACs induced by MARK/Par-1 kinases resulted directly from an increase in their nuclear export. FLIP experiments revealed a remarkable increase in the nucleocytoplasmic efflux of HDAC7 by both EMK and C-TAK1, resulting in less than 10% of the initial fluorescence left in the nucleus after 25 min (Fig. 7C).
FIG. 7.
EMK and C-TAK1 control nuclear export of class IIa HDACs and regulate their repressive activity. (A) Cos7 cells were transfected with expression vectors for GFP-HDAC7 and FLAG-tagged EMK or C-TAK1. The intracellular localization of HDAC7 was detected by direct immunofluorescence (GFP) while MARKs were revealed by indirect immunofluorescence using an AlexaFluor 568-labeled anti-FLAG antibody (FLAG). (B) Cos7 cells were transfected with expression vectors for GFP-HDAC4 or GFP-HDAC7 and constructs for EMK or C-TAK1. The subcellular distribution of the GFP-fused HDACs proteins was examined by confocal immunofluorescence microscopy. Bar histograms represent the mean percentages of cells showing predominant cytoplasmic staining. (C) A construct encoding GFP-HDAC7 was transfected into Cos7 cells, either with expressing vectors for EMK, C-TAK1, or a control plasmid (Empty). The cytoplasm of GFP-positive cells was repeatedly bleached, and FLIP measurements were performed as described in Materials and Methods. Analyses of the FLIP data from three independent experiments are shown as fit curves. (D) Do11.10 cells were transiently transfected with a luciferase reporter plasmid driven by the Nur77 promoter and the expression plasmids for the indicated proteins. Luciferase activities are presented relative to the basal luciferase activity of the reporter. Results are from five independent experiments, each performed in triplicate. (E) Total cellular lysates were prepared from Cos7 cells transiently expressing FLAG-tagged versions of EMK and C-TAK1. Cell lysates were examined by Western blotting with antisera for c-jun, FLAG, and actin.
In T cells, we have shown that HDAC7 associates with MEF2D to repress the Nur77 promoter and that this inhibitory action is relieved by T-cell receptor signaling which induces HDAC7 phosphorylation and cytoplasmic relocalization. (8). As EMK and C-TAK1 promote nuclear export of class IIa HDACs, both kinases would be expected to overcome the inhibitory activity of HDAC7 over the Nur77 promoter, even in the absence of T-cell receptor signaling. To address this question, we used the isolated Nur77 promoter in reporter assays. As expected, the transcriptional activity of the Nur77 promoter was strongly inhibited by HDAC7 (Fig. 7D). Overexpression of EMK or C-TAK1 totally reversed the repressive effect of wild-type HDAC7. Of note, EMK increased the transcriptional activity of Nur77 up to twofold above levels observed in the absence of HDAC7.
To further assess the functional consequences of phosphorylation of class IIa HDACs by MARK/Par-1 kinases, we examined the ability of EMK and C-TAK1 to activate c-jun expression, another class IIa HDAC-repressed gene (45). For this purpose, EMK and C-TAK1 were independently expressed in Cos7 cells, and levels of endogenous c-jun were examined by Western blotting. As expected, ectopic expression of EMK or C-TAK1 was associated with a marked increase in c-jun levels (Fig. 7E).
Taken together, these data demonstrate that the MARK/Par-1 kinases EMK and C-TAK1 are physiologically relevant kinases for class IIa HDACs and strongly support a novel role for these kinases in gene regulation.
EMK and C-TAK1 regulate phosphorylation and cytoplasmic localization of class IIa HDACs in the absence of extracellular stimuli.
We have previously shown that PKD efficiently phosphorylates Ser155 of HDAC7, even when the leucine residue at position −5 (Leu150) is replaced with an alanine (9). Interestingly, all C-TAK1 substrates identified so far have a leucine at position −5 of the targeted serine (Fig. 5B). While testing the importance of this leucine residue in EMK and C-TAK1 target recognition, we found out that the substitution of HDAC7 Leu150 to alanine totally abolished phosphorylation of Ser155 by both kinases (data available on request).
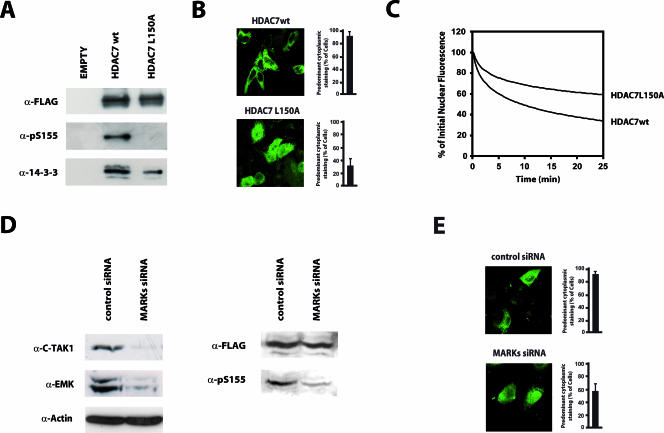
Based on these results, we generated an HDAC7 mutant specifically deficient for phosphorylation by EMK/C-TAK1 where Leu150 was mutated to alanine [HDAC7(L150A)]. We examined the in vivo phosphorylation of Ser155 in the context of this mutant using the phospho-specific antibody. As observed above (Fig. 6), HDAC7wt showed strong basal phosphorylation of Ser155 and robust association with 14-3-3 proteins (Fig. 8A). Interestingly, the L150A mutation totally inhibited phosphorylation of Ser155. In addition, the same mutation also reduced association with 14-3-3 proteins.
FIG. 8.
EMK and c-TAK1 phosphorylate Ser155 of HDAC7 in vivo. (A) FLAG-tagged versions of wild-type HDAC7 (HDAC7wt) or the HDAC7(L150A) mutant were transiently expressed in HEK293 cells, immunoprecipitated, and examined by Western blot analysis with antibodies directed against the FLAG epitope (α-FLAG), 14-3-3 family members (α-14-3-3), or HDAC7 phosphorylated at Ser155 (α-pS155). (B) HeLa cells were transfected with constructs encoding GFP fusion proteins of wild-type HDAC7 (HDAC7wt) or the HDAC7(L150A) mutant. Subcellular localization of the fluorescent proteins was observed by confocal microscopy. (C) Cos7 cells were transfected with constructs encoding GFP-tagged wild-type or L150A HDAC7. Cytoplamsic photobleaching and FLIP measurements were performed as described in Materials and Methods. Graphical analyses of the FLIP data from at least three independent experiments are shown as fit curves. (D) HeLa cells were transfected with FLAG-tagged HDAC7, along with pooled siRNA against EMK and C-TAK1 or with control siRNA. The next day, cells were transfected with a mix of siRNAs targeting an alternative sequence both in EMK and C-TAK1 or with control siRNA (see Materials and Methods). Seventy-two hours after the initial transfection, cells were harvested and lysed. Whole-cell lysates were analyzed by Western blotting with antibodies against EMK, C-TAK1, actin, the FLAG epitope or phosphorylated Ser155. (E) HeLa cells were transfected with an expression vector for GFP-HDAC7 along with pools of siRNAs directed against two different sequences of both EMK and C-TAK1 or control siRNA, as described in panel D. Sixty hours after the initial transfection, localization of GFP-HDAC7 was examined by confocal immunofluorescence microscopy. Bar histograms represent the mean percentages of cells showing predominant cytoplasmic staining.
Since the HDAC7(L150A) mutant is not phosphorylated on Ser155, it should consequently be impaired in its ability to exit the nucleus. To verify this hypothesis, we examined the subcellular localization of the L150A mutant in HeLa cells, where HDAC7 is very efficiently exported from the nucleus. Indeed, as shown above (Fig. 1C), wild-type HDAC7 was cytoplasmic in the majority of normally growing HeLa cells (Fig. 8B). By contrast, only 30% of HeLa cells expressing the L150A HDAC7 mutant showed predominant cytoplasmic staining (Fig. 8B). Interestingly, FLIP experiments revealed a clear difference between the abilities of wild-type HDAC7 and the L150A mutant to exit the nucleus. For wild-type HDAC7, half of the nuclear fluorescence was lost in about 10 min after bleaching the cytoplasm, and 35% of the initial nuclear fluorescence was left after 25 min (Fig. 8C). In contrast, the loss in nuclear fluorescence was much slower for HDAC7(L150A), which reached a plateau of ∼65% of its initial value after 25 min (Fig. 8C). These experiments show that the L150A HDAC7 mutant is greatly impaired in its ability to exit the nucleus, which results in an altered steady-state subcellular localization. These results thus strongly suggest that EMK and/or C-TAK1 specifically target Ser155 of HDAC7 to control its nuclear export.
To establish this hypothesis more firmly, we used RNAi to inhibit endogenous C-TAK1 and EMK activities in HeLa cells. A combination of siRNAs directed against both C-TAK1 and EMK reduced the endogenous levels of both kinases (Fig. 8D, left). Coincident with this reduction, we observed a substantial decrease in the phosphorylation of Ser155 in HDAC7 (Fig. 8D, right). As expected, knockdown of EMK and C-TAK1 also altered subcellular distribution of HDAC7. Indeed, when HeLa cells were cotransfected with GFP-HDAC7 and siRNAs against EMK and C-TAK, the proportion of cells showing a predominant cytoplasmic staining decreased significantly (Fig. 8E).
Altogether, these data strongly suggest that MARK/Par-1 kinases EMK and C-TAK1 phosphorylate class IIa HDACs on their most upstream 14-3-3 binding site and regulate their constitutive nuclear export in vivo.
The 14-3-3 binding sites are hierarchically phosphorylated in HDAC7.
The biological significance behind the specific constitutive phosphorylation of HDAC7 Ser155 (and corresponding HDAC4 Ser246, HDAC5 Ser259, and HDAC9 Ser220) by MARK/Par-1 kinases remained elusive. To address the role of Ser155 phosphorylation in the signal-independent nuclear efflux of HDAC7, we examined the dynamic nuclear export of an HDAC7 protein where Ser155 was mutated into alanine [HDAC7(S155A)]. Results from FLIP analysis were compared with data obtained with wild-type HDAC7 and the HDAC7ΔP mutant. Surprisingly, mutation of Ser155 alone had an effect comparable to mutating simultaneously the four serine residues and almost completely abolished constitutive nuclear export, as demonstrated by a constant postbleach relative nuclear fluorescence in HDAC7(S155A)-expressing cells (Fig. 9A). This observation demonstrates that Ser155 plays a dominant role in HDAC7 constitutive phosphorylation, association with 14-3-3, and nuclear export.
FIG. 9.
The 14-3-3 binding sites in HDAC7 are hierarchically phosphorylated. (A) Cos7 cells were transfected with constructs expressing GFP fusion proteins corresponding to wild-type HDAC7 (HDAC7wt), a mutant of HDAC7 in which the residues Ser155 was mutated to alanine alone [HDAC7(S155A)], or together with Ser181, Ser321, and Ser449 (HDAC7ΔP). Forty-eight hours posttransfection, the cytoplasm of fluorescent cells was bleached, and relative loss of fluorescence in the nuclear region was measured, as described in Materials and Methods. FLIP data are represented as nonlinear fit curves according to the legend at the right of the graph. (B) HEK293 cells expressing wild-type or S155A HDAC7 were labeled in vivo with [32P]orthophosphate. The labeled proteins were purified by immunoaffinity, digested with trypsin, and examined by HPLC analysis as described in Materials and Methods. (C) Expression constructs for FLAG-tagged wild-type HDAC7 (HDAC7wt) or mutant of HDAC7 in which the residues Ser155, Ser181, Ser321, or Ser449 were mutated to alanine independently [HDAC7(S155A), HDAC7(S181A), HDAC7(S321A), and HDAC7(S449A), respectively] or together (HDAC7ΔP) were transiently expressed in HEK293 cells. Cell extracts were examined by Western blotting with antibodies directed against the FLAG epitope (α-FLAG) or the phosphorylated forms of Ser155 (α-pS155) or Ser181 (α-pS181). (D) HEK293 cells were transiently transfected with expression vectors encoding FLAG-tagged versions of wild-type HDAC7 (HDAC7wt) or the L150A mutant [HDAC7(L150A)]. Total cellular extracts were analyzed by SDS-PAGE and subjected to immunoblotting using antibodies directed against either the FLAG epitope (α-FLAG) or HDAC7 phosphorylated at Ser155 (α-pS155) or Ser181 (α-pS181). (E) HEK293 cells expressing the L150A HDAC7 mutant were labeled in vivo with [32P]orthophosphate. The labeled protein was purified by immunoaffinity, digested with trypsin, and examined by HPLC analysis as described in Materials and Methods.
To further address the importance of Ser155, we examined the phosphorylation pattern of the HDAC7(S155A) mutant protein in vivo. For this purpose, wild-type and S155A mutant HDAC7 proteins were metabolically labeled with [32P] orthophosphate, affinity purified, and trypsin digested for HPLC analysis. As shown in Fig. 9B, wild-type HDAC7 was found to be constitutively phosphorylated on five major sites, among which the four serine residues previously implicated in subcellular trafficking of HDAC7, i.e., Ser155, Ser181, Ser321, and Ser449 were identified. Very unexpectedly, the S155A mutant exhibited a drastically different phosphorylation pattern, as the alanine mutation at Ser155 also resulted in a complete loss of phosphorylation at Ser181 (Fig. 9B).
We next tested whether phosphorylation of Ser181 could be also dependent on the two other 14-3-3 sites, Ser321 or Ser449. HDAC7 mutants, where each of the phosphorylatable serine residues was independently mutated to alanine [HDAC7(S155A), HDAC(S181A), HDAC7(S321A), and HDAC7(S449A)] were thus examined by Western blotting with antibodies specific for the phosphorylated forms of Ser155 and Ser181 (Fig. 9C) (data of antibody characterization available on request). As expected, phosphorylation of Ser155 was detected for all constructs except for HDAC7(S155A). Basal phosphorylation of Ser181 was undetectable after staurosporine treatment (data not shown) or in the HDAC7(S181A) mutant, confirming the specificity of the phospho-Ser181 antibody (Fig. 9C). In accordance with the HPLC data, phosphorylation of Ser181 was totally abolished when Ser155 was mutated into alanine (Fig. 9C). Phosphorylation of Ser181 was uniquely dependent on Ser155 as it was unaltered by the S321A or S449A mutations. These observations thus confirm that the presence and/or the phosphorylation of Ser155 are required for subsequent phosphorylation of Ser181 in vivo.
To discriminate between both hypotheses, we examined the phosphorylation levels of Ser181 in the HDAC7(L150A) mutant, where Ser155 is present but poorly phosphorylated (Fig. 8A) both by Western blotting and HLPC analysis (Fig. 9D and E, respectively). Interestingly, the L150A mutant also showed a concomitant reduction of Ser181 phosphorylation (Fig. 9D and E). These results demonstrate that basal phosphorylation of Ser155 is required for phosphorylation of Ser181 and raised the exciting possibility that class IIa HDACs may be regulated through a process of hierarchical phosphorylation.
DISCUSSION
Signal-dependent cytoplasmic relocalization of class IIa HDACs is achieved through phosphorylation of multiple serine residues by members of the CaMK and PKD families. In addition, some recent observations suggest that the subcellular localization of class IIa HDACs might also be constitutively regulated in a signal-independent manner (3, 15, 21). Our study identifies EMK and C-TAK1, two members of the MARK/Par-1 family, as new kinases interacting with the N-terminal adapter domain of class IIa HDACs. We have extended this finding by showing that EMK and C-TAK1 phosphorylate class IIa HDACs on one of their multiple 14-3-3 binding sites and alter their subcellular localization and repressive function in normally growing cells. Our study identified a new biological function for Par-1 kinases, as we provide direct experimental evidence that they can play a role in regulating gene transcription. An intriguing question that remains to be addressed is whether class IIa HDACs could play noncanonical roles in any Par-1 functions, such as regulating cell polarity, microtubules dynamic and stability, cell cycle progression and intracellular signaling (39).
Par-1 kinases substrate specificity.
The human MARK/Par-1 protein kinases include hPar-1a/C-TAK1 (MARK3/p78), hPar-1b/EMK (MARK2), hPar-1c (MARK1), and hPar-1d (MARK4/MARKL1). EMK and hPar-1c were originally identified based on their ability to phosphorylate Tau and the related MAP proteins MAP2 and MAP4 on their homologous KXGS motif (10). Recently, hPar-1d was also shown to phosphorylate the same KXGS motif (41). Strikingly, the KTVS motif around Ser155 of HDAC7 (conserved in other class IIa HDACs as KTAS) is very homologous to the KXGS consensus for hPar-1b, -c, and -d. Although it remains to be formally tested, it is logical to speculate that, in addition to EMK and C-TAK1, hPar-1c and hPar-1d could also phosphorylate class IIa HDACs.
Aside from other family members, C-TAK1/Par-1a exhibits specific substrate requirements. An extensive mutational analysis has defined φaXRXXSφXXXφa(where φ and φa are, respectively, a hydrophobic and a hydrophobic aliphatic residue) as its optimal substrate phosphorylation motif (30). From this study, the arginine residue at the −3 position relative to the phosphorylated serine, as well as hydrophobic amino acids at the +1 and +5 positions, was proven to be essential for efficient phosphorylation by C-TAK1. Surprisingly, none of these crucial amino acids is found around the serine residue phosphorylated by Par-1 kinases in class IIa HDACs (Ser246, Ser259, Ser155, and Ser220 of HDAC4, HDAC 5, HDAC7, and HDAC9, respectively) (Fig. 5B). Moreover, because they contain a hydrophobic residue at the +5 position, motifs around other phosphorylatable serines of HDAC7, Ser181, Ser321, and Ser449 (and corresponding residues in other class IIa HDACs) are very homologous to the optimal C-TAK1 consensus. Nonetheless, C-TAK1 does not phosphorylate (Fig. 5C and D) nor does it interact with (data not shown) any of these serine residues in vitro.
Previously identified substrates of C-TAK1 include the Cdc25C phosphatase (32, 35), the tyrosine phosphatase PTPH1 (51), the mitogen-activated protein kinase scaffolding protein KSR1, and plakophilin 2 (29, 30). Strikingly, for all known substrates of C-TAK1, the phosphorylated residue generates a 14-3-3 binding site. The identification of class IIa HDACs as C-TAK1 substrates thus provides additional evidence that C-TAK1 may function as a master regulator in the subcellular distribution of several proteins with diverse cellular functions. Identification of additional endogenous substrates for C-TAK1 should confirm this hypothesis.
Convergence of multiple families of protein kinases on class IIa HDACs.
The 14-3-3 binding sites of class IIa HDACs are efficiently phosphorylated by members of multiple families of protein kinases. In response to Ca2+ signaling, CaMKI, -II, and -IV have proven to be able to induce nuclear exclusion and cytoplasmic accumulation of class IIa HDACs in various cell types (49). Similarly, PKC signaling leads to the activation of PKD1, which directly phosphorylates the multiple signal-responsive serine residues on class IIa HDACs (9, 42). We have accumulated new evidence that PKD2 and PKD3, the two other PKD family members, can also do so (T. Seufferlein and F. Dequiedt, unpublished observations). Here, we show that EMK and C-TAK1, two members of the MARK/Par-1 family, can target one of the 14-3-3 binding sites in the N terminus of HDAC4 and HDAC7 (and presumably other class IIa members). It thus appears that the adapter domain of class IIa HDACs is targeted by multiple members of multiple protein kinase families. This has been illustrated in an elegant expression screen which was published while this article was in preparation (4). Looking for modulators of HDAC5 phosphorylation, the authors identified multiple protein kinases, including MARK2.
Experimentally, the convergence of multiple protein kinase families on the adapter domain of class IIa HDACs makes it difficult to fortify conclusions with convincing loss-of-function data. Knockdown of either EMK or C-TAK1 alone had no impact on the phosphorylation of Ser155 in HDAC7 (data not shown). As shown in Fig. 9D, only simultaneous knockdown of both EMK and C-TAK1 led to a reduced Ser155 phosphorylation. The existence of multiple class IIa HDACs kinases confirms that these enzymes are regulated by different signaling pathways and emphasizes the importance that class IIa HDACs may have in various genetic programs. In addition, it also highlights the fact that the functional relevance of each proposed HDAC kinase should be envisioned in a cellular/signaling context-dependent manner.
Multisite and hierarchical phosphorylation of class IIa HDACs.
Multisite phosphorylation often offers a sophisticated means to regulate protein functions. In most cases, each site impacts differently on a specific property of the protein, such as stability, cellular localization, catalytic activity, etc. The N terminus of class IIa HDACs contains multiple phosphorylation sites, with HDAC7 having four identified phosphorylatable serine residues within a 300-amino-acid region. Prior to the current study, these sites were thought to be phosphorylated indistinguishably by the same protein kinases in response to the same signals and equally important for class IIa HDACs nuclear export. In this context, why class IIa HDACs would have multiple phosphorylation sites in their N terminus remained obscure. Here, we establish the uniqueness and prominence of the most upstream phosphorylatable serine residue (Ser246, Ser259, and Ser155 in HDAC4, HDAC5, and HDAC7, respectively). First, we demonstrate that this site is uniquely and specifically phosphorylated by hPar-1 kinases in a constitutive manner. In addition, we establish the primary role of this residue in the constitutive nuclear export of class IIa HDACs. In exploring the functional relevance of these observations for HDAC7, we unexpectedly discovered that basal phosphorylation of Ser155 was a prerequisite to the phosphorylation of Ser181. For the first time, these findings demonstrate that class IIa HDACs undergo hierarchical phosphorylation of their 14-3-3 binding sites. Although it remains to be formally tested, it is tempting to speculate that similar hierarchical phosphorylations exist for other sites on HDAC7 and for other members of the class IIa family.
Par-1 kinases are constitutively active enzymes, and, accordingly, we observed constitutive phosphorylation of the most upstream phosphorylatable serine residue in class IIa HDACs. In this context, phosphorylation of the Par-1 target site has two important functions. First, it recruits 14-3-3 proteins and regulates the nucleocytoplasmic fluxes of the class IIa HDACs in the absence of any extracellular signaling. Second, phosphorylation of this particular site in HDAC7 allows subsequent phosphorylation of another 14-3-3 binding site, Ser181, which is also important for nuclear efflux (data not shown). These findings thus confer a central role to the most upstream phosphorylatable serine residue of class IIa HDACs. The precise mechanism by which phosphorylation of this residue would promote phosphorylation at other sites of class IIa HDACs remains unknown. Incorporation of a phosphate group at this site could alter conformation of class IIa HDACs and render other serine residues accessible. Alternatively, phosphorylation of Ser155 could also create a docking site for other, still unknown, kinases that require a priming phosphorylation in order to phosphorylate their substrate (e.g., glycogen synthase kinase 3). Finally, binding of 14-3-3 proteins at this serine residue, subsequently to its phosphorylation, could induce drastic conformational changes and have a similar effect. Mutants of class IIa HDACs that can still be phosphorylated at their most upstream serine residue, but deficient in 14-3-3 recruitment, should allow to discriminate between these options.
In this study, we provide multiple demonstrations that Ser155 phosphorylation is crucial for HDAC7 nuclear efflux. First, we show that phosphorylation of Ser155 coincides with cytoplasmic localization (Fig. 6C and D). In addition, mutation of Ser155 to alanine has an effect comparable to mutating the four serine residues simultaneously (Fig. 9A). However, phosphorylation of Ser155 has no impact on the phosphorylation of Ser321 or Ser449 of HDAC7 (Fig. 9B). These observations thus suggest that Ser155/Ser181 phosphorylation is necessary for nuclear export of HDAC7. Interestingly, HDAC7 nuclear localization signal (NLS) spans amino acids 160 to 168, exactly between Ser155 and Ser181. One speculative model would be that Ser155 would function as a “gatekeeper” (48) whose constitutive phosphorylation is necessary for binding of a 14-3-3 dimer but may not be sufficient for nuclear export. By a still unknown mechanism, phosphorylation of Ser155 would favor subsequent phosphorylation of Ser181 by a signal-responsive kinase or a still unidentified constitutively active kinase, depending on the cellular context. Dual phosphorylation of Ser155/Ser181 would simultaneously engage both monomeric subunits of a 14-3-3 dimer, which would prevent recognition of the NLS by importin α. Whether masking of the NLS by a 14-3-3 dimer would be sufficient for nuclear export or would necessitate signal-mediated phosphorylation of Ser321 and/or Ser449 remains unclear.
Our study provides experimental evidence that the multiple phosphorylation sites in class IIa HDACs display specific properties. In this context, combinatorial phosphorylation of these enzymes by multiple kinases would allow for a flexible and sophisticated control of their functions. Sequential or/and coordinated actions of the various protein kinases on the N terminus of class IIa HDACs would constitute a tightly regulated mechanism to rapidly, adequately, and reversibly induce expression of specific target genes in response to specific signals. Although the pieces are starting to fall into place, more efforts are required to fully understand regulation of class IIa HDACs by multisite phosphorylation.
Acknowledgments
This work was supported by the Fond National de la Recherche Scientific (F.N.R.S.), the Belgian Federation against Cancer, the Deutsche Forschungsgemeinschaft (SFB 518/B3 and SE 676/5-1), the Interuniversity Attraction Poles Programme, Belgian Science Policy, and the NIH (H.P.-W.). F.D. is Research Associate, M.M. is Research Fellow, and R.K. is Research Director of the F.N.R.S. H.P.-W. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Bachmann, M., H. Hennemann, P. X. Xing, I. Hoffmann, and T. Moroy. 2004. The oncogenic serine/threonine kinase Pim-1 phosphorylates and inhibits the activity of Cdc25C-associated kinase 1 (C-TAK1): a novel role for Pim-1 at the G2/M cell cycle checkpoint. J. Biol. Chem. 279:48319-48328. [DOI] [PubMed] [Google Scholar]
- 2.Bakin, R. E., and M. O. Jung. 2004. Cytoplasmic sequestration of HDAC7 from mitochondrial and nuclear compartments upon initiation of apoptosis. J. Biol. Chem. 279:51218-51225. [DOI] [PubMed] [Google Scholar]
- 3.Bolger, T. A., and T. P. Yao. 2005. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J. Neurosci. 25:9544-9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, S., S. Bezprozvannaya, S. Li, and E. N. Olson. 2005. An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc. Natl. Acad. Sci. USA 102:8120-8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, S., T. A. McKinsey, C. L. Zhang, J. A. Richardson, J. A. Hill, and E. N. Olson. 2004. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24:8467-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chawla, S., P. Vanhoutte, F. J. Arnold, C. L. Huang, and H. Bading. 2003. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J. Neurochem. 85:151-159. [DOI] [PubMed] [Google Scholar]
- 7.Davis, F. J., M. Gupta, B. Camoretti-Mercado, R. J. Schwartz, and M. P. Gupta. 2003. Calcium/calmodulin-dependent protein kinase activates serum response factor transcription activity by its dissociation from histone deacetylase, HDAC4. Implications in cardiac muscle gene regulation during hypertrophy. J. Biol. Chem. 278:20047-20058. [DOI] [PubMed] [Google Scholar]
- 8.Dequiedt, F., H. Kasler, W. Fischle, V. Kiermer, M. Weinstein, B. G. Herndier, and E. Verdin. 2003. HDAC7, a thymus-specific class II histone deacetylase, regulates Nur77 transcription and TCR-mediated apoptosis. Immunity 18:687-698. [DOI] [PubMed] [Google Scholar]
- 9.Dequiedt, F., J. Van Lint, E. Lecomte, V. Van Duppen, T. Seufferlein, J. R. Vandenheede, R. Wattiez, and R. Kettmann. 2005. Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis. J. Exp. Med. 201:793-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drewes, G., A. Ebneth, U. Preuss, E. M. Mandelkow, and E. Mandelkow. 1997. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89:297-308. [DOI] [PubMed] [Google Scholar]
- 11.Fischle, W., F. Dequiedt, M. Fillion, M. J. Hendzel, W. Voelter, and E. Verdin. 2001. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J. Biol. Chem. 276:35826-35835. [DOI] [PubMed] [Google Scholar]
- 12.Fischle, W., F. Dequiedt, M. J. Hendzel, M. G. Guenther, M. A. Lazar, W. Voelter, and E. Verdin. 2002. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9:45-57. [DOI] [PubMed] [Google Scholar]
- 13.Grozinger, C. M., and S. L. Schreiber. 2000. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA 97:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurov, J. B., T. S. Stappenbeck, C. M. Zmasek, L. S. White, S. H. Ranganath, J. H. Russell, A. C. Chan, K. M. Murphy, and H. Piwnica-Worms. 2001. Immune system dysfunction and autoimmune disease in mice lacking Emk (Par-1) protein kinase. Mol. Cell. Biol. 21:3206-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao, H.-Y., A. Verdel, C.-C. Tsai, C. Simon, H. Juguilon, and S. Khochbin. 2001. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276:47496-47507. [DOI] [PubMed] [Google Scholar]
- 16.Kirsh, O., J. S. Seeler, A. Pichler, A. Gast, S. Muller, E. Miska, M. Mathieu, A. Harel-Bellan, T. Kouzarides, F. Melchior, and A. Dejean. 2002. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemercier, C., A. Verdel, B. Galloo, S. Curtet, M.-P. Brocard, and S. Khochbin. 2000. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem. 275:15594-15599. [DOI] [PubMed] [Google Scholar]
- 18.Li, X., S. Song, Y. Liu, S.-H. Ko, and H.-Y. Kao. 2004. Phosphorylation of the histone deacetylase 7 modulates its stability and association with 14-3-3 proteins. J. Biol. Chem. 279:34201-34208. [DOI] [PubMed] [Google Scholar]
- 19.Linseman, D. A., C. M. Bartley, S. S. Le, T. A. Laessig, R. J. Bouchard, M. K. Meintzer, M. Li, and K. A. Heidenreich. 2003. Inactivation of the myocyte enhancer factor-2 repressor histone deacetylase-5 by endogenous Ca2+/calmodulin-dependent kinase II promotes depolarization-mediated cerebellar granule neuron survival. J. Biol. Chem. 278:41472-41481. [DOI] [PubMed] [Google Scholar]
- 20.Liu, F., M. Dowling, X.-J. Yang, and G. D. Kao. 2004. Caspase-mediated specific cleavage of human histone deacetylase 4. J. Biol. Chem. 279:34537-34546. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Y., W. R. Randall, and M. F. Schneider. 2005. Activity-dependent and -independent nuclear fluxes of HDAC4 mediated by different kinases in adult skeletal muscle. J. Cell Biol. 168:887-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, J., T. A. McKinsey, R. L. Nicol, and E. N. Olson. 2000. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. USA 97:4070-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manning, G., D. B. Whyte, R. Martinez, T. Hunter, and S. Sudarsanam. 2002. The protein kinase complement of the human genome. Science 298:1912-1934. [DOI] [PubMed] [Google Scholar]
- 24.McKinsey, T. A., C. L. Zhang, J. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2000. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 97:14400-14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 21:6312-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mejat, A., F. Ramond, R. Bassel-Duby, S. Khochbin, E. N. Olson, and L. Schaeffer. 2005. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat. Neurosci. 8:313-321. [DOI] [PubMed] [Google Scholar]
- 28.Miska, E. A., C. Karlsson, E. Langley, S. J. Nielsen, J. Pines, and T. Kouzarides. 1999. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller, J., S. Ory, T. Copeland, H. Piwnica-Worms, and D. K. Morrison. 2001. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol. Cell 8:983-993. [DOI] [PubMed] [Google Scholar]
- 30.Muller, J., D. A. Ritt, T. D. Copeland, and D. K. Morrison. 2003. Functional analysis of C-TAK1 substrate binding and identification of PKP2 as a new C-TAK1 substrate. EMBO J. 22:4431-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng, H. H., and A. Bird. 2000. Histone deacetylases: silencers for hire. Trends Biochem. Sci. 25:121-126. [DOI] [PubMed] [Google Scholar]
- 32.Ogg, S., B. Gabrielli, and H. Piwnica-Worms. 1994. Purification of a serine kinase that associates with and phosphorylates human Cdc25C on serine 216. J. Biol. Chem. 269:30461-30469. [PubMed] [Google Scholar]
- 33.Paroni, G., M. Mizzau, C. Henderson, G. Del Sal, C. Schneider, and C. Brancolini. 2004. Caspase-dependent regulation of histone deacetylase 4 nuclear-cytoplasmic shuttling promotes apoptosis. Mol. Biol. Cell 15:2804-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parra, M., H. Kasler, T. A. McKinsey, E. N. Olson, and E. Verdin. 2005. Protein dinase D1 phosphorylates HDAC7 and induces its nuclear export after T-cell receptor activation. J. Biol. Chem. 280:13762-13770. [DOI] [PubMed] [Google Scholar]
- 35.Peng, C. Y., P. R. Graves, S. Ogg, R. S. Thomas, M. J. Byrnes, 3rd, Z. Wu, M. T. Stephenson, and H. Piwnica-Worms. 1998. C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 protein binding. Cell Growth Differ. 9:197-208. [PubMed] [Google Scholar]
- 36.Petrie, K., F. Guidez, L. Howell, L. Healy, S. Waxman, M. Greaves, and A. Zelent. 2003. The histone deacetylase 9 gene encodes multiple protein isoforms. J. Biol. Chem. 278:16059-16072. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Shi, H., C. Asher, Y. Yung, L. Kligman, E. Reuveny, R. Seger, and H. Garty. 2002. Casein kinase 2 specifically binds to and phosphorylates the carboxy termini of ENaC subunits. Eur. J. Biochem. 269:4551-4558. [DOI] [PubMed] [Google Scholar]
- 39.Tassan, J. P., and X. Le Goff. 2004. An overview of the KIN1/PAR-1/MARK kinase family. Biol. Cell. 96:193-199. [DOI] [PubMed] [Google Scholar]
- 40.Thiel, G., M. Lietz, and M. Hohl. 2004. How mammalian transcriptional repressors work. Eur. J. Biochem. 271:2855-2862. [DOI] [PubMed] [Google Scholar]
- 41.Trinczek, B., M. Brajenovic, A. Ebneth, and G. Drewes. 2004. MARK4 is a novel microtubule-associated proteins/microtubule affinity-regulating kinase that binds to the cellular microtubule network and to centrosomes. J. Biol. Chem. 279:5915-5923. [DOI] [PubMed] [Google Scholar]
- 42.Vega, R. B., B. C. Harrison, E. Meadows, C. R. Roberts, P. J. Papst, E. N. Olson, and T. A. McKinsey. 2004. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 24:8374-8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vega, R. B., K. Matsuda, J. Oh, A. C. Barbosa, X. Yang, E. Meadows, J. McAnally, C. Pomajzl, J. M. Shelton, J. A. Richardson, G. Karsenty, and E. N. Olson. 2004. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119:555-566. [DOI] [PubMed] [Google Scholar]
- 44.Vertommen, D., M. Rider, Y. Ni, E. Waelkens, W. Merlevede, J. R. Vandenheede, and J. Van Lint. 2000. Regulation of protein kinase D by multisite phosphorylation. Identification of phosphorylation sites by mass spectrometry and characterization by site-directed mutagenesis. J. Biol. Chem. 275:19567-19576. [DOI] [PubMed] [Google Scholar]
- 45.Wang, A. H., N. R. Bertos, M. Vezmar, N. Pelletier, M. Crosato, H. H. Heng, J. Th'ng, J. Han, and X.-J. Yang. 1999. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol. 19:7816-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, A. H., M. J. Kruhlak, J. Wu, N. R. Bertos, M. Vezmar, B. I. Posner, D. P. Bazett-Jones, and X. J. Yang. 2000. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol. 20:6904-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White, J., K. Haskins, P. Marrack, and J. Kappler. 1983. Use of I region-restricted, antigen-specific T cell hybridomas to produce idiotypically specific anti-receptor antibodies. J. Immunol. 130:1033-1037. [PubMed] [Google Scholar]
- 48.Yaffe, M. B. 2002. How do 14-3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513:53-57. [DOI] [PubMed] [Google Scholar]
- 49.Yang, X.-J., and S. Gregoire. 2005. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol. Cell. Biol. 25:2873-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, C. L., T. A. McKinsey, S. Chang, C. L. Antos, J. A. Hill, and E. N. Olson. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, S.-H., R. Kobayashi, P. R. Graves, H. Piwnica-Worms, and N. K. Tonks. 1997. Serine phosphorylation-dependent association of the band 4.1-related protein-tyrosine phosphatase PTPH1 with 14-3-3beta protein. J. Biol. Chem. 272:27281-27287. [DOI] [PubMed] [Google Scholar]
- 52.Zhao, X., A. Ito, C. D. Kane, T.-S. Liao, T. A. Bolger, S. M. Lemrow, A. R. Means, and T.-P. Yao. 2001. The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J. Biol. Chem. 276:35042-35048. [DOI] [PubMed] [Google Scholar]