Abstract
Mitochondria constantly fuse and divide to adapt organellar morphology to the cell’s ever-changing physiological conditions. Little is known about the molecular mechanisms regulating mitochondrial dynamics. F-box proteins are subunits of both Skp1-Cullin-F-box (SCF) ubiquitin ligases and non-SCF complexes that regulate a large number of cellular processes. Here, we analyzed the roles of two yeast F-box proteins, Mfb1 and Mdm30, in mitochondrial dynamics. Mfb1 is a novel mitochondria-associated F-box protein. Mitochondria in mutants lacking Mfb1 are fusion competent, but they form aberrant aggregates of interconnected tubules. In contrast, mitochondria in mutants lacking Mdm30 are highly fragmented due to a defect in mitochondrial fusion. Fragmented mitochondria are docked but nonfused in Δmdm30 cells. Mitochondrial fusion is also blocked during sporulation of homozygous diploid mutants lacking Mdm30, leading to a mitochondrial inheritance defect in ascospores. Mfb1 and Mdm30 exert nonredundant functions and likely have different target proteins. Because defects in F-box protein mutants could not be mimicked by depletion of SCF complex and proteasome core subunits, additional yet unknown factors are likely involved in regulating mitochondrial dynamics. We propose that mitochondria-associated F-box proteins Mfb1 and Mdm30 are key components of a complex machinery that regulates mitochondrial dynamics throughout yeast’s entire life cycle.
INTRODUCTION
Mitochondria carry out a variety of different functions in eukaryotic cells. They supply the cell with ATP generated by oxidative phosphorylation, they are the site of many anabolic and catabolic pathways, they play an essential role in the assembly of Fe/S clusters, and they are key regulators of programmed cell death (Scheffler, 2000; Lill and Mühlenhoff, 2005; Youle and Karbowski, 2005). Depending on the cell’s physiological conditions, mitochondria frequently change their shape and intracellular position (Bereiter-Hahn, 1990; Yaffe, 1999; Griparic and van der Bliek, 2001; Westermann, 2002; Mozdy and Shaw, 2003; Scott et al., 2003; Chen and Chan, 2004). This dynamic behavior is important for a number of cellular processes. For example, mitochondrial fusion is required for sperm development in Drosophila (Hales and Fuller, 1997), for embryonic development in mice (Chen et al., 2003), and for maintenance of mitochondrial DNA in yeast (Chen and Butow, 2005). Conversely, mitochondrial division plays a crucial and evolutionarily highly conserved role in the apoptotic pathway (Jagasia et al., 2005; Youle and Karbowski, 2005). Although several proteins mediating mitochondrial motility, fusion and fission have been identified during the past several years (Okamoto and Shaw, 2005), little is known about regulatory components of mitochondrial dynamics.
The core machinery mediating mitochondrial membrane fusion in yeast consists of three proteins, namely, Fzo1, a conserved GTPase in the outer membrane (Hermann et al., 1998; Rapaport et al., 1998); the outer membrane protein Ugo1 (Sesaki and Jensen, 2001); and Mgm1, a conserved GTPase located in the intermembrane space (Wong et al., 2000). Ugo1 physically interacts with Fzo1 and Mgm1 and presumably coordinates their function (Wong et al., 2003; Sesaki and Jensen, 2004). Genetic and morphological evidence suggests that the mitochondrial fusion and fission machineries act antagonistically and operate in a perfectly balanced manner. Defects in cells lacking components of the fusion machinery can be relieved by deletion of genes encoding proteins of the fission machinery (Bleazard et al., 1999; Sesaki and Jensen, 1999). Wild-type cells growing logarithmically on glucose-containing medium show tightly balanced mitochondrial fusion and fission activities at a rate of 1.7 fusion and fission events per minute and cell (Jakobs et al., 2003). Thus, fusion and fission must be strictly coordinated to maintain a reticular mitochondrial network. A regulatory role in mitochondrial fusion has been proposed for Pcp1, a rhomboid-like protease in the mitochondrial inner membrane. Pcp1 generates two isoforms of Mgm1 by site-specific processing (Herlan et al., 2003; McQuibban et al., 2003; Sesaki et al., 2003). It has been suggested that Pcp1-dependent processing of Mgm1 adapts mitochondrial fusion activity to the ATP requirements of the cell (Herlan et al., 2004).
F-box proteins are particularly versatile regulators of a variety of cellular functions (Willems et al., 2004; Petroski and Deshaies, 2005). Typically, they serve as substrate adaptors in Skp1-Cullin-F-box (SCF) E3 ubiquitin ligases, where they are required to target proteins to ubiquitylation and degradation by the 26S proteasome. However, some F-box proteins may also function independently of the SCF complex and the 26S proteasome (Galan et al., 2001). In budding yeast, at least 21 proteins contain a discernible F-box motif (Willems et al., 2004). One of these proteins, Mdm30, is required for maintenance of fusion-competent mitochondria (Fritz et al., 2003). A subfraction of Mdm30 is associated with mitochondria (Fritz et al., 2003; Sickmann et al., 2003), and cells lacking Mdm30 contain aggregated or fragmented mitochondria, tend to lose their mitochondrial DNA, and fail to fuse mitochondria in vivo. The steady-state level of Fzo1 is dependent on the cellular level of Mdm30, suggesting that Mdm30 regulates mitochondrial fusion by directly or indirectly stimulating degradation of Fzo1 (Fritz et al., 2003). However, recent results by Neutzner and Youle (2005) suggest that the situation may be more complex. These authors showed that treatment of yeast cells with the mating pheromone α factor evokes a fragmentation of the mitochondrial network into small pieces. Mitochondrial fragmentation is accompanied by proteasome-dependent degradation of Fzo1 but does not require Mdm30 (Neutzner and Youle, 2005; Escobar-Henriques et al., 2006). These results point to the existence of multiple pathways for the regulation of the mitochondrial fusion machinery. To gain more insights into the role of F-box proteins in the regulation of mitochondrial behavior, we analyzed the functions of Mdm30 and Mfb1, a novel mitochondria-associated F-box protein, in vegetatively growing cells and during mating and sporulation.
MATERIALS AND METHODS
Plasmid Constructions
Standard methods were used for cloning procedures. The plasmid for expression of Mfb1 from the GAL1 promoter in yeast, pYES-MFB1, was generated by PCR amplification of the mitochondria-associated F-box protein 1 (MFB1) coding region from genomic yeast DNA using primers 5′-AAA AAG CTT ATG ACA TTA TTT TCT TGT AGT GTA C-3′ and 5′-AAA GGA TCC TTA TAT ATT AAA ATC GGT ATT AGC G-3′ and cloning into the HindIII/BamHI sites of vector pYES2.0 (Invitrogen, Groningen, The Netherlands). Plasmid pYES-MDM30 for expression of Mdm30 from the GAL1 promoter has been described previously (Fritz et al., 2003). Plasmids pVT100U-mtGFP, pYX113-mtGFP (Westermann and Neupert, 2000), pRS416-GAL1+PrF0ATP9-RFP (Mozdy et al., 2000), and pSJ55 (Jakobs et al., 2003) were used for expression of mitochondria-targeted green fluorescent protein (GFP) or mitochondria-targeted DsRed. Plasmids for expression of FLAG-tagged Mdm30 and an F-box–less variant thereof under control of the CUP1 promoter are described by Escobar-Henriques et al. (2006). A plasmid for expression of FLAG-tagged Mfb1 under control of the CUP1 promoter (pJDCEX2-MFB1) was constructed by PCR amplification of the MFB1 coding region using primers 5′-AAA AAG ATC TAT GAC ATT ATT TTC TTG TAG TGT AC-3′ and 5′-AAA ACC CGG GTT ATA TAT TAA AAT CGG TAT TAG GC-3′ and cloning into the BglII/XmaI sites of pJDCEX2-MDM30 (Escobar-Henriques et al., 2006). A plasmid for expression of a truncated Mfb1 protein lacking the 60 N-terminal amino acids (pJDCEX2-MFB1-ΔF-box) was constructed the same way using forward primer 5′-AAA AAG ATC TGT TAT CGG CTC AAA CGG GAA TAT G-3′.
Construction and Manipulation of Yeast Strains
Growth and manipulation of yeast strains was according to published procedures (Sherman, 1991; Sherman and Hicks, 1991; Gietz et al., 1992). All strains used in this study are isogenic to BY4741, BY4742, and BY4743 (Brachmann et al., 1998). Haploid deletion strains (Giaever et al., 2002) were obtained from EUROSCARF (Frankfurt, Germany), and strains expressing essential genes under control of a titratable promoter (Mnaimneh et al., 2004) were obtained from BioCat (Heidelberg, Germany). A strain expressing Mfb1-GFP (Huh et al., 2003) was obtained from Invitrogen. Mitochondria have a wild-type–like appearance in this strain, indicating that the Mfb1-GFP fusion protein is functional (Figure 1B; our unpublished observations). Homozygous diploid wild-type and Δmfb1 and Δmdm30 mutants expressing mitochondria-targeted GFP have been described previously (Dimmer et al., 2002). A haploid double mutant, Δmfb1/Δmdm30, was generated by mating of haploid deletion strains, sporulation, and tetrad dissection. Genotypes of haploid progeny were determined by PCR. Promoter shutoff in cdc4, cdc34, cdc53, ctf13, met30, and pre1 mutants carrying titratable promoter alleles (Mnaimneh et al., 2004) was as described previously (Altmann and Westermann, 2005). Expression of FLAG-tagged Mfb1 and Mdm30 proteins and ΔF-box variants thereof was induced by addition of 100 μM CuSO4 to the medium. Assay of mitochondrial fusion in vivo was according to published procedures (Nunnari et al., 1997; Fritz et al., 2003). Mitochondrial behavior during sporulation was observed essentially as described previously (Gorsich and Shaw, 2004) with minor modifications. Before sporulation, strains were incubated for 3 d on yeast extract-peptone-dextrose (YPD) plates at 30°C. Cells were allowed to form tetrads for 12 d at 30°C in liquid sporulation medium (1% potassium acetate; 20 μg/ml each of adenine, arginine, histidine, methionine, and tryptophan; and 30 μg/ml each of isoleucine, leucine, lysine, phenylalanine, threonine, and valine).
Figure 1.
Mfb1 is a novel mitochondria-associated F-box protein. (A) The F-box motifs of Mfb1 (amino acid residues 17–60) and Mdm30 (amino acid residues 16–59) are aligned with an F-box consensus sequence according to Willems et al. (2004). Favored sites in the consensus sequence are in uppercase letters, and moderately well tolerated residues are in lowercase letters. Residues matching the consensus sequence are in red. (B) Yeast cells expressing an Mfb1-GFP fusion protein under control of the MFB1 promoter were grown to log phase in glucose-containing medium (YPD), and mitochondria were stained with the membrane potential-sensitive dye rhodamine B hexyl ester. Yeast cells were analyzed by fluorescence and differential interference contrast (DIC) microscopy. Rhodamine fluorescence (mito) was analyzed first to avoid photoconversion of GFP. Bar, 1 μm.
Microscopy
Yeast cultures were grown at 30°C in liquid YPD medium to mid-logarithmic growth phase, if not indicated otherwise. Mitochondria were stained with plasmid-expressed mitochondria-targeted GFP or DsRed (see above) or rhodamine B hexyl ester (Invitrogen, Eugene, OR) as described previously (Dimmer et al., 2002). Filamentous actin, vacuoles, and endoplasmic reticulum (ER) were stained as described previously (Fritz et al., 2003). Epifluorescence microscopy was performed using an Axioplan 2 microscope (Carl Zeiss Lichtmikroskopie, Göttingen, Germany) equipped with a Plan-Neofluar 100×/1.30 Ph3 oil objective (Carl Zeiss Lichtmikroskopie). Images were recorded with an Evolution VF Mono Cooled monochrome camera (Intas, Göttingen, Germany) and processed with Image-Pro Plus 5.0 and Scope Pro4.5 software (Media Cybernetics, Silver Spring, MD).
Assay of Fzo1 Turnover after α Factor-induced Cell Cycle Arrest
To examine Fzo1 degradation upon α factor-induced cell cycle arrest (Neutzner and Youle, 2005), early log phase cells grown on YPD medium were treated with 20 mM sodium citrate and 10 μM α factor. At the indicated time points, cells corresponding to 3 optical density units were collected, lysed at alkaline pH, and trichloroacetic acid (TCA)-precipitated (Tatsuta and Langer, 2006), and protein extracts were analyzed by SDS-PAGE and immunoblotting using antisera against the GTPase domain of Fzo1 (kind gift from Jodi Nunnari, University of California, Davis, CA).
Assay of Mdm30 Binding to Mitochondria
Yeast cells expressing FLAG-tagged Mdm30 and Mdm30-ΔF-box were grown to logarithmic growth phase in SC medium lacking leucine to select for maintenance of the plasmid. The experiment was performed at intermediate Mdm30 expression levels, i.e., without the addition of copper to the medium. Total cell extracts were prepared as described above. Crude mitochondria were isolated by differential centrifugation, and soluble proteins were precipitated from the supernatant with TCA (Escobar-Henriques et al., 2006). Total cell proteins, mitochondrial proteins, and cytosolic proteins were analyzed by SDS-PAGE and immunoblotting.
Electron Microscopy
Yeast cells were prepared for electron microscopy according to published protocols (Bauer et al., 2001) with the exception that embedding was in Spurr’s resin (Spurr, 1969). Ultrathin sections (70 nm) were cut with a diamond knife (MicroStar, Huntsville, TX) on a Leica Ultracut UCT microtome (Leica Microsystems, Vienna, Austria) and mounted on pioloform-coated copper grids. Sections were viewed with a Zeiss CEM 902 A transmission electron microscope (Carl Zeiss, Oberkochen, Germany) at 80 kV. Micrographs were taken using SO-163 EM film (Eastman Kodak, Rochester, NY). Three-dimensional reconstructions were prepared from scanned films using IMOD software, version 3.5.5 (http://bio3d.colorado.edu/imod/; Kremer et al., 1996).
RESULTS
Mfb1 Is a Novel Mitochondria-associated F-Box Protein
The MFB1 gene (mitochondria-associated F-box protein 1; systematic name YDR219c) encodes a protein of 465 amino acid residues (≈53 kDa). Similar to Mdm30, the Mfb1 protein contains an F-box motif at its N terminus (Figure 1A). Outside the F-box, the Mfb1 amino acid sequence does not show significant homology to any other known protein domain or sequence motif. In a large-scale analysis aimed at the systematic identification of protein complexes by mass spectrometry, Mfb1 was found in a complex with Skp1 (Ho et al., 2002). A large-scale analysis aimed at the systematic subcellular localization of the yeast proteome suggested that Mfb1 is associated with mitochondria, because expression of an Mfb1-GFP fusion protein yielded a staining pattern resembling mitochondria (Huh et al., 2003). To prove a mitochondrial location of Mfb1, we stained live yeast cells expressing Mfb1-GFP under control of its endogenous promoter with a mitochondria-specific fluorescent dye, rhodamine B hexyl ester. Because Mfb1-GFP staining overlapped with mitochondrial staining (Figure 1B), we conclude that a major fraction of Mfb1 is associated with mitochondria.
Mfb1, Mdm30, and SCF Core Subunits Play Distinct and Separable Roles in Mitochondrial Distribution and Morphology
To investigate a role of Mfb1 in mitochondrial biogenesis and behavior, and to address the question whether its function overlaps with Mdm30, we constructed and analyzed mutant strains lacking or overexpressing the two mitochondria-associated F-box proteins in various combinations. Cells lacking Mfb1 showed a slight growth defect on the nonfermentable carbon source glycerol at elevated temperature (Figure 2A), which was less pronounced than the growth defect of cells lacking Mdm30 (Figure 2A; Fritz et al., 2003). The growth behavior of the Δmfb1/Δmdm30 double mutant was identical to that of the Δmdm30 single mutant (Figure 2A). We conclude that both Mfb1 and Mdm30 are important for maintenance of respiratory functions at elevated temperature, but yeast cells can tolerate the simultaneous loss of both proteins without the loss of vital functions.
Figure 2.
Mfb1, Mdm30, SCF complex, and proteasome subunits are required for normal growth and mitochondrial morphology. (A) Wild-type (WT), Δmfb1, Δmdm30, and Δmfb1/Δmdm30 (ΔΔ) cells were grown overnight in glucose-containing medium at 30°C. Then, 10-fold serial dilutions were spotted onto plates containing glucose (YPD) or glycerol (YPG) as carbon source. YPD plates were incubated for 2 d, and YPG plates were incubated for 3 d at the indicated temperatures. (B) Wild-type (WT), Δmfb1, Δmdm30, and Δmfb1/Δmdm30 (ΔΔ) cells expressing mitochondria-targeted GFP were grown to log phase in YPD or YPG medium and analyzed by DIC (left) and fluorescence (right) microscopy. Bar, 2 μm. (C) WT, cdc53, cdc34, and pre1 cells expressing mitochondria-targeted GFP were grown to log phase in YPD or YPG in the presence of doxycycline (+Dox) and analyzed as described above. Magnification is as in B.
To examine whether Mfb1 is required for normal mitochondrial distribution and morphology, we performed a comprehensive analysis of mitochondria in cells lacking Mfb1 and/or Mdm30. Logarithmically growing wild-type cells showed a characteristic tubular network, which is more ramified on the nonfermentable carbon source glycerol than on the fermentable carbon source glucose (Figure 2B). More than 80% of the cells in Δmfb1, Δmdm30, and Δmfb1/Δmdm30 cultures displayed aggregated mitochondria that were clumped together and deposited at random locations within the cells on glucose-containing medium (Figure 2B and Table 1). In contrast, on the nonfermentable carbon source glycerol, the majority of mutant cells contained fragmented mitochondria that were more or less evenly distributed in the cell (Figure 2B and Table 1). Because mitochondrial behavior observed by light microscopy seems very similar in Δmfb1 and Δmdm30 cells, we considered the possibility that Mfb1 and Mdm30 function in related pathways and might have overlapping substrate specificity. Thus, we tested whether overexpression of one component might rescue the loss of the other. The MFB1 and MDM30 genes were placed under control of the strong and inducible GAL1 promoter and expressed from a multicopy plasmid in wild type, Δmfb1, and Δmdm30 cells. Strains transformed with the empty vector served as controls. Δmfb1 cells expressing the MFB1 gene from the GAL1 promoter and Δmdm30 cells expressing the MDM30 gene from the GAL1 promoter showed wild-type–like mitochondria (Table 1), indicating that plasmid-expressed F-box proteins are functional and suggesting that overexpression of these genes does not have negative consequences for mitochondrial morphology. In contrast, Δmfb1 cells expressing the MDM30 gene from the GAL1 promoter and Δmdm30 cells expressing the MFB1 gene from the GAL1 promoter were indiscernible from the vector controls (Table 1). This suggests that loss of one mitochondria-associated F-box protein cannot be compensated by overexpression of the other. We conclude that Mfb1 and Mdm30 are both required for maintenance of a wild-type–like mitochondrial reticulum and that they have nonredundant functions.
Table 1.
Mitochondrial phenotypes during vegetative growth
| Strain/medium | Mitochondrial morphology (% of cells) | No. of cells scored | ||
|---|---|---|---|---|
| Wild-type–like | Aggregated | Fragmented | ||
| YPD | ||||
| WT | 91.1 | 5.1 | 3.8 | 315 |
| Δmfb1 | 9.4 | 84.7 | 5.9 | 374 |
| Δmdm30 | 10.4 | 88.4 | 1.2 | 346 |
| Δmfb1/Δmdm30 | 1.8 | 83.7 | 14.5 | 339 |
| YPG | ||||
| WT | 84.6 | 0.9 | 14.5 | 331 |
| Δmfb1 | 39.2 | 2.9 | 57.9 | 339 |
| Δmdm30 | 3.7 | 0.6 | 95.7 | 350 |
| Δmfb1/Δmdm30 | 1.3 | 1.3 | 97.4 | 312 |
| YPGal | ||||
| WT/pYES | 90.8 | 3.2 | 6.0 | 532 |
| WT/pYES-MFB1 | 87.5 | 3.0 | 9.5 | 305 |
| WT/pYES-MDM30 | 85.6 | 6.6 | 7.8 | 408 |
| Δmfb1/pYES-MFB1 | 82.8 | 10.5 | 6.7 | 313 |
| Δmdm30/pYES-MDM30 | 84.8 | 7.6 | 7.6 | 422 |
| Δmfb1/pYES | 38.9 | 56.0 | 5.1 | 553 |
| Δmfb1/pYES-MDM30 | 33.5 | 60.8 | 5.7 | 421 |
| Δmdm30/pYES | 6.8 | 86.4 | 6.8 | 308 |
| Δmdm30/pYES-MFB1 | 9.1 | 80.5 | 10.4 | 307 |
| YPD + Cu2+ | ||||
| Δmfb1 | 14.7 | 80.3 | 5.0 | 300 |
| Δmfb1/MFB1 | 69.5 | 27.2 | 3.3 | 305 |
| Δmfb1/MFB1ΔFbox | 78.5 | 20.5 | 1.0 | 302 |
| Δmdm30 | 14.3 | 80.0 | 5.7 | 300 |
| Δmdm30/MDM30 | 48.0 | 48.7 | 3.3 | 304 |
| Δmdm30/MDM30ΔFbox | 59.3 | 38.7 | 2.0 | 300 |
| YPD | ||||
| WT | 99.0 | 0 | 1.0 | 300 |
| cdc53 | 2.5 | 0.5 | 97 | 200 |
| cdc34 | 100.0 | 0 | 0 | 100 |
| pre1 | 97.5 | 0 | 2.5 | 200 |
| YPG | ||||
| WT | 91.5 | 0 | 8.5 | 200 |
| cdc53 | 2.5 | 0 | 97.5 | 200 |
| cdc34 | 78.0 | 0 | 22.0 | 100 |
| pre1 | 90.0 | 0 | 10.0 | 200 |
| YPD + Dox | ||||
| WT | 99.7 | 0.3 | 0 | 300 |
| cdc53 | 4.5 | 3.0 | 92.5 | 200 |
| cdc34 | 14.0 | 0 | 86.0 | 100 |
| pre1 | 24.7 | 0 | 75.3 | 300 |
| YPG + Dox | ||||
| WT | 90.0 | 0 | 10.0 | 200 |
| cdc53 | 3.0 | 2.0 | 95.0 | 100 |
| cdc34 | 24.0 | 0 | 76.0 | 100 |
| pre1 | 31.5 | 0 | 68.5 | 200 |
The F-box comprises a conserved binding site for Skp1 (Willems et al., 2004; Petroski and Deshaies, 2005). To test whether the presence of an intact F-box is essential for the proteins’ functions, we overexpressed truncated Mfb1 and Mdm30 proteins lacking their F-boxes from a CUP1 promoter (MFB1ΔFbox and MDM30ΔFbox). Surprisingly, these constructs rescued the mitochondrial morphology defects of the null mutants to a large extent (Table 1), suggesting that at least some of the proteins’ functions in mitochondrial dynamics do not require assembly into SCF complexes. It should be noted, however, that complementation of the mitochondrial phenotypes was seen under conditions that involved overexpression of the ΔF-box constructs from a strong promoter. Under these conditions, truncated, nonfunctional F-box proteins might be still able to bind to accumulated substrate proteins, such as Fzo1. Even though we consider it unlikely, we cannot exclude the possibility that this binding might inactivate excess Fzo1, or other substrate proteins, and thereby restore normal mitochondrial morphology in an indirect manner.
Several lines of evidence assign an important role to the ubiquitin/26S proteasome system in mitochondrial morphogenesis. For example, overexpression of a mutant ubiquitin variant unable to form polyubiquitin chains results in aberrant mitochondrial morphology (Fisk and Yaffe, 1999), and cells lacking functional proteasome subunits have severe mitochondrial defects (Rinaldi et al., 1998; Altmann and Westermann, 2005). However, the exact role of the proteasome in mitochondrial biogenesis is still unclear. We asked whether the mitochondrial phenotypes seen in F-box protein mutants resemble mitochondria in cells after depletion of Cdc53/Cullin and Cdc34, essential core components of SCF ubiquitin ligases, or Pre1, an essential subunit of the 20S proteasome core particle. Yeast mutants containing the CDC53, CDC34, or PRE1 gene under control of a promoter that can be shut off by the addition of doxycycline to the medium (Mnaimneh et al., 2004) were grown under inducing and repressing conditions on fermentable and nonfermentable carbon sources, and mitochondrial morphology was analyzed. Mitochondria were fragmented in cdc53 cells under all conditions and in pre1 and cdc34 cells under repressing conditions on either carbon source (Figure 2C). Surprisingly, aggregated mitochondria, the major phenotypic class seen in mutants lacking mitochondria-associated F-box proteins grown on YPD medium, could only very rarely be observed in cdc53, cdc34, and pre1 mutants (Figure 2C and Table 1). Thus, mitochondrial defects are clearly distinguishable in cdc53, cdc34, and pre1 mutants on the one hand and mfb1 and mdm30 mutants on the other hand. Together, our observations point to the involvement of other, yet unidentified factors, in F-box protein-dependent regulation of mitochondrial dynamics.
Mfb1 and Mdm30 Are the Only F-box Proteins Required for Maintenance of Mitochondrial Morphology
The yeast genome contains 21 genes potentially encoding F-box proteins (Willems et al., 2004). We asked whether in addition to MFB1 and MDM30 any of the remaining 19 genes is required for normal mitochondrial distribution and morphology. Mitochondria were stained with mitochondria-targeted GFP and examined by fluorescence microscopy in the following deletion mutants lacking nonessential genes: Δamn1, Δcos111, Δdia2, Δela1, Δgrr1, Δhrt3, Δrcy1, Δskp2, Δufo1, Δybr280c, Δydr131c, Δydr306c, Δyjl149w, Δylr224w, Δylr352w, and Δymr258c. Three genes encoding F-box proteins, CDC4, CFT13, and MET30, are essential for viability in yeast. Mitochondrial morphology was analyzed in strains carrying these genes under control of a doxycycline-repressible promoter (Mnaimneh et al., 2004). All of these mutant strains lacking F-box proteins showed wild-type–like mitochondria (Table 2and Supplemental Figure 1). We conclude that Mfb1 and Mdm30 are the only members of the F-box protein family that are essential for maintenance of normal mitochondrial morphology in yeast.
Table 2.
Role of yeast F-box proteins in mitochondrial morphology
| Strain/medium | Mitochondrial morphology (% of cells) | No. of cells scored | ||
|---|---|---|---|---|
| Wild-type–like | Aggregated | Fragmented | ||
| YPD | ||||
| WT | 91.1 | 5.1 | 3.8 | 315 |
| Δamn1(Δybr158w) | 100.0 | 0 | 0 | 100 |
| Δcos111(Δybr203w) | 70.0 | 25.0 | 5.0 | 100 |
| Δdia2(Δyor080w) | 100.0 | 0 | 0 | 100 |
| Δela1(Δynl230c) | 95.0 | 3.0 | 2.0 | 100 |
| Δgrr1(Δyjr090c) | 100.0 | 0 | 0 | 100 |
| Δhrt3(Δylr097c) | 100.0 | 0 | 0 | 100 |
| Δmdm30(Δylr368w) | 10.4 | 88.4 | 1.2 | 346 |
| Δmfb1(Δydr219c) | 9.4 | 84.7 | 5.9 | 374 |
| Δrcy1(Δyjl204c) | 95.0 | 5.0 | 0 | 100 |
| Δskp2(Δynl311c) | 95.0 | 0 | 5.0 | 100 |
| Δufo1(Δyml088w) | 74.0 | 0 | 26.0 | 100 |
| Δybr280c | 100.0 | 0 | 0 | 100 |
| Δydr131c | 98.0 | 0 | 2.0 | 100 |
| Δydr306c | 95.0 | 5.0 | 0 | 100 |
| Δyjl149w | 97.0 | 2.0 | 1.0 | 100 |
| Δylr224w | 100.0 | 0 | 0 | 100 |
| Δylr352w | 100.0 | 0 | 0 | 100 |
| Δymr258c | 100.0 | 0 | 0 | 100 |
| YPD | ||||
| WT | 99.0 | 0 | 1.0 | 300 |
| cdc4 (yfl009w) | 93.0 | 0 | 7.0 | 100 |
| ctf13 (ymr094w) | 100.0 | 0 | 0 | 100 |
| met30(yil046w) | 100.0 | 0 | 0 | 100 |
| YPD + Dox | ||||
| WT | 99.7 | 0.3 | 0 | 300 |
| cdc4 (yfl009w) | 67.0 | 12.0 | 21.0 | 100 |
| ctf13 (ymr094w) | 100.0 | 0 | 0 | 100 |
| met30(yil046w) | 91.0 | 4.0 | 5.0 | 100 |
To examine whether deletion of the MFB1 gene has pleiotropic effects on the morphology of other intracellular structures we analyzed vacuoles, ER, and filamentous actin by fluorescence microscopy of wild-type and Δmfb1 cells. Because these cellular structures seemed normal in Δmfb1 (Supplemental Figure 2) and Δmdm30 cells (Fritz et al., 2003), we conclude that organellar morphology defects in Δmfb1 and Δmdm30 mutants primarily affect mitochondria.
Mfb1 Is Not Required for Mitochondrial Fusion and Turnover of Fzo1
Mitochondrial fusion is blocked in Δmdm30 cells, presumably because the turnover of Fzo1 is inhibited and surplus Fzo1 protein accumulates in the cell (Fritz et al., 2003). To test whether Mfb1 is required for mitochondrial fusion, we assayed fusion by mating haploid cells of opposite mating types preloaded with different fluorescent mitochondrial matrix markers. At least 20 zygotes were analyzed per strain. Fluorescent labels immediately intermixed in 100% of the zygotes, indicating efficient mitochondrial fusion and content mixing both in wild-type and Δmfb1 cells (Figure 3A). Consistent with this finding, the steady-state level of Fzo1 in vegetatively growing cells is not elevated in Δmfb1 cells in comparison with the wild type, and deletion of the MFB1 gene in the Δmdm30 mutant does not elevate Fzo1 levels further in comparison with the Δmdm30 single mutant (Figure 3B). We conclude that Mfb1 is not required for mitochondrial fusion or turnover of Fzo1 during vegetative growth.
Figure 3.
Mfb1 is not required for mitochondrial fusion or turnover of Fzo1. (A) WT or Δmfb1 cells of opposite mating types containing mitochondria preloaded with GFP (mtGFP) or DsRed (mtDsRed) were mated, and zygotes were analyzed by fluorescence and DIC microscopy. DsRed fluorescence was analyzed first to avoid photoconversion of GFP. Bar, 5 μm. (B) WT, Δmfb1, Δmdm30, and Δmfb1/Δmdm30 cells were grown to log phase in YPD medium. Steady-state concentrations of Fzo1 in total cell extracts were analyzed by immunoblotting using Fzo1-specific antibodies (αFzo1). A cross-reacting protein (CR), which is invariantly expressed in all four strains, served as a loading control. (C) WT, Δmfb1, and Δmdm30 cultures were treated for the indicated time periods with mating pheromone α factor. Turnover of Fzo1 was analyzed by immunoblotting of total cell extracts as in B.
Interestingly, Neutzner and Youle (2005) reported that Mdm30 is not required for proteasome-dependent degradation of Fzo1 after G1 cell cycle arrest evoked by treatment of yeast wild-type cells with the mating pheromone α factor. These findings were genetically confirmed using yeast strains carrying mutations in genes encoding proteasome subunits (Escobar-Henriques et al., 2006). We considered the possibility that in G1 cell cycle-arrested cells Mfb1 might serve as an alternative F-box protein to target Fzo1 to degradation. To test this, wild-type, Δmfb1, and Δmdm30 cells were treated for different times with α factor, and turnover of Fzo1 was assayed by immunoblotting of cell extracts. Consistent with previous findings, we observed efficient degradation of Fzo1 in α factor-treated wild-type and Δmdm30 cells. However, efficient turnover of Fzo1 was also observed in Δmfb1 cells (Figure 3C). Hence, degradation of Fzo1 after mating pheromone-induced cell cycle arrest depends neither on Mdm30 nor on Mfb1.
Recruitment of Mdm30 to Mitochondria Is Independent of Fzo1
A major fraction of Mdm30 is associated with mitochondria (Fritz et al., 2003), and Mdm30 physically interacts with Fzo1 in an F-box–independent manner (Escobar-Henriques et al., 2006). Intriguingly, deletion of the MDM30 gene in a Δfzo1 background produces a synthetic growth defect, suggesting that Mdm30 may have other yet unknown target proteins in addition to Fzo1 (Fritz et al., 2003). To examine whether Mdm30 binds to mitochondria in the absence of its known binding partner, Fzo1, we fractionated Δfzo1 cells expressing FLAG-tagged Mdm30 (Mdm30FLAG) and FLAG-tagged Mdm30 lacking the F-box (ΔF-boxFLAG). On fractionation of yeast cells, we observed that Mdm30 cofractionated with mitochondria, even in the absence of Fzo1 (Figure 4). This suggests that Mdm30 has one or more mitochondrial binding partners in addition to Fzo1 and thus may have multiple target proteins on mitochondria.
Figure 4.
Binding of Mdm30 to mitochondria does not require Fzo1. Wild-type (FZO1) and Δfzo1 strains were transformed with plasmids expressing N-terminally FLAG-tagged full-length Mdm30 (Mdm30FLAG) and Mdm30 lacking its F-box (ΔF-boxFLAG) from the CUP1 promoter. Total cell extracts (T) and subcellular fractions were prepared from logarithmically growing cultures. Crude mitochondria contained in the pellet fraction (P) and cytosolic proteins contained in the supernatant (S) were isolated by differential centrifugation and analyzed by immunoblotting using antibodies against the FLAG epitope, the mitochondrial protein Tom40, and the cytosolic protein triose phosphate isomerase Tpi1.
Ultrastructure of Cells Lacking Mfb1 and Mdm30
Next, we examined the ultrastructure of wild-type, Δmfb1, Δmdm30, and Δmfb1/Δmdm30 cells by electron microscopy. To obtain high-resolution images of the three-dimensional organization of mitochondria, we prepared serial ultrathin sections (70 nm), analyzed them by transmission electron microscopy, and generated three-dimensional models (Figure 5, A and B). Although aggregated and clumped mitochondria of cells grown in glucose-containing medium looked very similar in all three mutant strains when observed by fluorescence microscopy (compare Figure 2B and Table 1), electron microscopic analyses revealed striking differences. When the three-dimensional structure of mitochondria was reconstructed from 31 consecutive sections (corresponding to ≈2170 nm) of a wild-type cell, three morphologically distinct organelles were observed within this volume (Figure 5A). After three-dimensional reconstruction of 27 sections (corresponding to ≈1890 nm) of a Δmfb1 cell, we found that all mitochondria within this volume were interconnected and formed one closed continuum consisting of clumped tubules (Figure 5A). In contrast, three-dimensional reconstruction of 30 sections (corresponding to ≈2100 nm) of a Δmdm30 cell revealed an aggregate of seven distinct organelles, and 18 sections (corresponding to ≈1260 nm) of a Δmfb1/Δmdm30 cell contained 16 small and morphologically separable organelles (Figure 5A). These analyses were repeated with at least one additional cell for each strain, yielding very similar results (our unpublished observations). All cells were harvested from logarithmically growing cultures, and for each strain at least one cell that clearly carried a bud was analyzed.
Figure 5.
Ultrastructure of cells lacking Mfb1 or Mdm30. (A) WT, Δmfb1, Δmdm30, and Δmfb1/Δmdm30 cells were grown to log phase in YPD medium, fixed with glutaraldehyde, treated with zymolyase to remove the cell wall, postfixed with osmium tetroxide, and en bloc stained with uranyl acetate. After embedding in Spurr’s resin, serial ultrathin sections (70 nm) were prepared and analyzed by transmission electron microscopy. The three-dimensional structure of mitochondria was reconstructed using IMOD software (Kremer et al., 1996). Morphologically separate organelles are displayed in different colors. The contour of the plasma membrane is shown by a white line for each section. The model of the wild-type cell was reconstructed from 31 consecutive sections, the Δmfb1 cell from 27 sections, the Δmdm30 cell from 30 sections, and the Δmfb1/Δmdm30 cell from 18 sections. (B) Three representative consecutive sections that were used for reconstruction of the three-dimensional models displayed in A. (C) Enlarged detail of a transmission electron micrograph of a Δmfb1/Δmdm30 cell (taken from the third section displayed in B). White arrows point to close contacts between morphologically separate nonfused mitochondria. It should be noted that yeast strains related to BY4743 generally contain only few cristae on glucose-containing medium. However, strains lacking Mfb1 or Mdm30 are able to form normal cristae (not visible here).
Intriguingly, mitochondria in Δmdm30 (our unpublished data) and Δmfb1/Δmdm30 cells (Figure 5C) were sometimes observed to be in close contact without fusion. This phenotype resembles clustered mitochondria seen in mammalian cells overexpressing Fzo1 homologues Mfn2 (Rojo et al., 2002) or Mfn1 (Santel et al., 2003). It is conceivable that surplus Fzo1 forms nonfunctional fusion complexes in strains lacking Mdm30. These complexes might tether neighboring organelles, as it has been observed for mammalian cells expressing mutant forms of Mfn2 (Eura et al., 2003) or truncated forms of Mfn1 (Koshiba et al., 2004). Interestingly, some electron-dense material is seen in the contact areas (Figure 5C, arrows), which might consist of accumulated Fzo1. A block of mitochondrial fusion in cells lacking Mdm30 leads to fragmentation of mitochondria by ongoing fission (Fritz et al., 2003). Because fusion is not blocked in Δmfb1 cells, mitochondria may fuse and form a continuum in cells lacking Mfb1 as long as Mdm30 is present. We conclude that mitochondrial aggregates are formed by different mechanisms in cells lacking Mfb1 and Mdm30.
Tubular, aggregated mitochondria in Δmfb1 cells that were clearly different from fragmented, aggregated mitochondria in Δmdm30 cells were also observed by Kondo-Okamoto et al. (2006). It should be noted that these researchers found that short tubules in Δmfb1 cells were less interconnected in the W303 genetic background than the structures we observed in BY4741-related Δmfb1 strains. We consider it likely that these differences are due to strain-specific effects.
Mdm30 Is Required for Remodeling of Mitochondria during Sporulation
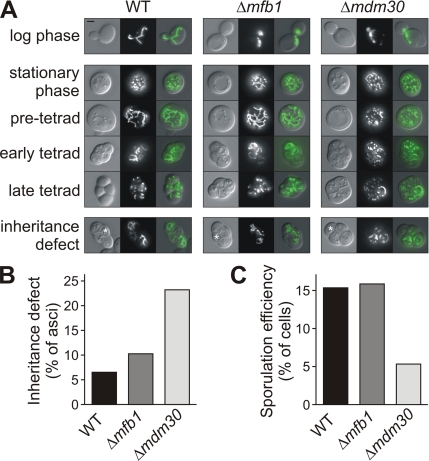
Mitochondria undergo extensive remodeling during sporulation (Miyakawa et al., 1984; Gorsich and Shaw, 2004). Interestingly, the MFB1 and MDM30 genes are induced during sporulation (Chu et al., 1998), raising the intriguing possibility that mitochondria-associated F-box proteins are required for progression through the mitochondrial remodeling program during meiosis and spore formation. To test this, we observed mitochondrial behavior during sporulation in diploid wild-type cells and homozygous diploid cells lacking Mfb1 or Mdm30. The tubular mitochondrial network of logarithmically growing wild-type cells fragments upon entry into stationary phase. Mitochondrial fragments fuse again in pretetrads to build a complex tubular network. Tubular mitochondria are then partitioned to the newly formed spores in early tetrads and fragment once again in late tetrads (Miyakawa et al., 1984; Gorsich and Shaw, 2004; Figure 6A and Table 3). Although the behavior of mitochondria during sporulation is virtually identical in wild-type cells and cells lacking Mfb1, formation of a tubular mitochondrial network in pretetrads is blocked in the absence of Mdm30, and mitochondria remain fragmented throughout the entire sporulation process (Figure 6A and Table 3).
Figure 6.
Mdm30 is required for mitochondrial remodeling and inheritance during sporulation. (A) Homozygous diploid WT, Δmfb1 and Δmdm30 cells expressing mitochondria-targeted GFP were grown to log phase (top) in YPD medium. To induce sporulation, strains were grown to stationary phase on YPD plates, transferred to sporulation medium, and incubated at 30°C for 12 d. At each stage of sporulation, representative cells were analyzed by fluorescence and DIC microscopy, as in Figure 2B. Right is a merged image of DIC and fluorescence micrographs. Mitochondrial inheritance defects were observed in early and late tetrads. Ascospores devoid of mitochondria are marked with an asterisk. Bar, 2 μm. (B) Percentage of asci is indicated that contained at least one spore devoid of mitochondria at the early or late tetrad stage (number of asci scored: WT, 413; Δmfb1, 422; and Δmdm30, 264). (C) Percentage of cells is indicated that had formed asci with two to four spores at the late tetrad stage (number of cells scored: 400 for each strain).
Table 3.
Mitochondrial phenotypes during sporulation
| Stage/strain | Mitochondrial morphology (% of cells) | No. of cells or asci scored | ||
|---|---|---|---|---|
| Tubular | Aggregated | Fragmented | ||
| Stationary phase | ||||
| WT | 13.0 | 0.6 | 86.4 | 828 |
| Δmfb1/Δmfb1 | 5.0 | 0.6 | 94.4 | 813 |
| Δmdm30/Δmdm30 | 1.5 | 1.3 | 97.2 | 800 |
| Pretetrad | ||||
| WT | 75.5 | 2.5 | 22.0 | 405 |
| Δmfb1/Δmfb1 | 62.8 | 3.0 | 34.2 | 404 |
| Δmdm30/Δmdm30 | 0.8 | 5.3 | 93.9 | 400 |
| Early tetrad | ||||
| WT | 91.6 | 3.5 | 4.9 | 286 |
| Δmfb1/Δmfb1 | 85.3 | 5.3 | 9.4 | 266 |
| Δmdm30/Δmdm30 | 4.1 | 14.2 | 81.7 | 169 |
| Late tetrads | ||||
| WT | 7.1 | 3.9 | 89.0 | 127 |
| Δmfb1/Δmfb1 | 6.4 | 1.3 | 92.3 | 156 |
| Δmdm30/Δmdm30 | 0 | 2.1 | 97.9 | 95 |
We asked whether the aberrant behavior of mitochondria in homozygous diploid Δmdm30 cells would result in a mitochondrial inheritance defect. To test this, spores devoid of mitochondria (Figure 6A, bottom) were counted in early and late tetrads. In wild-type cells, 6.5% of asci contained at least one spore without any visible mitochondrion. This number was slightly increased to 10.4% in Δmfb1 asci. Interestingly, the mitochondrial inheritance defect was much more severe in Δmdm30 asci, where 23.1% of the asci contained one or more spores devoid of mitochondria (Figure 6B). At the same time, sporulation efficiency was decreased in cells lacking Mdm30. Although 15.3% of wild-type cells and 15.8% of homozygous diploid Δmfb1 cells formed tetrads, only 5.3% of homozygous diploid Δmdm30 cells formed asci (Figure 6C). We conclude that Mdm30-dependent remodeling of mitochondria is important for mitochondrial inheritance during sporulation.
DISCUSSION
Even in a simple unicellular organism, such as yeast, mitochondria show a complex behavior. Volume, number, and morphology of mitochondria are readily adapted to the carbon source of the growth medium and oxygen supply (Stevens, 1981; Visser et al., 1995; Egner et al., 2002; Jakobs et al., 2003). During mitotic growth, mating, and sporulation, mitochondria are partitioned to newly formed cells by highly ordered inheritance mechanisms and dynamic remodeling processes (Miyakawa et al., 1984; Nunnari et al., 1997; Simon et al., 1997; Okamoto et al., 1998; Yang et al., 1999; Gorsich and Shaw, 2004). How are mitochondrial fusion and fission orchestrated during these events?
The complement of F-box proteins that is present in every eukaryotic cell is thought to have sweeping regulatory powers by virtue of its ability to directly link SCF substrates to signaling pathways (Willems et al., 2004). Consistently, our results assign a key role to F-box proteins in regulating mitochondrial dynamics. Mfb1 and Mdm30 are both required for proper mitochondrial distribution and morphology during mitotic growth. A comprehensive analysis of mutants of all known yeast F-box protein-encoding genes suggests that the remaining 19 members of this protein family are dispensable for maintenance of wild-type–like mitochondria. Consistent with this finding, large-scale analysis of protein localization in yeast (Huh et al., 2003) and analysis of the mitochondrial proteome (Sickmann et al., 2003) did not reveal any other mitochondria-associated F-box protein. Genetic and morphological evidence suggests that Mfb1 and Mdm30 function independently of each other and may have different target proteins. A Δmfb1/Δmdm30 double mutant does not show a more severe phenotype than a Δmdm30 single mutant; overexpression of one F-box protein does not compensate for the loss of the other; and deletion of the MDM30 gene leads to a block in fusion, whereas deletion of the MFB1 gene leads to clumped interconnected mitochondria. In addition to their role during mitotic growth, mitochondria-associated F-box proteins are important for mitochondrial dynamics during the execution of cell developmental programs. Consistent with a role in mitochondrial dynamics during development, both genes are several-fold induced during sporulation (Chu et al., 1998). Although a role of Mfb1 in regulating mitochondrial behavior during meiosis is still unknown, we consider it likely that Mdm30 modulates mitochondrial fusion activity because fusion of fragmented mitochondria to a tubular network is blocked in pretetrads lacking Mdm30 (Figure 6A). Because Mdm30 is also required for fusion of mitochondria during mating (Fritz et al., 2003), F-box proteins play an important role in mitochondrial dynamics throughout the entire life cycle of budding yeast.
A synthetic growth defect in Δfzo1/Δmdm30 double mutants (Fritz et al., 2003) and Fzo1-independent binding of Mdm30 to mitochondria (Figure 4) suggest that Mdm30 may have other yet unknown mitochondrial target proteins in addition to Fzo1. Interestingly, it has recently been reported that Mdm30 is a transcriptional coactivator of Gal4. This activity has been proposed to be required for activation of Gal4 target genes and growth on synthetic galactose medium (Muratani et al., 2005). However, we and others observed that Δmdm30 mutants are able to grow on galactose-containing media (Okamoto and Shaw, 2005), and mitochondria-targeted fluorescent proteins are efficiently expressed from the GAL1 promoter in Δmdm30 strains (Fritz et al., 2003). Thus, it is not clear whether Mdm30 is of general importance for expression of Gal4-dependent genes, and possible additional target proteins of mitochondria-associated F-box proteins remain to be identified.
Numerous lines of evidence point to an important role of the ubiquitin/26S proteasome system in mitochondrial biogenesis. Yeast mutants lacking functional subunits of ubiquitin ligases or the proteasome harbor aberrant mitochondria (Rinaldi et al., 1998; Fisk and Yaffe, 1999; Altmann and Westermann, 2005), and proteasome inhibitors (Neutzner and Youle, 2005) and mutations in proteasome subunit-encoding genes (Escobar-Henriques et al., 2006) reduce the rate of degradation of Fzo1 in α factor-arrested cells. Consistent with a specific role of ubiquitylation of mitochondrial proteins, the mitochondrial outer membrane harbors a deubiquitinating enzyme, Ubp16. However, neither deletion nor overexpression of the UBP16 gene produces any obvious phenotype, and mitochondrial morphology and inheritance are not affected in ubp16 mutants (Kinner and Kölling, 2003). What might be the role of Mfb1 and Mdm30 in mediating turnover of Fzo1 and other mitochondrial target proteins? Mdm30 has been shown to interact with Skp1 and Cdc53 in the yeast two-hybrid system (Uetz et al., 2000), recombinant Mdm30 assembles with Skp1, Cdc53, and Rbx1 into an SCF complex in vitro (Kus et al., 2004), and Mfb1 has been found in a complex with Skp1 in large-scale identification of yeast protein complexes by mass spectrometry (Ho et al., 2002) and in coimmunoprecipitation experiments (Kondo-Okamoto et al., 2006). However, Mfb1–Skp1 complexes apparently do not contain Cdc53 (Kondo-Okamoto et al., 2006). Intriguingly, we observed that the mitochondrial phenotype of yeast strains lacking Mfb1 and Mdm30 cannot be mimicked by depletion of SCF core complex or proteasome subunits and that the F-box motif of Mfb1 and Mdm30 is not essential for rescue of the mitochondrial morphology defect of the null mutants. Furthermore, proteasome-dependent degradation of Fzo1 in α factor-arrested cells does not require Mdm30 (Neutzner and Youle, 2005) or Mfb1 (Figure 3C), and Mdm30-mediated turnover of Fzo1 in vegetatively growing cells is independent of the proteasome (Escobar-Henriques et al., 2006). Thus, it seems that mitochondria-associated F-box proteins do not act as bona fide SCF ubiquitin ligases. Rather, they may exert at least some of their functions independently of the proteasome, as it has been demonstrated for Rcy1, an F-box protein involved in regulation of vesicular trafficking (Galan et al., 2001). Apparently, Mfb1 and Mdm30 are two important components of a complex network of substrate recognition factors and proteolytic systems mediating turnover of mitochondrial proteins. Many, yet unknown, factors remain to be identified.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Rita Grotjahn and Annette Suske for technical support and Koji Okamoto for communicating results before publication. This work was supported by grants of the Deutsche Forschungsgemeinschaft to B. W. (We 2174/2-4 and 4-1) and T. L. (SFB 635/P18) and an European Molecular Biology Organization fellowship to M.E.-H.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-01-0053) on June 21, 2006.
REFERENCES
- Altmann K., Westermann B. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:5410–5417. doi: 10.1091/mbc.E05-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C., Herzog V., Bauer M. F. Improved technique for electron microscope visualization of yeast membrane structure. Microsc. Microanal. 2001;7:530–534. doi: 10.1017.S1431927601010522. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int. Rev. Cytol. 1990;122:1–63. doi: 10.1016/s0074-7696(08)61205-x. [DOI] [PubMed] [Google Scholar]
- Bleazard W., McCaffery J. M., King E. J., Bale S., Mozdy A., Tieu Q., Nunnari J., Shaw J. M. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Chen H., Chan D. C. Mitochondrial dynamics in mammals. Curr. Top. Dev. Biol. 2004;59:119–144. doi: 10.1016/S0070-2153(04)59005-1. [DOI] [PubMed] [Google Scholar]
- Chen H., Detmer S. A., Ewald A. J., Griffin E. E., Fraser S. E., Chan D. C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. J., Butow R. A. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., Brown P. O., Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Dimmer K. S., Fritz S., Fuchs F., Messerschmitt M., Weinbach N., Neupert W., Westermann B. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:847–853. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner A., Jakobs S., Hell S. W. Fast 100 nm resolution 3D-microscope reveals structural plasticity of mitochondria in live yeast. Proc. Natl. Acad. Sci. USA. 2002;99:3370–3375. doi: 10.1073/pnas.052545099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Henriques M., Westermann B., Langer T. Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J. Cell Biol. 2006;173:645–650. doi: 10.1083/jcb.200512079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eura Y., Ishihara N., Yokota S., Mihara K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J. Biochem. 2003;134:333–344. doi: 10.1093/jb/mvg150. [DOI] [PubMed] [Google Scholar]
- Fisk H. A., Yaffe M. P. A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:1199–1208. doi: 10.1083/jcb.145.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz S., Weinbach N., Westermann B. Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast. Mol. Biol. Cell. 2003;14:2303–2313. doi: 10.1091/mbc.E02-12-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J. M., Wiederkehr A., Seol J. H., Haguenauer-Tsapis R., Deshaies R. J., Riezman H., Peter M. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol. 2001;21:3105–3117. doi: 10.1128/MCB.21.9.3105-3117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gietz D., Jean A. S., Woods R. A., Schiestl R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsich S. W., Shaw J. M. Importance of mitochondrial dynamics during meiosis and sporulation. Mol. Biol. Cell. 2004;15:4369–4381. doi: 10.1091/mbc.E03-12-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L., van der Bliek A. M. The many shapes of mitochondrial membranes. Traffic. 2001;2:235–244. doi: 10.1034/j.1600-0854.2001.1r008.x. [DOI] [PubMed] [Google Scholar]
- Hales K. G., Fuller M. T. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Herlan M., Bornhövd C., Hell K., Neupert W., Reichert A. S. Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J. Cell Biol. 2004;165:167–173. doi: 10.1083/jcb.200403022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlan M., Vogel F., Bornhövd C., Neupert W., Reichert A. S. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 2003;278:27781–27788. doi: 10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- Hermann G. J., Thatcher J. W., Mills J. P., Hales K. G., Fuller M. T., Nunnari J., Shaw J. M. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O’Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jagasia R., Grote P., Westermann B., Conradt B. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- Jakobs S., Martini N., Schauss A. C., Egner A., Westermann B., Hell S. W. Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p. J. Cell Sci. 2003;116:2005–2014. doi: 10.1242/jcs.00423. [DOI] [PubMed] [Google Scholar]
- Kinner A., Kölling R. The yeast deubiquitinating enzyme Ubp16 is anchored to the outer mitochondrial membrane. FEBS Lett. 2003;549:135–140. doi: 10.1016/s0014-5793(03)00801-9. [DOI] [PubMed] [Google Scholar]
- Kondo-Okamoto N., Ohkuni K., Kitagawa K., McCaffery J. M., Shaw J. M., Okamoto K. The novel F-box protein Mfb1p regulates mitochondrial connectivity and exhibits asymmetric localization in yeast. Mol. Biol. Cell. 2006;17:3756–3767. doi: 10.1091/mbc.E06-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T., Detmer S. A., Kaiser J. T., Chen H., McCaffery J. M., Chan D. C. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- Kremer J. R., Mastronarde D. N., McIntosh J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kus B. M., Caldon C. E., Andorn-Broza R., Edwards A. M. Functional interaction of 13 yeast SCF complexes with a set of yeast E2 enzymes in vitro. Proteins. 2004;54:455–467. doi: 10.1002/prot.10620. [DOI] [PubMed] [Google Scholar]
- Lill R., Mühlenhoff U. Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 2005;30:133–141. doi: 10.1016/j.tibs.2005.01.006. [DOI] [PubMed] [Google Scholar]
- McQuibban G. A., Saurya S., Freeman M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 2003;423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- Miyakawa I., Aoi H., Sando N., Kuroiwa T. Fluorescence microscopic studies of mitochondrial nucleoids during meiosis and sporulation in the yeast, Saccharomyces cerevisiae. J. Cell Sci. 1984;66:21–38. doi: 10.1242/jcs.66.1.21. [DOI] [PubMed] [Google Scholar]
- Mnaimneh S., et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Mozdy A., McCaffery J. M., Shaw J. M. Dnm1p GTPase-mediated mitochondrial fusion is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000;151:367–379. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A. D., Shaw J. M. A fuzzy mitochondrial fusion apparatus comes into focus. Nat. Rev. Mol. Cell Biol. 2003;4:468–478. doi: 10.1038/nrm1125. [DOI] [PubMed] [Google Scholar]
- Muratani M., Kung C., Shokat K. M., Tansey W. P. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell. 2005;120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Neutzner A., Youle R. J. Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:18598–18603. doi: 10.1074/jbc.M500807200. [DOI] [PubMed] [Google Scholar]
- Nunnari J., Marshall W. F., Straight A., Murray A., Sedat J. W., Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Perlman P. S., Butow R. A. The sorting of mitochondrial DNA and mitochondrial proteins in zygotes: preferential transmission of mitochondrial DNA to the medial bud. J. Cell Biol. 1998;142:613–623. doi: 10.1083/jcb.142.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Shaw J. M. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Petroski M. D., Deshaies R. J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Rapaport D., Brunner M., Neupert W., Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- Rinaldi T., Ricci C., Porro D., Bolotin-Fukuhara M., Frontali L. A mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces cell cycle arrest, overreplication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. Mol. Biol. Cell. 1998;9:2917–2931. doi: 10.1091/mbc.9.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo M., Legros F., Chateau D., Lombes A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J. Cell Sci. 2002;115:1663–1674. doi: 10.1242/jcs.115.8.1663. [DOI] [PubMed] [Google Scholar]
- Santel A., Frank S., Gaume B., Herrler M., Youle R. J., Fuller M. T. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J. Cell Sci. 2003;116:2763–2774. doi: 10.1242/jcs.00479. [DOI] [PubMed] [Google Scholar]
- Scheffler I. E. A century of mitochondrial research: achievements and perspectives. Mitochondrion. 2000;1:3–31. doi: 10.1016/s1567-7249(00)00002-7. [DOI] [PubMed] [Google Scholar]
- Scott S. V., Cassidy-Stone A., Meeusen S. L., Nunnari J. Staying in aerobic shape: how the structural integrity of mitochondria and mitochondrial DNA is maintained. Curr. Opin. Cell Biol. 2003;15:482–488. doi: 10.1016/s0955-0674(03)00070-x. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R. E. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H., Jensen R. E. UGO1 encodes an outer membrane protein required for mitochondrial fusion. J. Cell Biol. 2001;152:1123–1134. doi: 10.1083/jcb.152.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H., Jensen R. E. Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion. J. Biol. Chem. 2004;279:28298–28303. doi: 10.1074/jbc.M401363200. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Southard S. M., Hobbs A. E., Jensen R. E. Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem. Biophys. Res. Commun. 2003;308:276–283. doi: 10.1016/s0006-291x(03)01348-2. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sherman F., Hicks J. Micromanipulation and dissection of asci. Methods Enzymol. 1991;194:21–37. doi: 10.1016/0076-6879(91)94005-w. [DOI] [PubMed] [Google Scholar]
- Sickmann A., et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon V. R., Karmon S. L., Pon L. A. Mitochondrial inheritance: cell cycle and actin cable dependence of polarized mitochondrial movements in Saccharomyces cerevisiae. Cell Motil. Cytoskeleton. 1997;37:199–210. doi: 10.1002/(SICI)1097-0169(1997)37:3<199::AID-CM2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Spurr A. R. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Stevens B. In: Mitochondrial structure. In: The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Strathern E. W., Jones E. W., Broach J. R., editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1981. pp. 471–504. [Google Scholar]
- Tatsuta T., Langer T. Studying proteolysis within mitochondria. Curr. Topics Gen. 2006 doi: 10.1007/978-1-59745-365-3_25. (in press) [DOI] [PubMed] [Google Scholar]
- Uetz P., et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:601–603. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Visser W., van Spronsen E. A., Nanninga N., Pronk J. T., Gijs Kuenen J., van Dijken J. P. Effects of growth conditions on mitochondrial morphology in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek. 1995;67:243–253. doi: 10.1007/BF00873688. [DOI] [PubMed] [Google Scholar]
- Westermann B. Merging mitochondria matters. Cellular role and molecular machinery of mitochondrial fusion. EMBO Rep. 2002;3:527–531. doi: 10.1093/embo-reports/kvf113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B., Neupert W. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 2000;16:1421–1427. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Willems A. R., Schwab M., Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Wong E. D., Wagner J. A., Gorsich S. W., McCaffery J. M., Shaw J. M., Nunnari J. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 2000;151:341–352. doi: 10.1083/jcb.151.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E. D., Wagner J. A., Scott S. V., Okreglak V., Holewinske T. J., Cassidy-Stone A., Nunnari J. The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J. Cell Biol. 2003;160:303–311. doi: 10.1083/jcb.200209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M. P. The machinery of mitochondrial inheritance and behavior. Science. 1999;283:1493–1497. doi: 10.1126/science.283.5407.1493. [DOI] [PubMed] [Google Scholar]
- Yang H. C., Palazzo A., Swayne T. C., Pon L. A. A retention mechanism for distribution of mitochondria during cell division in budding yeast. Curr. Biol. 1999;9:1111–1114. doi: 10.1016/s0960-9822(99)80480-1. [DOI] [PubMed] [Google Scholar]
- Youle R. J., Karbowski M. Mitochondrial fission in apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.