Abstract
Chemotaxis of amoeboid cells is driven by actin filaments in leading pseudopodia and actin–myosin filaments in the back and at the side of the cell to suppress pseudopodia. In Dictyostelium, cGMP plays an important role during chemotaxis and is produced predominantly by a soluble guanylyl cyclase (sGC). The sGC protein is enriched in extending pseudopodia at the leading edge of the cell during chemotaxis. We show here that the sGC protein and the cGMP product have different functions during chemotaxis, using two mutants that lose either catalytic activity (sGCΔcat) or localization to the leading edge (sGCΔN). Cells expressing sGCΔN exhibit excellent cGMP formation and myosin localization in the back of the cell, but they exhibit poor orientation at the leading edge. Cells expressing the catalytically dead sGCΔcat mutant show poor myosin localization at the back, but excellent localization of the sGC protein at the leading edge, where it enhances the probability that a new pseudopod is made in proximity to previous pseudopodia, resulting in a decrease of the degree of turning. Thus cGMP suppresses pseudopod formation in the back of the cell, whereas the sGC protein refines pseudopod formation at the leading edge.
INTRODUCTION
Chemotaxis is a vital process in a variety of organisms, ranging from bacteria to vertebrates. Prokaryotes use chemotaxis to move toward high nutrient concentrations or away from unfavorable conditions, whereas in eukaryotes chemotaxis is also involved in embryogenesis, wound healing, and the immune response. Chemotaxis is achieved by coupling gradient sensing to basic cell movement. Prokaryotes are too small to sense spatial gradients and therefore rely on temporal changes of the chemoattractant concentration to achieve chemotaxis. They do this by adjusting their tumbling frequency in response to temporal changes of the chemoattractant concentration (Szurmant and Ordal, 2004; Wadhams and Armitage, 2004). Eukaryotic cells are typically large enough to be able to measure a spatial gradient. The difference in receptor occupation between each side of the cell leads to an internal polarization. A pseudopod is extended at the side with the highest receptor occupation and at the same time, pseudopod formation at all other sides is repressed, resulting in directional cell migration (Devreotes and Janetopoulos, 2003; Postma et al., 2004).
Dictyostelium is a eukaryotic organism that is widely used to study chemotaxis (Williams and Harwood, 2003; Parent, 2004; Affolter and Weijer, 2005). Starved Dictyostelium cells periodically secrete cAMP. Through relay of the cAMP signal by neighboring cells, concentric cAMP waves are generated. Starved Dictyostelium cells are chemotactically sensitive to cAMP and by movement in the direction of the origin of the cAMP waves, the cells are able to aggregate into groups of up to 100,000 cells. The chemotactic response of Dictyostelium is optimized for the dynamic cAMP waves that coordinate both aggregation and multicellular development. Cells show a much stronger chemotactic response to a cAMP wave, where the mean concentration increases over time, than to a static spatial gradient. Dictyostelium uses both spatial gradient sensing and the “bacterial-like” temporal gradient sensing to respond to these dynamic chemoattractant gradients (Futrelle, 1982; Varnum-Finney et al., 1987; Iijima et al., 2002; Xu et al., 2005).
The signal transduction pathways that are coupled to spatial gradient sensing and temporal gradient sensing rely on signal molecules with differential physical properties. Molecules that store spatial information have low diffusion rates to prevent the information from simply diffusing away. Because lipid molecules have very low diffusion constants (Almeida and Vaz, 1995), the phosphatidyl inositol phosphates that are formed during chemotaxis are excellent molecules to store spatial information (Funamoto et al., 2001; Wang et al., 2002). In contrast, molecules that carry temporal information benefit from high diffusion rates, which allow a rapid propagation of the signal throughout the cell. Small soluble molecules have high diffusion rates and are therefore most suitable as signal molecules for temporal signal transduction. One of the second messengers used during Dictyostelium chemotaxis is cGMP, which mediates the formation of myosin filaments in response to a cAMP stimulus (Mato and Malchow, 1978; Liu and Newell, 1988; Van Haastert and Kuwayama, 1997). Being a small molecule, cGMP has a very high diffusion constant of ∼300 μm2/min (Allbritton et al., 1992; Chen et al., 1999). It can be calculated that the average dispersion length of a cGMP molecule is 50 μm, which is about 5 times the size of a Dictyostelium cell (Postma and Van Haastert, 2001). This makes cGMP unsuitable to store spatial information but an excellent candidate to transduce temporal information. The predominant source of cGMP during development is soluble guanylyl cyclase (sGC) (Roelofs et al., 2001a; Roelofs and Van Haastert, 2002). With a molecular mass of 315 kDa, sGC is an unusually large protein. The protein is enriched in pseudopodia and localizes to the leading edge of the cell in a chemoattractant gradient (Veltman et al., 2005), where it exhibits very slow diffusion (D = 6 μm2/min; Ruchira Engel, unpublished data). With the observed localization and its slow diffusion, sGC has the potential to store spatial information. Thus, the sGC protein has opposite properties from its cGMP product, and the combination of sGC localization and transient cGMP formation allows the processing of both spatial and temporal information during chemotaxis. In this article, we explore the role of the localization of the sGC protein and formation of cGMP product during chemotaxis by generating two mutants of the sGC protein that uncouple localization of the protein and catalytic activity. Deletion of the N-terminal segment of sGC yields a protein that retains guanylyl cyclase activity but loses its localization to the leading edge. Cells expressing this protein exhibit excellent chemotaxis during the rising flank of a cAMP wave, but they lose their orientation in a stable spatial gradient. The second mutant contains a point mutation of a catalytic amino acid, yielding a protein that has no guanylyl cyclase activity but still localizes to the leading edge. Cells expressing this catalytically dead mutant show poor chemotaxis in a spatiotemporal gradient, but they improve chemotaxis in stable spatial gradients to the level of wild-type cells. These data suggest that localization of the sGC protein in pseudopodia and the leading edge retains spatial information and stabilizes the leading edge, whereas transient cGMP formation induces myosin filament formation and suppresses pseudopod formation in the back and at the sides of the cell.
MATERIALS AND METHODS
Strains and Culture Conditions
All cell strains were grown in HG5 medium (14.3 g/l oxoid pepton, 7.15 g/l bacto yeast extract, 1.36 g/l Na2HPO4·12H2O, 0.49 g/l KH2PO4, and 10.0 g/l glucose). When grown with selection, HG5 medium was supplemented with 10 μg/ml G418 or 50 μg/ml hygromycin. Previous to cGMP activity assays and confocal microscopy, cells were starved for 5 h by shaking in 17 mM phosphate buffer, pH 6.5 (PB), at a density of 107 cells/ml.
Cloning of the Expression Constructs
Plasmid pBK-CMV/sGC (Veltman et al., 2005) that contains the entire open reading frame (ORF) of sgc (GenBank AF361947) was used as starting material to construct the various mutants. The ORF of sgc is preceded by a PstI and BamHI site. The stop codon is immediately followed by a BamHI site. In the ORF used to construct the green fluorescent protein (GFP) fusion proteins, the endogenous stop codon was removed. To construct sGCΔN, we first obtained a small DNA fragment by PCR with full-length sgc DNA as template using the forward primer ctgcagggatccaaaatgggaaatacatcaccaatgtc together with a downstream, internal sgc primer, gttgcagaagctaataaacc; the DNA fragment contains base pairs 2632-3110 of sgc and is preceded by a start codon (bold in primer), a Kozak sequence, a BamHI site, and a PstI site. Subsequently, the N-terminal region of full-length sgc was excised from pBK-CMV/sGC by using PstI and KpnI. KpnI recognizes the internal sgc restriction site at base pair 3061. The fragment was replaced by the PCR fragment digested with the same enzymes. To obtain the sGCΔcat mutant, we used site-directed mutagenesis to replace the codon encoding amino acid 1106 from gat (aspartate) to gct (alanine). Mutant sGCΔNΔcat was obtained in the same way as the generation of sGCΔN, but using sGCΔcat as starting material instead. To obtain the expression constructs, the obtained mutant sGC constructs were excised with BamHI and cloned into either MB74 (Veltman et al., 2005) digested with BglII for nonfusion products or into MB74-GFP digested with BglII for GFP-fusion. Mutant sGCΔCΔcat was created as follows. The ORF of sGCΔcat was digested with BamHI and XbaI. The endogenous XbaI site is located at base pairs 4534, just C-terminal of the catalytic domain. This fragment was cloned into either pDM135 or MB74-GFP digested with BglII and SpeI. Plasmid pDM135 is identical to MB74 except that the SpeI site is immediately followed by a stop codon in all three reading frames. The resulting ORF encodes the first 1511 amino acids of sGCΔcat, with the addition of an extra serine residue at the C terminus just before the stop codon. All mutations were confirmed by sequencing.
Generation of the gc-Null Cell Strain
The gc-null cell strain was remade in an AX3 background cell strain. A knockout construct, termed pDM100, was created as follows. Plasmid pBK-CMV/sGC was digested with HindIII and EcoRV, removing the central 6.5 kb of the sgc gene. This fragment was replaced by a hygromycin resistance cassette that was obtained by digesting plasmid pHygTm(plus) (a kind gift of Jeff Williams, Developmental Biology, University of Dundee, Dundee, United Kingdom) with BstXI and HindIII. The BstXI site of the fragment was made blunt by a Klenow fill-in. A linear knockout fragment was obtained through PCR, using forward primer accaatcgaagaagtgcagg and reverse primer tgaacatcttcaccatcc on plasmid pDM100. gca-null cells (Roelofs et al., 2001b) were transfected with 5 μg of the purified PCR product. Cells were selected with 35 μg/ml hygromycin, and clonal transfectants were isolated. Proper disruption of the sgc gene in the obtained clones was confirmed with both PCR and Southern blotting.
cGMP Assays
In Vitro Guanylyl Cyclase Activity.
Starved cells were washed and resuspended in 10 mM Tris, pH 8.0, to a density of 2 × 108 cells/ml. All subsequent steps were performed at 4°C. One volume of cell suspension was mixed with 1 volume of lysis buffer yielding 15 mM Tris, 250 mM sucrose, and 3 mM EGTA, pH 8.0, and lysed through a 4-μm Nuclepore filter. Guanylyl cyclase assays were performed at 22°C with 50 μl of cell lysate in a total volume of 100 μl containing 15 mM Tris, pH 8.0, 250 mM sucrose, 0.5 mM GTP, 10 mM 1,4-dithiothreithol, 1.5 mM EGTA, and 2.5 mM MnCl2 (final concentrations). Reactions were terminated after 20, 40, and 60 s by addition of 40 μl of assay mixture to an equal volume of 3.5% perchloric acid. Samples were neutralized by adding 20 μl of 50% saturated KHCO3 and incubated for 5 min at room temperature to allow the CO2 to escape. Samples were then centrifuged 5 min at 500× g, and the cGMP levels of the supernatant were determined using the Biotrak [3H]cGMP assay kit (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) according to manufacturer’s instructions.
In Vivo cGMP Response.
Starved cells were washed in PB, resuspended to a density of 1 × 108 cells/ml in PB containing 2 mM caffeine, and aerated for at least 10 min. Cells were stimulated by adding 10 μl of cAMP solution to 40 μl of cell suspension, yielding a final concentration of 100 nM cAMP. The reaction was stopped by adding 40 μl of stimulated cell suspension to an equal volume of 3.5% perchloric acid at indicated time points. Samples were neutralized and assayed for cGMP content as described in the in vitro assay.
Chemotaxis Assay
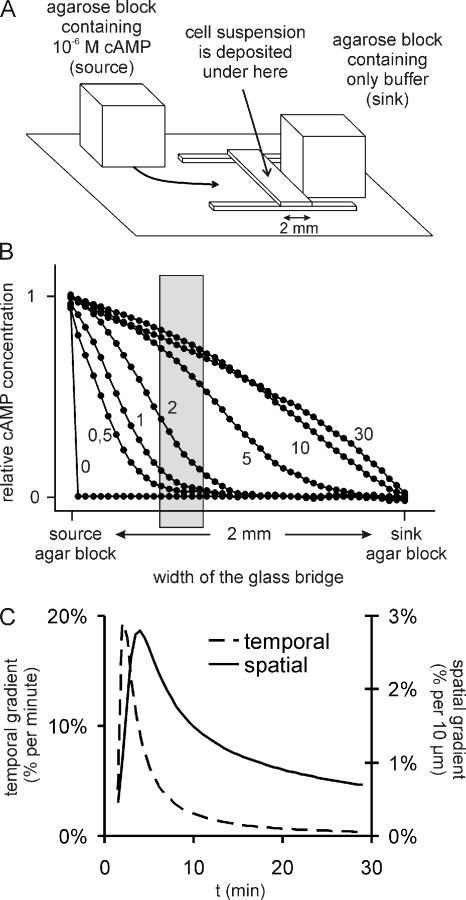
Vegetative cells were harvested, washed once in PB, settled as a monolayer on a glass slide under a small volume of PB, and allowed to starve. Cells were harvested at the onset of streaming. The starved cell suspension was pipetted under the glass bridge of a modified Zigmond chemotaxis chamber, and cells were allowed to settle down (Figure 2A; Zigmond, 1977). The chemotaxis chamber was constructed on a microscope slide, using glass strips with dimensions ∼2 × 24 mm, that were cut from the coverslip of a hemocytometer (24 × 24 mm, thickness of 0.15 mm). A bridge was made by placing one glass strip perpendicularly on top of two supporting glass strips. Blocks of agarose [1% (wt/vol) in PB] were used to create a cAMP gradient; these blocks were casted in a cuvette (10 × 10 mm). A block of agarose containing PB was placed alongside one edge of the bridge and a block of agarose containing 1 μM cAMP in PB was placed at the opposite side, making sure that both blocks make contact with the fluid under the bridge. The chemotaxis bridge was put in a chamber that was kept at saturated humidity to prevent the fluid under the bridge from evaporating. A gradient is formed across glass bridge by diffusion of the cAMP from the “source” block to the “sink” block. To determine the kinetics of gradient formation, the source agar block contained 50% saturated bromophenol blue, a dye with a molecular weight similar to that of cAMP. The distribution of dye concentration across the bridge was recorded at different times after assembly of the chamber using a digital camera.
Figure 2.
Chemotaxis setup and gradient formation. (A) Schematic drawing of the chemotaxis setup modeled after the Zigmond chamber. A droplet of cell suspension is deposited underneath the central glass bridge. An agarose block containing only buffer (sink) is placed on one side and an agarose block containing 10−6 M cAMP (source) is placed on the opposite side of the bridge. A cAMP gradient is formed across the bridge by diffusion of the cAMP from the source to the sink. (B) The formation of the cAMP gradient was deduced by measuring the diffusion across the bridge of bromophenol blue, a dye with a molecular weight similar to that of cAMP. The distribution of dye concentration across the bridge is plotted for different time points (indicated in minutes next to the graphs) after initialization of the gradient. Chemotaxis of cells is observed in an area of 350 × 270 μm at a distance of 700 μm from the source; this area is indicated by the gray box. (C) Deduced time course of the temporal and spatial cAMP gradients in the center of the area indicated by the gray box in Figure 3B. The temporal gradient is expressed as the percentage cAMP concentration increase per minute. The spatial gradient is expressed as the percentage cAMP concentration difference across 10 μm, which equals approximately the length of a Dictyostelium cell.
Chemotaxis of cells is recorded in an area of 350 × 270 μm at a distance of 700 μm from the source; this area is indicated by the gray box in Figure 2B. The cells were recorded for 35 min, starting immediately after assembly of the chemotaxis chamber. The recorded movie was analyzed as follows. In ImageJ (http://rsb.info.nih.gov/ij/), the contour and the position of each individual cell in the movie was determined every 30 s, yielding both a cell track and a series of coordinates for each cell. Using these coordinates, the chemotaxis index of every step was calculated (the ratio of its displacement in the direction of the gradient and its traveled distance), yielding a series of chemotaxis indices for each cell in the movie. To determine the degree of turning, the displacement of the cells during 1 min was calculated as a vector. The degree of turning is defined as the angle between subsequent vectors. At least four movies were analyzed for each cell strain, and ∼20 individual cells were analyzed per movie. The data shown are the average and SE of the mean, with n representing the number of cells analyzed.
Fluorescence of the expressed GFP and monomeric Kusabira-Orange (mKO) fusion constructs was visualized using a Zeiss LSM510 (Carl Zeiss, Jena, Germany) confocal fluorescence microscope fitted with a Plan-Neofluar 40× 1.3 numerical aperture oil immersion objective. The collected fluorescence data were quantified on a computer using the MatLab program (Mathworks, Natick, MA).
RESULTS
Localization and Activity of Two Mutant sGC Enzymes
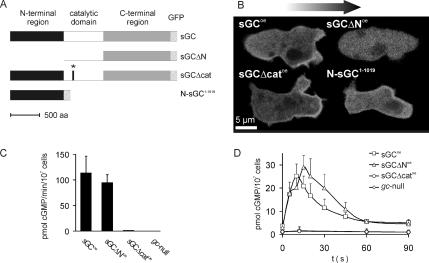
Two mutants of the Dictyostelium sGC were constructed (Figure 1A). In the first mutant, termed sGCΔN, the N-terminal 877 amino acids were deleted. In the second construct, termed sGCΔcat, a point mutation was introduced in the catalytic site. The aspartate at position 1106, which coordinates the magnesium ion during the cyclization reaction, was substituted for an alanine residue, rendering the enzyme inactive (Olson et al., 1998). Wild-type sGC and mutant sGC constructs were fused at the C terminus to GFP and expressed in gc-null cells. The Dictyostelium genome encodes two guanylyl cyclases, GCA and sGC. Both cyclases have been knocked out in the gc-null cell strain, and as a consequence, these cells are no longer able to synthesize cGMP. It has previously been shown that full-length sGC-GFP localizes to the leading edge of the cell during chemotaxis; the fluorescence intensity at the leading edge is about twofold higher than the fluorescence intensity in the cytosol (Veltman et al., 2005). The catalytically inactive mutant shows a similar, even somewhat more pronounced, anterior localization as wild-type sGC (Figure 1B and Supplemental Movie). In contrast, sGCΔN remains entirely cytosolic, suggesting that the N-terminal region targets the enzyme to the leading edge. A GFP-fusion protein, termed N-sGC1-1019, that consists of only the N-terminal region of sGC showed a similar localization as the full-length sGC. This indicates that the N-terminal region is not only required but also sufficient for anterior localization. In an attempt to identify a minimal fragment that is sufficient for the observed localization, we expressed another five GFP-fusion constructs that spanned amino acids 1-305, 306-680, 681-1019, 1-680, and 306-1019, respectively. However, none of these GFP-fusion proteins showed significant anterior localization (our unpublished data), indicating that the entire N-terminal region is required.
Figure 1.
Overview of the constructed sGC mutant, their cellular localization and activity. (A) Schematic overview and names of the sGC mutant proteins. (B) sGC mutant constructs from A were expressed in gc-null cells. Cells were starved for 5 h, placed in a chemoattractant gradient, and confocal images of the GFP fluorescence were acquired. The arrow points in the direction of movement. (C) In vitro guanylyl cyclase activity. Cells were lysed by pressing them through a 4-μm pore filter and lysates were incubated in assay mixture containing GTP. The reaction was stopped at various time points, and the amount of cGMP was determined using an isotope dilution assay. From these results, the rate of cGMP synthesis was calculated. (D) cGMP response of cells in vivo. Cells were stimulated at t = 0 with 10−7 M cAMP. The reaction was stopped at the indicated time points, and the amount of accumulated cGMP was determined.
Next, the guanylyl cyclase activity of the sGC mutant cell strains in vitro and the chemoattractant-stimulated cGMP response in vivo were measured, which are both absent in the parent gc-null cells (Figure 1C). A lysate of gc-null cells expressing the wild-type protein (sGCoe) showed a guanylyl cyclase activity of ∼100 pmol cGMP/min/107 cells (Figure 1C). The rate of cGMP synthesis of lysates of gc-null cells expressing N-terminally truncated sGC (sGCΔNoe) is almost identical to that of sGCoe cells. The in vivo cAMP-stimulated cGMP accumulation of starved sGCΔNoe cells is also comparable with that of sGCoe cells, indicating that the N-terminal region is not required for catalytic activity (Figure 1D). gc-null cells expressing catalytically inactive sGC (sGCΔcatoe), as expected, did not yield any detectable levels of cGMP in both the in vitro guanylyl cyclase assays and in vivo response assays.
The opposite characteristics of sGCΔN and sGCΔcat make up for a valuable set of mutants: sGCΔNoe cells show a proper temporal cGMP response to an extracellular cAMP concentration increase, but they show no spatial localization of the sGC protein in a cAMP gradient. In contrast, sGCΔcatoe cells do not synthesize any cGMP in response to a cAMP stimulus, but the protein still localizes to the leading edge of the cell in a gradient. Therefore, with these mutants we have effectively uncoupled the cGMP response and localization of the sGC protein.
Chemotaxis Efficiency in Dynamic and Static cAMP Gradients
Dictyostelium chemotaxis is optimized to respond to dynamic cAMP waves. Many signal transduction pathways that are involved in chemotaxis, including the cGMP pathway, are activated by an increase of the cAMP concentration and adapt when the concentration remains constant (Dinauer et al., 1980; Van Haastert and Van der Heijden, 1983). Therefore, we have devised a chemotaxis chamber that mimics the spatiotemporal gradient of the rising flank of the natural cAMP wave (see Materials and Methods and Figure 2). Cells were placed under the glass bridge of this modified Zigmond chemotaxis chamber. The cells are exposed to a spatial and temporal gradient between 1 and 10 min after application of the cAMP source, whereas after 15–35 min a nearly static spatial gradient is present (Figure 2C).
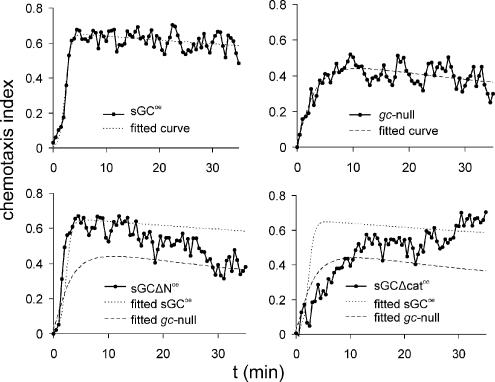
The coordinates of the cells were determined during 35 min at a 30-s interval immediately after application of the cAMP source. From these data, the chemotaxis index was calculated, which is the distance moved in the direction of the gradient divided by the total distance moved. The average chemotaxis index of each 30-s step is plotted in Figure 3. Cells expressing the wild-type protein (sGCoe) exhibit a sharp increase of the chemotaxis index around 3 min after initialization of the gradient chamber. This time corresponds with the maximal temporal increase of cAMP concentration. The chemotaxis index peaks at 0.64 and stays nearly constant for the remaining time of the experiment. Thus, the chemotaxis index that develops when cells experience both a temporal and spatial cAMP gradient persists when only a spatial gradient remains (see Table 1 for quantitative data). The kinetics and magnitude of the chemotaxis response of sGCoe cells, i.e., gc-null cells expressing the wild-type sGC protein, are similar to that of the response of wild-type AX3 cells (our unpublished data). The chemotaxis index of gc-null cells is significantly lower than that of cells expressing the sGC protein. The cells lack the sharp increase in response to the spatiotemporal gradient and reach a maximum chemotaxis index of only 0.43. Cells expressing sGCΔN, the sGC mutant that produces cGMP but does not localize to the leading edge, exhibit a very good chemotaxis response during the initial spatiotemporal gradient, being essentially identical to that of cells expressing the wild-type protein. However, in the static gradient the chemotaxis efficiency slowly declines until it reaches the value of gc-null cells at the end of the experiment (Table 1). Cells expressing sGCΔcat, the catalytically inactive sGC protein that localizes to the leading edge, show very poor chemotactic response to the initial spatiotemporal gradient, with a chemotaxis index that is even lower than that of gc-null cells. However, the chemotaxis index gradually increases and reaches the value of cells expressing the wild-type protein at the end of the experiment (Table 1).
Figure 3.
Chemotaxis efficiency of the sGC mutants. The chemotaxis efficiency of mutant sGC cell strains in the modified Zigmond chemotaxis chamber is plotted during the course of gradient formation. Cells experience both a spatial and a temporal gradient during the first 10 min, but the temporal component is slowly lost and a stable spatial gradient remains during the last 20 min. To facilitate comparison between the different cell strains, the fitted curve of the chemotaxis response of sGCoe cells (dotted line) and gc-null cells (dashed line) were included in the graphs of sGCΔNoe and sGCΔcatoe cells.
Table 1.
Chemotaxis efficiency and degree of turning of mutant sGC cell strains
| Cell strain | Spatiotemporal gradient |
Early spatial gradient |
Late spatial gradient |
No. of analyzed cells | |||
|---|---|---|---|---|---|---|---|
| Chemotaxis index | Directional change (°/min) | Chemotaxis index | Directional change (°/min) | Chemotaxis index | Directional change (°/min) | ||
| gc-null | 0.40 ± 0.04 | 35.7 ± 1.5 | 0.37 ± 0.04 | 37.4 ± 1.6 | 0.39 ± 0.05 | 38.4 ± 2.2 | 85 |
| sGC | 0.58 ± 0.03** | 28.6 ± 1.5** | 0.60 ± 0.04** | 27.2 ± 2.0** | 0.56 ± 0.06* | 31.3 ± 2.5* | 63 |
| sGCΔN | 0.56 ± 0.03** | 31.5 ± 1.2* | 0.57 ± 0.04** | 30.6 ± 1.2** | 0.42 ± 0.04 | 34.7 ± 1.4 | 115 |
| sGCΔcat | 0.35 ± 0.04 | 35.0 ± 1.7 | 0.43 ± 0.04 | 29.7 ± 1.3** | 0.58 ± 0.04** | 30.5 ± 1.8** | 104 |
| sGCΔNΔcat | 0.34 ± 0.05 | 34.7 ± 3.6 | 0.40 ± 0.06 | 36.3 ± 2.6 | 0.33 ± 005 | 41.4 ± 5.5 | 31 |
| sGCΔCΔcat | 0.41 ± 0.04 | 34.8 ± 3.0 | 0.29 ± 0.07 | 34.6 ± 4.4 | 0.27 ± 0.07 | 36.2 ± 4.0 | 29 |
The chemotaxis efficiency and degree of turning of mutant sGC cell strains in the modified Zigmond chemotaxis chamber in response to the spatiotemporal gradient (average of a 4-min window, centered around 6 min after initiation of the gradient), the early spatial gradient (average of a 5-min window at t = 15 min), and the late spatial gradient (average of a 5-min window at t = 30 min) are shown. The degree of turning is defined as follows. For each 1-min step, the displacement of the cells was calculated as a vector. The degree of turning is the angle between subsequent vectors. The data shown are the average and SE of the mean, with n representing the number of cells analyzed. The asterisk(s) indicates that the value is significantly different from the value of gc-null cells at *p < 0.05 or **p < 0.01; data without asterisk(s) are not significantly different at p > 0.05.
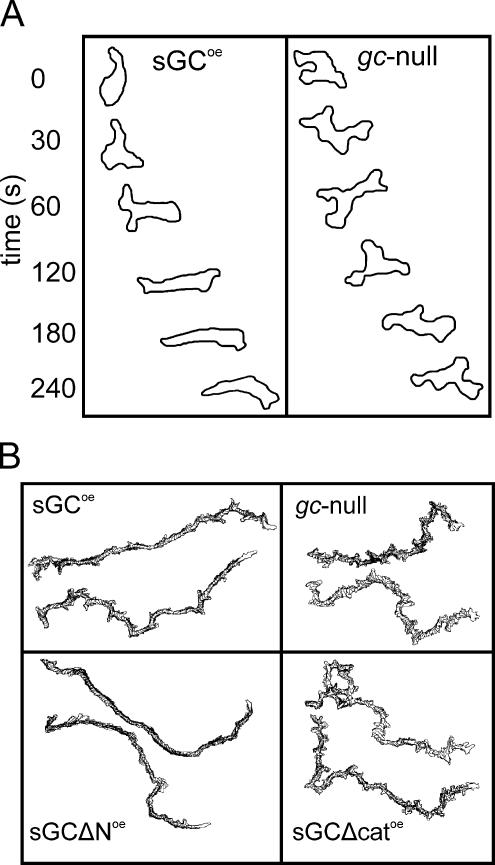
The results of Figure 3 show that the mutants exhibit a very different response to the spatiotemporal gradient compared with the static spatial gradient. In a spatiotemporal gradient, cells with a cGMP-plus phenotype (sGCoe and sGCΔNoe) have a much better chemotaxis response than cGMP-null cell strains (gc-null and sGCΔcatoe), and localization of the sGC protein has only minor effects. In contrast, in a stable spatial gradient, cells with anterior localization of sGC (sGCoe and sGCΔcatoe) have a much better chemotaxis response than cells without sGC localization (gc-null and sGCΔNoe), and cGMP is of less importance. To further investigate the function of cGMP, image sequences of a representative cGMP-plus cell (sGCoe) and cGMP-null cell (gc-null) during the spatiotemporal gradient are presented in Figure 4A. In the absence of a chemotactic gradient, cells from both strains extend multiple pseudopodia in random directions. As soon as the cAMP-wave arrives at the cell (top frame), both cells extend a pseudopod in the proper direction. This indicates that directional sensing is largely intact in cGMP-null cells. However, only cells with a proper cGMP response retract all remaining pseudopodia, resulting in an elongated cell shape and coordinated movement in the direction of the gradient, whereas cGMP-null cells continue to extend pseudopodia in the back of the cell, thereby compromising their chemotaxis efficiency. The cell tracks in Figure 4B shows that the smooth path with relatively few lateral pseudopodia in cGMP-plus cells is maintained throughout the course of the experiment.
Figure 4.
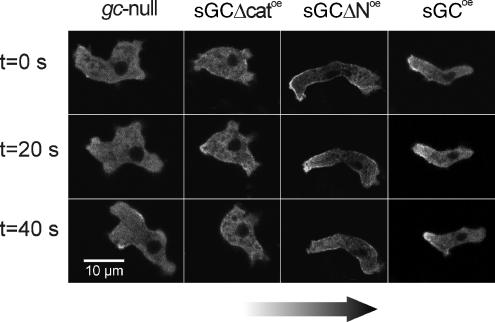
Cell shape of the sGC mutants during aggregation. (A) The contours of a representative sGCoe cell and gc-null cell during the spatiotemporal phase of the cAMP gradient. The top frame was taken ∼1 min after placing the agarose block with cAMP adjacent to the bridge, which is the time point when the cAMP wave arrives at the cell. Subsequent frames are depicted from top to bottom and were taken at indicated intervals. (B) The contour of each cell was determined every 30 s during chemotaxis of the sGC mutants in the modified Zigmond chamber. A stack of contours spanning at least 25 min of two representative cells of each strain is shown.
Cells expressing the catalytically inactive sGCΔcat lack the rapid reorientation toward the cAMP source during the initial spatiotemporal gradient. However, the cells eventually acquire a chemotaxis index that is as high as cells expressing wild-type sGC. To investigate the mechanism by which sGCΔcat attains this high chemotaxis index, we have determined many characteristics of sGCΔcat cells in late spatial gradients, such as speed, directional change, and persistence of movement. None of these were significantly different between gc-null and sGCΔcat cells, except for the directional change, which is 38.4°/min in gc-null cells and 30.5°/min in sGCΔcat cells (Table 1). Interestingly, in the early spatial gradient, 15 min after initiation of gradient formation, sGCΔcat cells still show poor chemotaxis but already have a significantly lower degree of turning than gc-null cells (Table 1). These results indicate that sGCΔcat cells still make lateral pseudopodia, but these do not become dominant. Instead, the leading edge makes less turns, thereby improving its orientation in the chemotactic gradient and allowing the cells to eventually acquire a chemotaxis index that is as high as cells expressing wild-type sGC.
Cells expressing the catalytically active but cytosolic sGCΔN have the opposite behavior. These cells show the rapid reorientation toward the cAMP source during the initial spatiotemporal gradient, but exhibits a high directional change in the late spatial gradient. The smooth cell tracks of sGCΔNoe throughout the experiment (Figure 4B) suggests that sGCΔNoe cells maintain the suppression of lateral pseudopodia in a stable spatial gradient but that the leading edge slowly loses orientation due to the high degree of turning.
To identify the parts of the sGC protein that are essential for the improved anterior orientation by the sGCΔcat protein, we expressed N-terminal and C-terminal deletions of sGCΔcat in gc-null cells. As expected, deletion of the N terminus (sGCΔNΔcat) yields a protein with homogenous cytoplasmic localization (our unpublished data). The chemotaxis data presented in Table 1 shows that these sGCΔNΔcatoe cells exhibit a similar chemotaxis response as gc-null cells. The C-terminal region of sGC is ∼1000 amino acids long and contains a well conserved AAA+ ATPase domain. Deletion of this C-terminal region from the catalytically inactive sGC (sGCΔCΔcat) yields a protein that still localizes to pseudopodia and the leading edge (our unpublished data), but no longer supports chemotaxis (Table 1). This suggests that the contribution of the sGC protein to chemotaxis in the spatial gradient requires both the N-terminal region which targets the protein to the pseudopodia and the C-terminal region containing the AAA+ ATPase domain.
Effects of sGC and cGMP on Myosin Distribution
In wild-type chemotaxing Dictyostelium cells, high levels of filamentous myosin are found in retracting pseudopodia and the posterior cell cortex (Clow and McNally, 1999). In vitro assays have shown that the cAMP-stimulated incorporation of myosin filaments into the Triton-insoluble cytoskeleton is absent in gc-null cells (Liu et al., 1993; Bosgraaf et al., 2002). To investigate the effects of the sGC protein and the cGMP product on myosin incorporation into the cytoskeleton, we coexpressed myosin-GFP in the various mutant sGC cell strains. Cells were placed under the bridge of the modified Zigmond chemotaxis chamber and allowed to migrate toward the cAMP. Cells expressing full-length sGC quickly become elongated in the cAMP gradient. In these elongated cells, a large amount of myosin is found along the posterior cell cortex (Figure 5). In contrast, in gc-null cells, most of the myosin remains cytosolic. A small myosin fraction is found in retracting pseudopodia, but the cells lack the pronounced incorporation of myosin along the posterior cell cortex. In accordance, the cells remain fairly nonpolar. Expression of the mutant sGCΔN protein, which no longer localizes to the leading edge but still synthesizes cGMP, rescues both the elongated cell shape and the myosin incorporation in the posterior cell cortex. In contrast, cells expressing the catalytically inactive sGCΔcat show a similar phenotype as gc-null cells; myosin remains mostly cytosolic and cells do not adopt an elongated cell shape, indicating that the regulation of myosin during chemotaxis is dependent only on cGMP and not on the localization of the sGC protein. The shape of sGCΔcat cells and the distribution of myosin-GFP as shown in Figure 5 do not change much during the chemotaxis assay. Thus, at 30 min after stimulation, when the sGCΔcat cells have a high chemotaxis index, myosin does not become enriched in the back of the cell, cells do not adopt an elongated cell shape, and cells still make pseudopodia at the back of the cell.
Figure 5.
Localization of myosin in sGC mutants. Myosin-GFP was expressed in gc-null, sGCΔcatoe, sGCΔNoe, and sGCoe cells. Cells were placed in the modified Zigmond chemotaxis chamber and were allowed to migrate toward the cAMP. Myosin-GFP fluorescence was visualized by confocal microscopy. The arrow points in the direction of migration.
Distribution of Filamentous Actin and sGC
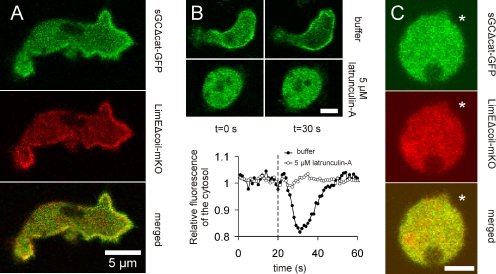
Filamentous actin is the major constituent of the cytoskeleton in extending pseudopodia. To investigate the relation between F-actin distribution and the localization of sGC during chemotaxis, the filamentous actin binding protein LimEΔcoil (Schneider et al., 2003) was C-terminally fused to mKO (Karasawa et al., 2004) and used as an F-actin probe in the various cell strains expressing the GFP-tagged sGC mutants. The cells were placed in a cAMP gradient and visualized by confocal microscopy. Most F-actin in the chemotaxing cells was detected in protruding pseudopodia at the leading edge. Interestingly, all sGC mutant proteins with an intact N-terminal domain (sGC-GFP, sGCΔcat-GFP [shown in Figure 6A] and N-sGC1-1019) were found to colocalize to a large extent with LimEΔcoil-mKO, indicating that sGC may interact with F-actin at the leading edge of the cell. To investigate whether the anterior localization of sGC is dependent on the presence of F-actin, we depolymerized all actin filaments by treating the cells with 5 μM latrunculin A, a molecular compound that traps actin in its monomeric state. Treated cells become spherical and, as expected, the distribution of LimEΔcoil-mKO is completely homogenous in these cells (Figure 6B). At the same time, distribution of the sGC mutant proteins in treated cells also becomes completely homogenous. It has previously been found that global stimulation of cells with cAMP greatly increases the amount of sGC in the cell cortex (Veltman et al., 2005). However, global stimulation of latrunculin A-treated cells no longer induces such a transient translocation of sGC to the cell cortex (Figure 6B). In addition, local stimulation by placing of a pipette with cAMP next to a latrunculin A-treated cell also no longer elicits a redistribution of sGC (Figure 6C). These results indicate that the localization of the sGC protein to the leading edge is dependent on the interaction of sGC with filamentous actin. In combination with the obtained chemotaxis data of the sGCΔcat mutant, this suggests that this interaction with the actin cytoskeleton is essential for the improved orientation that is observed in a stable cAMP gradient.
Figure 6.
Colocalization sGC and filamentous actin. (A) sGCΔcat-GFP was coexpressed with the filamentous actin binding protein LimEΔcoil-mKO in gc-null cells. The cells were placed in a cAMP gradient and allowed to migrate toward the cAMP. Localization of sGCΔcat-GFP is shown in the top panel, LimEΔcoil-mKO in the middle panel, and the overlay in the bottom panel. The arrow points in the direction of migration. (B) gc-null cells expressing the N-terminal localization domain of sGC fused to GFP (N-sGC1-1019) were globally stimulated with 10−6 M cAMP at t = 20 s, as indicated by the dashed line. Translocation of the localization domain to the cell cortex was monitored in the absence (open circles) and presence of 5 μM latrunculin A (solid circles) by measuring the depletion of the cytosolic fluorescence. Bar, 5 μm. (C) gc-null cells coexpressing N-sGC1-1019 and LimEΔcoil-mKO were treated with 5 μM latrunculin A. A pipette filled with 10−4 M cAMP was placed next to the cells, at the position indicated by the mark. The localization of N-sGC1-1019 is shown in the top panel, LimEΔcoil-mKO in the middle panel, and the overlay in the bottom panel. Bar, 5 μm.
DISCUSSION
Chemotaxis of amoeboid cells is driven by actin filaments in leading pseudopodia and actin–myosin filaments in the back and at the side of the cell to suppress pseudopodia. Several signal transduction components are involved in regulating the actin–myosin cytoskeleton during chemotaxis, including phosphatidylinositol 3-kinase, PAK, WASP, and Ras (Tuxworth et al., 1997; Chung and Firtel, 1999; Lim et al., 2005; Myers et al., 2005). Mutants with deletions of the encoding genes suggest that none of these signaling pathways is essential but that each contributes to chemotaxis to a different extent. In Dictyostelium, cGMP plays an important role during chemotaxis and is produced predominantly by a sGC that is homologous to mammalian soluble adenylyl cyclase. The sGC protein is enriched in extending pseudopodia and activated during chemotaxis. We show here that the sGC protein and the cGMP product have different functions during chemotaxis. Together, they are responsible for about one-third of the chemotactic response as indicated by the chemotaxis index of gc-null cells of 0.43 compared with 0.64 of gc-null cells expressing the wild-type sGC protein, or wild-type AX3 cells. The chemotaxis index of gc-null cells is similar to the chemotaxis defects observed in PI3K-null cells (Funamoto et al., 2001; Loovers et al., 2006) but much larger than the minimal defects seen in PLC-null (Drayer et al., 1994) and adenylyl cyclase-null cells (Pitt et al., 1992; Stepanovic et al., 2005). In Dictyostelium, cGMP is involved in the regulation of myosin. Stimulation of wild-type cells with cAMP leads to a transient incorporation of myosin filaments in the cell cortex (Liu and Newell, 1994). Mutant cell strains with disrupted cGMP signaling show a strong reduction of this response (Bosgraaf et al., 2002). In a cAMP gradient, myosin is mostly found in the back of the cell, where it increases the cortical tension and suppress pseudopod extension (Moores et al., 1996). We find that cGMP-null cells that are placed in a dynamic cAMP gradient incorporate almost no myosin in the posterior cell cortex during chemotaxis. Accordingly, mutants with disrupted cGMP signaling frequently extend pseudopodia at the back and sides of the cell, whereas cell strains with an elevated cGMP response acquire a strong axial polarity and have highly elongated cell shapes during chemotaxis. The myosin response and cell elongation are still intact in cells expressing the sGCΔN mutant, which shows a homogeneously cytosolic distribution but has a normal catalytic activity. This indicates that the cGMP-dependent myosin response is independent of the site where cGMP is synthesized.
We observe that the sGC protein associates with actin filaments in pseudopodia. The binding to filamentous actin is not essential for activation of sGC, because cells expressing the sGCΔN mutant that no longer associates to actin filaments in the leading edge still show a proper cGMP response to a cAMP stimulus. The role of the association of sGC to the actin cytoskeleton was investigated using a mutant with a point mutation in the catalytic site of the enzyme. The mutation renders the enzyme inactive and thereby blocks the ability of sGC to activate the cGMP signaling pathway. This catalytically inactive sGCΔcat protein is still present in pseudopodia at the leading edge. Expression of sGCΔcat in gc-null cells significantly enhances the chemotactic response in a static spatial chemoattractant gradient. This indicates that the sGC protein harbors additional functionality aside from cGMP synthesis. Expression of sGCΔcat leads to a decrease of the degree of turning of the cell from ∼38 to 30°/min. Cells expressing wild-type sGC also show the low degree of turning, but cells expressing the cytosolic sGCΔN protein do not. We propose that filamentous actin-associated sGC modulates pseudopod formation, such that a new pseudopod is formed preferentially at a site close to an sGC-rich, old pseudopod. On disruption of actin filaments with lantrunculin A, sGC becomes completely cytosolic. Local stimulation of these lantrunculin A-treated cells with a pipette filled with cAMP has been shown to induce phosphatidyl inositol-(3,4,5)-trisphosphate formation at the side of the cell close to the pipette (Parent et al., 1998). We did not observe any translocation of sGC in such stimulated lantrunculin A-treated cells, indicating that sGC localization is not a primary response to the cAMP gradient, but it is dependent on the formation of actin filaments at the side of highest chemoattractant concentration. In a spatial gradient the sGCΔcatoe cells exhibit a reduced degree of turning before the chemotaxis index increases compared with gc-null cells. This suggests that the improvement of chemotaxis by sGCΔcat is the consequence rather than the cause of the reduced degree of turning. Collectively, these data suggest that the reduced degree of turning by the sGC protein creates a more dominant leading edge, which allows a more efficient chemotactic response of sGCΔcatoe cells compared with cells lacking anteriorly localized sGC.
The additional function of the sGC protein can potentially be mapped to two different regions of the enzyme. The protein consists of 2843 amino acids. The central cyclase domain of ∼800 amino acids long is flanked on both sides by two regions of ∼1000 amino acids each, with a hitherto unknown function. The N-terminal region has a low complexity and shows no homology to any other sequences in the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST). We have now identified that the N-terminal region is essential and sufficient for targeting sGC to the actin cytoskeleton in pseudopodia. It is most likely that the C-terminal domain is responsible for mediating the observed effects of sGC on chemotaxis, because expression of catalytically inactive sGC that lacks the C-terminal domain does not improve the chemotactic response of gc-null cells. The sequence of the C-terminal domain of sGC has been well conserved throughout evolution with orthologues in bacterial kinases and soluble adenylyl cyclases. The domain contains a highly conserved P-loop ATPase motif that classifies sGC as a member of the AAA+ protein family (Leipe et al., 2004). Members of this family have been shown to use the energy released by the ATPase activity to generate mechanical force (Neuwald et al., 1999). The C-terminal domain of sGC also contains signatures of the tetratricopeptide repeat motif, which is known to be involved in establishing protein–protein interactions (Sikorski et al., 1990; D’Andrea and Regan, 2003). The presence of these domains suggests that the increased anterior polarity induced by the sGC protein may be due to the active rearrangement of protein–protein interactions or the recruitment of additional proteins to the leading edge. This suggests that the sGC protein can be regarded as an actin-binding protein with AAA–ATPase activity that modulates the local formation of pseudopodia.
Together, activation of sGC at the leading edge provides two very different but complementary sensory transduction pathways. Produced cGMP rapidly diffuses throughout the cell where it stimulates the incorporation of myosin in the cortex and inhibits pseudopod formation, whereas the sGC protein itself promotes the formation of a new pseudopod in the proximity of the old pseudopod. The sGC protein and cGMP product have opposite properties and functions. cGMP diffuses very fast by which it is expected to attain a spatially homogeneous concentration. Elevated cGMP levels are observed only after an increase of the chemoattractant concentration and adapt to constant concentrations (Van Haastert and Van der Heijden, 1983), indicating that cGMP senses the temporal component of the cAMP wave. In agreement with a function in temporal sensing, we observed that the sGCΔNoe mutant contributes to chemotaxis during the spatiotemporal part of the gradient but not during the stable spatial gradient. Alternatively, the sGC protein with its slow diffusion remains localized in pseudopodia in stable gradients, indicating that the sGC protein is involved in spatial sensing, which is strongly supported by the observation that sGCΔcat supports chemotaxis only in stable spatial gradients.
Several models have been proposed to explain how cells may read and respond to spatial gradients: local activation and global inhibition of pseudopod formation (Rappel et al., 2002; Ma et al., 2004), depletion of a cytosolic component to the leading edge (Postma and Van Haastert, 2001), or a bias of pseudopod formation by local activation at the front (Arrieumerlou and Meyer, 2005). Interestingly, the present observations on the function of cGMP and sGC provide essential elements for each of these models. Stimulation of pseudopod formation at the leading edge by sGC and inhibition of pseudopod formation in the back and at the side of the cell by cGMP represent the two components of the local activation and global inhibition model (Rappel et al., 2002; Ma et al., 2004). Recruitment of sGC to the leading edge also leads to a partial depletion of the limited amount of sGC from the cytosol. Models have been proposed that utilize the depletion of a cytosolic component to nonlinearly amplify the external chemoattractant gradient (Postma and Van Haastert, 2001). Finally, sGC reduces the degree of turning, supporting the local activation model. This model assumes that each receptor binding event within the leading edge triggers a local pseudopod extension and a small turn in the direction of migration. To achieve efficient chemotaxis, multiple steps are required where each subsequent step shows a slightly better orientation in the direction of the gradient. The relatively slow increase of chemotaxis efficiency of cells with anteriorly localized, catalytically inactive sGC in the spatial gradient is in excellent agreement with this model.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Leonard Bosgraaf for helping to develop the novel chemotaxis chamber.
Abbreviations used:
- mKO
monomeric Kusabira-Orange
- PB
phosphate buffer
- sGC
soluble guanylyl cyclase.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-05-0381) on June 21, 2006.
REFERENCES
- Affolter M., Weijer C. J. Signaling to cytoskeletal dynamics during chemotaxis. Dev. Cell. 2005;9:19–34. doi: 10.1016/j.devcel.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Allbritton N. L., Meyer T., Stryer L. Range of messenger action of calcium ion and inositol 1,4,5-trisphosphate. Science. 1992;258:1812–1815. doi: 10.1126/science.1465619. [DOI] [PubMed] [Google Scholar]
- Almeida P. F. F., Vaz W. L. C. Structure and Dynamics of Membranes. Amsterdam: Elsevier; 1995. Lateral diffusion in membranes; pp. 305–357. [Google Scholar]
- Arrieumerlou C., Meyer T. A local coupling model and compass parameter for eukaryotic chemotaxis. Dev. Cell. 2005;8:215–227. doi: 10.1016/j.devcel.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Bosgraaf L., Russcher H., Smith J. L., Wessels D., Soll D. R., Van Haastert P. J. M. A novel cGMP signalling pathway mediating myosin phosphorylation and chemotaxis in Dictyostelium. EMBO J. 2002;21:4560–4570. doi: 10.1093/emboj/cdf438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Nakamura T., Koutalos Y. Cyclic AMP diffusion coefficient in frog olfactory cilia. Biophys. J. 1999;76:2861–2867. doi: 10.1016/S0006-3495(99)77440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. Y., Firtel R. A. PAKa, a putative PAK family member, is required for cytokinesis and the regulation of the cytoskeleton in Dictyostelium discoideum cells during chemotaxis. J. Cell Biol. 1999;147:559–576. doi: 10.1083/jcb.147.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clow P. A., McNally J. G. In vivo observations of myosin II dynamics support a role in rear retraction. Mol. Biol. Cell. 1999;10:1309–1323. doi: 10.1091/mbc.10.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea L. D., Regan L. TPR proteins: the versatile helix. Trends Biochem. Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Devreotes P., Janetopoulos C. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J. Biol. Chem. 2003;278:20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- Dinauer M. C., Steck T. L., Devreotes P. N. Cyclic 3′,5′-AMP relay in Dictyostelium discoideum V. Adaptation of the cAMP signaling response during cAMP stimulation. J. Cell Biol. 1980;86:554–561. doi: 10.1083/jcb.86.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drayer A. L., Van der Kaay J., Mayr G. W., Van Haastert P. J. M. Role of phospholipase C in Dictyostelium: formation of inositol 1,4,5-trisphosphate and normal development in cells lacking phospholipase C activity. EMBO J. 1994;13:1601–1609. doi: 10.1002/j.1460-2075.1994.tb06423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamoto S., Milan K., Meili R., Firtel R. A. Role of phosphatidylinositol 3′ kinase and a downstream pleckstrin homology domain-containing protein in controlling chemotaxis in Dictyostelium. J. Cell Biol. 2001;153:795–810. doi: 10.1083/jcb.153.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futrelle R. P. Dictyostelium chemotactic response to spatial and temporal gradients. Theories of the limits of chemotactic sensitivity and of pseudochemotaxis. J. Cell Biochem. 1982;18:197–212. doi: 10.1002/jcb.1982.240180207. [DOI] [PubMed] [Google Scholar]
- Iijima M., Huang Y. E., Devreotes P. Temporal and spatial regulation of chemotaxis. Dev. Cell. 2002;3:469–478. doi: 10.1016/s1534-5807(02)00292-7. [DOI] [PubMed] [Google Scholar]
- Karasawa S., Araki T., Nagai T., Mizuno H., Miyawaki A. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem. J. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe D. D., Koonin E. V., Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 2004;343:1–28. doi: 10.1016/j.jmb.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Lim C. J., Zawadzki K. A., Khosla M., Secko D. M., Spiegelman G. B., Weeks G. Loss of the Dictyostelium RasC protein alters vegetative cell size, motility and endocytosis. Exp. Cell Res. 2005;306:47–55. doi: 10.1016/j.yexcr.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Liu G., Kuwayama H., Ishida S., Newell P. C. The role of cyclic GMP in regulating myosin during chemotaxis of Dictyostelium: evidence from a mutant lacking the normal cyclic GMP response to cyclic AMP. J. Cell Sci. 1993;106:591–595. doi: 10.1242/jcs.106.2.591. [DOI] [PubMed] [Google Scholar]
- Liu G., Newell P. C. Evidence that cyclic GMP regulates myosin interaction with the cytoskeleton during chemotaxis of Dictyostelium. J. Cell Sci. 1988;90:123–129. doi: 10.1242/jcs.90.1.123. [DOI] [PubMed] [Google Scholar]
- Liu G., Newell P. C. Regulation of myosin regulatory light chain phosphorylation via cyclic GMP during chemotaxis of Dictyostelium. J. Cell Sci. 1994;107:1737–1743. doi: 10.1242/jcs.107.7.1737. [DOI] [PubMed] [Google Scholar]
- Loovers H. M., Postma M., Keizer-Gunnink I., Huang Y. E., Devreotes P. N., Van Haastert P. J. Distinct roles of PI(3,4,5)P3 during chemoattractant signaling in Dictyostelium: a quantitative in vivo analysis by inhibition of PI3-kinase. Mol. Biol. Cell. 2006;17:1503–1513. doi: 10.1091/mbc.E05-09-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Janetopoulos C., Yang L., Devreotes P. N., Iglesias P. A. Two complementary, local excitation, global inhibition mechanisms acting in parallel can explain the chemoattractant-induced regulation of PI(3,4,5)P3 response in Dictyostelium cells. Biophys. J. 2004;87:3764–3774. doi: 10.1529/biophysj.104.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato J. M., Malchow D. Guanylate cyclase activation in response to chemotactic stimulation in Dictyostelium discoideum. FEBS Lett. 1978;90:119–122. doi: 10.1016/0014-5793(78)80311-1. [DOI] [PubMed] [Google Scholar]
- Moores S. L., Sabry J. H., Spudich J. A. Myosin dynamics in live Dictyostelium cells. Proc. Natl. Acad. Sci. USA. 1996;93:443–446. doi: 10.1073/pnas.93.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S. A., Han J. W., Lee Y., Firtel R. A., Chung C. Y. A Dictyostelium homologue of WASP is required for polarized F-actin assembly during chemotaxis. Mol. Biol. Cell. 2005;16:2191–2206. doi: 10.1091/mbc.E04-09-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Olson L. J., Ho B. Y., Cashdollar L. W., Drewett J. G. Functionally active catalytic domain is essential for guanylyl cyclase-linked receptor mediated inhibition of human aldosterone synthesis. Mol. Pharmacol. 1998;54:761–769. doi: 10.1124/mol.54.5.761. [DOI] [PubMed] [Google Scholar]
- Parent C. A. Making all the right moves: chemotaxis in neutrophils and Dictyostelium. Curr. Opin. Cell Biol. 2004;16:4–13. doi: 10.1016/j.ceb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Parent C. A., Blacklock B. J., Froehlich W. M., Murphy D. B., Devreotes P. N. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- Pitt G. S., Milona N., Borleis J., Lin K. C., Reed R. R., Devreotes P. N. Structurally distinct and stage-specific adenylyl cyclase genes play different roles in Dictyostelium development. Cell. 1992;69:305–315. doi: 10.1016/0092-8674(92)90411-5. [DOI] [PubMed] [Google Scholar]
- Postma M., Bosgraaf L., Loovers H. M., Van Haastert P. J. Chemotaxis: signalling modules join hands at front and tail. EMBO Rep. 2004;5:35–40. doi: 10.1038/sj.embor.7400051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma M., Van Haastert P. J. M. A diffusion-translocation model for gradient sensing by chemotactic cells. Biophys. J. 2001;81:1314–1323. doi: 10.1016/S0006-3495(01)75788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappel W. J., Thomas P. J., Levine H., Loomis W. F. Establishing direction during chemotaxis in eukaryotic cells. Biophys. J. 2002;83:1361–1367. doi: 10.1016/S0006-3495(02)73906-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs J., Meima M., Schaap P., Van Haastert P. J. M. The Dictyostelium homologue of mammalian soluble adenylyl cyclase encodes a guanylyl cyclase. EMBO. J. 2001a;20:4341–4348. doi: 10.1093/emboj/20.16.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs J., Snippe H., Kleineidam R. G., Van Haastert P. J. M. Guanylate cyclase in Dictyostelium discoideum with the topology of mammalian adenylate cyclase. Biochem. J. 2001b;354:697–706. doi: 10.1042/0264-6021:3540697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs J., Van Haastert P. J. M. Characterization of two unusual guanylyl cyclases from Dictyostelium. J. Biol. Chem. 2002;277:9167–9174. doi: 10.1074/jbc.M111437200. [DOI] [PubMed] [Google Scholar]
- Schneider N., Weber I., Faix J., Prassler J., Muller-Taubenberger A., Kohler J., Burghardt E., Gerisch G., Marriott G. A Lim protein involved in the progression of cytokinesis and regulation of the mitotic spindle. Cell Motil. Cytoskeleton. 2003;56:130–139. doi: 10.1002/cm.10139. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Boguski M. S., Goebl M., Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- Stepanovic V., Wessels D., Daniels K., Loomis W. F., Soll D. R. Intracellular role of adenylyl cyclase in regulation of lateral pseudopod formation during Dictyostelium chemotaxis. Eukaryot. Cell. 2005;4:775–786. doi: 10.1128/EC.4.4.775-786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H., Ordal G. W. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxworth R. I., Cheetham J. L., Machesky L. M., Spiegelmann G. B., Weeks G., Insall R. H. Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J. Cell Biol. 1997;138:605–614. doi: 10.1083/jcb.138.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert P. J., Van der Heijden P. R. Excitation, adaptation, and deadaptation of the cAMP-mediated cGMP response in Dictyostelium discoideum. J. Cell Biol. 1983;96:347–353. doi: 10.1083/jcb.96.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert P. J. M., Kuwayama H. cGMP as second messenger during Dictyostelium chemotaxis. FEBS Lett. 1997;410:25–28. doi: 10.1016/s0014-5793(97)00416-x. [DOI] [PubMed] [Google Scholar]
- Varnum-Finney B., Edwards K. B., Voss E., Soll D. R. Amebae of Dictyostelium discoideum respond to an increasing temporal gradient of the chemoattractant cAMP with a reduced frequency of turning: evidence for a temporal mechanism in ameboid chemotaxis. Cell Motil. Cytoskeleton. 1987;8:7–17. doi: 10.1002/cm.970080103. [DOI] [PubMed] [Google Scholar]
- Veltman D. M., Roelofs J., Engel R., Visser A. J., Van Haastert P. J. Activation of soluble guanylyl cyclase at the leading edge during Dictyostelium chemotaxis. Mol. Biol. Cell. 2005;16:976–983. doi: 10.1091/mbc.E04-08-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams G. H., Armitage J. P. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- Wang F., Herzmark P., Weiner O. D., Srinivasan S., Servant G., Bourne H. R. Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat. Cell Biol. 2002;4:513–518. doi: 10.1038/ncb810. [DOI] [PubMed] [Google Scholar]
- Williams H. P., Harwood A. J. Cell polarity and Dictyostelium development. Curr. Opin. Microbiol. 2003;6:621–627. doi: 10.1016/j.mib.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Xu X., Meier-Schellersheim M., Jiao X., Nelson L. E., Jin T. Quantitative imaging of single live cells reveals spatiotemporal dynamics of multistep signaling events of chemoattractant gradient sensing in Dictyostelium. Mol. Biol. Cell. 2005;16:676–688. doi: 10.1091/mbc.E04-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond S. H. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. J. Cell Biol. 1977;75:606–616. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.