Abstract
The endoplasmic reticulum (ER) is thought to play an important structural and functional role in phagocytosis. According to this model, direct membrane fusion between the ER and the plasma or phagosomal membrane must precede further invagination, but the exact mechanisms remain elusive. Here, we investigated whether various ER-localized SNARE proteins are involved in this fusion process. When phagosomes were isolated from murine J774 macrophages, we found that ER-localized SNARE proteins (syntaxin 18, D12, and Sec22b) were significantly enriched in the phagosomes. Fluorescence and immuno-EM analyses confirmed the localization of syntaxin 18 in the phagosomal membranes of J774 cells stably expressing this protein tagged to a GFP variant. To examine whether these SNARE proteins are required for phagocytosis, we generated 293T cells stably expressing the Fcγ receptor, in which phagocytosis occurs in an IgG-mediated manner. Expression in these cells of dominant-negative mutants of syntaxin 18 or D12 lacking the transmembrane domain, but not a Sec22b mutant, impaired phagocytosis. Syntaxin 18 small interfering RNA (siRNA) selectively decreased the efficiency of phagocytosis, and the rate of phagocytosis was markedly enhanced by stable overexpression of syntaxin 18 in J774 cells. Therefore, we conclude that syntaxin 18 is involved in ER-mediated phagocytosis, presumably by regulating the specific and direct fusion of the ER and plasma or phagosomal membranes.
INTRODUCTION
Phagocytosis is the coordinated process by which large foreign particles are internalized into a newly formed organelle, the phagosome. In professional phagocytes of the immune system, including macrophages, neutrophils, and dendritic cells, the internalization is triggered by activation of various types of cell surface receptors in the course of innate and adaptive immune responses. For example, in the case of immunoglobulin (Ig)-mediated phagocytosis, Fc receptors (FcγRs) cluster where they contact an Ig-opsonized solid surface; this induces actin polymerization, resulting in the formation of pseudopods that engulf the particle (Swanson and Hoppe, 2004). Subsequently, the phagosomes mature by fusing with other organelles of the endocytic pathway, leading to the formation of phagolysosomes (Downey et al., 1999; Vieira et al., 2002).
In macrophages, every membrane fusion event during phagocytosis is thought to be mediated by soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins (Coppolino et al., 2001). It is currently believed that the assembly of SNARE proteins leads to a tight connection between the vesicle and the target membrane, which initiates the opening of the fusion pore (Jahn et al., 2003). The SNARE complex forms an extended parallel four-helix bundle in order to fuse the two membranes (Sutton et al., 1998; Antonin et al., 2002). Three helices are extended from one membrane by proteins of the syntaxin and SNAP-25 families (Q-SNAREs) that contain a conserved glutamine residue at a central position called the “0” layer, and the remaining helix, extended from the opposite membrane, is derived from a protein of the VAMP or synaptobrevin family (an R-SNARE) that contains a conserved arginine residue at the same position (Fasshauer et al., 1998; Bock and Scheller, 1999; Jahn et al., 2003).
Syntaxin 7 and 13 are members of the Q-SNARE protein family that regulate endocytic membrane transport. Syntaxin 7 is involved in membrane fusion between late endosomes and lysosomes (Mullock et al., 2000; Nakamura et al., 2000; Ward et al., 2000), whereas syntaxin 13 is associated with recycling and early endosomes (Prekeris et al., 1998; Tang et al., 1998b). In the maturation process of the phagosome, both syntaxin 7 and syntaxin 13 are required at distinct steps for fusion of the phagosomes with either endosomes or lysosomes (Collins et al., 2002). Vesicle-associated membrane protein 3 (VAMP3, also called cellubrevin), a recycling endosome-localized R-SNARE protein (McMahon et al., 1993), is involved in local fusion between endocytic vesicles and the plasma membrane to form phagosomes, and is also required for focal membrane delivery to form the engulfing pseudopods (Bajno et al., 2000; Coppolino et al., 2001). In addition, the late endosome/lysosome-localized R-SNARE protein VAMP7 (also called TI-VAMP) (Galli et al., 1998) has been demonstrated to function in focal transport between the late endocytic vesicle and the plasma membrane during Fc receptor–mediated phagocytosis (Braun et al., 2004).
Recently, it has become evident that the endoplasmic reticulum (ER) plays important roles in phagocytosis. A series of studies by Desjardins and colleagues has shown that at least some phagosomal membranes are derived from the ER; these authors proposed that direct fusion of the ER with phagosomal membranes may be required to overcome the consumption of the cell surface membrane (Garin et al., 2001; Gagnon et al., 2002; Desjardins, 2003). Studies of antigen presentation have revealed that the ER also supplies MHC class I molecules to phagosomes for antigen cross-presentation in dendritic cells and macrophages (Guermonprez et al., 2003; Houde et al., 2003). In particular, two major ER proteins, calnexin and calreticulin, have been shown to be involved in the outgrowth of phagocytic cups in Dictyostelium discoideum. The analysis suggests that the Ca2+-binding capacity of these ER proteins may be essential to accommodate the machinery for particle uptake into phagosomes (Muller-Taubenberger et al., 2001). However, the precise molecular mechanism of this nonconventional heterotopic membrane fusion in phagocytosis remains largely unknown.
Considering that membrane fusion is mediated by a specific pair of SNARE proteins, the interaction between the ER and the plasma or phagosomal membrane should be regulated in part by ER-resident SNARE proteins. In this study, we set out to investigate how individual ER-related SNARE proteins are involved in the process. Syntaxin 18 is one of the best studied SNARE proteins associated with the ER (Hatsuzawa et al., 2000). This Q-SNARE and an abundant R-SNARE, Sec22b (Hay et al., 1997; Chao et al., 1999; Zhang et al., 1999), have been proposed to play a role in vesicular transport between the ER and Golgi (Zhang et al., 1999; Hatsuzawa et al., 2000). D12 (a mammalian homologue of yeast Use1p/Slt1p) is another Q-SNARE that associates with the ER and binds to both syntaxin18 and Sec22b (Okumura et al., 2006). Syntaxin18 is also known to associate with a wide variety of proteins, including ZW10, a spindle checkpoint protein (Hirose et al., 2004). Using J774 macrophages and a model cell system designed to trigger phagocytosis upon contact with Ig-opsonized particles, we show here that syntaxin18 selectively regulates the formation of phagosomes but not substrate tethering. Although some groups have recently contended that involvement of the ER in phagocytosis has not been proven (Groothuis and Neefjes, 2005; Touret et al., 2005), this study strongly supports the presence of syntaxin18–dependent ER membrane fusion step(s) in phagocytosis and also provides novel insights into the function of syntaxin18.
MATERIALS AND METHODS
Antibodies
Polyclonal antibodies to mouse D12 and Sec22b were raised against bacterially expressed His6-tagged D12 (amino acids 1–239) and Sec22b (amino acids 1–195). The produced antibodies were affinity-purified using antigen-coupled beads. The polyclonal syntaxin 18 antibody was prepared as described (Hatsuzawa et al., 2000). The polyclonal calnexin antibody was a kind gift from Dr. T. Yamashita (University of Iwate, Iwate, Japan). The polyclonal antiserum against EGFP was kindly provided by Dr. K. Akasaki (Fukuyama University, Fukuyama, Japan) and purified using GFP-coupled affinity beads. The purified antibody recognizes mVENUS as well as EGFP. The monoclonal c-Myc antibody was prepared from hybridoma cells (9E10; American Type Culture Collection, Manassas, VA). The remaining antibodies were purchased as follows: LAMP-1 (Santa Cruz Biotechnology, Santa Cruz, CA), ERp72 (Stressgen, Biotechnology, San Diego, CA), EEA1 (BD Transduction Laboratories, Lexington, KY), FLAG (Sigma-Aldrich, St. Louis, MO), and fluorescently labeled secondary antibodies (Molecular Probes Inc, Eugene, OR).
Cell Culture
J774 cells were obtained from the Riken Cell Bank (Tsukuba, Japan) and grown in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS). 293T cells were cultured in DMEM-high glucose supplemented with 10% FCS. All cell lines were cultured at 37°C under 5% CO2.
Preparation of IgG-opsonized Beads
Polystyrene latex beads (3.0 or 0.8 μm in diameter, Sigma-Aldrich) were washed in phosphate-buffered saline (PBS) and incubated in human IgG (Sigma-Aldrich) at 37°C for 4 h. After incubation, the beads were washed three times with PBS and further incubated with rabbit IgG against human-IgG (Chemicon International, Temecula, CA) at 37°C for 2 h. Opsonized beads were washed three times in PBS and resuspended in RPMI 1640 medium.
Isolation of Phagosomes
Latex beads (0.8 μm in diameter, dyed deep blue, Sigma-Aldrich) were or were not opsonized with IgGs. A 10% suspension of the beads was fed to J774 macrophages at a dilution of 1:200 in RPMI medium. The cells were given a 20-min pulse to ingest the beads, then washed with ice-cold PBS to remove extra beads, and homogenized either directly or after further incubation for 120, 240, or 360 min at 37°C. Phagosomes were then isolated by flotation in a discontinuous sucrose gradient, as described previously (Desjardins et al., 1994). To remove intact mitochondria and small ER debris from the phagosome fraction, the homogenate was treated with 10 mM ATP/4 mM MgCl2 for 15 min on ice before the sucrose density-gradient centrifugation (Gotthardt et al., 2002).
Expression Vectors and Establishment of Stable Transformants
For construction of the FcγRIIA expression vector, FcγRIIA cDNA was amplified by PCR from a MATCHMAKER human leukocyte cDNA library (BD Bioscience Clontech, Palo Alto, CA) using the primers 5′-CGGAATTCCAACTATGGAGACCCAAATG-3′ and 5′-CCGGGTACCGTTATTACTGTTGACATGG-3′. The FcγRIIA cDNA was cloned into the EcoRI/KpnI sites of pUC19 and sequenced with a DNA sequencer (ABI PRISM3100; Applied Biosystems, Foster City, CA) to confirm the sequences of the PCR products. The mammalian expression plasmid pFLAG-CMV-5a (Sigma-Aldrich) was used to express FcγRIIA fused to N-terminal FLAG.293T cells stably expressing FcγRIIA–FLAG were established by subcloning the FcγRIIA–FLAG cDNA into the appropriate cloning sites of a retrovirus expression vector, pCX4bsr, that was kindly donated by Dr. T. Akagi (Osaka BioScience Institute, Osaka, Japan; Akagi et al., 2000). The recombinant retrovirus was generated as described (Akagi et al., 2000) and used to infect 293T cells. The cells expressing FcγRIIA-FLAG (293T/RIIA-FLAG cells) were selected by culturing them in DMEM supplemented with 10% FCS in the presence of blasticidin S (final concentration, 10 μg/ml).
cDNAs of human VAMP3, VAMP7, and syntaxin 4 and of mouse D12 were obtained by PCR from human leukocyte or mouse MC9 cell cDNAs using synthesized primers based on the published nucleotide sequences. Plasmids encoding human syntaxins 2 and 3 were generously provided by Dr. K. Kasai (Gunma University, Maebashi, Japan). The expression plasmids for SNARE proteins and COPII proteins were constructed by subcloning cDNA fragments by PCR into pmVENUS-C1, pCX4puro with a C-terminal mVENUS, pmRFP-C1, pcDNA-myc, or pFLAG-CMV-2 (Sigma-Aldrich). To construct the pmVENUS-C1 vector, the codons for F47 and A206 in the pEYFP-C1 vector (BD Bioscience Clontech) were modified to L47 and K206, respectively, using the QuickChange protocol (Stratagene, La Jolla, CA) (Nagai et al., 2002; Zacharias et al., 2002). A cDNA for mRFP1 (mRFP) was a generous gift of Dr. R. Y. Tsien (University of California, San Diego, CA; Campbell et al., 2002). pcDNA-myc was derived from pcDNA3 (Invitrogen) to express a protein fused with the C-terminal Myc epitope. pCX4puro was also kindly donated by Dr. T. Akagi (Osaka BioScience Institute; Akagi et al., 2000). The J774 cell line expressing mVENUS-SNARE proteins (J774/mVENUS-syntaxin 18, J774/mVENUS-D12, and J774/mVENUS-Sec22b) were established by infection of the recombinant retrovirus generated from pCX4puro-mVENUS-SNARE proteins as described (Akagi et al., 2000) and culturing them in the presence of puromycin (final concentration 2 μM). To generate the pArf6 T27N-mRFP construct, the Arf6 T27N cDNA fragment containing the EcoRI/KpnI site was amplified by PCR from the pcDNA3-Arf6 T27N-HA kindly provided by Dr. K. Nakayama (Kyoto University, Kyoto, Japan; Hosaka et al., 1996) and was then subcloned into the pmRFP-N1 expression vector.
Phagocytosis Assay
293T/RIIA-FLAG cells were transiently transfected with the indicated constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Twelve hours after transfection, cells were incubated at 4°C with IgG-opsonized beads (0.1% suspension) for 20 min to allow attachment of the beads to the cells, washed three times with ice-cold PBS, and warmed to 37°C in a water bath to initiate phagocytosis in a synchronized manner. After a 40-min incubation, the cells were chilled on ice for 10 min to halt ingestion of the beads. Incompletely internalized beads were stained with Alexa 488–conjugated anti-rabbit IgG on ice, and then the cells were fixed with 4% paraformaldehyde. The number of internalized beads in the transfected cells was counted using a phase-contrast microscope. Fifty cells expressing each indicated construct were scored for attached and internalized beads. The association index is defined as the number of attached plus internalized beads per transfected cell. The phagocytosis index represents the percentage of internalized beads that were not stained with the Alexa 488–anti-rabbit IgG in 50 transfected cells. Data are expressed as a mean ± SE of three or four independent experiments (a total of 150 or 200 transfected cells were measured in each assay).
Texas Red–conjugated Zymosan A Assay
J774/mVENUS cells and J774/mVENUS-syntaxin 18 cells were plated at a density of 5 × 106 cells per 35-mm-diameter culture dish, each containing four 12-mm-diameter coverglasses, and cultured overnight. The culture medium was exchanged with fresh medium, and 30 min later 1.5 × 108 Texas Red–conjugated zymosan A particles (Molecular Probes) were added to the cells in each dish in the presence or absence of cytochalasin B at a final concentration of 10 μM. Coverglasses were withdrawn at the indicated time points and washed thoroughly in PBS to remove free particles, and then the cells were fixed with 4% paraformaldehyde. Fluorescence was measured on a Nikon ECLIPSE TE2000-U microscope using a Plan Fluor 20×/0.50 objective lens (Nikon, Tokyo, Japan). All image analysis was done using Metamorph Imaging System software (Universal Imaging, Downingtown, PA).
Luminol Beads Assay
J774 cells, J774/mVENUS cells, and J774/mVENUS-syntaxin 18 cells were plated at a density of 4 × 106 cells per 35-mm-diameter culture dish and cultured overnight. Four hours after the culture medium was exchanged with fresh medium, the cells were chilled on ice for 5 min and the medium was replaced with phenol red–free, CO2-independent MEM containing luminol-bound microbeads (1.5–2.0 μm in diameter, Kamakura Techno-Science, Kanagawa, Japan; Uchida et al., 1985), from which chemiluminescence was generated by reactive oxygen within phagosomes. After centrifugation at 2000 rpm for 2.5 min at 4°C, the cells were moved to a prewarmed chamber, and the chemiluminescence of each dish was quantified in a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA) by integrating the signal for 15 s every 1 min at a 50.1% sensitivity level. Results are given in relative light units (RLUs).
siRNA Experiment
Human syntaxin 18– and D12-specific duplex siRNAs with two-nucleotide overhangs at the 3′-ends of the sequences (syntaxin 18 siRNA: 5′-GUUUGAACAGGAAAAUCAGCGACUAAG-3′, which corresponds to positions 702–726 relative to the start codon, and D12 siRNA: 5′CCCAGAGUGUCAUCAAGAAGGACAAAG-3′, which corresponds to positions 569–593) were designed at iGENE Therapeutics (Tsukuba, Japan) and synthesized at Hokkaido System Science (Hokkaido, Japan). An siRNA duplex containing 47% GC content (catalogue number D-001206-09-05; Dharmacon, Lafayette, CO) was used as a control. 293T/RIIA-FLAG cells were transfected with siRNAs at a final concentration of 40 nM using Lipofectamine 2000 (Invitrogen). Twelve hours after transfection, control and syntaxin 18 siRNAs were transfected into the cells again under the same conditions. Twenty-four hours after the first transfection, the cells were split into three dishes for Western blot analysis, immunofluorescence characterization, and a phagocytosis assay.
Preparation of Cell Lysate
After transfection with a plasmid or an siRNA, the cells were suspended in extraction buffer (20 mM HEPES-KOH, pH 7.2, 100 mM KCl, 2 mM EDTA, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol, and a protease inhibitor cocktail; Nakarai Chemicals, Kyoto, Japan) and chilled on ice for 20 min. Cell lysates were collected from the supernatants after centrifugation at 13,000 rpm for 20 min. In the siRNA experiments, the protein concentration of cell lysates was determined using a BCA protein assay reagent kit (Pierce, Rockford, IL). Equal amounts of each cell lysate were separated by SDS-PAGE, followed by Western blotting.
Immunoprecipitation
Using Lipofectamine 2000 (Invitrogen), 293T cells were cotransfected with pcDNA-myc and pFLAG-CMV-2, for the expression of an N-terminal Myc fusion protein of syntaxin 18ΔTMD (Hatsuzawa et al., 2000) or N-terminal FLAG fusion of SNARE proteins including D12, Sec22b, VAMP3, VAMP7, and syntaxins 1–4. Twenty-four hours after transfection, the cells were harvested and lysed in extraction buffer as described above. The cell lysates were incubated with an anti-Myc monoclonal antibody (mAb; 9E10) for 30 min at 4°C. Protein G-Sepharose (Amersham Biosciences, Piscataway, NJ) was then added, and the mixture was allowed to incubate for 16 h at 4°C with gentle rotation. The beads were washed four times with extraction buffer, and immunoprecipitates were eluted with SDS-PAGE sample buffer. Five percent of the total lysate was mixed with 5× SDS-PAGE sample buffer and heated at 95°C for 5 min. The samples were analyzed with Western blotting using an anti-FLAG M2 antibody or an anti-Myc antibody.
Immunofluorescence
J774 cells expressing the indicated constructs were fed with latex beads of 3.0-μm diameter (Sigma-Aldrich) that were opsonized with IgGs for 20 min and then fixed with 4% paraformaldehyde for 20 min at room temperature. 293T/RIIA-FLAG cells were fixed with 4% paraformaldehyde for 20 min at room temperature to detect transiently or stably expressed proteins in the phagocytosis assay. Alternatively, they were fixed with methanol at −20°C for 7 min to detect endogenous syntaxin 18 in the siRNA experiment. Confocal images were captured on an LSM510meta laser scanning microscope using a plan-Apochromat 63× NA1.4 oil immersion objective (Zeiss, Oberkochen, Germany). The images were processed on PC computers using Adobe Photoshop (San Jose, CA). For the phagocytosis assay, conventional fluorescence microscopy and cell counting were performed with a BX60 microscope using a UPlanApo 40×/NA1.00 oil immersion objective (Olympus, Tokyo, Japan).
Electron Microscopy
J774/mVENUS-syntaxin 18 cells were incubated with 0.8-μm blue-dyed beads (Sigma-Aldrich) for 20 min, and then phagosome purification was performed as described above. Samples of the cell suspension before the purification procedure and of purified phagosomes were fixed in 4% paraformaldehyde and 0.02% glutaraldehyde in 100 mM HEPES buffer (pH 7.4) for 20 min. mVENUS-syntaxin 18 was detected with an anti-EGFP antibody and a goat anti-rabbit IgG coupled to 5-nm gold. The sample was dehydrated with ethanol and embedded in epoxy resin. The sections of the sample were examined with a JEM-1210 electron microscope (JEOL, Tokyo, Japan) at 80 kV.
SDS-PAGE and Western Blotting
SDS-PAGE and Western blotting were carried out as described previously (Hatsuzawa et al., 2000). Immobilon-P membranes (Millipore, Bedford, MA) were used for blotting. HRP-conjugated anti-rat IgG or anti-mouse IgG were used as secondary antibodies, and the blots were developed by enhanced chemiluminescence using ECL Western Blotting Detection Reagents (Amersham Biosciences) according to the manufacturer’s instructions.
RESULTS
The ER-localized SNARE Proteins Syntaxin 18, D12, and sec22b Are Present on Phagosomes in Macrophages
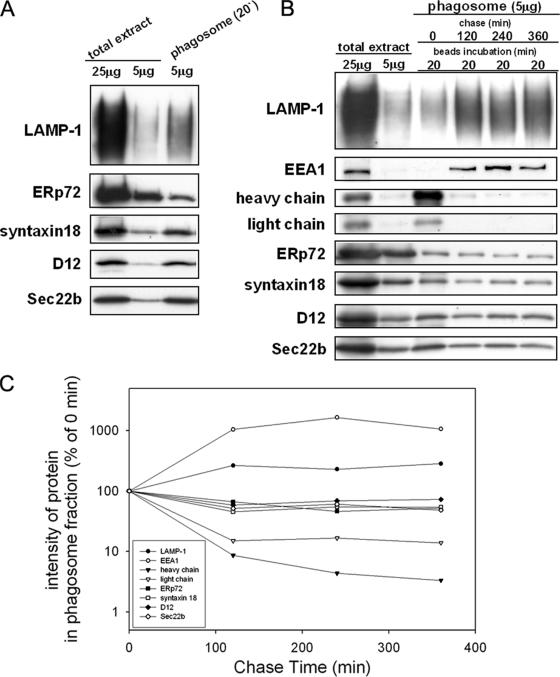
To define whether the known ER-localized SNARE proteins syntaxin 18, D12, and Sec22b are present on phagosomes, we purified phagosomes from the murine macrophage-like cell line J774 after the cells ingested latex beads (0.8 μm in diameter). Phagosomes purified 20 min after ingestion were subjected to Western blotting analysis. As shown in Figure 1, the phagocytic marker LAMP-1 was enriched in the phagosome fraction compared with the total extract, indicating the success of the phagosome purification. We found that the ER-localized SNARE proteins syntaxin 18, D12, and Sec22b were also markedly enriched in the phagosome fraction (Figure 1A). In contrast, the ER lumenal protein ERp72 was recovered in the phagosome fraction to a far lesser extent, consistent with previous results from a member of the same protein family, ERp57 (Guermonprez et al., 2003).
Figure 1.
ER-localized SNARE proteins are enriched in isolated phagosomes. (A) J774 macrophages were incubated in the presence of nonopsonized latex beads for 20 min, after which the phagosome fraction was isolated from the cells by sucrose density centrifugation as described in Materials and Methods. The total cell extract (25 μg and 5 μg) and the isolated phagosome fraction (5 μg) were analyzed by SDS-PAGE and subsequent Western blotting with the use of antibodies against LAMP-1 (a lysosome marker), ERp72 (an ER marker), and ER-localized SNARE proteins (syntaxin 18, D12, and Sec22b). (B) J774 cells were incubated in the presence of IgG-opsonized latex beads for 20 min and then washed to remove the unbound beads. After further incubation for the indicated time (120, 240, or 360 min), the phagosome fraction was isolated from the cells by sucrose density centrifugation. The total cell extract (25 and 5 μg) and the isolated phagosome fraction (5 μg) were analyzed by SDS-PAGE and subsequent Western blotting with various antibodies as indicated. EEA1 is an early endosome marker protein. The heavy and light chains were derived from the IgG used for opsonization of the beads. (C) The signal intensity on the Western blot (B) was quantified by densitometry using NIH ImageJ (developed at the U.S. National Institutes of Health) and expressed as a percentage of the signal intensity at time 0.
To determine whether the ER membranes are continuously supplied to phagosomes or are immediately removed, we examined the kinetics of various markers incorporated into phagosomes during their maturation. We used IgG-opsonized latex beads to trigger the formation of phagosomes, so that we could monitor the acquisition of the digestive activity of the phagosomes by monitoring the degradation of IgG fragments. As shown in Figure 1B, the heavy and light chains of the IgG attached to the surfaces of the latex beads were enriched in the first 20 min and then rapidly disappeared with further incubation. In contrast, the amounts of both LAMP-1 and EEA1, an early endosomal marker, continued to increase for up to 120 and 240 min, respectively (Figure 1C), indicating the continuous fusion of endosome-lysosomal compartments. Interestingly, the ER-localized SNARE proteins and ERp72 showed distinct kinetics, with their concentration remaining the same or only slightly reduced throughout the incubation up to 360 min (Figure 1C). These data suggest that fusion of the ER with phagosomes occurs at the early stages of invagination, most likely before the fusion with lysosomes.
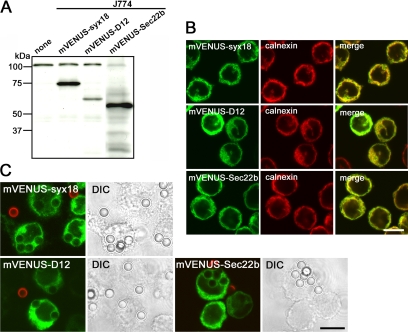
To investigate whether a set of ER-localized SNARE proteins was indeed recruited to phagosomes in J774 cells, we established cells stably expressing syntaxin 18, D12, and Sec22b, each of which was fused to monomeric VENUS fluorescent protein (mVENUS) at its N-terminus (Figure 2A). Indirect immunofluorescence experiments in these cells showed that the expressed proteins were colocalized with ER-resident markers, such as calnexin (Figure 2B). When these cells were incubated for 20 min at 37°C with IgG-opsonized beads (3.0 μm in diameter), all of the mVENUS-SNARE proteins appeared to be recruited to phagosomes (Figure 2C). Incompletely ingested beads that were not in closed vesicles were distinguished by staining the intact cell preparation with anti-rabbit IgG antibodies labeled with Alexa 594 on ice (red).
Figure 2.
mVENUS-SNARE proteins are recruited to phagosomes in J774 macrophages. (A) Western blotting analysis of total lysates from J774 cells stably expressing mVENUS-SNARE proteins. (B) J774 cells expressing mVENUS-SNARE proteins were fixed and stained with an antibody against an ER marker, calnexin. (C) J774 cells expressing mVENUS-SNARE proteins were incubated in the presence of IgG-opsonized latex beads for 20 min and then incompletely ingested beads were stained with Alexa 594–labeled anti-rabbit IgG antibodies (red) before fixation. Differential interference contrast (DIC) images indicate the location of the beads. Bars, 10 μm.
Overexpression of Syntaxin 18ΔTMD and D12ΔTMD Inhibits Fc Receptor–mediated Phagocytosis in 293T/RIIA-FLAG Cells
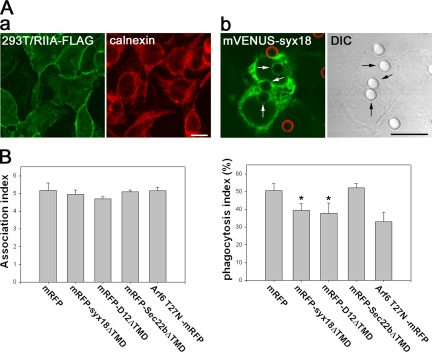
The results described above strongly suggest that in J774 macrophage cells, the ER-localized SNARE proteins participate in either phagosome formation or maturation. Therefore, we investigated whether the ER-localized SNARE proteins play a role in phagosome formation. Overexpression of the full-length SNARE protein or a truncated form lacking the transmembrane domain (TMD) often disturbs specific steps of membrane trafficking (Hay et al., 1997; Tang et al., 1998a; Hatsuzawa et al., 2000). We first attempted to transiently overexpress the cytoplasmic domain of SNARE proteins (SNAREΔTMD) N-terminally fused to monomeric red fluorescent protein (mRFP) in J774 cells. Because we were unable to obtain a sufficiently high efficiency of transfection in J774 cells, we instead established 293T cells stably expressing the FcγRIIA receptor C-terminally tagged with FLAG (293T/RIIA-FLAG cells). Examination by immunofluorescence confirmed that the majority of FcγRIIA-FLAG was localized on the cell surface (Figure 3Aa). As shown in Figure 3Ab, 293T/RIIA-FLAG cells were able to engulf IgG-opsonized latex beads, as had been observed with professional phagocytes. Consistent with the results in J774 cells, when mVENUS-syntaxin 18 was transiently expressed in the 293T cells, the florescent protein was detected in membranes of phagosomes as well as in the tubular/reticular ER-like structure, but not on the cell surface (Figure 3Ab).
Figure 3.
Overexpression of the cytoplasmic portions of syntaxin 18 and D12 inhibits Fc receptor–mediated phagocytosis in 293T cells expressing FcγRIIA-FLAG. (A) (a) 293T cells stably expressing FcγRIIA-FLAG (293T/RIIA-FLAG cells) were fixed and double-stained with antibodies against FLAG and calnexin. (b) 293T/RIIA-FLAG cells were transfected with a plasmid expressing mVENUS-syntaxin 18. After 12 h, the cells were incubated in the presence of IgG-opsonized latex beads for 30 min, and then incompletely ingested beads were stained with Alexa 594–labeled anti-rabbit IgG antibodies (red) before fixation. The DIC image indicates the location of the beads. Arrows point to internalized IgG-opsonized beads. Bars, 10 μm. (B) 293T/RIIA-FLAG cells were transfected with the indicated constructs (the N-terminally mRFP-tagged cytoplasmic portions of SNARE proteins and the C-terminally mRFP-tagged T27N mutant of Arf6). After 12 h, the cells were incubated in the presence of IgG-opsonized latex beads for 40 min, and then incompletely ingested beads were stained with Alexa 488–labeled anti-rabbit IgG antibodies before fixation. The association index (left panel) and the phagocytosis index (right panel) represent the number of beads attached and ingested per single transfected cell and the percentage of beads that were ingested, respectively. Fifty transfected cells were used for each measurement. Data presented are the mean ± SE of four independent experiments (*p < 0.05 vs. mRFP).
We first verified that 293T/RIIA-FLAG cells would be useful for the evaluation of a dominant-negative effect on phagocytosis. It has been shown previously that overexpression of an Arf6 GTP binding-deficient mutant (Arf6 T27N) strongly inhibits Fc receptor–mediated phagocytosis in professional phagocytes (Zhang et al., 1998; Uchida et al., 2001; Niedergang et al., 2003). We therefore examined whether or not the Arf6 T27N mutant caused the same effects in 293T/RIIA-FLAG cells. The ability of cells to phagocytose was evaluated by calculating the fraction of completely ingested beads relative to all the beads associated with the cells (the phagocytosis index). When the phagocytosis index was determined in the cells overexpressing C-terminally mRFP-tagged Arf6 T27N (Arf6 T27N-mRFP) 12 h after transfection, inhibition was clearly observed (Figure 3B). Overexpression of mRFP alone had no effect (Figure 3B).
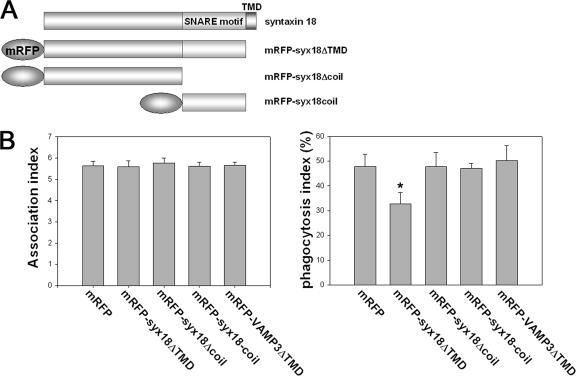
Using mRFP and Arf6 T27N-mRFP as a negative and positive control, respectively, we next determined whether overexpression of the ΔTMD form of ER-localized SNARE proteins affected phagosome formation. As shown in the right panel of Figure 3B, overexpression of both mRFP-syntaxin 18ΔTMD and mRFP-D12ΔTMD significantly reduced the number of beads that had been ingested by the cells by 12 h after transfection, although the extent of the inhibition was slightly less than that of Arf6 T27N- mRFP. In contrast, overexpression of mRFP-Sec22bΔTMD had no effect on phagocytosis in this analysis, although in a second analysis using the luminol beads system, mVENUS-Sec22bΔTMD did inhibit phagocytosis (Supplementary Figure S1). The association of beads with cells per se was not significantly affected by overexpression of any of the mRFP-tagged proteins (Figure 3B, left). Because it was conceivable that overexpression of ER-localized SNARE proteins might have affected the cell surface expression of FcγRIIA-FLAG, we quantitated the cell surface signal of FcγRIIA-FLAG by staining the intact cells with an anti-FLAG M2 mAb; however, the expression level of the receptor was unchanged in any case for at least 12 h after transfection (unpublished data). These results suggest that syntaxin 18 and D12 participate in phagosome formation during Fc receptor–mediated phagocytosis; in contrast, Sec22b may have a role in phagosome formation only under limited circumstances (Becker et al., 2005).
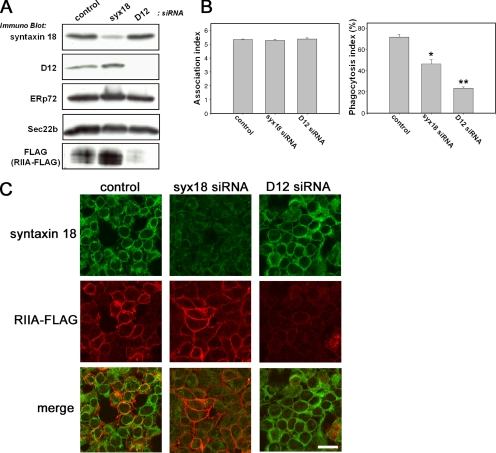
siRNA-induced Knockdown of Syntaxin 18 Expression Inhibits Fc Receptor–mediated Phagocytosis
Next, to determine whether both syntaxin 18 and D12 mediate the phagocytosis of IgG-opsonized beads, we made use of siRNAs directed against syntaxin 18 and D12 in order to silence the expression of these endogenous proteins in 293T/RIIA-FLAG cells. Compared with control siRNA transfection, transfection of syntaxin 18 siRNA specifically decreased its expression without affecting the expression of other ER resident proteins (Figure 4A). Using D12 siRNA, we also detected the marked reduction of D12 expression and observed no effect on syntaxin 18, ERp72, or Sec22b expression (Figure 4A). The number of beads associated with the cells in our phagocytosis assay was not reduced by transfection of the cells with any of the siRNAs (Figure 4B, left). Similarly, surface expression of transiently transfected VSVG-EGFP was not affected by the silencing (our unpublished data). However, as shown in the right panel of Figure 4B, Fc receptor–mediated phagocytosis was significantly inhibited by the transfection of syntaxin 18 siRNA, and transfection of D12 siRNA reduced the phagocytosis index even more strongly. However, in this case, it was conceivable that the observed inhibition might have been caused by a failure of the cell to express FcγRIIA-FLAG on the surface. Previously, we reported that syntaxin 18 plays an important role in protein transport between the ER and the Golgi (Hatsuzawa et al., 2000), and we have recently shown that D12 is involved in post-Golgi transport (Okumura et al., 2006). This prompted us to investigate the cell surface expression of FcγRIIA-FLAG in 293T/RIIA-FLAG cells transfected with siRNA. The immunofluorescence of the cells transfected with syntaxin 18 siRNA showed reduced staining of syntaxin 18 without affecting the fluorescent signals of anti-FLAG antibodies (Figure 4C, middle column), suggesting that siRNA-induced knockdown of syntaxin 18 directly inhibits Fc receptor–mediated phagocytosis. In contrast, when D12 was knocked down, FcγRIIA-FLAG was almost absent (Figure 4C, right) as confirmed with Western blotting (Figure 4A). However, the residual FcγRIIA-FLAG appeared to be sufficient to bind to IgG-opsonized beads because no differences were observed in the association index between control and D12 siRNA (Figure 4B, left). These results indicate that the D12 siRNA-induced inhibition of phagocytosis could be mainly due to down-regulation of FcγRIIA-FLAG expression.
Figure 4.
siRNA-induced knockdown of syntaxin 18 expression inhibits Fc receptor–mediated phagocytosis in 293T/RIIA-FLAG cells. (A) 293T/RIIA-FLAG cells transfected with a nonspecific siRNA as a control or transfected with syntaxin 18 siRNA or D12 siRNA were analyzed. A mixture of total cell lysates from three independently transfected cell cultures was analyzed by SDS-PAGE and subsequent Western blotting using the indicated antibodies. (B) 293T/RIIA-FLAG cells were transfected with the indicated siRNAs. Thirty-six hours after the first siRNA transfection, the cells were transfected with an mRFP plasmid to mark expression-competent cells. After 12 h, the cells were incubated in the presence of IgG-opsonized latex beads for 40 min, and incompletely ingested beads were stained with Alexa 488–labeled anti-rabbit IgG antibodies before fixation. The association index (left panel) represents the number of beads attached and ingested per single transfected cell and the phagocytosis index (right panel) represents the percentage of beads ingested; the indices were calculated from 50 mRFP-expressing cells per condition. Data presented are the mean ± SE of three independent experiments (*p < 0.05 and **p < 0.005, compared with control). (C) 293T/RIIA-FLAG cells were transfected with the indicated siRNAs. Forty-eight hours after the first siRNA-transfection, the cells were fixed and double-stained with antibodies against syntaxin 18 (top column) and FLAG (middle column). Bar, 10 μm.
The Entire Cytoplasmic Region of Syntaxin 18 Is Needed for Inhibition of Phagocytosis
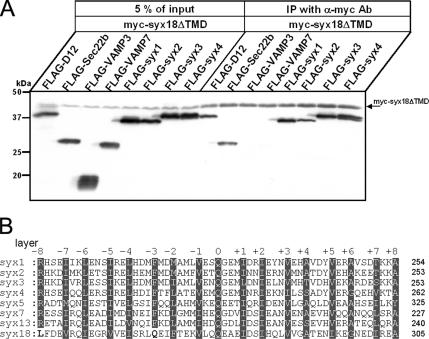
Syntaxin 18, like other syntaxins, possesses a predicted coiled-coil region, which is called the SNARE motif (see Figure 8B) and is thought to be important for fusion events (Hatsuzawa et al., 2000). We therefore investigated whether expression of the SNARE motif of syntaxin 18 had dominant-negative effects on phagocytosis in 293T/RIIA-FLAG cells. When mRFP-tagged syntaxin 18Δcoil, which lacks the SNARE motif, or mRFP-syntaxin 18coil, consisting of the SNARE motif alone (Figure 5A), was transfected into the cells, we found no difference in the number of beads associated with the cells (Figure 5B, left). Overexpression of mRFP-syntaxin 18ΔTMD significantly reduced phagocytosis; in contrast, neither overexpression of mRFP-syntaxin 18coil nor mRFP-syntaxin 18Δcoil had an inhibitory effect (Figure 5B, right). Overexpression of mRFP-VAMP3ΔTMD also did not inhibit phagocytosis in our system, although VAMP3, a SNARE protein of endosomes, has been reported to be involved in phagocytosis (Bajno et al., 2000; Coppolino et al., 2001). This discrepancy seems to be caused by a difference in the assay system because overexpressing VAMP3ΔTMD did repress phagocytosis in a luminol beads assay (see Materials and Methods and Supplementary Figure S1). These data suggest that the SNARE motif of syntaxin 18 is not sufficient for its function in phagocytosis.
Figure 8.
Syntaxin 18ΔTMD is capable of interacting with the plasma membrane syntaxins. (A) 293T cells were cotransfected with the Myc-tagged syntaxin 18ΔTMD and the indicated FLAG-tagged constructs. Twenty-four hours after transfection, cells were harvested and lysed in a buffer containing 20 mM HEPES/KOH, 100 mM KCl, 2 mM EDTA, 1 mM DTT, 1 mM PMSF, and 1% Triton X-100. Myc-syntaxin 18 ΔTMD (myc-syx18ΔTMD) was immunoprecipitated from lysates with anti-Myc antibodies. After washes, immunoprecipitates were separated on SDS-PAGE, and the antigens were detected by Western blotting using anti-Myc and anti-FLAG antibodies. (B) Sequence alignment of the Qa-SNARE motif region of Homo sapiens syntaxin family proteins. The positions of the heptad repeat layers (−8 to +8) are indicated. Highly homologous residues of conserved layers are indicated with white letters on a gray background. The bold black residue letters denote the amino acid residues in the −8 and −3 layer positions of syntaxin 18 that are inconsistent with the conserved residues of other syntaxins.
Figure 5.
The entire cytoplasmic region of syntaxin 18 is responsible for inhibition of Fc receptor–mediated phagocytosis in 293T/RIIA-FLAG cells. (A) Schematic representation of syntaxin 18 and the mRFP-syntaxin 18 constructs. mRFP was fused to the N-terminus of all constructs. The SNARE-motif and TMD represent a coiled-coil and a transmembrane domain, respectively. (B) 293T/RIIA-FLAG cells were transfected with the mRFP-tagged syntaxin 18 variants or the N-terminally mRFP-tagged VAMP3 ΔTMD, which lacks the transmembrane domain. After 12 h, the cells were incubated with IgG-opsonized latex beads for 40 min and then the incompletely ingested beads were stained with Alexa 488–labeled anti-rabbit IgG antibodies before permeabilization. The association index (left panel) and the phagocytosis index (right panel) were calculated from 50 transfected cells per condition. Data presented are the mean ± SE of four independent experiments (*p < 0.01, as compared with mRFP).
COPII-coated Vesicles Are Not Involved in Syntaxin 18–mediated Phagocytosis
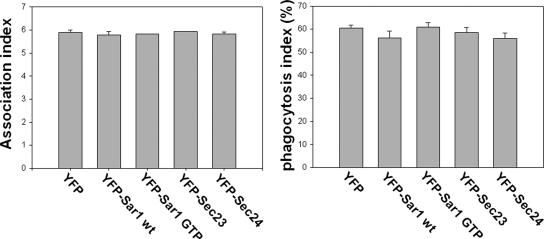
The results described above indicate that syntaxin 18 is involved in phagosome formation; however, it is still possible that syntaxin 18 may mediate fusion between COPII-coated vesicles detached from the ER and the plasma membrane, rather than directly between the ER and plasma or phagosomal membranes. To investigate this possibility, we performed the phagocytosis assay using 293T/RIIA-FLAG cells transiently expressing the COPII components Sar1, Sec23, and Sec24 tagged with yellow fluorescent protein (YFP; Barlowe, 2002; Sato, 2004). As shown in Figure 6, the expression of a GTP-bound form of Sar1, the Sar1 H79G mutant (YFP-Sar1-GTP), whose expression inhibits fusion of COPII-coated vesicles by locking the GTP-GDP cycles of the coats (Aridor et al., 1995), did not decrease the phagocytosis index or the number of associated beads compared with the expression of YFP alone or of wild-type YFP-Sar1. Similar results were obtained when YFP-Sec23 or YFP-Sec24 were overexpressed (Figure 6). These results suggest that processes mediated by COPII-coated vesicles are not involved in phagosome formation.
Figure 6.
Inhibition of COPII-mediated transport had no influence on phagocytosis. 293T/RIIA-FLAG cells were transfected with the N-terminally YFP-tagged COPII components as indicated. After 12 h, the cells were incubated with IgG-opsonized latex beads for 40 min, and then the incompletely ingested beads were stained as in Figure 5. The association index (left panel) and the phagocytosis index (right panel) were calculated from 50 transfected cells per condition. Data presented are the mean ± SE of three independent experiments.
Syntaxin 18 Is also Required for Fc Receptor–independent Phagocytosis in J774 Macrophages
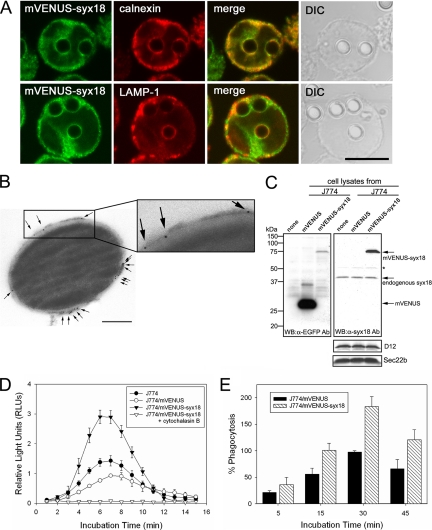
To gain further insights into the function of syntaxin 18 during phagocytosis, we examined the involvement of syntaxin 18 in Fc receptor–independent phagocytosis using J774 cells. To facilitate the analysis, we utilized J774 cells stably expressing mVENUS-syntaxin 18. Western blotting analysis showed that mVENUS-syntaxin 18 was expressed at levels about threefold higher than endogenous syntaxin 18 in J774/mVENUS-syntaxin 18 cells (Figure 7C). When nonopsonized latex beads (3.0 μm in diameter) were fed to the cells and incubated for 20 min, the phagosomes were clearly demarcated by mVENUS-syntaxin 18 as well as LAMP-1 (Figure 7A). Interestingly, whereas almost all phagosomes were labeled with mVENUS-syntaxin 18, LAMP-1 staining was observed in only a subpopulation of the phagosomes. Considering the kinetic results shown in Figure 1, B and C, these LAMP-1–negative phagosomes may represent immature phagolysosomes (Supplementary Figure S2). The presence of syntaxin 18 on the phagosomal membranes was also confirmed by electron microscopic (EM) analysis. We purified phagosomes from J774 cells incubated with nonopsonized latex beads (0.8 μm in diameter, dyed blue) for 20 min and performed immunolabeling with an anti-EGFP antibody followed by incubation with a goat anti-rabbit IgG coupled to 5-nm gold. The result clearly revealed that mVENUS-syntaxin 18 was indeed integrated nonuniformly into the phagosomal membrane surrounding the bead (Figure 7B and Supplementary Figure S3).
Figure 7.
mVENUS-syntaxin 18 is localized on phagosomal membranes surrounding nonopsonized latex beads and enhances phagocytosis of luminol-bound microbeads in J774 macrophages. (A) J774 cells expressing mVENUS-syntaxin 18 (J774/mVENUS-syx18 cells) were incubated in the presence of nonopsonized latex beads (3.0 μm diameter) for 20 min before fixation. Cells were then stained with antibodies against calnexin or LAMP-1. Bar, 10 μm. (B) Electron microscopic (EM) analysis of mVENUS-syntaxin 18 recruitment on purified phagosomes. Formed phagosomes ingested 0.8-μm diameter blue-dyed latex beads for 20 min and were then purified by sucrose gradient centrifugation from J774/mVENUS-syx18 cells. The purified phagosomes were labeled with an anti-EGFP antibody and a goat anti-rabbit antibody coupled to 5-nm gold (arrows indicate some of the gold particles) as described in Materials and Methods (also see Supplementary Figure S3). Bar, 200 nm. (C) Western blotting analysis of total lysates from J774 cells stably expressing mVENUS and mVENUS-syntaxin 18. Western blotting was carried out using antibodies against EGFP and syntaxin 18. The expression of mVENUS-syntaxin 18 was about threefold higher than that of endogenous syntaxin 18 in J774/mVENUS-syntaxin 18 cells. Asterisk (*) denotes nonspecific band recognized by anti-syntaxin 18 antibodies. The expression levels of D12 and Sec22b in these cells were not different. (D) J774, J774/mVENUS, and J774/mVENUS-syntaxin 18 cells were incubated with luminol-bound microbeads. Chemiluminescence from cells that ingested the beads was measured on a TD-20/20 luminometer for 15 s every 1 min, up to 15 min. These data indicate the rate of phagocytosis in the cells. When J774/mVENUS-syntaxin 18 cells were incubated with the beads in the presence of cytochalasin B (final 10 μM), no significant signal was detected. Data presented are the mean ± SE of six independent experiments. (E) J774/mVENUS and J774/mVENUSsyntaxin 18 cells were incubated with Texas Red-conjugated zymosan A prespectively, as articles. At the indicated time points, samples were washed in PBS to remove free particles and fixed and then processed for fluorometric analyses as described in Materials and Methods (also see Supplementary Figure S5). Arbitrary fluorescence units were obtained by subtracting the average fluorescence intensity in the presence of cytochalasin B from that in the absence of cytochalasin B and then normalizing to the maximal value obtained for J774/mVENUS cells within the same experiment. Data presented are the mean ± SE of three independent experiments.
We next analyzed whether mVENUS-syntaxin 18 expressed in J774 cells could up-regulate the rate of phagocytosis. For this, we quantified phagocytosis using luminol-bound microbeads (1.5–2.0 μm in diameter) as described in Materials and Methods. As shown in Figure 7D, J774/mVENUS-syntaxin 18 cells had a significantly higher rate of phagocytosis compared with wild-type J774 cells or cells expressing mVENUS alone, indicating that mVENUS-syntaxin 18 is required for progression of phagocytosis, presumably by facilitating membrane fusion between the ER and the plasma membrane during phagocytosis. There was no difference in the cell shape and size between J774/mVENUS-syntaxin 18 cells J774/mVENUS cells (Supplementary Figure S4). In addition, we performed another analysis (Texas Red–conjugated zymosan A assay; see Materials and Methods) to detect phagocytosis directly rather than by measuring the activity of reactive oxygen intermediates after phagocytosis as in the luminol beads assay. In this assay system, cells overexpressing mVENUS-syntaxin 18 took up more labeled zymosan particles than control (J774/mVENUS) cells (Figure 7E and Supplementary Figure S5). Interestingly, the effect of mVENUS-syntaxin 18 was already observed at the earliest time point of measurement, and the enhancement remained nearly constant at the later phases, although the number of zymosan taken up continued to be increased (Figure 7E). These results clearly indicate that syntaxin 18 is functionally involved in Fc receptor–independent phagocytosis, particularly at the very early stage.
Direct Interaction of Syntaxin 18 with Plasma Membrane Syntaxins
Syntaxins 1–4 are mainly localized in the plasma membrane and are known to function in several exocytic pathways. It has been reported that syntaxins 2–4, but not syntaxin 1, are expressed in J774 macrophages and are present on their phagosomes (Hackam et al., 1996). Additionally, the endocytic pathway–related SNARE proteins VAMP3 and VAMP7 have been shown to be involved in the early steps of phagocytosis (Bajno et al., 2000; Braun et al., 2004). These facts raise the possibility that these SNARE proteins participate in ER-mediated phagocytosis as cognate SNARE partners of syntaxin 18. To address this possibility, we performed immunoprecipitation experiments as described in Materials and Methods. Lysates of 293T cells coexpressing Myc-tagged syntaxin 18ΔTMD (myc-syx18ΔTMD) and one of the FLAG-tagged SNARE proteins syntaxin 1, 2, 3, 4; VAMP3; VAMP7; D12; or Sec22b were incubated with anti-Myc antibodies, and the immunoprecipitates were analyzed by SDS-PAGE followed by Western blotting with anti- Myc and anti-FLAG M2 antibodies. FLAG-D12 and FLAG-Sec22b, which are known to bind to syntaxin 18, were used as positive controls. FLAG-syntaxins 1–4 coimmunoprecipitated with Myc-syntaxin 18ΔTMD, but FLAG-VAMP3 and -VAMP7 did not (Figure 8A). These results suggest that syntaxin 18 is capable of interacting with plasma membrane syntaxins in vitro, and this interaction may be required for membrane fusion between the ER and the plasma membrane during phagocytosis.
DISCUSSION
During phagocytosis, the ER is thought to supply membranes to engulf large foreign particles and to recruit MHC class I molecules to phagosomes by direct fusion with the plasma membrane and/or phagosomes (Gagnon et al., 2002; Guermonprez et al., 2003; Houde et al., 2003). Proteomic and EM analyses have revealed that phagosomes contain ER resident proteins, and the ER membranes are directly in contact with either the plasma membranes or phagosomes (Garin et al., 2001; Gagnon et al., 2002). However, the molecular mechanism of their membrane fusion has never been elucidated. In this study, we have described the direct involvement of ER-localized SNARE proteins in phagocytosis. In the yeast Saccharomyces cerevisiae, the ER-localized SNARE proteins Ufe1p, Sec22p, Sec20p, and Use1p/Slt1p form a SNARE complex required for vesicular transport between the ER and the Golgi (Burri et al., 2003; Dilcher et al., 2003). In addition to the previously known mammalian orthologues syntaxin 18 and Sec22b, and BNIP1 and D12 (also called p31) were recently identified as mammalian orthologues of Sec20p and Use1p/Slt1p, respectively (Hirose et al., 2004; Nakajima et al., 2004; Okumura et al., 2006). BNIP1, unlike other ER-localized SNARE proteins, has been shown to be mainly required for apoptotic cell death and homotypic ER membrane fusion rather than for vesicle trafficking (Nakajima et al., 2004). We therefore focused on syntaxin 18, Sec22b, and D12 and investigated their localization and function during ER-mediated phagocytosis.
Identification of the ER-localized SNARE proteins syntaxin 18, D12, and Sec22b by subcellular fractionation or direct visualization using mVENUS tagging suggested that these proteins may be involved in ER-mediated phagocytosis as a part of the fusion machinery. To prove this, we used 293T/RIIA-FLAG cells because it has been observed previously that expression of FcγRIIA in CHO, COS-1, and ts20 cells can give them the ability to phagocytose IgG-opsonized particles (Indik et al., 1995; Bajno et al., 2000; Booth et al., 2002; Collins et al., 2002). 293T cells were chosen so that we could efficiently and easily express high levels of exogenously transfected plasmids. We observed that dominant-negative expression of soluble mutants of syntaxin 18 and D12 lacking the transmembrane domain, but not the Sec22b mutant, significantly suppressed the rate of phagocytosis by an amount similar to Arf6 T27N, which has been reported to cause inhibition of Fc receptor–mediated phagocytosis (Uchida et al., 2001; Niedergang et al., 2003; Figure 3B). Similarly, silencing of syntaxin 18 and D12 by siRNA led to reduced phagocytotic efficiency in 293T/RIIA-FLAG cells. However, we found that knockdown of D12 expression also drastically reduced the protein levels of FcγRIIA-FLAG as well as of some other proteins (Okumura et al., 2006), and it is therefore not clear at the present time whether D12 actually functions in fusion events during phagocytosis. In addition, J774/mVENUS-syntaxin 18 cells, in which the expression of mVENUS-syntaxin 18 is about threefold higher than that of endogenous syntaxin 18, phagocytosed luminol-bound microbeads and Texas Red–labeled zymosan particles at increased rates, regardless of IgG-opsonization, indicating that the expressed mVENUS-syntaxin 18 positively contributes to phagocytosis. These results strongly suggest that syntaxin 18 is a functional participant in the fusion events between the ER and the plasma or phagosomal membranes during the general phagocytic process, particularly at the very early stage.
More recently, Sec22b (also called ERS24) has been reported to function selectively in phagocytosis triggered by IgG-opsonized large particles (3.0 μm in diameter) in J774 macrophages (Becker et al., 2005). In contrast, we were unable to detect involvement of Sec22b in phagocytosis when using our 293T/RIIA-FLAG system. However, we do not exclude the possibility of a functional role for Sec22b even in our system because we did not examine the effect of loading antibody against Sec22b as they did. However, in a luminol beads assay, mVENUS-Sec22bΔTMD as well as mVENUS-VAMP3ΔTMD inhibited phagocytosis in J774 cells (Supplementary Figure S1C), suggesting that the role of Sec22b may depend in part of the environment. This result confirms previous reports (Bajno et al., 2000; Becker et al., 2005) and suggests that syntaxin 18 may play a more general role in phagocytosis.
The plasma membrane SNARE proteins syntaxins 2–4 are expressed and localized on phagosomal membranes in J774 cells, although syntaxin 1 is not (Hackam et al., 1996). Furthermore, the endosomal SNARE proteins VAMP3 and VAMP7 are recruited underneath the site of particle attachment on the plasma membrane during early phagocytosis and then engage in phagosome formation in RAW264.7 cells (Bajno et al., 2000; Braun et al., 2004). These findings led us to investigate whether these SNARE proteins bind to syntaxin 18. The results clearly showed that syntaxin 18 has the ability to bind to syntaxins 2–4 but not VAMP3 and VAMP7 (Figure 8), suggesting that the plasma membrane–localized syntaxins quickly form a SNARE complex with syntaxin 18 at the site where foreign particles are attached on the plasma membrane. This may be surprising because syntaxin 18 has been classified as a member of the Qa-SNARE family, like the plasma membrane–localized (syntaxins 1–4), the Golgi-localized (syntaxin 5), and the endocytic pathway–related (syntaxin 7 and 13) syntaxins (Figure 8B). However, the SNARE motif of syntaxin 18 does not completely satisfy the criteria for Qa-SNARE classification (Antonin et al., 2000). Generally, a phenylalanine residue fills the −3 layer position of Qa-SNAREs, whereas syntaxin 18 has a glutamine residue in this position. Furthermore, a leucine residue is found at the position for the −8 layer in syntaxin 18 instead of the arginine residue that is conserved in most Qa-SNAREs (Figure 8B). Both of these layers are known to be responsible for asymmetric interactions with R- and Qb-/Qc-SNAREs (Fasshauer et al., 1998). We propose that syntaxin 18 is an unusual Qa-SNARE in that it could collaborate with plasma membrane–localized syntaxins (Qa-SNAREs), as can D12 (Qb- or Qc-SNARE; Dilcher et al., 2003; Okumura et al., 2006) and Sec22b (R-SNARE).
We have further found that ER-localized SNARE proteins are involved in phagocytosis from the very early stages and continue to be involved for long periods during phagosome formation, consistent with previous observations of syntaxin 2–4 and 7 but not of syntaxin 13 (Hackam et al., 1996; Collins et al., 2002). As proposed for syntaxin 7, this feature may indicate their direct involvement in phagosome maturation per se. Alternatively, the ER-localized SNARE proteins may be used in an exocytosis process of phagosomes, as reported for syntaxin 3 in J774 macrophages (Di et al., 2002). Our observations that syntaxin 18 is involved in phagosome formation regardless of IgG-opsonization of foreign particles are consistent with previous results on ER-mediated phagocytosis in J774 macrophages and dendritic cells (Gagnon et al., 2002; Guermonprez et al., 2003; Houde et al., 2003), suggesting that syntaxin 18 is part of the general machinery for ER-mediated phagocytosis. However, it seems to be clear that the ER plays no role in the phagocytosis of either the Leishmania donovani parasite in neutrophils (Gagnon et al., 2002) or IgG-opsonized red blood cells in RAW264.7 macrophages (Braun et al., 2004; Henry et al., 2004). In the latter case, indeed, it is focal exocytosis of endocytic vesicles mediated by VAMP3 and/or VAMP7 that is required for phagocytosis. Assuming that the ER membranes are used to supply membranes during phagocytosis, it is rather mysterious why such a system does not require the ER. At the least, fusion of the ER with phagosomal membranes or the cell surface has to be tightly regulated, because the fusion should not disturb the environment of the ER that is suitable for oxidative folding. The zipper model of receptor-mediated phagocytosis (Swanson and Baer, 1995), including that mediated by the Fc receptor, may explain the maintenance of the ER environment. Alternatively, various SNARE binding proteins, such as Munc18–2, 18–3 (18c), or tomosyn (Araki et al., 1997; Riento et al., 2000; Hatsuzawa et al., 2003; Widberg et al., 2003), may strictly regulate the fusion process. It may be a tradeoff for the cells whether or not they use the ER for phagocytosis.
During the preparation of this article, the contribution of the ER to phagosome formation was assessed by Grinstein and colleagues using several independent methods (Touret et al., 2005). Because they did not obtain evidence proving direct fusion between the ER and the plasma membrane, they finally advocated that the plasma membrane rather than the ER is the main source of membrane in the early stage of phagosome formation (Touret et al., 2005). In contrast, Desjardins and colleagues concluded that the ER is involved in early phagosome formation because, for example, ER molecules are more abundant on early phagosomes than on late phagosomes (Gagnon et al., 2005). Although the ER-mediated phagocytosis model remains a subject of controversy, we think that our findings in this study strongly support this model.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mayumi Takeuchi and Hiroko Nakano for excellent technical assistance. We are grateful to Drs. Kazuhisa Nakayama, Kazuo Kasai, Tatsuro Yamashita, Kenji Akasaki, Tsuyoshi Akagi, and Roger Y. Tsien for kindly providing materials and Dr. Satoshi Waguri for helpful discussion. This work was supported in part by a Grant-in-Aid (16044237) from the Japan Society for Promotion of Science and the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Abbreviations used:
- ER
endoplasmic reticulum
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- ECL
enhanced chemiluminescence
- COP
coat protein
- TMD
transmembrane domain
- VSVG-EGFP
enhanced green fluorescent protein-tagged glycoprotein of vesicular stomatitis virus ts045.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-12-1174) on June 21, 2006.
References
- Akagi T., Shishido T., Murata K., Hanafusa H. v-Crk activates the phosphoinositide 3-kinase/AKT pathway in transformation. Proc. Natl. Acad. Sci. USA. 2000;97:7290–7295. doi: 10.1073/pnas.140210297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W., Fasshauer D., Becker S., Jahn R., Schneider T. R. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol. 2002;9:107–111. doi: 10.1038/nsb746. [DOI] [PubMed] [Google Scholar]
- Antonin W., Holroyd C., Fasshauer D., Pabst S., Von Mollard G. F., Jahn R. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 2000;19:6453–6464. doi: 10.1093/emboj/19.23.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki S., Tamori Y., Kawanishi M., Shinoda H., Masugi J., Mori H., Niki T., Okazawa H., Kubota T., Kasuga M. Inhibition of the binding of SNAP-23 to syntaxin 4 by Munc18c. Biochem. Biophys. Res. Commun. 1997;234:257–262. doi: 10.1006/bbrc.1997.6560. [DOI] [PubMed] [Google Scholar]
- Aridor M., Bannykh S. I., Rowe T., Balch W. E. Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J. Cell Biol. 1995;131:875–893. doi: 10.1083/jcb.131.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajno L., Peng X. R., Schreiber A. D., Moore H. P., Trimble W. S., Grinstein S. Focal exocytosis of VAMP3-containing vesicles at sites of phagosome formation. J. Cell Biol. 2000;149:697–706. doi: 10.1083/jcb.149.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C. COPII-dependent transport from the endoplasmic reticulum. Curr. Opin. Cell Biol. 2002;14:417–422. doi: 10.1016/s0955-0674(02)00348-4. [DOI] [PubMed] [Google Scholar]
- Becker T., Volchuk A., Rothman J. E. Differential use of endoplasmic reticulum membrane for phagocytosis in J774 macrophages. Proc. Natl. Acad. Sci. USA. 2005;102:4022–4026. doi: 10.1073/pnas.0409219102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock J. B., Scheller R. H. SNARE proteins mediate lipid bilayer fusion. Proc. Natl. Acad. Sci. USA. 1999;96:12227–12229. doi: 10.1073/pnas.96.22.12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth J. W., Kim M. K., Jankowski A., Schreiber A. D., Grinstein S. Contrasting requirements for ubiquitylation during Fc receptor-mediated endocytosis and phagocytosis. EMBO J. 2002;21:251–258. doi: 10.1093/emboj/21.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V., Fraisier V., Raposo G., Hurbain I., Sibarita J. B., Chavrier P., Galli T., Niedergang F. TI-VAMP/VAMP7 is required for optimal phagocytosis of opsonised particles in macrophages. EMBO J. 2004;23:4166–4176. doi: 10.1038/sj.emboj.7600427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri L., Varlamov O., Doege C. A., Hofmann K., Beilharz T., Rothman J. E., Sollner T. H., Lithgow T. A SNARE required for retrograde transport to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2003;100:9873–9877. doi: 10.1073/pnas.1734000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao D. S., Hay J. C., Winnick S., Prekeris R., Klumperman J., Scheller R. H. SNARE membrane trafficking dynamics in vivo. J. Cell Biol. 1999;144:869–881. doi: 10.1083/jcb.144.5.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R. F., Schreiber A. D., Grinstein S., Trimble W. S. Syntaxins 13 and 7 function at distinct steps during phagocytosis. J. Immunol. 2002;169:3250–3256. doi: 10.4049/jimmunol.169.6.3250. [DOI] [PubMed] [Google Scholar]
- Coppolino M. G., Kong C., Mohtashami M., Schreiber A. D., Brumell J. H., Finlay B. B., Grinstein S., Trimble W. S. Requirement for N-ethylmaleimide-sensitive factor activity at different stages of bacterial invasion and phagocytosis. J. Biol. Chem. 2001;276:4772–4780. doi: 10.1074/jbc.M007792200. [DOI] [PubMed] [Google Scholar]
- Desjardins M. ER-mediated phagocytosis: a new membrane for new functions. Nat. Rev. Immunol. 2003;3:280–291. doi: 10.1038/nri1053. [DOI] [PubMed] [Google Scholar]
- Desjardins M., Celis J. E., van Meer G., Dieplinger H., Jahraus A., Griffiths G., Huber L. A. Molecular characterization of phagosomes. J. Biol. Chem. 1994;269:32194–32200. [PubMed] [Google Scholar]
- Di A., Krupa B., Bindokas V. P., Chen Y., Brown M. E., Palfrey H. C., Naren A. P., Kirk K. L., Nelson D. J. Quantal release of free radicals during exocytosis of phagosomes. Nat. Cell Biol. 2002;4:279–285. doi: 10.1038/ncb771. [DOI] [PubMed] [Google Scholar]
- Dilcher M., Veith B., Chidambaram S., Hartmann E., Schmitt H. D., Fischer von Mollard G. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 2003;22:3664–3674. doi: 10.1093/emboj/cdg339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey G. P., Botelho R. J., Butler J. R., Moltyaner Y., Chien P., Schreiber A. D., Grinstein S. Phagosomal maturation, acidification, and inhibition of bacterial growth in nonphagocytic cells transfected with FcgammaRIIA receptors. J. Biol. Chem. 1999;274:28436–28444. doi: 10.1074/jbc.274.40.28436. [DOI] [PubMed] [Google Scholar]
- Fasshauer D., Sutton R. B., Brunger A. T., Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon E., Bergeron J. J., Desjardins M. ER-mediated phagocytosis: myth or reality? J. Leukoc. Biol. 2005;77:843–845. doi: 10.1189/jlb.0305129. [DOI] [PubMed] [Google Scholar]
- Gagnon E., Duclos S., Rondeau C., Chevet E., Cameron P. H., Steele-Mortimer O., Paiement J., Bergeron J. J., Desjardins M. Endoplasmic reticulum–mediated phagocytosis is a mechanism of entry into macrophages. Cell. 2002;110:119–131. doi: 10.1016/s0092-8674(02)00797-3. [DOI] [PubMed] [Google Scholar]
- Galli T., Zahraoui A., Vaidyanathan V. V., Raposo G., Tian J. M., Karin M., Niemann H., Louvard D. A novel tetanus neurotoxin-insensitive vesicle-associated membrane protein in SNARE complexes of the apical plasma membrane of epithelial cells. Mol. Biol. Cell. 1998;9:1437–1448. doi: 10.1091/mbc.9.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garin J., Diez R., Kieffer S., Dermine J. F., Duclos S., Gagnon E., Sadoul R., Rondeau C., Desjardins M. The phagosome proteome: insight into phagosome functions. J. Cell Biol. 2001;152:165–180. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt D., Warnatz H. J., Henschel O., Bruckert F., Schleicher M., Soldati T. High-resolution dissection of phagosome maturation reveals distinct membrane trafficking phases. Mol. Biol. Cell. 2002;13:3508–3520. doi: 10.1091/mbc.E02-04-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groothuis T. A., Neefjes J. The many roads to cross-presentation. J. Exp. Med. 2005;202:1313–1318. doi: 10.1084/jem.20051379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermonprez P., Saveanu L., Kleijmeer M., Davoust J., Van Endert P., Amigorena S. ER-phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature. 2003;425:397–402. doi: 10.1038/nature01911. [DOI] [PubMed] [Google Scholar]
- Hackam D. J., Rotstein O. D., Bennett M. K., Klip A., Grinstein S., Manolson M. F. Characterization and subcellular localization of target membrane soluble NSF attachment protein receptors (t-SNAREs) in macrophages. Syntaxins 2, 3, and 4 are present on phagosomal membranes. J. Immunol. 1996;156:4377–4383. [PubMed] [Google Scholar]
- Hatsuzawa K., Hirose H., Tani K., Yamamoto A., Scheller R. H., Tagaya M. Syntaxin 18, a SNAP receptor that functions in the endoplasmic reticulum, intermediate compartment, and cis-Golgi vesicle trafficking. J. Biol. Chem. 2000;275:13713–13720. doi: 10.1074/jbc.275.18.13713. [DOI] [PubMed] [Google Scholar]
- Hatsuzawa K., Lang T., Fasshauer D., Bruns D., Jahn R. The R-SNARE motif of tomosyn forms SNARE core complexes with syntaxin 1 and SNAP-25 and down-regulates exocytosis. J. Biol. Chem. 2003;278:31159–31166. doi: 10.1074/jbc.M305500200. [DOI] [PubMed] [Google Scholar]
- Hay J. C., Chao D. S., Kuo C. S., Scheller R. H. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell. 1997;89:149–158. doi: 10.1016/s0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- Henry R. M., Hoppe A. D., Joshi N., Swanson J. A. The uniformity of phagosome maturation in macrophages. J. Cell Biol. 2004;164:185–194. doi: 10.1083/jcb.200307080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose H., et al. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 2004;23:1267–1278. doi: 10.1038/sj.emboj.7600135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka M., Toda K., Takatsu H., Torii S., Murakami K., Nakayama K. Structure and intracellular localization of mouse ADP-ribosylation factors type 1 to type 6 (ARF1-ARF6) J. Biochem. (Tokyo) 1996;120:813–819. doi: 10.1093/oxfordjournals.jbchem.a021484. [DOI] [PubMed] [Google Scholar]
- Houde M., Bertholet S., Gagnon E., Brunet S., Goyette G., Laplante A., Princiotta M. F., Thibault P., Sacks D., Desjardins M. Phagosomes are competent organelles for antigen cross-presentation. Nature. 2003;425:402–406. doi: 10.1038/nature01912. [DOI] [PubMed] [Google Scholar]
- Indik Z. K., Park J. G., Hunter S., Schreiber A. D. The molecular dissection of Fc gamma receptor mediated phagocytosis. Blood. 1995;86:4389–4399. [PubMed] [Google Scholar]
- Jahn R., Lang T., Sudhof T. C. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- McMahon H. T., Ushkaryov Y. A., Edelmann L., Link E., Binz T., Niemann H., Jahn R., Sudhof T. C. Cellubrevin is a ubiquitous tetanus-toxin substrate homologous to a putative synaptic vesicle fusion protein. Nature. 1993;364:346–349. doi: 10.1038/364346a0. [DOI] [PubMed] [Google Scholar]
- Muller-Taubenberger A., Lupas A. N., Li H., Ecke M., Simmeth E., Gerisch G. Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO J. 2001;20:6772–6782. doi: 10.1093/emboj/20.23.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullock B. M., et al. Syntaxin 7 is localized to late endosome compartments, associates with Vamp 8, and Is required for late endosome-lysosome fusion. Mol. Biol. Cell. 2000;11:3137–3153. doi: 10.1091/mbc.11.9.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Hirose H., Taniguchi M., Kurashina H., Arasaki K., Nagahama M., Tani K., Yamamoto A., Tagaya M. Involvement of BNIP1 in apoptosis and endoplasmic reticulum membrane fusion. EMBO J. 2004;23:3216–3226. doi: 10.1038/sj.emboj.7600333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Yamamoto A., Wada Y., Futai M. Syntaxin 7 mediates endocytic trafficking to late endosomes. J. Biol. Chem. 2000;275:6523–6529. doi: 10.1074/jbc.275.9.6523. [DOI] [PubMed] [Google Scholar]
- Niedergang F., Colucci-Guyon E., Dubois T., Raposo G., Chavrier P. ADP ribosylation factor 6 is activated and controls membrane delivery during phagocytosis in macrophages. J. Cell Biol. 2003;161:1143–1150. doi: 10.1083/jcb.200210069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura A. J., Hatsuzawa K., Tamura T., Nagaya H., Saeki K., Okumura F., Nagao K., Nishikawa M., Yoshimura A., Wada I. Involvement of a novel Q-SNARE, D12, in quality control of the endomembrane system. J. Biol. Chem. 2006;281:4495–4506. doi: 10.1074/jbc.M509715200. [DOI] [PubMed] [Google Scholar]
- Prekeris R., Klumperman J., Chen Y. A., Scheller R. H. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J. Cell Biol. 1998;143:957–971. doi: 10.1083/jcb.143.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riento K., Kauppi M., Keranen S., Olkkonen V. M. Munc18-2, a functional partner of syntaxin 3, controls apical membrane trafficking in epithelial cells. J. Biol. Chem. 2000;275:13476–13483. doi: 10.1074/jbc.275.18.13476. [DOI] [PubMed] [Google Scholar]
- Sato K. COPII coat assembly and selective export from the endoplasmic reticulum. J. Biochem. (Tokyo) 2004;136:755–760. doi: 10.1093/jb/mvh184. [DOI] [PubMed] [Google Scholar]
- Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Swanson J. A., Baer S. C. Phagocytosis by zippers and triggers. Trends Cell Biol. 1995;5:89–93. doi: 10.1016/s0962-8924(00)88956-4. [DOI] [PubMed] [Google Scholar]
- Swanson J. A., Hoppe A. D. The coordination of signaling during Fc receptor-mediated phagocytosis. J. Leukoc. Biol. 2004;76:1093–1103. doi: 10.1189/jlb.0804439. [DOI] [PubMed] [Google Scholar]
- Tang B. L., Low D. Y., Hong W. Hsec22c: a homolog of yeast Sec22p and mammalian rsec22a and msec22b/ERS-24. Biochem. Biophys. Res. Commun. 1998a;243:885–891. doi: 10.1006/bbrc.1998.8194. [DOI] [PubMed] [Google Scholar]
- Tang B. L., Tan A. E., Lim L. K., Lee S. S., Low D. Y., Hong W. Syntaxin 12, a member of the syntaxin family localized to the endosome. J. Biol. Chem. 1998b;273:6944–6950. doi: 10.1074/jbc.273.12.6944. [DOI] [PubMed] [Google Scholar]
- Touret N., et al. Quantitative and dynamic assessment of the contribution of the ER to phagosome formation. Cell. 2005;123:157–170. doi: 10.1016/j.cell.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Uchida H., Kondo A., Yoshimura Y., Mazaki Y., Sabe H. PAG3/Papalpha/KIAA0400, a GTPase-activating protein for ADP-ribosylation factor (ARF), regulates ARF6 in Fcgamma receptor-mediated phagocytosis of macrophages. J. Exp. Med. 2001;193:955–966. doi: 10.1084/jem.193.8.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T., Kanno T., Hosaka S. Direct measurement of phagosomal reactive oxygen by luminol-binding microspheres. J. Immunol. Methods. 1985;77:55–61. doi: 10.1016/0022-1759(85)90183-8. [DOI] [PubMed] [Google Scholar]
- Vieira O. V., Botelho R. J., Grinstein S. Phagosome maturation: aging gracefully. Biochem. J. 2002;366:689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. M., Pevsner J., Scullion M. A., Vaughn M., Kaplan J. Syntaxin 7 and VAMP-7 are soluble N-ethylmaleimide-sensitive factor attachment protein receptors required for late endosome-lysosome and homotypic lysosome fusion in alveolar macrophages. Mol. Biol. Cell. 2000;11:2327–2333. doi: 10.1091/mbc.11.7.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widberg C. H., Bryant N. J., Girotti M., Rea S., James D. E. Tomosyn interacts with the t-SNAREs syntaxin4 and SNAP23 and plays a role in insulin-stimulated GLUT4 translocation. J. Biol. Chem. 2003;278:35093–35101. doi: 10.1074/jbc.M304261200. [DOI] [PubMed] [Google Scholar]
- Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Cox D., Tseng C. C., Donaldson J. G., Greenberg S. A requirement for ARF6 in Fcgamma receptor-mediated phagocytosis in macrophages. J. Biol. Chem. 1998;273:19977–19981. doi: 10.1074/jbc.273.32.19977. [DOI] [PubMed] [Google Scholar]
- Zhang T., Wong S. H., Tang B. L., Xu Y., Hong W. Morphological and functional association of Sec22b/ERS-24 with the pre-Golgi intermediate compartment. Mol. Biol. Cell. 1999;10:435–453. doi: 10.1091/mbc.10.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.