Abstract
Members of the Ste20 and NDR protein kinase families are important for normal cell differentiation and morphogenesis in various organisms. We characterized POD6 (NCU02537.2), a novel member of the GCK family of Ste20 kinases that is essential for hyphal tip extension and coordinated branch formation in the filamentous fungus Neurospora crassa. pod-6 and the NDR kinase mutant cot-1 exhibit indistinguishable growth defects, characterized by cessation of cell elongation, hyperbranching, and altered cell-wall composition. We suggest that POD6 and COT1 act in the same genetic pathway, based on the fact that both pod-6 and cot-1 can be suppressed by 1) environmental stresses, 2) altering protein kinase A activity, and 3) common extragenic suppressors (ropy, as well as gul-1, which is characterized here as the ortholog of the budding and fission yeasts SSD1 and Sts5, respectively). Unlinked noncomplementation of cot-1/pod-6 alleles indicates a potential physical interaction between the two kinases, which is further supported by coimmunoprecipitation analyses, partial colocalization of both proteins in wild-type cells, and their common mislocalization in dynein/kinesin mutants. We conclude that POD6 acts together with COT1 and is essential for polar cell extension in a kinesin/dynein-dependent manner in N. crassa.
INTRODUCTION
Factors that determine and modulate cell polarity have been the subject of extensive investigations in a variety of experimental organisms (Drubin and Nelson, 1996; Nelson, 2003), with the most progress having been made in the unicellular yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe (Bähler and Peter, 2000; Pruyne and Bretscher, 2000a, 2000b; Pruyne et al., 2004). The mechanisms by which polarity is established in filamentous fungi have remained largely obscure, but it is likely that the fundamental principles leading to the initial polarization of the cell are conserved among unicellular organisms (Bähler and Peter, 2000; Wendland, 2001), filamentous fungi (Galagan et al., 2003; Borkovich et al., 2004; Harris and Momany, 2004), and animals (Hall, 1998). But in contrast to baker's yeast, where growth becomes isotropic soon after bud emergence, the growth of filamentous fungi must stay highly polar to produce a tip-growing hypha that can extend at astonishing rates of more than 1 μm/s (Lopez-Franco et al., 1994; Seiler and Plamann, 2003; Harris et al., 2005). Thus, filamentous fungi present good model systems to study how this highly polar shape is maintained over long distances, the way in which new branch points are initiated and how their spatial relationship is regulated.
In recent years, protein kinases of the NDR Ser/Thr protein kinase family have emerged as being important for normal cell differentiation and polar morphogenesis in various organisms, yet their specific functions are still elusive (Tamaskovic et al., 2003; Hergovich et al., 2006). In Drosophila melanogaster, the NDR kinases Tricornered and Warts are required for control of the extent and direction of cell proliferation as well as for neuronal morphogenesis (Justice et al., 1995; Xu et al., 1995; Geng et al., 2000; Emoto et al., 2004). The Caenorhabditis elegans homolog SAX1 regulates aspects of neuronal cell shape and has been proposed to be involved in cell spreading, neurite initiation, and dendritic tiling (Zallen et al., 2000; Gallegos and Bargmann, 2004). Verde et al. (1998) have shown that the fission yeast gene orb6 is required to maintain cell polarity during interphase. The budding yeast kinase Cbk1p is involved in cell separation and modulates cell shape (Racki et al., 2000; Bidlingmaier et al., 2001). A number of recent large-scale screens have identified several proteins that interact with Cbk1p (Ito et al., 2001; Du and Novick, 2002; Ho et al., 2002), establishing the idea that Cbk1p and other interacting proteins may represent the core components of a conserved complex required for polarized morphogenesis. Further work in both yeasts as well as in animal cells has resulted in an emerging network, which includes the NDR kinase and its binding partner and activator MOB2, which are regulated through a Ste20 type kinase that interacts with a MO25- as well as a FURRY-like scaffolding protein (Nelson et al., 2003; Kanai et al., 2005; Stegert et al., 2005; Hergovich et al., 2006).
The founding member of the NDR family, the kinase COT1 of the filamentous fungus Neurospora crassa, is required for hyphal tip elongation (Collinge and Trinci, 1974; Collinge et al., 1978; Yarden et al., 1992), and the temperature-sensitive cot-1 mutant ceases hyphal elongation after being shifted to restrictive temperature. This is accompanied by a massive induction of new hyphal tip formation, creating the typical barbed-wired morphology of cot-1 cells. A similar branching and growth-termination phenotype has been observed in neuronal cells of sax-1 and fry mutants in C. elegans and D. melanogaster (Geng et al., 2000; Zallen et al., 2000), suggesting an evolutionarily conserved function of NDR kinases in the formation of branched cellular structures. This may be linked to changes in a general stress-sensing response, similar to that reported for the mammalian NDR-related myotonic dystrophy kinase (Mounsey et al., 1995; Chahine and George, 1997; Kushnir et al., 1997). Evidence for this includes suppression of the cot-1 phenotype by osmotic and other environmental stresses as well as by altering cAMP-dependent kinase (protein kinase A [PKA]) activity levels in the temperature-shifted cultures (Gorovits and Yarden, 2003).
Another large emerging group of kinases that have been implicated in various signaling pathways are the Ste20 kinases (Dan et al., 2001; Bokoch, 2003). Originally defined by S. cerevisiae Ste20p, an upstream kinase of the mitogen-activated protein kinase pathway, the Ste20 group of kinases is divided into the p21-activated (PAK) kinases and several germinal center kinase (GCK) subfamilies. The true PAKs, originally characterized as the primary downstream effectors of Rac/Cdc42-type GTPases, are defined by a C-terminal kinase domain and an N-terminally located Cdc42/Rac interacting/binding (CRIB) motif that mediates binding of the small G-protein to the kinase and its subsequent activation. The GCK group differs from the PAKs in that the kinase domain is located N-terminally, they lack typical CRIB domains, and their noncatalytic domains are highly variable. In contrast to the PAKs, the function of the GCKs is much less defined, but they have been implicated in stress response, proliferation, and apoptosis (Dan et al., 2001; Bokoch, 2003).
Despite the relevance of an apically growing tip cell for members of the fungal kingdom, the key components that are specifically required for tip extension and branch-point specification are poorly understood, with COT1 being the best-characterized player to date. An important link between cytoskeleton assembly and function and COT1 activity has been established by the analysis of cot-1 suppressor mutants, which are defective in the microtubule-dependent motor protein complex dynein/dynactin (Plamann et al., 1994; Bruno et al., 1996b), but the underlying molecular mechanisms remain unclear. Several other complementation groups have been identified, which result in cot-1-like growth and defective hyphal elongation when these genes are mutated (Seiler and Plamann, 2003). Here, we report the characterization of POD6 (NCU02537.2), a novel member of the GCK family of Ste20 kinases that influences cell morphology in a manner highly similar to COT1 and acts in concert, and perhaps in cooperation, with this NDR kinase. We also present evidence that POD6 is essential for cellular extension in a motor-protein–dependent manner in N. crassa.
MATERIALS AND METHODS
Strains, Media, and Growth Conditions
General procedures and media used in the handling of N. crassa have been described (Davis and DeSerres, 1970) or are available through the Fungal Genetic Stock Center (www.fgsc.net), and the N. crassa strains used are listed in Table 1. Strains were grown in either liquid or solid (supplemented with 1.5% agar) Vogel's minimal media with 1.5% (wt/vol) sucrose, unless otherwise stated. When required, H2O2 (7 mM), NaCl (0.5–1.2 M), sorbitol (0.5–1.2 M), ethanol (7.5%, vol/vol), or KT5720 (75 μM), all purchased from Sigma (St. Louis, MO), were added to the growth medium. DNA-mediated transformation of N. crassa spheroplasts was carried out as described (Vollmer and Yanofsky, 1986). The gul-1 deletion strain was provided by the Neurospora knock out project (H. V. Colot, G. Park, Ringelberg, S. Curilla, C. Crew, K. A. Borkovich, and J. C. Dunlap, unpublished results).
Table 1.
Neurospora crassa strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| Wild type | 74-OR23-1A | FGSC 987 |
| pod-6(31-21) | pod-6(31-21) | Seiler and Plamann (2003) |
| cot-1(1) | cot-1(C102t) | FGSC 4066 |
| pod-6(31-21);cot-1(1) | pod-6(31-21) cot-1(C102t) | This study |
| HP1 | benR his-3+ fpaS pan-2− + benS his-3− fpaR pan-2+ | Nargang et al. (1995) |
| Δpod-6 | benR his-3+ fpaS pan-2− hph::pod-6Δ + benS his-3− fpaR pan-2+ pod-6+ | This study |
| ro-1 | ro-1(B15) | FGSC 146 |
| ro-3 | ro-3(R2354) | FGSC 3 |
| ro-10 | ro-10(AR7) | FGSC 3619 |
| Nkin | nkin(RIP-1) | Seiler et al. (1997) |
| gul-1803 | gul-1(CA1) | FGSC 803 |
| gul-1803;cot-1(1) | cot-1(C102t) gul-1(CA1) | FGSC 1962 |
| gul-1803;pod-6(31-21) | cot-1(C102t) gul-1(CA1) | This study |
| Δgul-1 | hph::gul-1Δ | FGSC 11288 |
| Δgul-1;cot-1(1) | hph::gul-1Δ cot-1(C102t) | This study |
| Δgul-1;pod-6(31-21) | hph::gul-1Δ pod-6(31-21) | This study |
| myc::cot-1 | cot-1(C102t) myc::cot-1(EC) | This study |
| myc::cot-1(1);ro-1 | cot-1(C102t) myc::cot-1(EC) ro-1(B15) | This study |
| myc::cot-1(1);nkin | cot-1(C102t) myc::cot-1(EC) nkin(RIP-1) | This study |
| pod-6(31-21);ro-1 | pod-6(31v21) ro-1(B15) | This study |
| pod-6(31-21);nkin | pod-6(31-21) nkin(RIP-1) | This study |
| cr-1 | cr-1(B47) | FGSC #826 |
| pod-6(31-21);cr-1 | pod-6(31-21) cr-1(B47) | This study |
| cot-1(1);cr-1 | cot-1(C102t) cr-1(B47) | This study |
| mcb(14-4) | mcb(14-4) | Seiler and Plamann (2003) |
| pod-6(31-21);mcb(14-4) | pod-6(31-21) mcb(14-4) | This study |
| cot-1(1);mcb(14-4) | cot-1(C102t) mcb(14-4) | This study |
| mcb(14-4); Δgul-1 | mcb(14-4) hph::gul-1Δ | This study |
pod-6 was isolated by identification of cosmid X1F7 from the Orbach/Sachs library (Orbach, 1984), which rescued the growth defects of pod-6(31-21). To identify pod-6, the cosmid was cut independently with several restriction endonucleases, and the digested DNAs were cotransformed with a hygromycin-resistance-selectable marker to identify enzymes that cut within and outside of pod-6. The information of the cosmid sequence boundaries, the available genome sequence, and the pattern of inactivating restriction-enzyme cuts allowed the identification of an ORF that was likely to complement pod-6(31-21). Confirmation was obtained by PCR with primers 5-TTC TAT ATC GGA TCG ATA ACA TGA-3 and 5-ATA GGT AAG CTT TCA TCG AGA TCG-3 in order to amplify this segment of genomic DNA from a wild-type strain and from pod-6(31-21) to complement the mutant. This approach also verified that we had cloned the gene that harbored the original mutation and not a multicopy suppressor. gul-1 was amplified using primers 5-CCA CAT GAC GAC GGA CAT TAA GG-3 and 5-CTC CTC TCA GGT CCA AGG CCC AG-3.
To construct the resistance cassette for the generation of a Δpod-6 strain, the Aspergillus nidulans gpdA promoter was obtained as an 888-base pair SacI/NcoI fragment from pSM1 (Poggeler et al., 2003), fused via NcoI with the 581-base pair nourseothricin resistance gene (NatR) from pAG25 (Goldstein and McCusker, 1999) that was amplified with the primers 5-ACC CCA TGG CCA TGA CCA CTC TTG ACG AC-3 and 5-AGG GAA TTC TCA GGG GCA GGG CAT GC-3 and introduced together via SacI/EcoRI into pBluescript SK+ (Stratagene, La Jolla, CA). The nourseothricin concentration was adjusted to 20 μg/ml to select for transformants.
The deletion cassette was obtained by plasmid gap repair in S. cerevisiae (Orr-Weaver and Szostak, 1983), with pRS416 (Sikorski and Hieter, 1989) as the yeast vector cut with XbaI and XhoI and the following primers to generate the NatR cassette and the 5′ and 3′ flanking regions of pod-6 by PCR with cosmid pXIF7 as template: 5-GCG AGC GGC AGG CGC TCT ACA TGA GCA TGC CCT GCC CCT GAG GGA GGT AGG GTC TTG-3, 5-TGG AAT TGT GAG CGG ATA ACA ATT TCA CAC AGG AAA CAG CGC ATG TGC GGG TGG GTA ATG-3, 5-ATT AAG TTG CGT AAC GCC AGG GTT TTC CCA GTC ACG ACG CTA CAG CAC TTG TGA TGG TGC-3, 5-CTC CGC ATG CCA GAA AGA GTC ACC GGT CAC TGT ACA GAG CTG ACT GCC CCC GCA AGG CC-3, 5-TAC TAC CAA CAG TCT CCT CGG GTC CTT GCG GGG GCA GTC AGC TCT GTA CAG TGA CCG GT-3, and 5-TTC TGA GAC AAA TAA CAT CCC GTT ACA AGA CCC TAC CTC CCT CAG GGG CAG GGC ATG CT-3.
Homologous recombination events in N. crassa were verified by phenotypic analysis of transformants grown on 400 μM p-fluorophenylalanine (fpa) and 200 μg/ml histidine or 5 μg/ml benomyl and 200 μg/ml pantothenic acid and by the complementation of the growth defect of potential Δpod-6 strains with a pod-6-containing amplicon using the same primers as for the complementation of pod-6(31-21).
An MYC-tagged version of COT1 was constructed, utilizing plasmid pOY18, containing the cot-1 wild-type allele, capable of complementing the cot-1 temperature-sensitive mutant (Yarden et al., 1992), along with plasmid pCM2-MT containing a 6MYC tag sequence (Cheng et al., 2001). The 6MYC tag sequence was previously PCR-amplified from pCM2-MT, while introducing an NsiI restriction site at both ends of the product, and ligated into a pDrive vector (Qiagen, Hilden, Germany) to create pME8 (Efrat, Gorovits, and Yarden, unpublished results). In this study, pME8 was used as a template for PCR amplification using the primers 5-GGT ATG CAT CGT TCG AAA GCT-3 and 5-TTG AAT TCG AAG CCT CAT GCA T-3, which introduced an NspV restriction site (underlined) at the PCR product ends, thereby facilitating the cloning of the amplicon into the NspV site in pOY18 and creating pCZ18. Correct integration of the MYC tag sequence at the 5′ end of the cot-1 gene coding region was verified by sequencing with the primer 5-TCT GGT TGT TGT TGG CAT TG-3. A 3.3-kb XbaI fragment from pMP6 containing a hygromycin-resistance cassette was finally introduced into the unique XbaI site of pCZ218 creating pCZ218/hygR. To verify proper activity of the fused MYC::COT1 protein, this construct was used to complement the growth defects of cot-1(1). Western-blot analysis using anti-MYC antibodies verified the presence of MYC-tagged COT1 with the expected molecular mass.
Immunological Methods
For the preparation of polyclonal anti-POD6 antibodies, a POD6 fragment containing amino acids 421–675 was expressed in Escherichia coli BL21(DE3) as a pQE60-based His6-fusion protein (Stratagene) and purified via the Ni-NTA purification system (Qiagen). Polyclonal antibodies were generated by Pineda Antikörper Service (Berlin), and the serum was affinity-purified using the original antigen.
N. crassa mycelial samples were frozen in liquid nitrogen, suspended in 50 mM phosphate buffer, pH 7.0, 150 mM KCl, 1 mM DTT, and protein inhibitors and centrifuged, and the supernatant was used for further experiments. Proteins were separated by 7.5 or 10% SDS-PAGE. Western blotting was performed according to standard procedures. For immunoprecipitation (IP), protein G-Sepharose beads (Amersham Biosciences, Piscataway, NJ) were incubated on a rotation device with an excess of monoclonal anti-MYC antibody (clone 9E10) in binding buffer (50 mM phosphate buffer, pH 7.0, 150 mM KCl, 1 mM DTT, 0.2% wt/vol NP40, 1 mM EDTA, and protein inhibitors) for 2 h at 4°C. After the unbound antibodies were removed, the treated beads were resuspended in binding buffer with 400–500 μg protein extract and incubated for 3 h at 4°C. Immunoprecipitated protein was eluted with 100 mM glycine, boiled in Sample Buffer before SDS-PAGE, blotted, and probed with α-MYC or α-POD6 antibodies.
Microscopy
Immunolocalization for N. crassa hyphae was conducted following a protocol adapted from Minke et al. (1999). Fresh conidia were spread on a small piece of cellulose filter (GN-6, Gelman Sciences, Ann Arbor, MI) placed on the surface of a sucrose agar plate, and incubated until small mycelia formed. Filters were plunge-frozen in liquid propane and transferred to fixative (3% formaldehyde in 100% ethanol precooled to −80°C). Samples were maintained at −80°C for at least 2 d and then slowly transferred to room temperature (2 h at −20°C, 2 h at 4°C). Filters were rehydrated in a series of ethanol:buffer (100 mM phosphate, pH 7.0) solutions starting at a ratio of 90:10 and ending at 10:90. After several rinses in phosphate buffer, the samples were incubated for 0.5–5 min in 2 mg/ml lysing enzymes (Sigma) in 100 mM potassium citrate, pH 6.0, 20 mM EGTA to digest the cell wall. After several washes with phosphate buffer, filters were incubated in 1% BSA to block nonspecific binding of antibody to the filter. Samples were immersed in the primary antibody for at least 8 h, washed several times in phosphate buffer and incubated in the secondary antibody for ≥8 h, and visualized using standard rhodamine and FITC filter sets. For light microscopy, samples were viewed with a Zeiss Axioscope microscope (Oberkochen, Germany) and the Improvision Openlab 4.04 software (Lexington, MA). For the colocalization of MYC::COT1 and POD6, 200-μm hyphal stacks were acquired and volume-deconvoluted using Openlab 4.04. Documentation was performed with a Nikon DXM1200F digital camera (Melville, NY) or an Olympus SZX12 stereomicroscope (Hamburg, Germany), and a Kappa PS30 camera (Gleichen, Germany).
RESULTS
POD6 Belongs to the Germinal Center Family of the Ste20 Group of Kinases
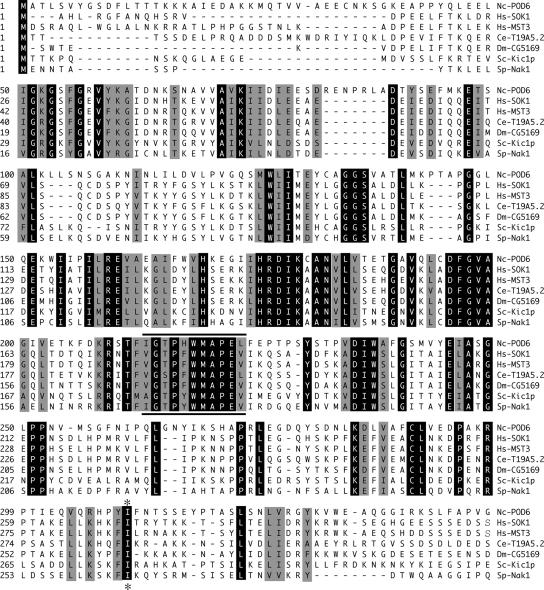
To identify critical components that contribute to polarized growth in N. crassa, we recently developed a large-scale screen for the isolation of conditional mutants defective in hyphal morphogenesis and isolated seven independent alleles of pod-6 (polarity defective-6; Seiler and Plamann, 2003). During the complementation of pod-6, we noticed that the N-terminal two thirds of the automatically annotated ORF NCU02537.2 were sufficient to complement pod-6. Further inspection of NCU02537.2 by BLAST analyses suggested that two ORFs were incorrectly fused during the annotation by the introduction of an erroneous intron, omitting the stop codon of the first ORF (pod-6) and joining it with a second ORF (probably an ortholog of the translation initiation factor SUA5). RT-PCR and sequencing experiments of cDNA with POD6-specific primers were used to confirm this prediction. Thus, pod-6 encodes a 928-amino acid protein containing an N-terminal kinase domain with the highest sequence similarity to members of the Ste20 group of kinases and a C-terminal region with no characteristic sequence motifs (Figure 1). Based on the homology of the kinase domain, its N-terminal localization and the lack of defined sequence motifs in the C-terminus, POD6 belongs to the GCK-III subfamily of eukaryotic Ste20 kinases. The known vertebrate members of this subfamily, SOK1, MST3, and MST4, have been implicated in the regulation of stress response, apoptosis, and proliferation, but the molecular mechanisms involved are unknown (Dan et al., 2001, 2002; Lin et al., 2001; Qian et al., 2001). The most closely related budding and fission yeast kinases are Kic1p and Nak1, respectively. Both kinases have been recently reported as part of a morphogenetic network that also contains the NDR kinases Cbk1p and Orb6, respectively, that is important for coordinating polarized growth with daughter cell specific transcription and cell cycle progression (Nelson et al., 2003; Kanai et al., 2005; Leonhard and Nurse, 2005).
Figure 1.
POD6 is a GC kinase. The catalytic kinase domains of N. crassa POD6, S. cerevisiae Kic1p, S. pombe Nak1, human MST3 and SOK1, and two uncharacterized GCK-III kinases from D. melanogaster and C. elegans (Dan et al., 2001) were aligned using the CLUSTAL W alignment algorithm. Identical and conserved corresponding amino acids are shaded dark and light, respectively, the PAK signature motif “GTPFWMAPE” is underlined and the position of the isoleucine-lysine substitution of pod-6(31-21) is marked with an asterisk.
POD6 Is Essential for Hyphal Tip Extension
When wild-type N. crassa conidia (asexual spores) germinate, they rehydrate and begin to grow isotropically. Growth soon becomes polarized, and usually one hyphal tip is generated. Continued polarized growth results in unidirectional extension of the straight primary hypha, and new hyphal tips are subsequently generated by branching from subapical compartments at intervals of 30–150 μm. Continuous hyphal elongation and branching results in the formation of spreading colonies.
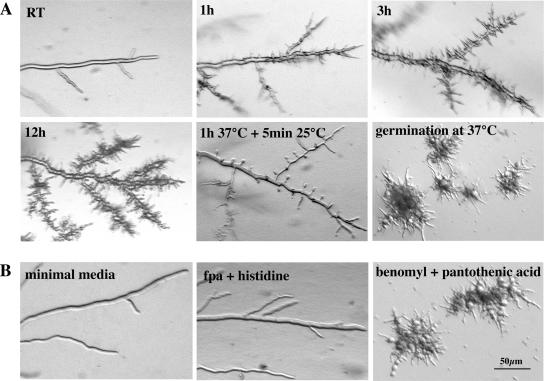
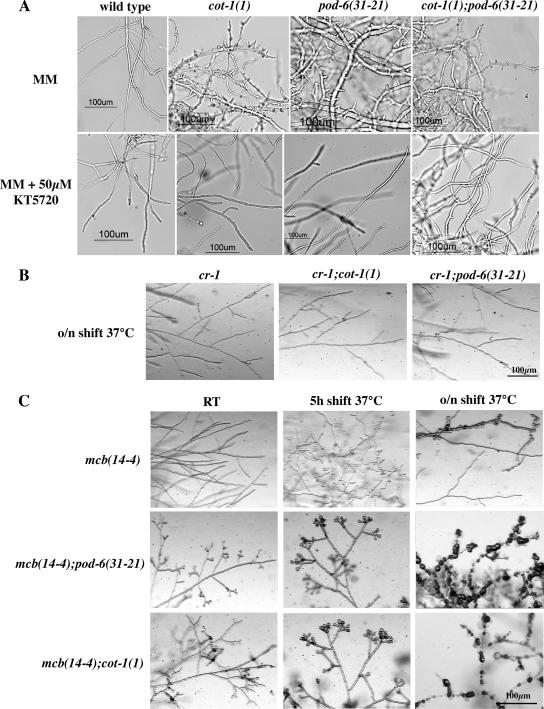
To determine the morphological changes conferred by pod-6, we conducted a microscopic analysis of pod-6 grown under permissive and restrictive temperatures (Figure 2A). As no major morphological differences were observed among the seven isolated pod-6 alleles, we focused our analyses on the pod-6(31-21) allele. When grown on agar plates at 25°C, no differences in hyphal elongation or branching frequency were detected between pod-6(31-21) and the wild type. However, within 45 min of transferring pod-6(31-21) to 37°C, we observed a rapid cessation of tip extension, whereas the temperature shift had no effect on the morphology of wild type (note that the agar plate served as a strong temperature buffer in these shift experiments). In contrast to a wild-type apex, which is a dome-shaped structure, the tips of pod-6(31-21) have a characteristic pointed/needle-like shape after the shift to restrictive growth conditions. This cessation of tip extension was simultaneously accompanied by the appearance of numerous subapical branches 20–50 μm in length that also stopped growth with a pointed tip. After prolonged incubation at restrictive temperature, these branches produced secondary and tertiary branches, and the hyphae began to swell up in a bulbous and apolar manner. Transfer of such a culture back to a permissive temperature resulted in all tips resuming normal growth rates, diameter, and morphology within 2–5 min. Germination of pod-6(31-21) at restrictive temperature resulted in the formation of compact colonies with multiple 10–50-μm-long germ tubes with pointed tips, which also produced secondary and tertiary branches. These experiments indicate that POD6 is essential for tip extension, but is also required to inhibit excessive branch formation in subapical regions of the hypha. Sequencing of pod-6(31-21) revealed a single amino acid substitution of the conserved isoleucine 310 (ATA) at the C-terminus of the kinase domain by a lysine (AAA; Figure 1).
Figure 2.
POD6 is essential for hyphal tip extension. (A) pod-6(31-21) was grown on minimal media plates at 25°C and shifted for 1, 3, and 12 h to restrictive temperature to illustrate the cessation of tip extension with pointed, needle-like tips and the progressive hyperbranching of the mutant. Hyphae that were shifted to the restrictive temperature and then shifted back to the permissive temperature resumed growth at all generated tips within 5 min (center panel). When germinated at the restrictive temperature, pod-6 exhibited a compact, hyperbranched, colonial morphology. (B) Growth of the HP1 pod-6+ + HP1 Δpod6 heterokaryotic strain on minimal medium, medium containing fpa and histidine and medium containing benomyl and pantothenic acid (which forces the knockout nucleus to predominate), resulting in morphological defects identical to pod-6(31-21) germinated at or shifted to restrictive temperature.
To examine the phenotype of a pod-6 deletion strain, we constructed a null mutant by using the sheltered disruption method (Nargang et al., 1995), which takes advantage of the fact that N. crassa is a multinucleated cell. In brief, the pod-6 gene was deleted via homologous recombination in the heterokaryotic strain HP1, thereby allowing the generation of mutants in genes that are essential or important for growth. The resulting mutant harbors two kinds of nuclei: one with a null allele of pod-6 and one with a wild-type copy. These nuclei contain selectable markers that allow a shift in the nuclear ratio within the heterokaryotic cells. Growth on media containing fpa and histidine favors the propagation of the wild-type nucleus. In contrast, growth of heterokaryotic cells on media containing benomyl and pantothenic acid forces the knockout nucleus to predominate and thus leads to the depletion of POD6 and morphological defects that are indistinguishable from pod-6(31-21) germinated at or shifted to restrictive temperatures (Figures 2B). These results indicate that pod-6(31-21) is a temperature-sensitive loss-of-function allele of pod-6. Both mutants indicate an essential role for POD6 during the extension of the hyphal tip and in controlling the number and position of subapical branches.
POD6 and COT1 Act in the Same Genetic Pathway in Parallel to the PKA Pathway
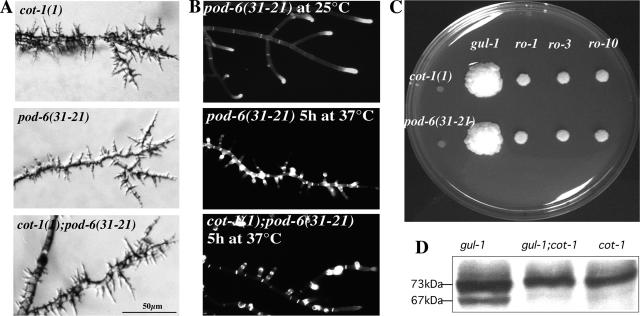
The morphological defects of pod-6(31-21) and of Δpod-6 were strikingly similar to those of cot-1(1) and led us to hypothesize that POD6 and COT1 may have related functions. This was supported by the observation that several recessive alleles of cot-1 and pod-6 showed unlinked noncomplementation. This genetic phenomenon suggests that the two proteins are either part of independent pathways, which are equally necessary for the successful completion of a process or that the two proteins interact physically. To further analyze the relation between COT1 and POD6, double mutants were generated (Figure 3A). cot-1(1), pod-6(31-21), and cot-1(1);pod-6(31-21) mutants showed identical phenotypes at restrictive temperature. This provides further evidence of their involvement in a common genetic pathway and, possibly, of a physical interaction between the two proteins.
Figure 3.
COT1 and POD6 act in the same genetic pathway. (A) A cot-1;pod-6 double mutant displayed the same morphological defects as the two parental strains. All three strains were grown on minimal media plates shifted to restrictive temperature for 5 h after growth at permissive temperature. (B) Calcofluor White stained primarily the hyphal apex of mutant strains grown at permissive temperature but mutants shifted to restrictive temperature for 5 h showed an abnormal and patchy label throughout the hyphae, including strong septal staining. (C) Suppressor analysis of cot-1(1) and pod-6(31-21) showed that gul-1 as well as components of the dynein/dynactin complex partially suppress the cot-1(1) or pod-6(31-21) growth defects (ro-1: dynein heavy chain mutant, ro-3: p150glued mutant; ro-10: 24-kDa subunit of dynactin). (D) Western-blot analysis of cell extracts probed with anti-COT1 antibody indicated that suppression of the cot-1(1) phenotype at restrictive temperature is independent of 67-kDa COT1 presence.
The cot-1(1) mutation has been shown to result in increased cell-wall thickness at restrictive temperature (Collinge et al., 1978; Gorovits et al., 2000), suggesting a defect in cell-wall metabolism. We therefore tested chitin deposition by staining hyphae with Calcofluor White. Chitin is the primary component of the inner layers of the hyphal cell wall and is therefore accessible to the dye primarily at the hyphal tips. At permissive temperature, hyphal tips (and to a minor extent also septa) of pod-6(31-21) and also cot-1(1) are strongly labeled in a manner identical to the wild type (Figure 3B). Within 5 h at 37°C, pod-6(31-21), cot-1(1), and the pod-6(31-21);cot-1(1) double mutant showed extensive label in a patchy, subapical manner throughout the hyphae, including strong septal staining, indicating excessive chitin deposition in all mutants, and further suggesting a functional connection between POD6 and COT1. Furthermore, it appears that the high calcofluor stain at the tip observed in mutants grown at the permissive temperature is nearly absent when the strains are cultured at the restrictive temperature, indicating decreased chitin synthesis at these nonelongating tips.
If POD6 and COT1 have related functions, they may share the same extragenic suppressors. Therefore, we tested whether components of the dynein/dynactin complex could partially complement the growth defects of pod-6, similar to the mechanism described for cot-1(1) (Plamann et al., 1994; Bruno et al., 1996b). We compared growth rates of ro-1;pod-6(31-21), ro-3;pod-6(31-21), and ro-10;pod-6(31-21) with pod-6(31-21) grown at restrictive temperatures and found that these mutations partially suppress the pod-6(31-21) phenotype (Figure 3C; for hyphal phenotypes of the double mutants, see Supplementary Figure 1).
In addition to dynein/dynactin mutations, gul-1 has also been described as a dominant modifier of cot-1 (Terenzi and Reissig, 1967). Although the identity of the affected gene has not yet been determined, its close linkage with am-1 and ace-5 (Perkins et al., 2001) and the available genome sequence led us to suspect that GUL1 may be encoded by NCU01197.2. To test this, we amplified wild-type and mutant gul-1803 alleles by PCR and transformed cot-1(1) and pod-6(31-21) with both amplicons. While transformation with gul-1+ resulted in compact cot-1(1) or pod-6(31-21) colonies, strains transformed with the mutant gul-1803 allele partially suppressed the growth defects of cot-1(1) and pod-6(31-21), similar to the original cot-1;gul-1 strain. This indicated that NCU01197.2 codes for GUL1 and that gul-1 is a common dominant suppressor of both cot-1(1) and pod-6(31-21). Database searches indicated that GUL1 is an evolutionarily conserved protein. Ssd1p and STS5, the budding and fission yeast orthologues, respectively, are naturally polymorphic proteins and have been implicated in the maintenance of cell-wall integrity, RNA binding, and the TOR pathway, as well as in protein phosphatase-associated functions, but the underlying molecular mechanisms are entirely unknown (Matsusaka et al., 1995; Toda et al., 1996; Evans and Stark, 1997; Uesono et al., 1997). To characterize the function of GUL1 in apical tip extension, we compared the phenotypes of the dominant gul-1803 and a gul-1 deletion strain. Morphological characteristics of both strains suggested a role of GUL1 in the establishment of a functional tip both during germination as well as during the formation of new branches (Supplementary Figure 1; Table 2).
Table 2.
Morphological characteristics of wild type and gul-1 mutants
| Wild type | Δgul-1 | gul-1803 | |
|---|---|---|---|
| Growth rate (cm/d; n = 3) | 6.2 ± 0.2 | 5.6 ± 0.3 | 5.5 ± 0.2 |
| Conidial germination rate after 5 h (%; n = 150) | 83 | 54 | 61 |
| Distance between branch points (μm; n = 80) | 338 ± 115 | 125 ± 43 | 153 ± 57 |
According to Gorovits and Yarden (2003), the cot-1(1) defect can be suppressed by regaining partial COT1 activity in mutants of the dynein/dynactin (ropy) complex. We therefore tested gul-1803 and gul-1803;cot-1(1) cultures at 37°C for the presence/absence of the 67-kDa COT1 band and determined that the dominant mutation in gul-1803 eliminated the need for the 67-kDa COT1 form (Figure 3D), indicating that suppression by the dynein/dynactin mutations is different from that involving gul-1. Bypassing the need for COT1 was also observed when the mutants were treated with various environmental stresses that alter PKA activity (see below), suggesting a potential link between the proposed functions of GUL1 and the PKA pathway. The consistency of the effects that common genetic suppressors have on cot-1 and pod-6 morphology strongly support that POD6 and COT1 act in a common genetic pathway.
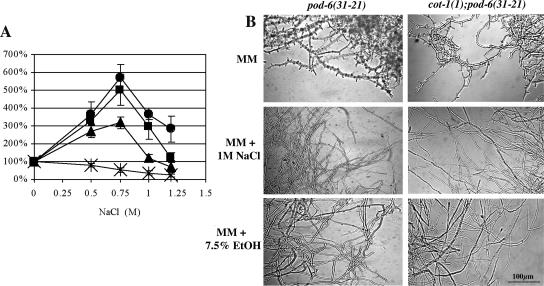
In addition to the suppression by extragenic mutations, cot-1(1) has been shown to undergo environmental stress-related suppression (Gorovits and Yarden, 2003). Therefore, we cultured pod-6(31-21) as well as pod-6(31-21);cot-1(1) under similar environmental stresses. When grown in the presence of NaCl, a significant increase in radial growth of pod-6(31-21) as well as pod-6(31-21);cot-1(1) was observed (Figure 4A). Furthermore, when either NaCl or ethanol were added to the medium, suppression of these strains was observed (Figure 4B). Similar results were obtained when the strains were cultured in the presence of sorbitol or H2O2. A common component of fungal stress response is the PKA-dependent signaling pathway (Thevelein, 1994; Gustin et al., 1998), and the stress-related suppression of cot-1(1) was accompanied by reduced levels of PKA activity (Gorovits and Yarden, 2003). Thus, we inhibited PKA by amending pod-6(31-21), cot-1(1) and the double mutant grown at restrictive temperature with the PKA-specific inhibitor KT5720 and observed partial suppression of the mutant phenotypes in a manner similar to that observed in the stressed cultures (Figure 5A), indicating that lowering PKA activity can bypass the POD6/COT1 defects. This is further supported by the analysis of double mutants between cot-1(1)/pod-6(31-21) and cr-1, an adenylate cyclase mutant with reduced levels of cAMP that can be complemented by adding cAMP (Terenzi et al., 1974). These double mutants showed normal tip growth at 37°C (Figure 5B). In contrast, when we generated double mutants using a temperature-sensitive allele in the regulatory subunit of PKA that displayed increased levels of PKA activity at restrictive temperature (Bruno et al., 1996a; Seiler and Plamann, 2003) by crossing cot-1(1)/pod-6(31-21) with mcb, we observed a synthetic effect (Figure 5C). Unlike the single mutants, mcb(14-4);pod-6(31-21) and mcb(14-4);cot-1(1) showed slight growth abnormalities at permissive temperatures. After shifting to 37°C, the synthetic defects were even more pronounced, resulting in branched and swollen hyphal tips and marked septation within 5 h in the double mutants, whereas mcb(14-4) displayed only minor abnormalities, such as some swollen tips and branches. Overnight incubation at 37°C resulted in cell lysis and subsequent death of double mutant cultures, whereas mcb(14-4) displayed a mixture of normally growing hyphae and chains of spherical cells. As changes in cell polarity in cot-1 and pod-6 are emphasized when PKA activity levels are altered, we concluded that COT1/POD6 and PKA act in parallel pathways to regulate cell polarity in a positive or negative manner, respectively.
Figure 4.
Environmental suppression of pod-6(31-21) and cot-1(1); pod-6(31-21). (A) NaCl amendment in the growth medium suppresses the cot-1, pod-6 and double mutant phenotype. Relative growth (expressed as percent of nonamended control) was determined on the basis of radial growth of N. crassa strains in the presence of different concentrations of NaCl. Cultures were incubated at 34°C. The results are the average of at least three independent results. Error bars, SE. *, wild type; ●, cot-1(1); ▴, pod-6(31-21); ■, cot-1(1);pod-6(31-21). (B) Pictures of liquid cultures supplemented with either 1 M NaCl or 7.5% ethanol were taken 8 h after they were shifted from permissive to restrictive temperatures.
Figure 5.
PKA and POD6/COT1 act in parallel pathways. (A) Effect of amending liquid growth media with the PKA inhibitor KT5720. Suppression of the growth defect was monitored after shifting the cultures to restrictive temperatures for 6 h. (B) The growth defect of cot-1(1) and pod-6(31-21) was suppressed in the cr-1 mutant background, whereas (C) mcb;cot-1/pod-6 double mutants are synthetically lethal. For B and C, the strains were grown on minimal media plates supplemented with 1% yeast extract and shifted for the indicated times.
POD6 and COT1 Are Physically Associated and Depend on Opposing Microtubule-dependent Motor Proteins for Correct Localization
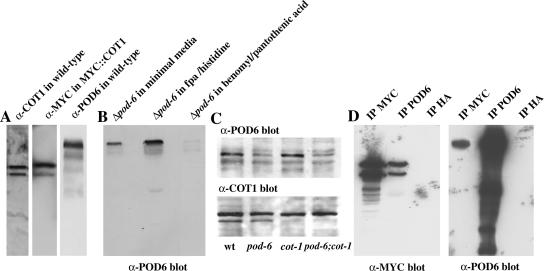
To examine whether POD6 physically interacts with COT1, we generated and affinity-purified antisera against POD6 that recognized a single polypeptide of ca. 100-kDa in wild-type extracts and generated a strain expressing a MYC::COT1 fusion protein (Figure 6A). The POD6 signal was strongly reduced in extracts of the heterokaryotic Δpod-6 strain grown on benomyl and pantothenic acid, indicating that the anti-POD6 antibodies are specific for POD6 (Figure 6B). Reduced amounts of POD6 were also observed in pod-6(31-21) shifted to restrictive temperature, indicating that a reduction in POD6 protein levels accompanies the temperature shift. Nevertheless, we observed no change in protein abundance of either kinase in the other mutant grown at restrictive temperature (Figure 6C). cot-1 was tagged N-terminally with a 6MYC tag under the control of its own promoter. This construct fully complemented cot-1(1) and ensured full activity of the modified protein. Anti-MYC, anti-POD6, and, as a control, anti-HA antibodies were used for IP experiments from extracts of the MYC::COT1 strain (Figure 6D). The anti-POD6 IP recovered a markedly higher portion of POD6 from the extracts when compared with the amount of coprecipitated COT1, whereas anti-MYC IP recovered an abundant quantity of COT1 and coprecipitated much less of POD6. These results indicate that under the conditions tested, low but significant portions of COT1 and POD6 were associated with each other.
Figure 6.
COT1 and POD6 interact. (A) COT1, POD6, and MYC::COT1 were detected in extracts prepared from wild-type cells or MYC::COT1 cells, respectively, to illustrate the specificity of the antibodies being used. (B) Extracts of the nuclear-ratio-modulated heterokaryotic Δpod-6 strain grown in appropriate media (see Materials and Methods) were adjusted to equal amounts of soluble protein and probed to determine the effect of the predominance of Δpod-6 nucleus (in the presence of benomyl and pantothenic acid) on POD6 abundance. (C) Reduced amounts of POD6 were also observed in pod-6(31-21) shifted to restrictive temperature for 5 h, but no alteration in the expression level of either kinase in the other mutant grown at restrictive temperature was observed. (D) Cell extracts of MYC::COT1 cells were prepared, precipitated with α-MYC, α-POD6 or, as control, α-HA antibodies, separated by SDS-PAGE and probed with α-MYC or α-POD6 antibodies.
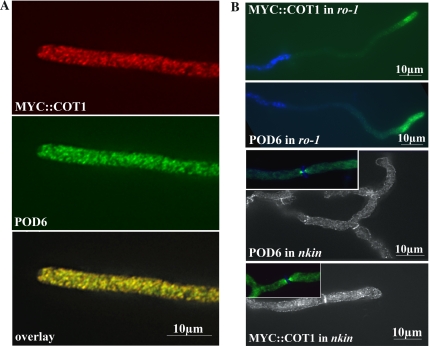
To determine the cellular distribution of MYC::COT1 and POD6 in N. crassa hyphae, we used the anti-MYC and anti-POD6 antibodies, respectively (Figure 7A). Both proteins exhibit a vesicular-reticulate distribution throughout the hypha that is, to a large extent, overlapping. In conjunction with the co-IP data, indicating a low but significant level of interaction, we concluded that COT1 and POD6 may be part of one or more dynamic protein complexes in the cell, but that the proteins are not necessarily associated with each other at all times. This punctate localization pattern and our previous results that both mutants can be suppressed by components of the dynein/dynactin motor complex prompted us to analyze the dependence of POD6 and COT1 on motor protein function (Figure 7B). In ro-1 hyphae, POD6 was not evenly distributed, but rather accumulated in clusters at the hyphal apex. In contrast, we observed the opposite distribution in conventional kinesin mutants with both kinases accumulating subapically at the septae. Finally, we also tested the interdependence of POD6 and COT1 for localization in the respective mutants shifted to restrictive temperatures. In neither case were significant alterations in the kinase distribution patterns detected (unpublished data).
Figure 7.
COT1 and POD6 colocalize and are dependent on microtubule-dependent motor proteins for their correct localization. MYC::COT1 and POD6 were colocalized in growing wild-type hyphae and exhibited punctate/reticulate staining that was evenly distributed throughout the hypha. Note that the Spitzenkörper region at the extreme hyphal apex is unlabeled. (B) This punctate distribution of myc::COT1 and POD6 was altered in dynein heavy chain and conventional kinesin mutants, with myc::COT1 and POD6 accumulating at the hyphal tip in the minus-end-directed (−) ro-1 mutant and in subapical areas/septae in the plus-end-directed (+) nkin mutant. In ro-1, the nuclei are counterstained with DAPI to illustrate that the accumulation at the tip is not due to their clustering, whereas in nkin, septal association of the kinases is indicated by Calcufluor White staining.
DISCUSSION
Our characterization of pod-6(31-21) and Δpod-6 revealed the same morphogenetic defects as those previously characterized in the cot-1(1) mutant. These include the rapid cessation of tip extension, subsequent hyperbranching, and altered cell-wall organization. In addition, we showed that pod-6(31-21);cot-1(1) double mutants exhibit phenotypic characteristics identical to the parental strains and that mutations in both genes share common extragenic suppressors, which include mutations in components of the dynein/dynactin motor protein complex (ropy) as well as mutations in the conserved polymorphic protein GUL1. These results strongly suggest that POD6 and COT1 function in the same genetic pathway. This conclusion is further supported by our localization and IP results, which provide evidence that POD6 and COT1 can physically associate with each other in the cell and in vitro. Finally, we showed that the correct distribution of both kinases is dependent on the two opposing motor proteins: conventional kinesin and cytoplasmic dynein.
Although our data clearly indicate that COT1 and POD6 can, at least to some extent, physically interact with one another and act in the same genetic pathway, the hierarchical relationship between COT1 and POD6 have yet to be elucidated. Neither kinase's expression level was altered in the other mutant grown at restrictive temperature. Furthermore, overexpression of one kinase, driven by the strong modified cpc-1 promoter in the other mutant did not alter the mutant phenotypes either (unpublished data). In addition, no codependence for localization was observed, suggesting that COT1 and POD6 operate in a network or in parallel, rather than in a linear genetic path. This view is supported by the observation that pod-6(31-21);cot-1(1) double mutants displayed a synthetic growth defect when cultivated at semirestrictive temperatures, i.e., between 30 and 32°C. Similarly, Nelson et al. (2003) found no codependence of Cbk1p localization on Kic1p in S. cerevisiae.
The morphological defects of both pod-6 and cot-1 can be partially suppressed by various environmental stresses, which have been shown to decrease PKA activity and bypass the requirement for a functional COT1 or POD6. Based on the fact that in contrast to mutations in dynein/dynactin components, mutant gul-1803 eliminated the need for the 67-kDa COT1 isoform, it is tempting to speculate that gul-1 suppression is linked with the environment- and PKA-activity-dependent suppression mechanism. This is supported by S. cerevisiae ssd1 mutants, which can suppress the absence of Bcy1p (the yeast PKA regulatory subunit; Sutton et al., 1991), as well as by the fact that the S. pombe homolog sts5 has been implicated in the maintenance of cell polarity involving stress-signaling pathways (Toda et al., 1996). Nevertheless, we did not find any observable genetic interactions between gul-1 and the PKA pathway in Δgul-1;mcb(14-4) double mutants in N. crassa (unpublished data), suggesting a functional link of GUL1 with COT1/POD6 and not PKA. This is supported by the fact that budding yeast SSD1 was isolated in large scale two-hybrid and affinity purification approaches as a direct interaction partner of Cbk1p (Racki et al., 2000; Ho et al., 2002). Furthermore, mutations that compromise SSD1 function or deletion of SSD1 result in viable Cbk1 pathway deletions (Jorgensen et al., 2002).
Based on our analysis, it is conceivable that both kinases function to promote tip elongation, whereas at the same time curb excessive branch formation in subapical regions of the hypha. This is further supported by a previous study demonstrating that COT1 distribution is not restricted to the hyphal tip and can be found in association with the plasma membrane along the entire hyphal filament (Gorovits et al., 2000). Inhibiting PKA activity by amending the medium with KT5720 or in pod-6/cot-1;cr-1 double mutants resulted in partial suppression of the pod-6/cot-1 defects. pod-6/cot-1;mcb double mutants displayed a synthetic phenotype, further suggesting that POD6/COT1 and PKA act in parallel pathways that regulate polarity formation in a positive or negative manner, respectively. A common downstream target of both pathways may involve the link between PKA activity and actin organization. Disruption of the N. crassa actin cytoskeleton with actin depolymerizing drugs or mutants affecting the actin organization result in increased septation and the generation of spherical cells (Heath et al., 2000; Silverman-Gavrila and Lew, 2001; Seiler and Plamann, 2003). This phenotype strongly resembles the phenotype of the latrunculin A-hypersensitive, and PKA-hyperactive mutant, mcb. PKA has also been shown to regulate actin polymerization in budding yeast (Ho and Bretscher, 2001). Nonetheless, the cellular responses of yeast and N. crassa are the opposite. Although activation of PKA in yeast leads to increased polarization and pseudohyphal differentiation (Pan and Heitman, 1999), activation of PKA in N. crassa results in apolar growth (Bruno et al., 1996a; Seiler and Plamann, 2003). Similar to the observation in the filamentous fungus, PKA in animal cells has been shown to promote a round cell shape during cytokinesis and to phosphorylate monomeric actin to prevent actin-fiber formation (Ohta et al., 1987; Prat et al., 1993). The fact the animal NDR kinase function has been implied in regulating actin organization (Geng et al., 2000; Zallen et al., 2000; Emoto et al., 2004; Stork et al., 2004) along with the stated effect of PKA on the cytoskeleton in a variety of organisms supports the possibility that the fungal components function in a similar manner. It is conceivable that COT1/POD6 may positively modulate actin dynamics in parallel to the negatively acting PKA pathway. Thus, in N. crassa, polymerized actin is essential for polar tip extension (Heath et al., 2000), but subapical lowering or modification of the F-actin pool would act as a cue for the initiation of new branches. Probably the best indication for regulation of the actin cytoskeleton through GC kinases comes from work in fission yeast. Huang et al. (2005) have shown that the GC kinase Nak1 directly interacts with the actin-binding proteins Hob1 (an Rvs167/amphiphysin homolog that stimulates actin polymerization through Wsp1), and Leonhard and Nurse (2005) determined that F-actin localization at cell tips is Nak1 dependent.
Interestingly, in none of the organisms studied has the function of GC-III and NDR kinases yet been associated with the microtubule cytoskeleton. However, where determined, their cellular localization is always punctate (Verde et al., 1998; Nelson et al., 2003; Gallegos and Bargmann, 2004; Stork et al., 2004; He et al., 2005; Kanai et al., 2005), suggesting a vesicular localization and therefore also motor-dependent transport. Conventional kinesin and dynein drive apical and retrograde-directed transport in N. crassa, respectively (Seiler et al., 1997, 1999). The observed accumulation of COT1 and POD6 in the two motor mutants in apical or distal regions (corresponding to microtubule + and − ends, respectively; Konzack et al., 2005; Schuchardt et al., 2005) indicates that the equal distribution of both kinases in wild type is a result of the activity of the two opposing motor proteins. An explanation for the suppression of the kinase mutant phenotypes by dynein/dynactin mutations may be that the reduced retrograde transport rate in dynein mutants and the subsequent accumulation of mutant kinases in apical areas could result in higher levels of partially functional protein at the hyphal tip. The observed enrichment of kinases in a 10–20-μm-broad tip region and not specifically within the hyphal apex of ro-1 may be explained by altered microtubule dynamics in addition to retrograde transport defects in dynein mutants, which has been reported for all systems studied (e.g., Shaw et al., 1997; Han et al., 2001).
Earlier observations demonstrated that COT1 could be found in various subcellular compartments, including the cytoplasm and nuclei, and in association with the plasma membrane (Gorovits et al., 2000). Since then, other NDR kinases have also been shown to be present in the mentioned compartments (Chen and Dickman, 2002; Hergovich et al., 2005, 2006). The punctate label observed in the present study suggests that COT1 and POD6 may also be associated with internal membrane structures—a distribution pattern that may not be as evident in thin sections analyzed by electron microscopy. Nevertheless, some immunogold label was also associated with internal membrane structures in the mentioned study, suggesting that, depending on the strains, antibodies, and labeling techniques used, the kinase localization may shift from primarily plasma membrane to primarily vesicle associated. In nkin mutants, an increased cortical/plasma membrane localization of POD6 and COT1 was found, suggesting that altering membrane dynamics can influence kinase localization. Furthermore, the strong association of the kinases with septae in the kinesin mutant may indicate a function of both proteins during septum formation in wild-type cells too, similar to what has been reported for S. pombe ORB6 and NAK1 (Verde et al., 1998; Leonhard and Nurse, 2005). This is in line with the altered ultrastructure of septae in cot-1 hyphae shifted to restrictive temperatures (Gorovits et al., 2000).
The variety of phenotypic alterations such as an altered cell-wall structure, cytoskeletal organization, and transport defects associated with dysfunctional COT1/POD6 and their homologues in other organisms (Jorgensen et al., 2002; Nelson et al., 2003; Kanai et al., 2005; Leonhard and Nurse, 2005) strongly suggests that these kinases have multiple cellular functions, probably requiring additional components. In fact, several genetic screens in S. cerevisiae have identified proteins that could potentially interact with Cbk1p, the yeast homolog of COT1 (Ito et al., 2001; Du and Novick, 2002; Ho et al., 2002; Kurischko et al., 2005; Voth et al., 2005). Analyses in fission yeast have identified additional proteins that interact with the ORB6 (Verde et al., 1998; Hou et al., 2003; Wiley et al., 2003). In N. crassa, co-IP experiments have provided evidence for a potential physical interaction between COT1 and the type 2B phosphatase calcineurin (Gorovits et al., 1999). It is therefore likely that additional proteins can interact with POD6/COT1 or their homologues. The added phenotypic complexity of filamentous fungi and cells of higher eukaryotes provides an opportunity to dissect the contribution of different COT1-interacting proteins to the establishment and maintenance of cell polarity. Given the similarity between NDR and GC-III kinases in filamentous fungi and animals together with the described differences in COT1 complex regulation between yeasts and N. crassa/animals, they may serve as useful models to separate signals required for branch formation and tip extension in neuronal cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yi Liu for providing us with plasmid pCM2-MT. Sequence data of pod-6 have been deposited under GenBank accession number DQ336953. This joint research project was financially supported by the German Bundesland of Lower Saxony and the Volkswagen Foundation, Hannover, Germany (S.S. and O.Y.) and by the Deutsche Forschungsgemeinschaft (DFG) through the DFG Research Center of Molecular Physiology of the Brain (CMPB) to S.S., the DFG Schwerpunkt Zellpolarität (SPP1111) to S.S. and N.V., and The Israel Science Foundation to O.Y.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-01-0072) on July 5, 2006.
REFERENCES
- Bähler J., Peter M. Cell polarity in yeast. In: Drubin D. G., editor. Cell Polarity. Oxford: Oxford University Press; 2000. pp. 21–77. [Google Scholar]
- Bidlingmaier S., Weiss E. L., Seidel C., Drubin D. G., Snyder M. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:2449–2462. doi: 10.1128/MCB.21.7.2449-2462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokoch G. M. Biology of the p21-activated kinases. Annu. Rev. Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- Borkovich K. A., et al. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno K. S., Aramayo R., Minke P. F., Metzenberg R. L., Plamann M. Loss of growth polarity and mislocalization of septa in a Neurospora mutant altered in the regulatory subunit of cAMP-dependent protein kinase. EMBO J. 1996a;15:5772–5782. [PMC free article] [PubMed] [Google Scholar]
- Bruno K. S., Tinsley J. H., Minke P. F., Plamann M. Genetic interactions among cytoplasmic dynein, dynactin, and nuclear distribution mutants of Neurospora crassa. Proc. Natl. Acad. Sci. USA. 1996b;93:4775–4780. doi: 10.1073/pnas.93.10.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar-Ton That C., Rossier C., Barja F., Turian G., Ross U.-P. Induction of multiple germ tubes in Neurospora crassa by antitubulin agents. Eur. J. Cell Biol. 1988;46:68–79. [PubMed] [Google Scholar]
- Chahine M., George A. L., Jr. Myotonic dystrophy kinase modulates skeletal muscle but not cardiac voltage-gated sodium channels. FEBS Lett. 1997;412:621–624. doi: 10.1016/s0014-5793(97)00869-7. [DOI] [PubMed] [Google Scholar]
- Chen C. B., Cickmann M. B. Colletotrichum trifolii TB3 kinase, a COT1 homolog, is light inducible and becomes localized in the nucleus during hyphal elongation. Eukaryot. Cell. 2002;1:626–633. doi: 10.1128/EC.1.4.626-633.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., Yang Y., Heintzen C., Liu Y. Coiled-coil domain-mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. EMBO J. 2001;20:101–108. doi: 10.1093/emboj/20.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge A. J., Fletcher M. H., Trinci A.P.J. Physiological and cytology of septation and branching in a temperature-sensitive colonial mutant (cot-1) of Neurospora crassa. Trans. Br. Mycol. Soc. 1978;71:107–120. [Google Scholar]
- Collinge A. J., Trinci A. P. Hyphal tips of wild type and spreading colonial mutants of Neurospora crassa. Arch. Microbiol. 1974;99:353–368. doi: 10.1007/BF00696249. [DOI] [PubMed] [Google Scholar]
- Dan I., Ong S. E., Watanabe N. M., Blagoev B., Nielsen M. M., Kajikawa E., Kristiansen T. Z., Mann M., Pandey A. Cloning of MASK, a novel member of the mammalian germinal center kinase III subfamily, with apoptosis-inducing properties. J. Biol. Chem. 2002;277:5929–5939. doi: 10.1074/jbc.M110882200. [DOI] [PubMed] [Google Scholar]
- Dan I., Watanabe N. M., Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- Davis R. D., DeSerres F. J. Genetic and microbiological research techniques for Neurospora crassa. Meth. Enzymol. 1970;17:79–143. [Google Scholar]
- Drubin D. G., Nelson W. J. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Du L. L., Novick P. Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:503–514. doi: 10.1091/mbc.01-07-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K., He Y., Ye B., Grueber W. B., Adler P. N., Jan L. Y., Jan Y. N. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Evans D. R., Stark M. J. Mutations in the Saccharomyces cerevisiae type 2A protein phosphatase catalytic subunit reveal roles in cell wall integrity, actin cytoskeleton organization and mitosis. Genetics. 1997;145:227–241. doi: 10.1093/genetics/145.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J. E., et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Gallegos M. E., Bargmann C. I. Mechanosensory neurite termination and tiling depend on SAX-2 and the SAX-1 kinase. Neuron. 2004;44:239–249. doi: 10.1016/j.neuron.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Geng W., He B., Wang M., Adler P. N. The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics. 2000;156:1817–1828. doi: 10.1093/genetics/156.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Gorovits R., Propheta O., Kolot M., Dombradi V., Yarden O. A mutation within the catalytic domain of COT1 kinase confers changes in the presence of two COT1 isoforms and in Ser/Thr protein kinase and phosphatase activities in Neurospora crassa. Fungal Genet. Biol. 1999;27:264–274. doi: 10.1006/fgbi.1999.1152. [DOI] [PubMed] [Google Scholar]
- Gorovits R., Sjollema K. A., Sietsma J. H., Yarden O. Cellular distribution of COT1 kinase in Neurospora crassa. Fungal Genet. Biol. 2000;30:63–79. doi: 10.1006/fgbi.2000.1198. [DOI] [PubMed] [Google Scholar]
- Gorovits R., Yarden O. Environmental suppression of Neurospora crassa cot-1 hyperbranching: a link between COT1 kinase and stress sensing. Eukaryot. Cell. 2003;2:699–707. doi: 10.1128/EC.2.4.699-707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin M. C., Albertyn J., Alexander M., Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G., Liu B., Zhang J., Zuo W., Morris N. R., Xiang X. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr. Biol. 2001;11:719–724. doi: 10.1016/s0960-9822(01)00200-7. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Harris S. D., Momany M. Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 2004;41:391–400. doi: 10.1016/j.fgb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Harris S. D., Read N. D., Roberson R. W., Shaw B., Seiler S., Plamann M., Momany M. Polarisome meets spitzenkorper: microscopy, genetics, and genomics converge. Eukaryot. Cell. 2005;4:225–229. doi: 10.1128/EC.4.2.225-229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Fang X., Emoto K., Jan Y. N., Adler P. N. The tricornered Ser/Thr protein kinase is regulated by phosphorylation and interacts with furry during Drosophila wing hair development. Mol. Biol. Cell. 2005;16:689–700. doi: 10.1091/mbc.E04-09-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath I. B., Gupta G., Bai S. Plasma membrane-adjacent actin filaments, but not microtubules, are essential for both polarization and hyphal tip morphogenesis in Saprolegnia ferax and Neurospora crassa. Fungal Genet. Biol. 2000;30:45–62. doi: 10.1006/fgbi.2000.1203. [DOI] [PubMed] [Google Scholar]
- Hergovich A., Bichsel S. J., Hemmings B. A. Human NDR kinases are rapidly activated by MOB proteins through recruitment to the plasma membrane and phosphorylation. Mol. Cell Biol. 2005;25:8259–8272. doi: 10.1128/MCB.25.18.8259-8272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., Stegert M. R., Schmitz D., Hemmings B. A. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 2006;4:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- Ho J., Bretscher A. Ras regulates the polarity of the yeast actin cytoskeleton through the stress response pathway. Mol. Biol. Cell. 2001;12:1541–1555. doi: 10.1091/mbc.12.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hou M. C., Wiley D. J., Verde F., McCollum D. Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J. Cell Sci. 2003;116:125–135. doi: 10.1242/jcs.00206. [DOI] [PubMed] [Google Scholar]
- Huang T. Y., Renaud-Young M., Young D. Nak1 interacts with Hob1 and Wsp1 to regulate cell growth and polarity in Schizosaccharomyces pombe. J. Cell Sci. 2005;118:199–210. doi: 10.1242/jcs.01608. [DOI] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Nelson B., Robinson M. D., Chen Y., Andrews B., Tyers M., Boone C. High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics. 2002;162:1091–1099. doi: 10.1093/genetics/162.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice R. W., Zilian O., Woods D. F., Noll M., Bryant P. J. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kanai M., Kume K., Miyahara K., Sakai K., Nakamura K., Leonhard K., Wiley D. J., Verde F., Toda T., Hirata D. Fission yeast MO25 protein is localized at SPB and septum and is essential for cell morphogenesis. EMBO J. 2005;24:3012–3025. doi: 10.1038/sj.emboj.7600782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzack S., Rischitor P. E., Enke C., Fischer R. The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans. Mol. Biol. Cell. 2005;16:497–506. doi: 10.1091/mbc.E04-02-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurischko C., Weiss G., Ottey M., Luca F. C. A role for the Saccharomyces cerevisiae regulation of Ace2 and polarized morphogenesis signaling network in cell integrity. Genetics. 2005;171:443–455. doi: 10.1534/genetics.105.042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir T., Knubovets T., Itzchak Y., Eliav U., Sadeh M., Rapoport L., Kott E., Navon G. In vivo 23Na NMR studies of myotonic dystrophy. Magn. Reson. Med. 1997;37:192–196. doi: 10.1002/mrm.1910370209. [DOI] [PubMed] [Google Scholar]
- Leonhard K., Nurse P. Ste20/GCK kinase Nak1/Orb3 polarizes the actin cytoskeleton in fission yeast during the cell cycle. J. Cell Sci. 2005;118:1033–1044. doi: 10.1242/jcs.01690. [DOI] [PubMed] [Google Scholar]
- Lin J. L., Chen H. C., Fang H. I., Robinson D., Kung H. J., Shih H. M. MST4, a new Ste20-related kinase that mediates cell growth and transformation via modulating ERK pathway. Oncogene. 2001;20:6559–6569. doi: 10.1038/sj.onc.1204818. [DOI] [PubMed] [Google Scholar]
- Lopez-Franco R., Bartnicki-Garcia S., Bracker C. E. Pulsed growth of fungal hyphal tips. Proc. Natl. Acad. Sci. USA. 1994;91:12228–12232. doi: 10.1073/pnas.91.25.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan M. S., et al. Structure and genomic sequence of the myotonic dystrophy (DM kinase) gene. Hum. Mol. Genet. 1993;2:299–304. doi: 10.1093/hmg/2.3.299. [DOI] [PubMed] [Google Scholar]
- Matsusaka T., Hirata D., Yanagida M., Toda T. A novel protein kinase gene ssp1+ is required for alteration of growth polarity and actin localization in fission yeast. EMBO J. 1995;14:3325–3338. doi: 10.1002/j.1460-2075.1995.tb07339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke P. F., Lee I. H., Plamann M. Microscopic analysis of Neurospora ropy mutants defective in nuclear distribution. Fungal Genet. Biol. 1999;28:55–67. doi: 10.1006/fgbi.1999.1160. [DOI] [PubMed] [Google Scholar]
- Mounsey J. P., Xu P., John J. E., 3rd, Horne L. T., Gilbert J., Roses A. D., Moorman J. R. Modulation of skeletal muscle sodium channels by human myotonic protein kinase. J. Clin. Invest. 1995;95:2379–2384. doi: 10.1172/JCI117931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargang F. E., Kunkele K. P., Mayer A., Ritzel R. G., Neupert W., Lill R. ‘Sheltered disruption’ of Neurospora crassa MOM22, an essential component of the mitochondrial protein import complex. EMBO J. 1995;14:1099–1108. doi: 10.1002/j.1460-2075.1995.tb07093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B., Kurischko C., Horecka J., Mody M., Nair P., Pratt L., Zougman A., McBroom L. D., Hughes T. R., Boone C., Luca F. C. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell. 2003;14:3782–3803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W. J. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422:766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y., Akiyama T., Nishida E., Sakai H. Protein kinase C and cAMP-dependent protein kinase induce opposite effects on actin polymerizability. FEBS Lett. 1987;222:305–310. doi: 10.1016/0014-5793(87)80391-5. [DOI] [PubMed] [Google Scholar]
- Orbach M. J. A cosmid with a HyR marker for fungal library construction and screening. Gene. 1984;150:159–162. doi: 10.1016/0378-1119(94)90877-x. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. USA. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D., Radford A., Sachs M. S. The Neurospora Compendium. San Diego: Academic Press; 2001. [Google Scholar]
- Plamann M., Minke P. F., Tinsley J. H., Bruno K. S. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J. Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggeler S., Masloff S., Hoff B., Mayrhofer S., Kuck U. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr. Genet. 2003;43:54–61. doi: 10.1007/s00294-003-0370-y. [DOI] [PubMed] [Google Scholar]
- Prat A. G., Bertorello A. M., Ausiello D. A., Cantiello H. F. Activation of epithelial Na+ channels by protein kinase A requires actin filaments. Am. J. Physiol. 1993;265:C224–C233. doi: 10.1152/ajpcell.1993.265.1.C224. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A. Polarization of cell growth in yeast. J. Cell Sci. 2000a;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 2000b;113:365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Qian Z., Lin C., Espinosa R., LeBeau M., Rosner M. R. Cloning and characterization of MST4, a novel Ste20-like kinase. J. Biol. Chem. 2001;276:22439–22445. doi: 10.1074/jbc.M009323200. [DOI] [PubMed] [Google Scholar]
- Racki W. J., Becam A. M., Nasr F., Herbert C. J. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 2000;19:4524–4532. doi: 10.1093/emboj/19.17.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme M., Reynaga-Pena C. G., Gierz G., Bartnicki-Garcia S. What determines growth direction in fungal hyphae? Fungal Genet.Biol. 1998;24:101–109. doi: 10.1006/fgbi.1998.1074. [DOI] [PubMed] [Google Scholar]
- Schuchardt I., Assmann D., Thines E., Schuberth C., Steinberg G. Myosin-V, Kinesin-1, and Kinesin-3 cooperate in hyphal growth of the fungus Ustilago maydis. Mol. Biol. Cell. 2005;16:5191–5201. doi: 10.1091/mbc.E05-04-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S., Nargang F. E., Steinberg G., Schliwa M. Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 1997;16:3025–3034. doi: 10.1093/emboj/16.11.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S., Plamann M. The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell. 2003;14:4352–4364. doi: 10.1091/mbc.E02-07-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S., Plamann M., Schliwa M. Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr. Biol. 1999;9:779–785. doi: 10.1016/s0960-9822(99)80360-1. [DOI] [PubMed] [Google Scholar]
- Shaw S. L., Yeh E., Maddox P., Salmon E. D., Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman-Gavrila L. B., Lew R. R. Regulation of the tip-high [Ca2+] gradient in growing hyphae of the fungus Neurospora crassa. Eur. J. Cell Biol. 2001;80:379–390. doi: 10.1078/0171-9335-00175. [DOI] [PubMed] [Google Scholar]
- Stegert M. R., Hergovich A., Tamaskovic R., Bichsel S. J., Hemmings B. A. Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol. Cell. Biol. 2005;25:11019–11029. doi: 10.1128/MCB.25.24.11019-11029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork O., Zhdanov A., Kudersky A., Yoshikawa T., Obata K., Pape H. C. Neuronal functions of the novel serine/threonine kinase Ndr2. J. Biol. Chem. 2004;279:45773–45781. doi: 10.1074/jbc.M403552200. [DOI] [PubMed] [Google Scholar]
- Sutton A., Immanuel D., Arndt K. T. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaskovic R., Bichsel S. J., Hemmings B. A. NDR family of AGC kinases—essential regulators of the cell cycle and morphogenesis. FEBS Lett. 2003;546:73–80. doi: 10.1016/s0014-5793(03)00474-5. [DOI] [PubMed] [Google Scholar]
- Terenzi H. F., Flawia M. M., Torres H. N. A Neurospora crassa morphological mutant showing reduced adenylate cyclase activity. Biochem. Biophys. Res. Commun. 1974;58:990–996. doi: 10.1016/s0006-291x(74)80241-x. [DOI] [PubMed] [Google Scholar]
- Terenzi H. F., Reissig J. L. Modifiers of the cot gene in Neurospora: the gulliver mutants. Genetics. 1967;56:321–329. doi: 10.1093/genetics/56.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein J. M. Signal transduction in yeast. Yeast. 1994;10:1753–1790. doi: 10.1002/yea.320101308. [DOI] [PubMed] [Google Scholar]
- Toda T., Niwa H., Nemoto T., Dhut S., Eddison M., Matsusaka T., Yanagida M., Hirata D. The fission yeast sts5+ gene is required for maintenance of growth polarity and functionally interacts with protein kinase C and an osmosensing MAP-kinase pathway. J. Cell Sci. 1996;109:2331–2342. doi: 10.1242/jcs.109.9.2331. [DOI] [PubMed] [Google Scholar]
- Uesono Y., Toh-e A., Kikuchi Y. Ssd1p of Saccharomyces cerevisiae associates with RNA. J. Biol. Chem. 1997;272:16103–16109. doi: 10.1074/jbc.272.26.16103. [DOI] [PubMed] [Google Scholar]
- Verde F., Wiley D. J., Nurse P. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl. Acad. Sci. USA. 1998;95:7526–7531. doi: 10.1073/pnas.95.13.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer S. J., Yanofsky C. Efficient cloning of genes of Neurospora crassa. Proc. Natl. Acad. Sci. USA. 1986;83:4869–4873. doi: 10.1073/pnas.83.13.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth W. P., Olsen A. E., Sbia M., Freedman K. H., Stillman D. J. ACE2, CBK1, and BUD4 in budding and cell separation. Eukaryot. Cell. 2005;4:1018–1028. doi: 10.1128/EC.4.6.1018-1028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland J. Comparison of morphogenetic networks of filamentous fungi and yeast. Fungal Genet. Biol. 2001;34:63–82. doi: 10.1006/fgbi.2001.1290. [DOI] [PubMed] [Google Scholar]
- Wiley D. J., Marcus S., D’Urso G., Verde F. Control of cell polarity in fission yeast by association of Orb6p kinase with the highly conserved protein methyltransferase Skb1p. J. Biol. Chem. 2003;278:25256–25263. doi: 10.1074/jbc.M209703200. [DOI] [PubMed] [Google Scholar]
- Xu T., Wang W., Zhang S., Stewart R. A., Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Yarden O., Plamann M., Ebbole D. J., Yanofsky C. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 1992;11:2159–2166. doi: 10.1002/j.1460-2075.1992.tb05275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J. A., Peckol E. L., Tobin D. M., Bargmann C. I. Neuronal cell shape and neurite initiation are regulated by the Ndr kinase SAX-1, a member of the Orb6/COT-1/warts serine/threonine kinase family. Mol. Biol. Cell. 2000;11:3177–3190. doi: 10.1091/mbc.11.9.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.