Abstract
A reverse transcription-seminested PCR (RT-snPCR) assay was developed for the detection and identification of enterovirus (EV) RNA in clinical specimens. Three conserved protein motifs were identified by aligning the VP3 and VP1 sequences of prototype EV strains. Consensus degenerate primers were designed from a conserved VP3 motif and a distal VP1 motif for the first PCR. Consensus-degenerate hybrid oligonucleotide primers were designed from an internal VP1 motif and used with the same distal VP1 motif for the second, seminested PCR step. The primers were designed for broad target specificity and amplified all recognized and proposed EV serotypes and other antigenic variant strains tested. The VP1 RT-snPCR assay was slightly more sensitive than our in-house EV 5′ nontranslated region RT-snPCR assay, detecting as few as 10 RNA copies per reaction. As an example application, the VP1 RT-snPCR assay was used to identify EVs in clinical specimens. A product of the expected size was successfully amplified and sequenced from cerebrospinal fluid; serum; stool suspensions; and nasopharyngeal, eye, and rectal swab specimens, allowing unambiguous identification of the infecting virus in all cases. The VP1 sequences derived from the RT-snPCR products allow rapid phylogenetic and molecular epidemiologic analysis of strains circulating during the EV season and comparison with EV sequences from past seasons or from different locations around the world.
Enteroviruses (EVs) (genus Enterovirus, family Picornaviridae) are among the most common viruses infecting humans. Most infections are asymptomatic or result in only mild symptoms, such as nonspecific febrile illness or mild upper respiratory symptoms (common cold). However, enteroviruses can also cause a wide variety of other clinical illnesses, including acute hemorrhagic conjunctivitis, aseptic meningitis, undifferentiated rash, acute flaccid paralysis, myocarditis, and neonatal sepsis-like disease (23).
Molecular diagnostic tests for the detection of EVs in clinical specimens usually target highly conserved sites in the 5′ nontranslated region (5′-NTR), allowing detection of all members of the genus (27). Many enteroviruses do not grow readily in cell culture; hence, these diagnostic tests based on PCR are frequently more sensitive than traditional methods. Because it is more sensitive and much faster than culture, providing reliable results in a clinically relevant time frame, PCR is rapidly becoming the new “gold standard” for enterovirus detection (5, 25, 26, 32). Despite their generally better detection sensitivities, however, these tests are genus specific and provide an EV-positive or EV-negative result, but they cannot be used to identify the serotype.
The sequence of the EV VP1 capsid gene correlates with the serotype determined by antigenic methods (14) and is an ideal target for EV detection tests that can both detect and identify EVs. To take advantage of the excellent correlation between the VP1 sequence and the enterovirus serotype, we developed primers with broad target specificities to facilitate sequencing of a portion of VP1 as a molecular typing system (12, 13, 19). Since then, other investigators have developed analogous methods targeting different portions of VP1 (3, 4, 11, 31). In this system, nucleotide sequences which are at least 75% identical (85% amino acid identity) are considered to represent strains of the same serotype (12-14, 19, 20). As a result, this approach has been widely accepted as a surrogate for antigenic typing for the identification of EV isolates (1, 2, 6-9, 15-18, 21, 22, 24).
Even though the detection of virus from original clinical specimens by targeting the 5′-NTR has achieved very high sensitivities and sequencing of the VP1 region provides a practical means of establishing the identity of an enterovirus, identification of the serotype directly from clinical specimens has been considerably more problematic. This is primarily because the virus titer is typically very low in original specimens; as a result, nonspecific amplification may outcompete virus-specific amplification. Several reverse transcription (RT)-nested or RT-seminested PCR (RT-snPCR) methods have been developed to address these issues (4, 10, 31). However, these methods either target only a subset of EV serotypes (31) or rely on highly degenerate, inosine-containing primers in both PCR steps to broaden the target specificity to include all serotypes (4, 10), which can adversely affect sensitivity. The use of degenerate, inosine-containing primers often results in nonspecific amplification of host cell nucleic acids and may obscure the virus-specific product (30). We have adapted a primer design strategy that reduces nonspecific amplification and that was originally applied to the amplification and identification of new homologs of cellular genes and to the discovery of novel herpesviruses (28, 30). Our RT-snPCR method achieves high sensitivity, amplifies all known human EV serotypes, and permits the detection of EVs in clinical specimens as well as the identification of the serotype by amplicon sequencing.
MATERIALS AND METHODS
Virus isolates and model templates.
To evaluate the specificity of the VP1 RT-snPCR, we assembled panels of virus isolates that included reference strains of all 64 recognized EV serotypes (see Table 2), reference strains of 22 proposed new serotypes (see Table 2), and 87 recent clinical isolates (see Table 3).
TABLE 2.
Enterovirus reference strains amplified by VP1 RT-snPCRa
| Serotype | Strain | Serotype | Strain | |
|---|---|---|---|---|
| CVA1 | Tompkins | E16 | Harrington | |
| CVA2 | Fleetwood | E17 | CHHE-29 | |
| CVA3 | Olson | E18 | Metcalf | |
| CVA4 | High Point | E19 | Burke | |
| CVA5 | Swartz | E20 | JV-1 | |
| CVA6 | Gdula | E21 | Farina | |
| CVA7 | AB-IV | E24 | De Camp | |
| CVA8 | Donovan | E25 | JV-4 | |
| CVA9 | Griggs | E26 | Coronel | |
| CVA10 | Kowalik | E27 | Bacon | |
| CVA11 | Belgium-1 | E29 | JV-10 | |
| CVA12 | Texas-12 | E30 | Bastianni | |
| CVA13 | Flores | 30 | Frater | |
| CVA14 | G-14 | E30 | Giles | |
| CVA15 | G-9 | E30 | PR-17 | |
| CVA16 | G-10 | E31 | Caldwell | |
| CVA17 | G-12 | E32 | PR-10 | |
| CVA18 | G-13 | E33 | Toluca-3 | |
| CVA19 | 8663 | EV68 | Fermon | |
| CVA20 | IH-35 | EV69 | Toluca-1 | |
| CVA21 | Kuykendall | EV70 | J670/71 | |
| CVA22 | Chulman | EV71 | BrCr | |
| CVA24 | Joseph | EV73 | CA55-1988 | |
| CVA24 | EH24 | EV74 | 10213 | |
| CVA24 | DN-19 (E34) | EV75 | 10362 | |
| CVB1 | Conn-5 | EV76 | 10369 | |
| CVB2 | Ohio-1 | EV79 | 10384 | |
| CVB3 | Nancy | EV80 | 10387 | |
| CVB4 | JVB | EV81 | 10389 | |
| CVB5 | Faulkner | EV82 | 10390 | |
| CVB6 | Schmitt | EV83 | 10392 | |
| E1 | Farouk | EV84 | 10603 | |
| E1 | Bryson (E8) | EV85 | 10353 | |
| E2 | Cornelis | EV86 | 10354 | |
| E3 | Morrisey | EV87 | 10396 | |
| E4 | Du Toit | EV88 | 10398 | |
| E4 | Shropshire | EV89 | 10395 | |
| E4 | Pesacek | EV90 | 10399 | |
| E5 | Noyce | EV91 | 10406 | |
| E6 | D;Amori | EV92 | 10408 | |
| E6 | Cox | EV96 | 10358 | |
| E6 | Burgess | EV97 | 10355 | |
| E6 | Charles | EV100 | 10500 | |
| E7 | Wallace | EV10 | 110361 | |
| E9 | Hill | PV1 | Mahoney | |
| E11 | Gregory | PV1 | Sabin | |
| E11 | Silva | PV2 | Lansing | |
| E12 | Travis | PV2 | Sabin | |
| E13 | Del Carmen | PV2 | Lansing | |
| E14 | Tow | PV3 | Leon | |
| E15 | CH96-51 | PV3 | Sabin |
Reference strains for EV77 to EV78 and EV93 to EV95 were not tested because they are not publicly available. Some other numbers are missing due to reclassification (i.e., coxsackievirus A serotype 23 [CVA23] is a variant of echovirus 9 [E9], E8 is a variant of E1, E34 is a variant of CVA24, E10 is reovirus 1 [genus Orthoreovirus, family Reoviridae], E28 is human rhinovirus 1A [genus Rhinovirus, family Picornaviridae], and EV72 is human hepatitis A virus [genus Hepatovirus, family Picornaviridae]). The “newer” serotypes (EV73 to EV76, EV79 to EV92, EV96, EV97, EV100, and EV101) were identified and classified by molecular means rather than by antigenic means (15, 18, 21; M. S. Oberste, unpublished data). PV, poliovirus.
TABLE 3.
Clinical isolates amplified by VP1 RT-snPCR
| Serotype | No. of isolates | Yr(s) of isolation |
|---|---|---|
| CVA14 | 2 | 1992-1994 |
| CVA16 | 4 | 1984-1995 |
| CVA20 | 2 | 1983 |
| CVA21 | 6 | 1986-1996 |
| CVA24 | 1 | 1984 |
| CVA9 | 2 | 1993-1996 |
| CVB2 | 3 | 1982-1995 |
| CVB3 | 6 | 1988-1997 |
| CVB4 | 1 | 1984 |
| CVB5 | 4 | 1983-1993 |
| E3 | 3 | 1986-1988 |
| E4 | 2 | 1985-1988 |
| E5 | 1 | 1996 |
| E6 | 3 | 1991-1998 |
| E7 | 3 | 1993-1998 |
| E9 | 3 | 1992-1995 |
| E11 | 5 | 1988-1998 |
| E12 | 1 | 1988 |
| E13 | 2 | 1986-1995 |
| E18 | 6 | 1985-1997 |
| E21 | 1 | 1994 |
| E24 | 1 | 1983 |
| E25 | 6 | 1984-1994 |
| E29 | 1 | 1988 |
| E30 | 4 | 1991-1995 |
| E33 | 1 | 1998 |
| EV71 | 7 | 1988-1995 |
| EV75 | 4 | 1985-1987 |
| HRV2 | 1 | 1990 |
| HRV31 | 1 | 1988 |
RNA extraction.
Stool suspensions were prepared by adding 5 ml of phosphate-buffered saline, 1 g of glass beads (Corning Inc., Corning, NY), and 0.5 ml of chloroform to 1 g of stool sample, shaking the mixture vigorously for 20 min in a mechanical shaker, and centrifuging at 1,500 × g for 20 min at 4°C (33). For rectal swab samples, the fluid was centrifuged at 13,000 × g for 1 min at room temperature to remove the solids, and the supernatant was transferred to a fresh tube. For fecal specimens (10% stool suspensions or clarified rectal swab supernatants), 140 μl of the specimen extract was combined with an equal volume of Vertrel XF (Miller-Stephenson Chemical Co., Danbury, CT), shaken vigorously, and then centrifuged at 13,000 × g for 1 min at room temperature. The aqueous phase was transferred to a fresh tube. Other specimen types (including cerebrospinal fluid; serum; virus isolates; and supernatants from nasopharyngeal, oropharyngeal, and conjunctival swab samples) were processed without pretreatment. Twenty micrograms of proteinase K (Roche Applied Science, Indianapolis, IN) was added to 140 μl of each liquid specimen or fecal extract, and the mixture was then incubated for 30 min at 37°C. Nucleic acid was extracted from the digested specimen with a QIAamp Viral RNA mini kit (QIAGEN, Inc., Valencia, CA), which was used according to the manufacturer's instructions. The eluted RNAs were passively dried in a benchtop desiccator under vacuum. The dried RNA was resuspended in 16 μl of sterile nuclease-free water and stored at −20°C until use.
RT-PCR and sequencing.
Synthesis of cDNA was carried out in a 10-μl reaction mixture containing 5 μl of RNA, 100 μM each deoxynucleoside triphosphate (dNTP; Amersham Biosciences, Piscataway, NJ), 2 μl of 5× reaction buffer (Invitrogen, Carlsbad, CA), 0.01 M dithiothreitol, 1 pmol each cDNA primer (primers AN32, AN33, AN34, and AN35; Table 1), 20 U of RNasin (Promega Corp., Madison, WI), and 100 U of SuperScript II reverse transcriptase (Invitrogen). Following incubation at 22°C for 10 min, 45°C for 45 min, and 95°C for 5 min, the entire 10 μl RT reaction mixture was then used in the first PCR (final volume, 50 μl) (PCR1), consisting of 5 μl of 10× PCR buffer (Roche Applied Science), 200 μM each dNTP, 50 pmol each of primers 224 and 222 (Table 1), and 2.5 U of Taq DNA polymerase (Roche Applied Science), with 40 cycles of amplification (95°C for 30 s, 42°C for 30 s, 60°C for 45 s). One microliter of the first PCR was added to a second PCR (PCR2) for seminested amplification. PCR2 contained 40 pmol each of primers AN89 and AN88 (Table 1), 200 μM each dNTP, 5 μl of 10× FastStart Taq buffer (Roche Applied Science), and 2.5 U of FastStart Taq DNA polymerase (Roche Applied Science) in a final volume of 50 μl. The FastStart Taq polymerase was activated by incubation at 95°C for 6 min prior to 40 amplification cycles of 95°C for 30 s, 60°C for 20 s, and 72°C for 15 s. The reaction products were separated and visualized on 1.2% agarose gels containing 0.5 μg ethidium bromide per ml and were purified from the gel by using a QIAquick gel extraction kit (QIAGEN). Slight variations in the sizes of the PCR products (∼350 to 400 bp) were observed due to VP1 gene length differences in the different serotypes, as described previously (12-14, 19). The resulting DNA templates were sequenced with a BigDye Terminator v1.1 ready reaction cycle sequencing kit on an ABI Prism 3100 automated sequencer (both from Applied Biosystems, Foster City, CA) by using primers AN89 and AN88 or primers AN232 and AN233 (Table 1).
TABLE 1.
Primers used for cDNA synthesis, PCR amplification, and sequencing
| Primer | Sequence | Amino acid motif | Gene | Locationa |
|---|---|---|---|---|
| AN32 | GTYTGCCA | WQT | VP1 | 3009-3002 |
| AN33 | GAYTGCCA | WQS | VP1 | 3009-3002 |
| AN34 | CCRTCRTA | YDG | VP1 | 3111-3104 |
| AN35 | RCTYTGCCA | WQS | VP1 | 3009-3002 |
| 224 | GCIATGYTIGGIACICAYRT | AMLGTH(I/L/M) | VP3 | 1977-1996 |
| 222 | CICCIGGIGGIAYRWACAT | M(F/Y)(I/V)PPG(A/G) | VP1 | 2969-2951 |
| 292 | MIGCIGYIGARACNGG | (Q/T)A(A/V)ETG | VP1 | 2612-2627 |
| AN89 | CCAGCACTGACAGCAGYNGARAYNGGb | PALTA(A/V)E(I/T)G | VP1 | 2602-2627 |
| AN88 | TACTGGACCACCTGGNGGNAYRWACATb | M(F/Y)(I/V)PPGGPV | VP1 | 2977-2951 |
| AN232 | CCAGCACTGACAGCAb | PALTA | VP1 | 2602-2616 |
| AN233 | TACTGGACCACCTGGb | PGGPV | VP1 | 2977-2963 |
| AN230 | AATTAACCCTCACTAAAGGGAGAAGATATTATACTCAYTGGc | RYYTHW | VP3 | 1993-2010 |
| AN231 | GTCAGCTGGGTTTATNCCRTA | YGINPAD | VP1 | 3069-3049 |
The locations of all primers except AN230 and AN231 are those relative to the genome of PV1 Mahoney (GenBank accession number J02281); the locations for AN230 and AN231 are relative to those of the genome of EV68 Fermon (GenBank accession number AY426531).
AN232 is the nondegenerate “clamp” portion of AN89, and AN233 is the nondegenerate clamp portion of AN88. Within the AN88 and AN89 sequences, these clamp regions are indicated in italic type.
The T3 RNA polymerase promoter sequence is underlined.
Sequence analysis.
The amplicon sequences were compared with the VP1 sequences of EV reference strains, including at least one representative of each recognized serotype, by script-driven sequential pairwise comparison with the program Gap (Wisconsin Sequence Analysis Package, version 10.2; Accelrys, Inc., San Diego, CA), as described previously (15, 18, 19). In cases where the result was not unequivocal (highest score less than 75% or second-highest score greater than 70%), the deduced amino acid sequences were compared by a similar method.
Assay sensitivity.
Sensitivity was tested by two methods. The sensitivity relative to the results of cell culture infectivity was measured by using a stock of the EV serotype 68 (EV68) prototype strain Fermon whose titer had been determined. Serial 10-fold dilutions of the EV68-Fermon stock were made in Hank's balanced salt solution, and RNA from 100 μl of each dilution was extracted with the QIAamp Viral RNA mini kit. RNAs representing from 104 cell culture infectious dose 50% end-point units (CCID50) to 10−3 CCID50 per 5 μl were tested with the VP1 RT-snPCR assay.
Absolute sensitivity was measured by using an in vitro-transcribed synthetic RNA standard derived from EV68-Fermon. To make the synthetic RNA standard, RT-PCR primers were designed to flank the VP3-VP1 RT-snPCR assay cDNA product. The sense primer AN230 contains the 23-base T3 RNA polymerase promoter at the 5′ end, and it was used with the antisense primer AN231 (Table 1) in a two-step RT-PCR. cDNA was made with SuperScript II reverse transcriptase (Invitrogen), according to the instructions provided with the kit, by using 10 pmol AN231 to prime the cDNA. PCR was performed with FastStart Taq (Roche Applied Science) by using the manufacturer's 10× buffer with MgCl2, 2 μl of cDNA, 200 μM each dNTP, and 20 pmol each of primers AN230 and AN231 in a final reaction volume of 50 μl. The thermocycler program consisted of 40 cycles of 95°C for 30 s, 55°C for 40 s, and 72°C for 40 s. The PCR product was purified by using a High Pure PCR product purification kit (Roche Applied Science), according to the manufacturer's instructions. The purified PCR product was quantitated spectrophotometrically, and 1 μg of PCR product was used as the template for in vitro RNA transcription with a MEGAscript high yield transcription kit (Ambion, Inc., Austin, TX), according to the manufacturer's protocol. The VP3-VP1 single-stranded, positive-sense standard RNA product (VP3-VP1 sRNA; 1,082 nucleotides) was digested with DNase I to remove the template DNA and was then purified with the QIAamp Viral RNA mini kit (QIAGEN). The manufacturer's instructions were followed, except that no carrier tRNA was added to the kit's lysis buffer. The purified VP3-VP1 sRNA was quantitated spectrophotometrically, and the concentration was calculated in units of RNA molecules per microliter. Two separate lots of the VP3-VP1 sRNA were synthesized and diluted to contain from 104 copies to 1 copy per 5 μl and then tested in two separate experiments with the VP1 RT-snPCR assay.
The sensitivity of the VP1 RT-snPCR assay was also directly compared to that of our published 5′-NTR RT-snPCR assay (10) by serially diluting RNA extracted from a recent EV68 clinical isolate and running both the VP1 and the 5′-NTR RT-snPCR assays in parallel by using the same diluted RNA preparations.
Nucleotide sequence accession numbers.
The sequences for the original clinical specimens described here have been deposited in the GenBank sequence database (accession numbers AY903638 to AY903644).
RESULTS
Primer design.
To achieve the sensitivity necessary for the amplification and identification of EVs in original clinical specimens, we previously developed an RT-snPCR approach, using a primer (primer 224) in VP3 paired with a primer (primer 222) in VP1 for the first round of amplification and primers (primers 292 and 222) within VP1 (19) for the second, seminested PCR amplification (10) (Table 1; Fig. 1). This primer combination unfortunately sometimes results in an undesirable accumulation of nonspecific amplification products (data not shown), probably due to the use of highly degenerate inosine-containing primers, which require relatively low annealing temperatures for both PCR steps. To eliminate this problem, we designed a new set of internal primers to replace primers 292 and 222 using the consensus degenerate hybrid oligonucleotide primer (CODEHOP) approach (28, 30). CODEHOP primers contain a consensus degenerate core region of 9 to 12 bases at the 3′ end that is designed from conserved motifs within aligned amino acid sequences. The 5′ portion of a CODEHOP primer comprises a nondegenerate consensus clamp region that is typically 15 to 25 bases long. The degenerate core provides for broad target specificity, while the clamp increases the stability of the primer-template helix and allows the use of higher annealing temperatures to reduce nonspecific amplification. The optimal design strategy for the nondegenerate consensus clamp requires the analysis of codon usage for any conserved amino acid residues that occur within the clamp region. The predominant codon, if any, is incorporated into the primer sequence. For positions with multiple amino acid residues in the nondegenerate clamp, an arbitrary codon can be chosen or a specific codon can be selected to manipulate the G+C content of the primer.
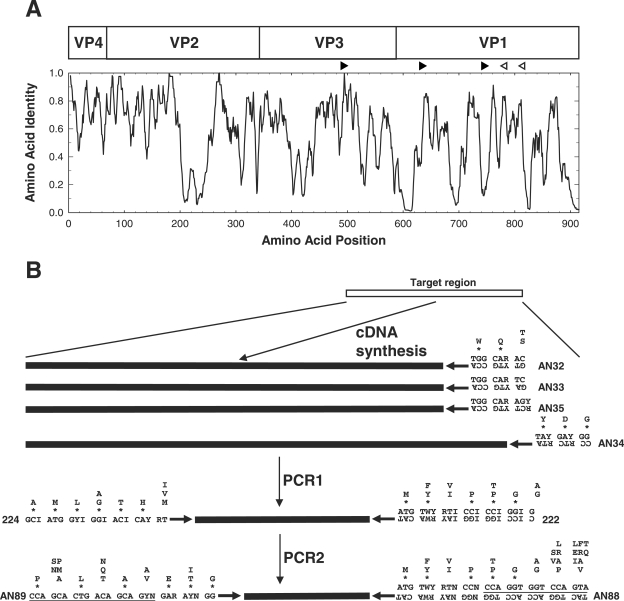
FIG. 1.
Schematic representation of the locations of the primers used in the CODEHOP VP1 RT-snPCR. (A) Similarity plot of the aligned capsid amino acid sequences of 64 enterovirus prototype strains. Sequence identity scores were calculated within each 6-residue window, and the window was progressively moved across the alignment in 1-residue increments, with the identity score plotted versus the position at the center of the window. The positions of the four mature EV capsid proteins, VP4, VP2, VP3, and VP1, are shown at the top. The orientations and approximate positions of the cDNA primers (open arrowheads) and PCR primers (filled arrowheads) are shown directly above the plot. (B) Amino acid motifs used in primer design and schematic representation of the steps in the CODEHOP VP1 RT-snPCR assay. Consensus amino acid motifs are shown. Asterisks indicate that the residue directly above the asterisk is present at that position in at least 90% of EV prototype strains; when only a single residue is shown, it is present in all prototype strains. Primer sequences are shown directly below the amino acid motif sequences. Ambiguity codes are as follows: R, A or G; Y, C or T; W, A or T; N, A, C, G, or T; I, inosine.
Consensus degenerate primers for the cDNA and PCR1 steps were designed from conserved amino acid motifs in the aligned capsid sequences of the 64 serotype prototype strains (Fig. 1). The four cDNA primers were designed to anneal to conserved sites downstream of the reverse PCR primer; these sites encode the motifs WQT (primer AN32), WQS (primers AN33 and AN35), and YDG (primer AN34) (Table 1; Fig. 1). PCR1 forward primer 224 was designed to target the site in VP3 encoding the highly conserved motif AMLGTH(I/L/M) (Table 1; Fig. 1), while PCR1 reverse primer 222 targets a conserved motif M(F/Y)(I/V)PPG(A/G), near the middle of VP1 (Table 1; Fig. 1) (12, 19). These two motifs are conserved among all EV serotypes and many human rhinoviruses. The degenerate primer design approach was used for the PCR1 primers to broaden the target specificity of amplification, allowing amplification of all EV serotypes and increasing the absolute concentration of virus-specific product to be used as the template for PCR2. The presence of inosine residues in positions of fourfold codon degeneracy reduces the overall degeneracy of the PCR1 primers but also results in the decreased thermostability of the primer-template helix. As a result, low-stringency annealing conditions are required for PCR1.
The internal primers used in PCR2 were designed without the use of inosine residues by using the CODEHOP strategy and with target sites encoding conserved motifs in VP1. Our original protocol used forward internal primer 292, which targets the conserved motif (Q/T)A(A/V)ETG, paired with reverse primer 222 (Table 1; Fig. 1). The new forward PCR2 primer, primer AN89, is 3′ coterminal with primer 292, but it targets a longer motif [PALTA(A/V)E(I/T)G] and incorporates an additional degeneracy at position −6 to allow annealing to codons that encode either isoleucine or threonine in the penultimate codon of the motif. Primer AN88 is 3′ coterminal with primer 222 and targets the motif M(F/Y)(I/V)PPGGPV (Table 1; Fig. 1). The consensus clamp and increased length both probably contribute to the increased thermostabilities of primers AN88 and AN89 compared with those of primers 292 and 222.
Assay evaluation.
To ensure that the new primers are broadly reactive and have the same ability to amplify VP1 sequences of all EV serotypes as our previous primers, we tested a panel of reference strains representing all serotypes (101 strains total), as well as a panel of 87 recent clinical isolates of 29 different types. All 64 EV serotype reference strains, the 22 proposed new serotypes, and 15 additional reference strains for some serotypes were successfully amplified and sequenced by the CODEHOP VP1 RT-snPCR procedure (Table 2). All 87 clinical isolates tested were also successfully amplified, sequenced, and identified by comparing the nucleic acid sequence to the sequences in our EV reference strain VP1 sequence database (Table 3). In all cases, the serotype based on the VP1 RT-snPCR amplicon was identical to the serotype previously determined by neutralization or by VP1 sequencing with different primers and conventional PCR (12, 13, 19).
Assay sensitivity.
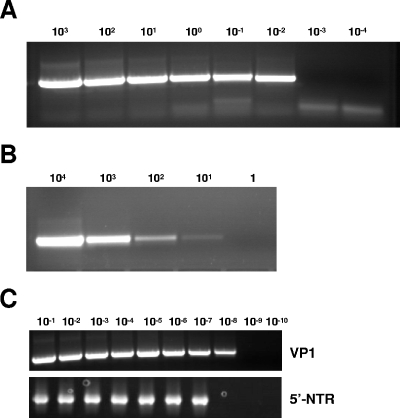
Due to the absence of a gold standard for an analytical sensitivity measure that is applicable across all serotypes of enteroviruses, sensitivity was measured in two different ways so that the sensitivity could be compared to those obtained by previously described assays. The VP1 RT-snPCR assay detected RNA extracted from as little as 0.01 CCID50 per 5 μl of EV68-Fermon (Fig. 2A), indicating that the assay is at least 100-fold more sensitive than cell culture for this strain of virus (since 1 CCID50 defines the cell culture end point). As few as 10 copies of the in vitro-transcribed VP3-VP1 sRNA produced a detectable gel band in two independent experiments, indicating a low limit of absolute sensitivity (Fig. 2B). The diluted EV68 clinical isolate RNA was amplified from the 10−1 to 10−7 dilutions with the 5′-NTR RT-snPCR assay and from the 10−1 to 10−8 dilutions with the VP1 RT-snPCR assay (Fig. 2C), indicating that the VP1 assay has sensitivity at least equal to or possibly 10-fold better than that of the 5′-NTR assay.
FIG. 2.
Sensitivity of VP1 RT-snPCR and comparison of the sensitivity with that of the 5′ nontranslated region RT-snPCR. (A) Amplification of RNA extracted from 10-fold serial dilutions of an EV68 virus stock; (B) amplification of 10-fold serial dilutions of VP3-VP1 sRNA; (C) comparison of VP1 RT-snPCR (top) with 5′-NTR RT-snPCR (bottom) using 10-fold serial dilutions.
Application to clinical specimens.
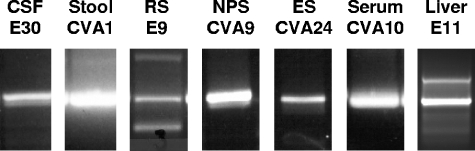
To illustrate the application of the VP1 RT-snPCR method, RNA was extracted from original clinical specimens obtained from patients with a number of different enteroviral illnesses and specimen types. The specimens (and associated illnesses) included cerebrospinal fluid (aseptic meningitis), stool (aseptic meningitis), rectal swab (febrile rash), nasopharyngeal swab (mild upper respiratory illness), conjunctival swab (acute hemorrhagic conjunctivitis), serum (febrile rash), and postmortem liver tissue (neonatal sepsis-like illness). A specific product was amplified by VP1 RT-snPCR from each of these extracted RNA templates (Fig. 3). In each assay, the tested RNA represents the equivalent of approximately 45 μl of original specimen fluid or 2 mg of stool sample. Following gel purification, the EV present in each specimen was identified by amplicon sequencing and comparison to a database of EV VP1 sequences. All of the amplification products yielded clean, readable sequences, including those with weak or multiple bands (e.g., rectal swab and liver tissue specimens). In this example, the EVs identified were E30 (cerebrospinal fluid), CVA1 (stool), E9 (rectal swab), CVA9 (nasopharyngeal swab), CVA24 (conjunctival swab), CVA10 (serum), and E11 (liver tissue).
FIG. 3.
VP1 RT-snPCR amplification of RNA extracted directly from original clinical specimens. For each reaction, 50 μl of each seminested PCR2 product was analyzed and gel purified by electrophoresis on a 1.5% agarose gel containing 0.5 μg ethidium bromide per milliliter. The specimens tested were cerebrospinal fluid (CSF), stool, rectal swab (RS), nasopharyngeal swab (NPS), eye (conjunctival) swab (ES), serum, and postmortem liver tissue.
DISCUSSION
Because of the degeneracy of the genetic code and the high degree of EV sequence diversity, it is more efficient to conceive of molecular diagnostic targets in the coding region as conserved amino acid sequences from which oligonucleotide primer sequences may be derived by back-translation. Codon degeneracy and sequence diversity, even within a serotype, necessitate the use of degenerate primers or multiple primer pairs to amplify all EV serotypes. Even with the use of multiple nondegenerate or minimally degenerate primers, it is difficult to design “pan-EV” primers that can be used for VP1 sequencing and molecular serotyping. To address this problem, our strategy has been to design degenerate primers that contain deoxyinosine residues at positions of fourfold codon degeneracy (e.g., using GGI to specify a glycine codon, which is normally GGN) (12, 13, 15, 16, 18, 19, 21). For example, the primers that we currently use to identify EV cell culture isolates (primers 292 and 222) target the conserved VP1 amino acid motifs (QT)A(AV)ETG and M(FY)(IV)PPG(AG), respectively (19). These primers amplify RNA templates from all human EV serotypes, most nonhuman EVs, and some human rhinoviruses (12, 15, 16, 18, 19, 21). Casas et al. have used a similar approach, using degenerate, deoxyinosine-containing primers to target a different part of VP1 (4).
Several PCR assays have been developed to allow EV serotype identification directly from original clinical specimens by nested amplification and sequencing of VP1 (4, 10, 31). The nested amplification format increases the sensitivity substantially, rivaling that of nested 5′-NTR assays (4, 10), so that nested VP1 assays can be used for both detection and identification. However, nonspecific amplification can sometimes occur when degenerate primers are used in a nested PCR assay, particularly when nucleic acids isolated from tissues or other high-complexity specimens are amplified. The CODEHOP primer design strategy has the advantage that most or all primers are able to initiate DNA synthesis in early PCR rounds by virtue of annealing of the degenerate core sequence to the template, which is further stabilized by base pairing by at least some of the residues in the nondegenerate clamp sequence. The clamp sequence is present in all newly synthesized molecules during subsequent rounds of amplification, enabling virtually all primer molecules to participate in amplification. The inclusion of the nonconserved consensus clamp sequences in primers AN88 and AN89 did not affect the amplification efficiency for viruses that contained mismatches in this region (data not shown), even when stringent annealing conditions were used. This is probably because the degenerate core sequence is a good match for virtually all EVs and because at least a few of the bases in the clamp are able to form base pairs and contribute to the stability of the primer-template helix in the initial rounds of PCR. Once the primers have been incorporated into the PCR products, the primers and templates match perfectly, allowing highly efficient amplification. Since its initial description (30), the CODEHOP approach has been used to amplify previously unknown gene homologs in a wide variety of plants, animals, and bacteria, as well as herpesviruses and retroviruses (29). To our knowledge, this work represents the first application of the CODEHOP strategy to the amplification of an RNA virus and the first application of the method to primary virus diagnostics. By eliminating inosine from the primers used in the second, nested PCR, our VP1 CODEHOP PCR has increased specificity versus nonviral templates and an increased sensitivity for viral targets, while it maintains the broad viral target specificity of the methods that use inosine-containing primers (4, 10).
As an example of the potential clinical/public health application of our method, we used the VP1 RT-snPCR assay to identify EVs in a diverse array of clinical specimens (Fig. 3). EVs were successfully amplified and amplicons were sequenced from cerebrospinal fluid; stool; serum; liver tissue; and rectal, nasopharyngeal, and eye swab specimens. Amplicons from all of these specimen sources yielded high-quality sequences that provided clear serotype identification of the infecting viruses. We have subsequently applied this approach to the testing of routine diagnostic specimens in parallel with more conventional real-time 5′-NTR detection assays. The approach achieved sensitivity comparable to those of conventional real-time 5′-NTR detection assays and provided virus identification without the use of cell culture (unpublished data).
Our CODEHOP VP1 RT-snPCR is a sensitive assay of broad target specificity that can successfully amplify template RNA from all known EV serotypes. Its sensitivity is superior to that of 5′-NTR RT-snPCR assays, with product detection performed with stained gels (Fig. 2), probably because of the high degree of RNA secondary structure in the 5′-NTR, which prevents complete template melting and efficient annealing of the primers. In addition to EV identification, molecular epidemiologic analysis of the sequences derived from the VP1 RT-snPCR products can also furnish clues to virus transmission pathways during epidemiologic field investigations. Bypassing cell culture in the early phases of an outbreak investigation can help to focus limited resources and allow rapid identification of epidemiologically linked cases. When more detailed virus characterization is desired, an informed choice can be made as to which specimens should be inoculated into cell culture and, in many cases, which cell lines should be used to maximize the efficiencies of these more labor-intensive methods.
REFERENCES
- 1.Barrios Olivera, J. A., L. Sarmiento Pérez, O. Valdés, P. Mas Lago, and R. Palomera Puentes. 2003. Aplicación de la secuenciación de VP1 a la identificación de enterovirus humanos. Rev. Cubana Med. Trop. 55:133-137. [PubMed] [Google Scholar]
- 2.Bolanaki, E., C. Kottaridi, P. Markoulatos, L. Margaritis, and T. Katsorchis. 2005. Nucleotide analysis and phylogenetic study of the homology boundaries of coxsackie A and B viruses. Virus Genes 31:307-320. [DOI] [PubMed] [Google Scholar]
- 3.Caro, V., S. Guillot, F. Delpeyroux, and R. Crainic. 2001. Molecular strategy for ‘serotyping’ of human enteroviruses. J. Gen. Virol. 82:79-91. [DOI] [PubMed] [Google Scholar]
- 4.Casas, I., G. F. Palacios, G. Trallero, D. Cisterna, M. C. Freire, and A. Tenorio. 2001. Molecular characterization of human enteroviruses in clinical samples: comparison between VP2, VP1, and RNA polymerase regions using RT nested PCR assays and direct sequencing of products. J. Med. Virol. 65:138-148. [PubMed] [Google Scholar]
- 5.Hamilton, M. S., M. A. Jackson, and D. Abel. 1999. Clinical utility of polymerase chain reaction testing for enteroviral meningitis. Pediatr. Infect. Dis. J. 18:533-537. [DOI] [PubMed] [Google Scholar]
- 6.Kottaridi, C., E. Bolanaki, and P. Markoulatos. 2004. Amplification of echoviruses genome regions by different RT-PCR protocols—a comparative study. Mol. Cell. Probes 18:263-269. [DOI] [PubMed] [Google Scholar]
- 7.Kottaridi, C., E. Bolanaki, N. Siafakas, and P. Markoulatos. 2005. Evaluation of seroneutralization and molecular diagnostic methods for echovirus identification. Diagn. Microbiol. Infect. Dis. 53:113-119. [DOI] [PubMed] [Google Scholar]
- 8.Manayani, D. J., R. V. Shaji, G. J. Fletcher, T. Cherian, N. Murali, N. Sathish, T. Solomon, C. Gnanamutha, and G. Sridraran. 2002. Comparison of molecular and conventional methods for typing of enteroviral isolates. J. Clin. Microbiol. 40:1069-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manzara, S., M. Muscillo, G. La Rosa, C. Marianelli, P. Cattani, and G. Fadda. 2002. Molecular identification and typing of enteroviruses isolated from clinical specimens. J. Clin. Microbiol. 40:4554-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nix, W. A., M. Berger, M. S. Oberste, B. R. Brooks, D. McKenna-Yasek, R. H. Brown, R. P. Roos, and M. A. Pallansch. 2004. Failure to detect enterovirus genome in the spinal cord of ALS patients using a sensitive, semi-nested RT-PCR method. Neurology 62:1372-1377. [DOI] [PubMed] [Google Scholar]
- 11.Norder, H., L. Bjerregaard, and L. O. Magnius. 2001. Homotypic echoviruses share aminoterminal VP1 sequence homology applicable for typing. J. Med. Virol. 63:35-44. [PubMed] [Google Scholar]
- 12.Oberste, M. S., K. Maher, M. R. Flemister, G. Marchetti, D. R. Kilpatrick, and M. A. Pallansch. 2000. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J. Clin. Microbiol. 38:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oberste, M. S., K. Maher, D. R. Kilpatrick, M. R. Flemister, B. A. Brown, and M. A. Pallansch. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oberste, M. S., K. Maher, S. M. Michele, M. Uddin, G. Belliot, and M. A. Pallansch. 2005. Enteroviruses 76, 89, 90, and 91 represent a novel group within the species human enterovirus A. J. Gen. Virol. 86:445-451. [DOI] [PubMed] [Google Scholar]
- 16.Oberste, M. S., K. Maher, and M. A. Pallansch. 2002. Molecular phylogeny and classification of the simian picornaviruses. J. Virol. 76:1244-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oberste, M. S., K. Maher, A. J. Williams, N. Dybdahl-Sissoko, B. A. Brown, M. T. Gookin, S. Peñaranda, N. G. Mishrik, M. Uddin, and M. A. Pallansch. 2006. Species-specific RT-PCR amplification of human enteroviruses: a tool for rapid species identification of uncharacterized enteroviruses. J. Gen. Virol. 87(Pt 1):119-128. [DOI] [PubMed] [Google Scholar]
- 18.Oberste, M. S., S. M. Michele, K. Maher, D. Schnurr, D. Cisterna, N. Junttila, M. Uddin, J.-J. Chomel, C.-S. Lau, W. Ridha, S. al-Busaidy, H. Norder, L. Magnius, and M. A. Pallansch. 2004. Molecular identification and characterization of two proposed new enterovirus serotypes, EV74 and EV75. J. Gen. Virol. 85:3205-3212. [DOI] [PubMed] [Google Scholar]
- 19.Oberste, M. S., W. A. Nix, K. Maher, and M. A. Pallansch. 2003. Improved molecular identification of enteroviruses by RT-PCR and amplicon sequencing. J. Clin. Virol. 26:375-377. [DOI] [PubMed] [Google Scholar]
- 20.Oberste, M. S., and M. A. Pallansch. 2005. Enterovirus molecular detection and typing. Rev. Med. Microbiol. 16:163-171. [Google Scholar]
- 21.Oberste, M. S., D. Schnurr, K. Maher, S. al-Busaidy, and M. A. Pallansch. 2001. Molecular identification of new picornaviruses and characterization of a proposed enterovirus 73 serotype. J. Gen. Virol. 82:409-416. [DOI] [PubMed] [Google Scholar]
- 22.Palacios, G., I. Casas, A. Tenorio, and C. Freire. 2002. Molecular identification of enterovirus by analyzing a partial VP1 genomic region with different methods. J. Clin. Microbiol. 40:182-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pallansch, M. A., and M. S. Oberste. 2003. Molecular detection and characterization of human enteroviruses, p. 245-257. In A. Matsumori (ed.), Cardiomyopathies and heart failure: biomolecular, infectious and immune mechanisms. Kluwer Academic Publishers, Boston, Mass.
- 24.Rakoto-Andrianarivelo, M., D. Rousset, R. Razafindratsimandresy, and F. Delpeyroux. 2002. Nouvelle méthode de typage moléculaire des entérovirus humains: caractérisation des souches malgaches “non sérotypables.” Arch. Instit. Pasteur Madagascar 68:55-58. [PubMed] [Google Scholar]
- 25.Read, S. J., K. J. M. Jeffery, and C. R. M. Bangham. 1997. Aseptic meningitis and encephalitis: the role of PCR in the laboratory. J. Clin. Microbiol. 35:691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson, C. C., M. Willis, A. Meagher, K. E. Gieseker, H. Rotbart, and M. P. Glodé. 2002. Impact of rapid polymerase chain reaction results on management of pediatric patients with enteroviral meningitis. Pediatr. Infect. Dis. J. 21:283-286. [DOI] [PubMed] [Google Scholar]
- 27.Romero, J. R. 1999. Reverse-transcription polymerase chain reaction detection of the enteroviruses. Arch. Pathol. Lab. Med. 123:1161-1169. [DOI] [PubMed] [Google Scholar]
- 28.Rose, T. M. 2005. CODEHOP-mediated PCR—a powerful tool for the identificcation and characterization of viral genomes. Virol. J. 2:20-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose, T. M., J. G. Henikoff, and S. Henikofff. 2003. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 31:3763-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thoelen, I., P. Lemey, I. Van der Donck, K. Beuselink, A. M. Lindberg, and M. Van Ranst. 2003. Molecular typing and epidemiology of enteroviruses identified from an outbreak of aseptic meningitis in Belgium during the summer of 2000. J. Med. Virol. 70:420-429. [DOI] [PubMed] [Google Scholar]
- 32.Vuorinen, T., R. Vainionpää, and T. Hyypiä. 2003. Five years' experience of reverse-transcriptase polymerase chain reaction in daily diagnosis of enterovirus and rhinovirus infections. Clin. Infect. Dis. 37:452-455. [DOI] [PubMed] [Google Scholar]
- 33.World Health Organization. 2001. Manual for the virological investigation of polio WHO/EPI/GEN/97.01. World Health Organization, Geneva, Switzerland.