Abstract
Mutants of Candida albicans, Candida glabrata, and Saccharomyces cerevisiae with disruptions in the ACE2 gene and C. glabrata and S. cerevisiae swi5 disruption mutants were tested for virulence in a murine challenge model of disseminated yeast infection. All mutants showed a clumping phenotype, but clumping was minimized in challenge inocula by inclusion of chitinase in the growth medium. In animals rendered temporarily neutropenic by cyclophosphamide treatment, the C. glabrata ace2 null mutant was confirmed as hypervirulent: it led to early terminal illness and kidney, brain, and lung fungal burdens substantially and significantly larger than those in controls. The C. glabrata swi5 null mutant did not lead to terminal illness but generated significantly larger brain and lung burdens than those in controls. The C. albicans ace2 null mutant was very slightly attenuated and the S. cerevisiae ace2 and swi5 null mutants were substantially attenuated relative to their parental control strains. The phenotype of aggressive hypervirulence, unique to disruption of the C. glabrata ACE2 gene among the strains tested, was not seen when the C. glabrata ace2 strain was tested in immunologically intact mice. The different effects seen with these mutants rule out the clumping phenotype as the explanation for hypervirulence in the C. glabrata ace2 mutant. The absence of C. glabrata ace2 hypervirulence in healthy mice may be a tool for definitive future study of host-parasite cross talk in microbial opportunism.
In the generally nonpathogenic yeast Saccharomyces cerevisiae, two transcription factors, Ace2p and Swi5p, with 37% identity, regulate the expression of several genes during the late M1 and early G1 stages of the mitotic cell cycle (5, 17). Targets of Ace2p only are predominantly expressed in the daughter cell, and they include chitinase (encoded by CTS1) and other proteins required for separation from the mother cell (4, 5, 23). Deletion of ACE2 results in increased pseudohyphal growth and invasion of agar (11, 16). Swi5p alone regulates the expression of the HO endonuclease, which initiates mating-type switching (5, 15).
Ace2p and Swi5p are both conserved in species closely related to S. cerevisiae, including the opportunistic pathogenic yeast Candida glabrata (6). The Candida albicans genome, however, contains only a single gene orthologue (C. albicans ACE2), whose product is equidistant from both S. cerevisiae proteins (10). Deletion of ACE2 from C. albicans results in gross attenuation of virulence of the fungus in a mouse model of disseminated infection (10). In contrast, deletion of C. glabrata ACE2 results in hypervirulence of the mutant strain in neutropenic mice (9) and in alterations in levels of more than 60 proteins (19). Because these two Candida species have evolved to exist as human commensals, it is possible that centrally important regulatory factors such as the Ace2p family may have adapted to play a role in the regulation of molecules essential for survival in the mammalian milieu. For both species, the ace2 mutants show a phenotype in vitro which is similar to that of S. cerevisiae deletion mutants: the yeast cells grow as clumped forms, and the expression of several cell wall proteins is reduced (9).
We sought to determine if the enhanced virulence caused by deleting ACE2 is unique to C. glabrata. Phylogenetically, C. glabrata lies closer to S. cerevisiae than to C. albicans (3, 20), which raises the possibility that deleting ACE2 or SWI5 from S. cerevisiae may also lead to enhanced virulence for mice. We therefore compared the effects of specific disruption of SWI5 in S. cerevisiae and C. glabrata and of ACE2 in S. cerevisiae, C. glabrata, and C. albicans on virulence for immunocompetent and temporarily neutropenic mice, using identical conditions for preparation of the inocula of mutant and control strains.
MATERIALS AND METHODS
Yeast isolates and mutants.
The C. glabrata ACE2 (HLS121), C. glabrata ace2 (HLS122), C. albicans ace2 (MK106), C. albicans SC5314, and C. albicans CAI-4 (with URA3 from CIp10 [14] integrated at the RPS1 locus) strains have all been described for animal experiments in previous publications (1, 9, 10, 12). The CAI-4 Ura+ strain shows virulence in the mouse model which is indistinguishable from that of its parent (SC5314) and a URA3 heterozygous mutant (CAF-2) (12). The pathogenic S. cerevisiae isolate YJM145 (8, 13) was the parental strain for ACE2 and SWI5 deletions.
Culture media.
YEPD comprised Bacto peptone (Becton Dickinson, Cowley, Oxford, United Kingdom) at 20 g/liter, yeast extract (Becton Dickinson) at 10 g/liter, and glucose at 20 g/liter. Sabouraud broth comprised mycological peptone (Oxoid, Basingstoke, United Kingdom) at 10 g/liter and glucose at 40 g/liter. NGY comprised Neopeptone (Becton Dickinson) at 1 g/liter, glucose at 5 g/liter, and yeast extract at 1 g/liter. SPO1 buffer comprised potassium acetate at 10 g/liter, yeast extract at 1 g/liter, and glucose at 0.5 g/liter.
To produce challenge yeast inocula free of cell clumps for animal experiments, two alternative approaches were investigated. For the first approach, the test strains were grown for 18 h in NGY at 30°C with constant rotation at 20 rpm. Yeast cells were centrifuged at 2,500 × g for 5 min, resuspended in sterile saline, and then treated with chitinase (Sigma, Poole, Dorset, United Kingdom) (9). For the second approach, chitinase was included at 2 U/ml in the NGY medium for the duration of growth.
Construction of ACE2 and SWI5 null mutants of S. cerevisiae.
A 1.3-kb fragment from the KanMX gene cassette was amplified from pFA6-kanMX2 (21), using primers BMDACE2-F and BMDACE2-R for ACE2 and KOS-1 and KOS-2 for SWI5 (Table 1). Both primer pairs were designed to remove the entire open reading frame of ACE2 or SWI5. The deletion cassettes were transformed into S. cerevisiae YJM145 (13) by the TRAFO method (7).
TABLE 1.
Oligonucleotide primers used for ACE2 and SWI5 deletions in S. cerevisiae
| Primer | Sequencea |
|---|---|
| BMDACE2-F | ATGGATAACGTTGTAGATCCGTGGTATATAAATCCCAGCTGAAGCTTCGTACGC |
| BMDACE2-R | TCAGAGAGCATCAGTTTCGTTTGAAAGGGTGCGGTTGCATAGGCCACTAGTGGATCTG |
| ACE2-TEST-F | CTCAAGCAACAGTTAAAGTGC |
| ACE2-TEST-R | CCACCTCTTCTTGCTCTTCTA |
| KAN-2 | TGCGCCTGAGCGAGACGA |
| KOS-1 | ACAATATAGATACCACCATATAACTCGTTCTTGGAGCTAGCAGCTGAAGCTTCGTACGC |
| KOS-2 | AAAGTGTTCATACTATATTATTTACTTAGATTATACATAAGCATAGGCCACTAGTGGATCTG |
| KOS-3 | AGAGCGAATCAAATATCGCAG |
| KOS-5 | CATAATCCCCGTTCCATTCC |
Underlined sequences were derived from the pFA6-kanMX2 multiple cloning site or from the C. glabrata HIS3 gene.
Disruption of ACE2 or SWI5 in confirmed G418-resistant transformants was achieved with diagnostic PCR, using the primers ACE2-TEST-F, ACE2-TEST-R, and KAN-2 or KOS-3, KOS-5, and KAN-2 (Table 1), respectively. To disrupt both alleles of ACE2 or SWI5 in the diploid S. cerevisiae isolate YJM145, suitable heterozygous deletion transformants were induced to sporulate in SPO1 buffer at 25°C for 4 to 6 days. Tetrads were dissected with the aid of a Singer MSM microscope. Since YJM145 is a homothallic isolate, haploid spores spontaneously undergo mating-type switching and immediately form diploids. Homozygous deletions (derived from two of four tetrads) were verified by PCR as described above. Single representative ACE2 (BMEY-5) and SWI5 (YCK2) knockouts were chosen for subsequent analysis.
Construction of SWI5 null mutant of C. glabrata.
The C. glabrata HIS3 gene was amplified using primers SWI5-HIS-F and SWI5-HIS-R. These primers include 60 bp of SWI5 flanking sequences and were designed to delete the entire open reading frame. The purified deletion cassette was used to transform C. glabrata ΔH1 (22). Thirty histidine prototrophs were selected and analyzed by PCR. Primers SWI5-FT and SWI5-RT were used to distinguish SWI5 wild-type cells (generating a PCR product of 2.4 kb) from swi5Δ mutant cells (generating a PCR product of 1.5 kb). The primer combination of SWI5-FT and HIS-T (generating a product of 1.3 kb) confirmed that the SWI5 locus was replaced with HIS3. Three colonies were detected that had undergone the desired recombination event. One was selected and named CK1-1.
Mouse model.
All animal experimentation was done in accordance with UK Home Office regulations and was approved by both the Home Office and an institutional ethical review committee. Female BALB/c mice (Harlan, United Kingdom) with a weight range from 17 to 23 g were maintained in groups of up to 12 animals per cage. The mice were supplied with food and water ad libitum. To induce temporary neutropenia, mice were injected intraperitoneally with cyclophosphamide at 150 mg/kg of body weight 3 days before challenge and on the day of challenge. Cell densities were measured with a hemocytometer, and yeast cells, treated with chitinase during growth or after growth, were centrifuged and resuspended a second time to produce a concentration that allowed delivery of the challenge inoculum in a volume not exceeding 100 μl. Viable counts in the inocula were determined by dilution plating of the suspensions.
Mice were challenged intravenously via the lateral tail vein. The size of the challenge was determined from published experimentation with C. albicans (10, 12) and C. glabrata (9) and in pilot experiments with small groups of mice for S. cerevisiae. Mice were weighed individually every day and were humanely terminated when body weight fell >20% below the weight at the time of infection or the animals showed signs of serious illness. Dates of demise or termination of animals were used to determine survival after challenge. At the time of death, the left kidney and the brain were dissected with aseptic precautions and homogenized in 0.5-ml volumes of sterile water, and tissue burdens were determined by viable counting on Sabouraud agar. In some experiments, lung burdens were also determined. The detection limits were 2.3, 2.2, and 2.1 log CFU/g tissue for lungs, brains, and kidneys, respectively. Negative tissues were recorded as having burdens 0.5 log lower than these figures for inclusion in statistical analyses.
Data analysis.
Differences in survival data from Kaplan-Meier curves were assessed statistically by the log rank test. Differences in tissue burdens were analyzed with the nonparametric Mann-Whitney U test. In all cases, a difference was regarded as significant when the P value associated with the statistic was <0.05.
RESULTS
Influence of chitinase treatment on infection outcomes.
No significant differences were found in survival curves or mean kidney fungal burdens determined for groups of six cyclophosphamide-treated mice challenged with C. glabrata suspensions treated with chitinase after growth or grown with chitinase added to the medium (Table 2). Microscopic examination of challenge inocula of both Candida species showed no difference in appearance of the cells: both methods of chitinase treatment effectively reduced cell clumps to pairs or occasional tetrads. For all subsequent experiments, inocula were grown in the presence of chitinase.
TABLE 2.
Effects of chitinase treatment applied to fungal cell suspensions after growth and during growth on the outcome of intravenous challenge experiments with micea
| Fungal strain | Time of chitinase treatment | Challenge dose (CFU/g) | MST (days)b | MKB (log CFU)c |
|---|---|---|---|---|
| C. glabrata HLS121 | After growth | 2.0 × 106 | 28.0 ± 0.0 | 5.4 ± 0.6 |
| During growth | 2.0 × 106 | 24.0 ± 9.8 | 6.4 ± 1.6 | |
| C. glabrata HLS122 | After growth | 1.5 × 106 | 2.0 ± 0.0 | 5.6 ± 0.7 |
| During growth | 1.5 × 106 | 2.0 ± 0.0 | 6.0 ± 0.8 | |
| C. albicans SC5314 | After growth | 1.6 × 104 | 4.0 ± 2.4 | 5.8 ± 0.9 |
| During growth | 1.6 × 104 | 3.0 ± 0.9 | 6.7 ± 0.4 |
All experiments were done with groups of six mice. Mice challenged with C. glabrata isolates were pretreated with cyclophosphamide to induce temporary neutropenia.
MST, mean survival time.
MKB, mean kidney burden of fungi.
Virulence for mice of ace2 and swi5 mutants of C. albicans, C. glabrata, and S. cerevisiae.
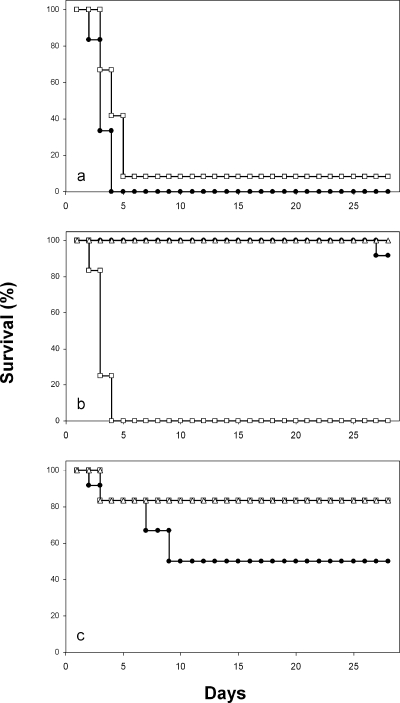
Temporarily neutropenic mice infected with C. albicans CAI-4 containing CIp10 survived with a similar curve to that for mice infected with the ace2 null strain MK106. The small difference was statistically significant by log rank analysis (P < 0.001) (Fig. 1a) but was the consequence of prolonged survival of a single animal. The difference in mean survival times (Table 3) was only 4 days. Tissue burdens of viable C. albicans (Table 3) for mice challenged with the two strains indicated that mean kidney and lung burdens were slightly but significantly (P < 0.05) smaller in animals infected with the ace2 null mutant than in mice infected with the CAI-4 control. In terms of both survival and two of three sets of tissue burden data, the ace2 null mutant, given at the same level of challenge as its parental strain, showed a very minor though statistically significant level of virulence attenuation. The significant differences remained even when data for the MK106-infected mouse were excluded from analysis.
FIG. 1.
Survival curves for temporarily neutropenic mice challenged with (a) C. albicans CAI-4 plus CIp10 (filled circles) and C. albicans MK106 (ace2 null) (open squares); (b) C. glabrata HLS121 (control) (filled circles), C. glabrata HLS122 (ace2 null) (open squares), and C. glabrata CK1-1 (swi5 null) (open triangles); and (c) S. cerevisiae YJM145 (filled circles), S. cerevisiae BMEY-5 (ace2 null) (open squares), and S. cerevisiae YCK2 (swi5 null) (open triangles).
TABLE 3.
Summary of virulence parameters for temporarily neutropenic mice challenged intravenously with three fungal species and their ace2 or swi5 null mutants
| Fungal strain | Challenge dose (CFU/g body wt) | No. of animals tested | Mean survival time (days) ± SD | Mean burden (log CFU/g) ± SD
|
||
|---|---|---|---|---|---|---|
| Kidneys | Brain | Lungs | ||||
| C. albicans CAI-4 + CIp10 | 1.5 × 104 | 12 | 2.1 ± 0.7 | 6.7 ± 0.3 | 4.9 ± 0.6 | 4.7 ± 0.7 |
| C. albicans MK106 (ace2 null) | 1.6 × 104 | 12 | 6.0 ± 7.0 | 6.0 ± 1.0 | 4.5 ± 0.7 | 3.7 ± 0.8 |
| C. glabrata HLS121 (control) | 1.2 × 106 | 12 | 27.9 ± 0.3 | 3.4 ± 1.1 | 2.0 ± 0.5 | 2.5 ± 0.5 |
| C. glabrata HS122 (ace2 null) | 1.6 × 105 | 12 | 3.0 ± 0.6 | 4.8 ± 0.4 | 5.9 ± 0.3 | 6.0 ± 0.4 |
| C. glabrata CK1-1 (swi5 null) | 1.3 × 105 | 12 | 28.0 ± 0.0 | 4.0 ± 0.7 | 3.7 ± 0.3 | 4.0 ± 0.7 |
| S. cerevisiae YJM145 (parent) | 4.2 × 105 | 6 | 9.7 ± 9.5 | 5.5 ± 1.9 | 4.8 ± 1.7 | 5.3 ± 1.0 |
| S. cerevisiae BMEY-5 (ace2 null) | 4.2 × 105 | 6 | 23.8 ± 10.2 | 5.0 ± 1.8 | 3.2 ± 0.8 | 5.9 ± 1.1 |
| S. cerevisiae YCK2 (swi5 null) | 4.2 × 105 | 6 | 23.8 ± 10.2 | 3.4 ± 1.2 | 3.5 ± 0.7 | 5.5 ± 0.3 |
All but one of the mice challenged with the C. glabrata control strain (HLS121) survived until 28 days after challenge, unlike mice infected with the ace2 null strain (HLS122), which all died by day 4 (Fig. 1b) (P < 0.001 versus control), even though the challenge dose was 10 times lower than that of the control strain, thus confirming the hypervirulence of this mutant. All mice challenged with the C. glabrata swi5 null strain (CK1-1) at the same dose as that of HLS122 survived for 28 days after challenge, essentially the same result as that for the control strain (Fig. 1b). Data for tissue burdens of C. glabrata (Table 3) confirmed the hypervirulence of the ace2 null mutant, which was already evident from the survival rates (Fig. 1b). For HLS122, the mean fungal burdens in kidneys (P = 0.002), brains (P < 0.001), and lungs (P < 0.001) were all significantly larger than those of the control strain HLS121, even though the challenge dose was approximately 10 times lower. These results paralleled the short survival times for mice challenged with this strain. For the swi5 mutant, also given as a challenge at a 10-fold lower dose than that of the control strain, the mean burdens in brains and lungs were significantly larger than those of controls (P < 0.001), while the mean kidney burden was not significantly different from that of controls. The C. glabrata swi5 null mutant's mean brain and lung burdens were also significantly lower than the corresponding means for the ace2 null mutant.
Groups of mice challenged intravenously with 1.0 × 105 to 1.6 × 105 yeast cells of wild-type S. cerevisiae (pathogenic isolate YJM145) or the ace2 (BMEY5) or swi5 (YCK2) null mutant all survived to day 28 and had negative cultures for kidney, brain, and lung burdens at postmortem (details not shown). With a higher challenge dose of 4.2 × 105 yeast cells/g, S. cerevisiae YJM145 led to fatal disease in 50% of a group of six mice (Fig. 1c), while five of six mice survived the same dose of the ace2 and swi5 null mutants (Fig. 1c). These survival differences were significant by the log rank test (P = 0.039). None of the tissue burden differences between S. cerevisiae control and mutant strains reached statistical significance, even though mean kidney and brain burdens were >10-fold smaller in animals challenged with the swi5 null mutant than in control animals.
Effects of C. albicans and C. glabrata ace2 null mutants in immunologically normal BALB/c mice.
Because our previous work with the C. albicans ace2 null mutant was done with DBA/2 mice, not the BALB/c mouse strain used in the present study, we challenged six unmodified BALB/c mice with the MK106 (ace2 null) mutant grown in the presence of chitinase at a dose of 1.9 × 104 CFU/g. All mice survived to 28 days postchallenge. Five of six kidney samples and all six brain samples were negative for C. albicans at postmortem examination. These data confirm the attenuation of the ace2 mutant seen in DBA/2 mice.
We challenged three pairs of healthy BALB/c mice with the C. glabrata ace2 and swi5 null mutants at doses of 3 × 104, 3 × 105, and 3 × 106 CFU/g. At the two lowest doses of the ace2 null mutant, all mice survived to day 28. At 3 × 106 CFU/g, one of the mice was terminated 19 days after challenge, and the other survived to day 28. Kidneys were negative for C. glabrata for both animals challenged with 3 × 104 CFU/g; one animal challenged with 3 × 105 CFU/g had a positive kidney result, with 4.7 log CFU/g, and the mean kidney burden (± standard deviation) for the two mice challenged with 3.3 × 106 CFU/g was 4.5 ± 0.1 log CFU/g. All animals survived to day 28 with all three doses of the swi5 null mutant. The mean kidney burdens for the pairs of mice were 3.5 ± 0.1, 3.9 ± 0.1, and 4.0 ± 0.4 log CFU/g for doses of 3 × 104, 3 × 105, and 3 × 106 CFU/g, respectively. No evidence suggestive of hypervirulence was obtained for either mutant in these experiments with immunologically intact mice.
DISCUSSION
We undertook this study to determine if deletion of ACE2 or SWI5 from S. cerevisiae leads to hypervirulence in the immunocompromised mouse model, in a manner analogous to that after deletion of ACE2 from C. glabrata (9), since these two fungal species are quite closely related and C. glabrata may have lost its mating ability relatively recently (2). However, the results of our experiments show that the consequence of ACE2 and SWI5 deletion in S. cerevisiae is a reduction in mouse virulence from the already low virulence of the parental strain. We used a segregant from a clinical isolate of S. cerevisiae (YJM145) that was previously shown to colonize the organs of infected mice (8). The ACE2 and SWI5 genes were replaced with dominant drug resistance markers that have no effect on virulence (8). Our S. cerevisiae challenge dose was more than twice as high as that used with the hypervirulent ace2 null mutant of C. glabrata and approximately 30 times higher than the challenge dose used for C. albicans strains, so it is unlikely that any change in the direction of hypervirulence in the S. cerevisiae ace2 and swi5 null mutants would have been missed in our experiments.
The results for the swi5 null strain of C. glabrata make an interesting comparison with those for the C. glabrata ace2 null strain. Only the latter demonstrated hypervirulence by causing severe illness in infected mice that appeared as reduced survival. The mean brain and lung burdens of the C. glabrata swi5 mutant at postmortem were substantially (1.5 log) and significantly (P < 0.001) larger than those in mice challenged with a 10 times higher dose of the C. glabrata control strain, but kidney burdens did not differ (Table 3). These mean burdens were also smaller, at similar orders of magnitude and the same statistical significance, than the brain and lung burdens in mice challenged at the same level with the ace2 null mutant. By these criteria, the swi5 null mutant has only moderate virulence effects in temporarily neutropenic mice. The terminal illness resulting from C. albicans infection is due to the development of sepsis, with kidney pathology being a primary contributor to terminal events and the kidney burden being directly correlated with renal failure (18). From our data, it seems reasonable to conclude that the aggressive hypervirulence of the C. glabrata ace2 null mutant results from the development of large kidney burdens combined with unusually large burdens in the brain. The contribution of lung burdens to gross symptoms may be less important or negligible, since the S. cerevisiae strains tested also achieved large postmortem lung burdens without leading to overt symptomatology. Compared with the control strain, the C. glabrata swi5 null mutant generates high brain burdens but does not achieve renal dysfunction sufficient to lead to terminal sepsis. Since the ace2 and swi5 null mutants of all three fungal species have a clumping phenotype, yet obvious differences were found in the pathological consequences of their infections, our data support the interpretation that the C. glabrata ace2 mutant's increased virulence is not due to the clumpy growth phenotype but rather is a specific result of some other attribute associated with ACE2 inactivation.
Our aim was to compare the virulence of ace2 and swi5 null mutants of three fungal species under conditions that were as identical as possible. This included preparing all challenge inocula identically, with chitinase included in the growth medium to minimize the clumping phenotype of the mutants, and conducting all tests in temporarily neutropenic mice. We adjusted challenge doses species by species: the wild-type strains of C. glabrata and S. cerevisiae are inherently less virulent than the C. albicans wild type, so identical challenge inocula were not possible in the experimental design if we wished to detect possible attenuation in the mutants of the less virulent species. The results of the comparison show that in the immunosuppressed mouse host, the C. albicans ace2 null mutant is slightly attenuated in virulence, the C. glabrata ace2 null mutant is notably hypervirulent, the C. glabrata swi5 null mutant gives larger lung and brain burdens than those of the control, but without consequences for survival, and the S. cerevisiae ace2 and swi5 null mutants are attenuated.
It is perhaps worth emphasizing that challenges with all three strains of S. cerevisiae in the immunosuppressed animals led to lung burdens that were of the same order of magnitude as those achieved by the hypervirulent C. glabrata ace2 null mutant (Table 2) and larger than the burdens seen with either of the C. albicans strains tested. It is unclear why cyclophosphamide-treated mice should be less effective at removing C. glabrata and S. cerevisiae from the lungs than they are with C. albicans. The effect might be a technical artifact—a consequence of C. albicans forming hyphae in the lungs which artificially reduce viable counts or of CFU counts being affected by the clumping phenotype—and quantitative PCR may be needed to obtain a more accurate estimate of pulmonary fungal burdens.
The C. albicans ace2 null mutant was previously shown to be highly attenuated in mouse virulence compared with its parent (10). In the previous study, immunologically unaltered DBA/2 mice were used as hosts. This mouse strain is inherently more susceptible to C. albicans challenge than are the BALB/c mice used in the present experiments (12), so we retested the mutant at an appropriate intravenous challenge dose in immunologically normal BALB/c mice. The data replicated the nearly total attenuation of the null mutant in an intact host. However, in our main experiments, where the parent and null mutant strains of three fungal species were compared in mice rendered neutropenic at the time of challenge, only a low degree of attenuation was found with the C. albicans ace2 null construct (Fig. 1a; Table 3), emphasizing the importance of host status in susceptibility to C. albicans challenge. Our tests with the C. glabrata ace2 and swi5 mutants in a small number of immunologically unaltered mice gave no evidence of the hypervirulence phenomenon. We concluded that the positive and negative virulence differences seen in challenge experiments of the type we have described are very highly influenced by the host immune status. The differences we have found in this study provide a strong basis for detailed investigation of the differences in immune recognition of and response to C. glabrata strains.
The observation that hypervirulence was seen only in temporarily neutropenic hosts is particularly notable in the context of studies already done with the C. glabrata ace2 mutant. A search for gene products that might account for hypervirulence revealed more than 60 proteins which are altered significantly in quantity in response to the presence or absence of Ace2p (19). This result confirms the central regulatory role played by Ace2p in the growth and development of C. glabrata, but it also suggests that the hypervirulence noted for the mutant in the immunosuppressed mouse model may result from changes in expression of more than one gene. If transcript profiling in vivo can be achieved with current models, it may become possible to pinpoint the most significant genes that lead to hypervirulence in immunosuppression.
We undertook these experiments in collaboration to establish the comparative virulence of the mutants in a single experimental program. Our individual laboratories work separately with the different species involved and are currently testing individual hypotheses for the virulence differences seen. The very different consequences of ACE2 disruption between C. albicans and C. glabrata show that these species are handled differently by murine immune defenses. The absence of C. glabrata ace2 mutant hypervirulence in normal mice and the relative attenuation of the swi5 mutant in normal and immunosuppressed mice may be tools for definitive future studies of microbial opportunism.
Acknowledgments
We thank John McCusker for providing the clinical isolate of S. cerevisiae and Brenda McEvoy for help in constructing the deletion strains.
This work was supported by the Health Research Board, The Wellcome Trust (072420 and 076954), and the BBSRC (BBS/B/10331).
Editor: A. Casadevall
REFERENCES
- 1.Brand, A., D. M. MacCallum, A. J. P. Brown, N. A. R. Gow, and F. C. Odds. 2004. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Mol. Microbiol. 3:900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler, G., C. Kenny, A. Fagan, C. Kurischko, C. Gaillardin, and K. H. Wolfe. 2004. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc. Natl. Acad. Sci. USA 101:1632-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai, J., I. N. Roberts, and M. D. Collins. 1996. Phylogenetic relationships among members of the ascomycetous yeast genera Brettanomyces, Debaryomyces, Dekkera and Kluyveromyces deduced by small-subunit rRNA gene sequences. Int. J. Syst. Bacteriol. 46:542-549. [DOI] [PubMed] [Google Scholar]
- 4.Colman-Lerner, A., T. E. Chin, and R. Brent. 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107:739-750. [DOI] [PubMed] [Google Scholar]
- 5.Doolin, M.-T., A. L. Johnson, L. H. Johnston, and G. Butler. 2001. Overlapping and distinct roles of the duplicated transcription factors Ace2p and Swi5p. Mol. Microbiol. 40:422-432. [DOI] [PubMed] [Google Scholar]
- 6.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 430:35-44. [DOI] [PubMed] [Google Scholar]
- 7.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein, A. L., and J. H. McCusker. 2001. Development of Saccharomyces cerevisiae as a model pathogen: a system for the genetic identification of gene products required for survival in the mammalian host environment. Genetics 159:499-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamran, M., A. M. Calcagno, H. Findon, E. Bignell, M. D. Jones, P. Warn, P. Hopkins, D. W. Denning, G. Butler, T. Rogers, F. A. Muhlschlegel, and K. Haynes. 2004. Inactivation of transcription factor gene ACE2 in the fungal pathogen Candida glabrata results in hypervirulence. Eukaryot. Cell 3:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly, M. T., D. M. MacCallum, S. D. Clancy, F. C. Odds, A. J. P. Brown, and G. Butler. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol. Microbiol. 53:969-983. [DOI] [PubMed] [Google Scholar]
- 11.King, L., and G. Butler. 1998. Ace2p, a regulator of Cts1 (chitinase) expression, affects pseudohyphal production in Saccharomyces cerevisiae. Curr. Genet. 34:183-191. [DOI] [PubMed] [Google Scholar]
- 12.MacCallum, D. M., and F. C. Odds. 2005. Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses 48:151-161. [DOI] [PubMed] [Google Scholar]
- 13.McCusker, J. H., K. V. Clemons, D. A. Stevens, and R. W. Davis. 1994. Genetic characterization of pathogenic Saccharomyces cerevisiae isolates. Genetics 136:1261-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murad, A. M. A., P. R. Lee, I. D. Broadbent, C. J. Barelle, and A. J. P. Brown. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325-327. [DOI] [PubMed] [Google Scholar]
- 15.Nasmyth, K., G. Adolf, D. Lydall, and A. Seddon. 1990. The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SW15 nuclear entry. Cell 62:631-647. [DOI] [PubMed] [Google Scholar]
- 16.O'Conallain, C., M. T. Doolin, C. Taggart, F. Thornton, and G. Butler. 1999. Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol. Gen. Genet. 262:275-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon, I., J. Barnett, N. Hannett, C. T. Harbison, N. J. Rinaldi, T. L. Volkert, J. J. Wyrick, J. Zeitlinger, D. K. Gifford, T. S. Jaakkola, and R. A. Young. 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106:697-708. [DOI] [PubMed] [Google Scholar]
- 18.Spellberg, B., A. S. Ibrahim, J. E. Edwards, and S. G. Filler. 2005. Mice with disseminated candidiasis die of progressive sepsis. J. Infect. Dis. 192:336-343. [DOI] [PubMed] [Google Scholar]
- 19.Stead, D., H. Findon, Z. K. Yin, J. Walker, L. Selway, P. Cash, B. A. Dujon, C. Hennequin, A. J. P. Brown, and K. Haynes. 2005. Proteomic changes associated with inactivation of the Candida glabrata ACE2 virulence-moderating gene. Proteomics 5:1838-1848. [DOI] [PubMed] [Google Scholar]
- 20.Tavanti, A., A. D. Davidson, N. A. R. Gow, M. C. J. Maiden, and F. C. Odds. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43:284-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 22.Weig, M., K. Haynes, T. R. Rogers, O. Kurzai, M. Frosch, and F. A. Muhlschlegel. 2001. A GAS-like gene family in the pathogenic fungus Candida glabrata. Microbiology 147:2007-2019. [DOI] [PubMed] [Google Scholar]
- 23.Weiss, E. L., C. Kurischko, C. Zhang, K. Shokat, D. G. Drubin, and F. C. Luca. 2002. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158:885-900. [DOI] [PMC free article] [PubMed] [Google Scholar]