Abstract
To survive in a host environment, microbial pathogens must sense local conditions, including nutrient availability, and adjust their growth state and virulence functions accordingly. No comprehensive investigation of growth phase-related gene regulation in Bordetella pertussis has been reported previously. We characterized changes in genome-wide transcript abundance of B. pertussis as a function of growth phase and availability of glutamate, a key nutrient for this organism. Using a Bordetella DNA microarray, we discovered significant changes in transcript abundance for 861 array elements during the transition from log phase to stationary phase, including declining transcript levels of many virulence factor genes. The responses to glutamate depletion exhibited similarities to the responses induced by exit from log phase, including decreased virulence factor transcript levels. However, only 23% of array elements that showed at least a fourfold growth phase-associated difference in transcript abundance also exhibited glutamate depletion-associated changes, suggesting that nutrient limitation may be one of several interacting factors affecting gene regulation during stationary phase. Transcript abundance patterns of a Bvg+ phase-locked mutant revealed that the BvgAS two-component regulatory system is a key determinant of growth phase- and nutrient limitation-related transcriptional control. Several adhesin genes exhibited lower transcript abundance during stationary phase and under glutamate restriction conditions. The predicted bacterial phenotype was confirmed: adherence to bronchoepithelial cells decreased 3.3- and 4.4-fold at stationary phase and with glutamate deprivation, respectively. Growth phase and nutrient availability may serve as cues by which B. pertussis regulates virulence according to the stage of infection or the location within the human airway.
During the course of infection, bacterial pathogens encounter a variety of challenging conditions in the host, including scarcity of carbon, nitrogen, iron, and other nutrients; fluctuations in pH; oxidative stress; and adverse conditions produced by the host innate and adaptive immune responses. In order to succeed, microorganisms must be able to modify their physiological state and virulence phenotype in accordance with changing features within the host, in a reciprocal and dynamic fashion. Growth phase-associated regulation is inherently related to pathogenicity, for in order to survive, bacteria must alter their growth rate and metabolic activity appropriately, according to cues in the host environment. Entry into stationary phase, defined as the moment when the bacterial growth rate begins to decline, is accompanied by development of cross-resistance to multiple stressors, promoting survival (34). In addition, studies have identified correlations between bacterial virulence and growth phase (22, 38).
No comprehensive study of growth phase-associated regulation has been performed with the obligate human pathogen Bordetella pertussis, the causative agent of whooping cough. The most well-characterized regulatory system in B. pertussis and in the closely related respiratory pathogens B. bronchiseptica and B. parapertussis is the BvgAS two-component phosphorelay system (3, 7). Growth at 37°C in low concentrations of nicotinic acid (NA) and sulfate leads to the Bvg+ phase, in which BvgAS activates expression of most known virulence factors. When the bordetellae are grown at low temperatures (below 26°C) or in the presence of millimolar concentrations of NA or sulfate, BvgAS is inactivated, resulting in the Bvg− phase, in which transcription of most virulence factor genes is repressed and the expression of other factors is induced. Intermediate phases, called Bvgi (Bvg intermediate), occur transiently during shifts between the Bvg+ and Bvg− states and during growth in concentrations of NA or sulfate below those that induce the Bvg− phase (10). Bvg− phase-locked mutants of B. bronchiseptica, which has a broad mammalian host range and is capable of survival ex vivo, show improved survival when they are subjected to nutrient deprivation (8). In addition to controlling virulence factor expression, BvgAS also regulates transcription of a number of metabolic pathway genes in B. bronchiseptica but not in B. pertussis (10).
Although Bvg-mediated responses of B. pertussis under in vitro conditions have been studied extensively, little is known about how conditions within the human airway and the organism's growth state affect control of virulence factor expression or other key cellular functions in B. pertussis, either via the Bvg system or by other mechanisms. We examined the global transcriptional profile of B. pertussis as it varied with growth phase and in response to nutrient limitation, which is postulated to be a critical stressor during stationary phase in the host environment. We hypothesized that virulence genes would be among the genes exhibiting varying transcript abundance levels as a function of growth phase and nutrient availability, in keeping with the organism's need to coordinate virulence functions with its environment and resulting physiologic state. Through use of a Bvg+ phase-locked mutant, we examined the involvement of BvgAS in these gene expression programs. We also investigated how a crucial B. pertussis virulence phenotype, adherence, varies according to growth phase and nutritional status.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Tohama I (16) and GMT-1 (23) are wild-type clinical isolates from the 1950s and 1990s, respectively. Tohama I is the B. pertussis strain for which a complete genome sequence has been reported (29). Tohama I-Sc3 is a Bvg+ phase-locked mutant derived by a single-nucleotide substitution in bvgS that renders its product constitutively active (28). Tohama I ΔfhaB was derived by complete replacement of fhaB with a chloramphenicol resistance gene. The design and method used for construction of this mutation were not expected to cause disruption of the promoter for the downstream FHA and fimbria accessory genes, although transcription of these genes may still be affected by alterations in DNA structure. Strains were grown at 37°C on Bordet Gengou (BG) agar supplemented with 15% sheep blood (BD Biosciences, San Jose, CA) or in modified Stainer-Scholte liquid medium (SSM) (14) at 37°C with shaking. Beta-hemolysis on BG agar was verified following growth in liquid cultures to ensure that bacteria remained Bvg+ and were not spontaneous Bvg− mutants.
Growth phase time course experiments.
Mid-log-phase cultures were diluted in SSM to obtain a starting optical density at 600 nm (OD600) of 0.03 to 0.05, in duplicate or triplicate. Growth was monitored via measurement of OD600, and culture aliquots were collected at multiple times for counting of CFU, measurement of the DNA concentration, and RNA isolation.
Glutamate limitation experiments.
Duplicate mid-log-phase cultures were washed twice and resuspended at a starting OD600 of 0.5 in three medium preparations: SSM, SSM made with one-half the usual amount of supplementary sodium glutamate in the standard medium, and SSM made with none of the supplementary sodium glutamate in the standard medium (resulting in a final glutamate concentration of 2.6% due to the glutamate content of Casamino Acids in the medium). The glutamate concentrations in these medium preparations were 8.65, 4.44, and 0.225 g/liter, respectively. Growth was monitored via measurement of OD600 at 2-h intervals, and samples were obtained for RNA isolation 6 h after the cultures were started.
Measurement of DNA concentration.
Culture aliquots (0.5 ml) were centrifuged at 16,000 × g for 60 s, the supernatants were removed, and the cell pellets were frozen at −80°C. DNA was extracted by a boiling-lysis method as previously described (18), with the modification that cells were incubated at 80°C for 10 min. DNA concentrations were determined using a PicoGreen dsDNA quantitation kit (Molecular Probes, Eugene, OR).
RNA isolation and microarray hybridization.
Culture aliquots (1 ml) were collected by the same method that was used to collect aliquots for measurement of the DNA concentration. RNA was isolated, labeled with Cy5, and cohybridized along with a Cy3-labeled genomic DNA reference mixture to a Bordetella microarray representing 97.4% of the B. pertussis Tohama I open reading frames (ORFs), 97.9% of the B. parapertussis 12822 ORFs, and 98.5% of the B. bronchiseptica RB50 ORFs, as previously described (5). Arrays were scanned using a GenePix 4000B scanner and the GenePix Pro 5.0 software (Axon Instruments, Union City, CA).
Microarray analysis.
Using the GenePix Pro 6.0 software (Axon Instruments), spots were screened visually to exclude the low-quality spots, and background-subtracted Cy5/Cy3 intensity ratios were calculated. Data were filtered to include only spots containing more than 30 pixels and having a mean background-subtracted Cy3 signal greater than 150 U. Cy5/Cy3 ratios were averaged for replicate spots, log transformed, and normalized by calculating the median log ratio and subtracting the value obtained from each data point.
For the analysis of growth phase-associated regulation in Tohama I and GMT-1, Significance Analysis of Microarrays (SAM) (version 1.0) (39) was performed for the subset of B. pertussis array elements whose data passed filtering criteria for at least 14 of 19 arrays. For this analysis, combined data from the two strains were assigned to two groups, log phase and stationary phase, in order to identify transcripts in both strains with significant differences in abundance between the two growth phases. The transition from log phase to stationary phase was defined for each strain as the time at which a majority of genes showed a change in transcript abundance; this point was determined to occur at 35 h of growth for Tohama I and at 29 h of growth for GMT-1.
Hierarchical clustering of data was performed with Cluster (version 2.11.0.1) (11), using only array elements whose data passed filtering criteria for at least 80% of the arrays. Transcript abundance was considered to vary significantly according to growth phase or glutamate availability if there was a difference of at least 2 between the maximum and minimum log values across the data set (fourfold variation). Results were displayed using Treeview (version 1.60) (11). Gene annotation was performed as described previously (29), using functional categories defined according to MultiFun (33). The headings “Unknown proteins, no known homologs,” “Some information, but not classifiable,” and “Conserved in organism other than Escherichia coli” were combined into the category “Hypothetical and conserved hypothetical.” Our category “Metabolism of small molecules” includes the MultiFun headings “Amino acid biosynthesis,” “Biosynthesis of cofactors, carriers,” “Central intermediary metabolism,” “Degradation of small molecules,” “Energy metabolism, carbon,” “Fatty acid biosynthesis,” and “Nucleotide biosynthesis.” “Chromosome replication and cell division” includes both headings with these names. The statistical significance of overrepresentation of functional categories within sets of genes with increased or decreased transcript abundance was calculated using the binomial distribution with a Bonferroni correction. A P value less than 0.05 was defined as indicative of significance.
Bacterial adhesion assays.
BEAS-2B cells, a line of transformed human bronchial epithelial cells (1), were seeded in 12-well plates (8 × 103 cells per 12-mm well) in Eagle’s minimal essential medium supplemented with 10% fetal bovine serum (EMEM-10% FBS) and grown for 3 days to preconfluence (4 × 104 to 1 × 105 cells per well), and then bacteria were inoculated at a multiplicity of infection of 30, using Tohama I or Tohama I-Sc3. For assays examining differences in adhesion between logarithmic-phase and stationary-phase cultures, bacterial inocula were prepared from cultures at mid-log phase or early stationary phase. For assays determining differences in adhesion between cultures grown under glutamate-replete conditions and cultures grown under glutamate-limited conditions, aliquots were collected following growth for 7 to 8 h in SSM with 100% or 2.6% of the usual glutamate concentration. Tohama I ΔfhaB grown in SSM to mid-log phase was used as a control in each assay. Bacterial inocula were washed twice in phosphate-buffered saline (PBS) and once in EMEM-10% FBS before resuspension at the desired concentration in EMEM-10% FBS. Dilutions of bacterial inocula were spread on BG blood agar plates to determine CFU counts for verifying the multiplicity of infection. After bacterial inoculation, tissue culture plates were centrifuged at 134 × g for 2 min and then incubated at 37°C in 5% CO2 for 30 min. Wells were washed four times with PBS to remove nonadherent bacteria, and then BEAS-2B cells were lysed using 3% (wt/vol) saponin in PBS, a treatment determined not to affect bacterial viability (data not shown). Dilutions of the resulting bacterial suspensions were spread on BG agar plates for determination of CFU counts, which were used to calculate proportions of adherent bacteria, expressed as percentages of the original inoculum. Results were analyzed for significance using the Student's t test, and a P value less than 0.05 was defined as indicating significance. To verify that the adhesion assays reflected adherence primarily to the eukaryotic cells rather than to the tissue culture plates, a control assay was performed, in which adhesion of Tohama I and Tohama I ΔfhaB, grown to mid-log phase in SSM, to tissue culture wells without BEAS-2B cells was measured. Only 0.074% of the original inoculum of Tohama I cells and 0.086% of Tohama I ΔfhaB cells adhered to these wells (the values are averages for 16 wells). The similar and extremely low numbers of adherent cells for the two strains suggest that adhesion to the tissue culture plate surface contributed little to the overall levels of adherence measured in the presence of eukaryotic cells.
Microarray accession numbers.
Microarray data have been deposited in ArrayExpress under accession numbers E-TABM-99, E-TABM-100, and E-TABM-101.
RESULTS
Growth phase-associated patterns of gene transcript abundance in B. pertussis.
To assess variation in patterns of bacterial gene expression across different phases of growth, genome-wide transcript abundance was measured in Tohama I and GMT-1 during mid-log phase, late log phase, the transition from log phase to stationary phase, and stationary phase. Two well-studied strains were examined in order to appreciate more universal growth-associated changes rather than the changes unique to a particular strain. Growth phases were defined by serial measurement of the OD600 of duplicate cultures of each strain. Because optical density measurements may not reflect cell number accurately due to factors such as clumping and cannot distinguish between culturable and nonculturable cells, DNA concentrations and CFU counts were also determined at each time. DNA concentrations correlated well with optical densities (Fig. 1A). Although GMT-1 and Tohama I reached similar maximum optical densities, duplicate GMT-1 cultures exhibited decreases in CFU counts during stationary phase (Fig. 1A), suggesting that there was greater resistance to cultivation or decreased survival in stationary phase.
FIG. 1.
Growth-phase associated patterns of transcript abundance. (A) Optical densities, DNA concentrations, and CFU counts during growth of Tohama I and GMT-1 from mid-log phase to stationary phase. The data are representative of similar results obtained with duplicate cultures. (B) Transcript patterns of array elements whose levels were found to vary significantly (by SAM) between log phase and stationary phase. Rows correspond to array elements. The two major expression classes, with some representative loci, are indicated on the right. Data are mean centered for each array element. Data were averaged from duplicate cultures, when available. In some cases, a gene is represented by multiple array elements. Gray indicates missing data. The elapsed time since the start of culture is shown at the bottom. (C) Functional categorization of genes that are regulated according to growth phase. Data are expressed as the percentage of genes that show increased or decreased transcript abundance (a significant change between log phase and stationary phase as identified by SAM) among all Tohama I array elements in each category.
The transcriptional profiles of both strains revealed a prominent shift during the transition from log phase to stationary phase (Fig. 1B), with 144 array elements showing an increase in transcript level and 717 array elements showing a decrease in transcript level (identified using SAM with a false discovery rate of 1%) (see Table S1 in the supplemental material). GMT-1 exhibited the same pattern as Tohama I, but the pattern was offset at an earlier point in the growth curve.
Genes encoding folding and ushering proteins were significantly overrepresented among the genes whose transcript levels were increased later during growth (Fig. 1C and Table 1). This group included the heat shock protein genes dnaJ, dnaK, and cbpA, which also exhibit increased expression during stationary phase in other bacterial species (6, 19, 38), as well as a gene encoding a putative GrpE chaperone (BP2501).
TABLE 1.
Functional categories overrepresented among array elements whose transcript abundance changed significantly in association with growth phasea
| Category | No. of array elements (% of total)
|
|
|---|---|---|
| Tohama I genome | Array elements whose transcript abundance changed in association with growth phase | |
| Increased transcript abundance | ||
| Folding and ushering proteins | 24 (0.6) | 5 (3.5) |
| Decreased transcript abundance | ||
| Transport/binding proteins | 413 (10.4) | 115 (16.0) |
| Macromolecule synthesis and modification | 208 (5.2) | 56 (7.8) |
| Virulence factors | 81 (2.0) | 41 (5.7) |
| Ribosome constituents | 53 (1.3) | 24 (3.3) |
| Chromosome replication and cell division | 22 (0.55) | 11 (1.5) |
P < 0.05.
Genes involved in global regulatory functions were also highly represented, although not significantly so, among the genes whose transcript levels were higher in stationary phase. The genes encoding named transcriptional regulators included the SOS gene repressor lexA, as well as rpoH, which encodes an RNA polymerase sigma factor that has been implicated in induction of heat shock proteins and survival during carbon starvation in E. coli (15). Thus, like other gram-negative organisms (34), B. pertussis may possess a stationary-phase program that includes development of increased resistance to multiple stressors, such as heat shock and starvation.
Although their differential representation was not statistically significant, genes in the “Adaptation” functional category also comprised a relatively high proportion of the genes displaying rising transcript levels during the transition to stationary phase. This group included the following genes involved in iron acquisition: three putative siderophore receptor genes, bfrB, bfrC, and bfrH; bhuR, encoding the outer membrane heme receptor; and hmsH, encoding a putative hemin storage protein (see Table S1 in the supplemental material). Transcript levels of hurI, encoding a regulatory protein in the heme acquisition system, and fecI, encoding a regulatory molecule in a citrate-dependent iron transport system, also increased. The concentrations of free iron were not measured in these experiments, but the B. pertussis stationary-phase program may include entry into an iron-scavenging mode, either in direct response to iron restriction or because iron scarcity tends to accompany other conditions that induce the stationary phase.
Within the set of genes discovered to have lower transcript abundance in stationary phase than in log phase, a significantly overrepresented functional category of interest was “Virulence factors.” This group included genes for known adhesins or adhesin-related proteins, including prn, encoding pertactin, and tcfA, encoding tracheal colonization factor (see Table S1 in the supplemental material). Other adhesin genes, classified in the MultiFun category “Cell envelope,” also exhibited decreased transcript levels with the transition to stationary phase; these genes included the fimbrial gene fim2 and the putative adhesin genes fhaL and fimD/fhaE. Other cell envelope-associated virulence factor genes with this transcription pattern included five members of the wlb locus involved in synthesis of lipopolysaccharide (LPS). The pertussis toxin subunit and transport genes also displayed decreased transcript abundance in stationary phase; these genes included ptxABCD, ptlA, and ptlB, as well as ptlC, ptlEFG, and ptlI (the GenBank-derived descriptions used for the latter genes in Table S1 in the supplemental material refer only to their ORF numbers, BP3790, BP3793 to BP3795, and BP3792, respectively, rather than their gene names). The transcript abundance changes for ptxE and pltH (BP3796) exhibited the same downward pattern but were not determined to be significant using SAM. Only the transcript levels for ptlD (BP3791) showed no particular trend in association with growth phase. Multiple autotransporter genes shared a decreasing transcript abundance expression pattern as well; these genes included vag8 and the protease gene sphB1, which catalyzes maturation of the FHA precursor protein. In addition, bvgS, the Bvg transmembrane sensor kinase gene, was among the regulatory protein genes exhibiting decreased transcript abundance during stationary phase.
The functional category most overrepresented among genes having decreased transcript levels with growth was “Transport/binding proteins.” Eleven bsc type III secretion system genes, which, although intact in B. pertussis, appear to be posttranscriptionally blocked (26), exhibited lower transcript abundance in stationary phase. Surprisingly, 15 amino acid transporter genes also displayed this expression pattern, and these genes included glnP and glnQ, encoding components of the glutamine transporter, and livF, livG, and livH, encoding components of two branched-chain amino acid transport systems. Studies of E. coli have illustrated that survival in stationary phase is enhanced by induction of nutrient-scavenging systems (12), including amino acid acquisition machinery (42). B. pertussis, however, appears not to increase amino acid scavenging as part of its adaptations during stationary phase.
Several genes exhibited patterns consistent with a predicted decrease in cell replication and division and in overall protein synthesis (19) during stationary phase. Five members of the mur locus for peptidoglycan synthesis, for example, had lower transcript levels in stationary phase, as did ftsA, ftsE, and ftsZ, which are involved in cell division, and mreC and mreD, which are involved in determination of cell shape. Two functional categories comprising a relatively large proportion of the genes with decreased transcript levels were “Ribosome constituents” and “Macromolecule synthesis and modification proteins,” which included several genes encoding DNA and RNA polymerase subunits.
Transition from log phase to stationary phase.
Since the characterization of growth phase-associated changes in gene transcript abundance revealed major changes occurring around the transition from log phase to stationary phase, we sought to characterize this period of transition in greater detail. We assessed transcript levels in Tohama I at intervals of 1 to 2 h, starting at mid-log phase and continuing until 3 h after the transition from log phase to stationary phase. A cutoff of at least fourfold variation in expression across the time course was applied to identify genes with significant variations in transcript abundance during this phase of the growth curve.
Eight hundred seventy-eight array elements exceeded this threshold and fell into three broad expression patterns: genes with increasing transcript abundance starting in late log phase, genes whose transcript abundance increased starting at the transition from log phase to stationary phase, and genes with decreasing transcript abundance starting in mid-log phase (Fig. 2A; see Table S2 in the supplemental material). A small proportion showed increasing transcript abundance primarily in late log phase or no obvious transcriptional pattern. As in the experiments described above, the genes displaying increased transcript levels with growth from log phase to stationary phase included genes encoding proteins involved in resistance to heat shock and starvation and in metabolism of small molecules, including iron acquisition and use. Two genes of the wcb putative capsule biosynthesis locus showed increased transcript abundance starting in late log phase. Whether B. pertussis is capable of producing capsule is unclear (29), but induction of this locus just prior to stationary phase might reflect a protective mechanism under stressful conditions. The genes having decreased transcript abundance levels from mid-log phase to stationary phase again included several genes encoding virulence factors, proteins participating in cell division, and transporter proteins.
FIG. 2.
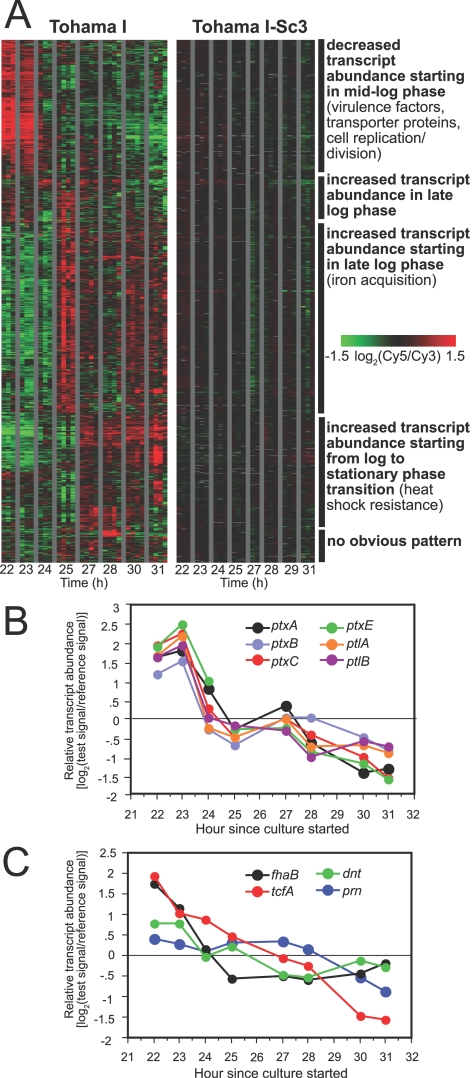
Details of the gene transcript abundance changes at the transition from log phase to stationary phase. (A) Growth phase-associated gene expression patterns assessed in Tohama I and in Tohama I-Sc3 at 1- to 2-h intervals from mid-log phase until 3 h after the transition from log phase to stationary phase. Rows correspond to array elements. The array elements shown are those exhibiting a minimum fourfold variation in transcript levels with growth. The major expression classes, with some representative loci, are indicated on the right. Data are mean centered across each array element for each strain. Columns between gray bars show data from duplicate or trip-licate cultures for each strain and, when available, from duplicate independent RNA preparations from one of the cultures. In some licate cultures for each strain and, when available, from duplicate independent RNA preparations from one of the cultures. In some cases, a gene is represented by multiple array elements. Gray indicates missing data. The elapsed time since the culture was started is shown at the bottom. (B) Variation in relative transcript abundance, expressed as log ratios of test signal intensity to reference signal intensity, for the pertussis toxin operon. For genes represented by multiple array elements, the data shown are averages of values for all of the array elements passing quality filters. (C) Variation in relative transcript abundance, expressed as log ratios of test signal intensity to reference signal intensity, for selected other virulence factor genes. For genes represented by multiple array elements, the data shown are averages of values for all of the array elements passing quality filters.
Transcriptional profile details not appreciated in the initial examination of growth phase-associated changes in transcript abundance were elucidated. Transcript levels for iron acquisition genes, for example, including bhuR, bhuU, hurI, and hurR of the heme uptake system and four members of the alcABCDE locus for biosynthesis of the siderophore alcaligin, increased starting in late log phase, whereas transcript levels for heat shock protein genes increased starting at the transition from log phase to stationary phase. This pattern is consistent with the observation that bacteria may first proceed through a “hunger” response, scavenging for increasingly limited nutrients, before they enter a stationary-phase program that emphasizes stress resistance when nutrient scarcity or other environmental conditions completely prohibit growth (12).
Downward changes in transcript abundance for genes encoding virulence factors followed distinct temporal patterns. All of the genes comprising the pertussis toxin operon exhibited a steep decline in transcript levels from mid-log to late-log phase, followed by a less marked decrease during the transition from log phase to stationary phase (Fig. 2B). In contrast, the transcript abundance of other virulence factors exhibited a more heterogeneous pattern (Fig. 2C). The transcript levels for fhaB, for instance, dropped sharply from mid-log to late-log phase and then remained fairly constant, whereas the levels for prn were stable through late log phase and then decreased. The levels for tcfA showed yet another pattern, with a gradual decrease from mid-log phase to stationary phase. Our findings imply that B. pertussis exercises finely tuned control of transcript abundance in accordance with its growth phase. The changes in transcript abundance may reflect different patterns in the repression of transcriptional initiation or in the degradation of transcripts or both.
Role of the BvgAS system in growth phase-related patterns of transcript abundance.
Because BvgAS is the best-characterized regulatory system in the bordetellae and many Bvg-regulated virulence factors displayed variations in transcript abundance with growth, we examined whether BvgAS participates in growth phase-associated transcriptional control. Using the same method that was used for its wild-type parent strain, the gene expression profile of the Bvg+ constitutive phase-locked mutant Tohama I-Sc3 was evaluated at hourly intervals from mid-log to stationary phase. The Bvgc mutant strain exhibited strikingly few transcript abundance changes (Fig. 2A); no array elements exhibited at least a fourfold change in transcript abundance across the time course, even though the growth pattern of this strain was similar to that of Tohama I, as measured by optical density (Fig. 3A). A Bvg system capable of modulation is thus evidently necessary for the growth phase-associated changes in transcript abundance described above.
FIG. 3.
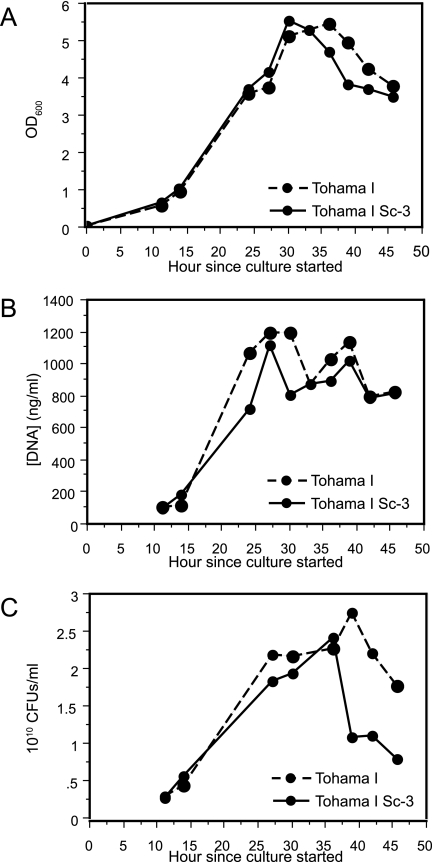
Role of BvgAS in stationary-phase survival: optical densities (A), DNA concentrations (B), and CFU counts (C) during growth of Tohama I and Tohama I-Sc3 from mid-log phase to several hours after the transition from log phase to stationary phase. The data are representative of similar results obtained from duplicate cultures.
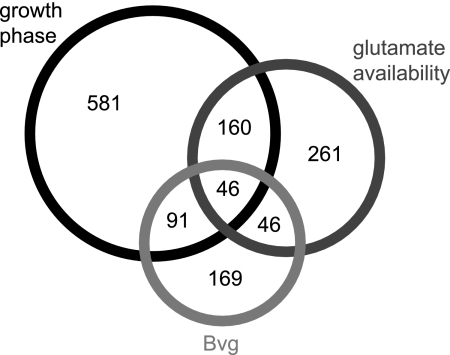
Despite the apparent importance of BvgAS in growth phase-related control, its regulatory role does not seem to be straightforward or exclusive. In a recent study we defined the Bvg regulon comprehensively through comparison of phase-locked mutants and through analysis of transcript abundance changes in response to the classical modulators NA and sulfate (10). Comparison of the array elements that vary at least fourfold with growth from log phase to stationary phase in Tohama I to the 352 array elements determined in the previous study to be Bvg regulated in B. pertussis (10) revealed only 137 array elements common to both sets (Fig. 4). The remaining array elements exhibiting alterations in transcript abundance with growth from log phase to stationary phase, therefore, seem to be subject to other regulatory systems acting in concert with BvgAS.
FIG. 4.
Overlap between array elements regulated in association with growth phase, according to nutrient limitation, and by the Bvg system. The values indicate the number of array elements in each group.
Role of BvgAS in stationary-phase survival.
Given the paucity of growth-phase associated transcriptional changes in the presence of a constitutively active BvgAS system, we measured growth parameters of Tohama I and Tohama I-Sc3 several hours after entry into stationary phase in order to assess whether BvgAS affects stationary-phase survival. All measurements were performed with duplicate cultures of each strain. Optical density (Fig. 3A) and DNA concentration (Fig. 3B) measurements for both strains showed similar temporal patterns. A difference was observed, however, in the proportion of culturable cells in cultures of the two strains, with Tohama I-Sc3 achieving a lower CFU count and experiencing an earlier and more rapid decline than Tohama I (Fig. 3C). Hence, while a Bvg system capable of modulation is apparently not essential for reaching maximum cell mass, it does seem to be required for optimal maintenance of culturability in stationary phase.
Response of B. pertussis to glutamate limitation.
A variety of conditions can induce bacterial cultures to enter stationary phase, including starvation (19), oxidative stress (27), and acid stress (32). Of these stimuli, nutrient scarcity is one factor that may be relevant to host-pathogen interactions since bacterial nutrient availability is likely to be low in the intact human airway (36). We examined the response of B. pertussis to nutrient deprivation by determining its transcriptional profile during growth in glutamate-depleted media. B. pertussis generally does not use carbohydrates as carbon sources, perhaps due to deficiencies in its glycolytic pathway (29); amino acids are its preferred energy source (20). It has an absolute requirement for only a few amino acids, and of those, glutamate appears to be among the amino acids most rapidly depleted during in vitro growth (20). SSM contains as its primary carbon source a mixture of amino acids that is enriched for glutamate and proline, and glutamate appears to be the major carbon source for B. pertussis (29). Glutamate restriction was therefore chosen as a likely relevant and informative condition for studying nutrient limitation effects in B. pertussis.
Tohama I was cultured in SSM containing 2.6%, 51%, or 100% of the usual glutamate concentration in order to compare the effects of severe and moderate glutamate limitation with the effects of glutamate-replete conditions. Cells were collected for RNA isolation 6 h after inoculation of the media since this was the time that corresponded with a major shift in gene transcript levels in response to glutamate depletion (in preliminary experiments) (data not shown). This time was also the time at which the optical density of the cultures reached a plateau in the severely glutamate-depleted culture yet still indicated the log phase of the cultures grown in SSM with 51% or 100% of the usual glutamate concentration (Fig. 5A), thus helping to separate effects associated purely with glutamate limitation from effects likely due to the multiple factors inducing the stationary phase in the growth phase experiments.
FIG. 5.
Patterns of gene transcript abundance as a function of glutamate availability. (A) Optical densities, DNA concentrations, and CFU counts during growth of Tohama I and Tohama I-Sc3 in standard media and media containing 51% and 2.6% of the usual glutamate concentration. The data are representative of similar results obtained with duplicate cultures. (B) Gene transcript abundance patterns in Tohama I and Tohama I-Sc3 for duplicate cultures of each strain after growth for 6 h in each of the three medium formulations. Rows correspond to array elements. The array elements shown are those exhibiting at least fourfold variation in transcript level as a function of glutamate availability. The major expression classes, with some representative loci, are indicated on the right. Data are mean centered across each array element for each strain. In some cases, a gene is represented by multiple array elements. Gray indicates missing data. (C) Functional categorization of genes that are regulated according to glutamate availability. Data are expressed as the percentage of genes that exhibit increased or decreased transcript abundance (a minimum fourfold change as a function of glutamate availability) among all Tohama I array elements in each category.
Analysis of microarray data revealed 513 array elements with at least fourfold changes in transcript abundance according to glutamate availability and yielded expression patterns that could be separated into three major groups: increased transcript levels only with severe glutamate restriction, increased transcript levels with both moderate and severe glutamate restriction, and decreased transcript levels with severe glutamate restriction (Fig. 5B; see Table S3 in the supplemental material). While the transcriptional profile of glutamate-depleted cells exhibited similarities with that of cells in stationary-phase cultures, as described in greater detail below, it also had unique features. Three hundred seven, or about 60% of the array elements that were discovered to have altered transcript levels under glutamate restriction conditions, did not vary in transcript abundance in accordance with the growth phase (Fig. 4).
Among the genes showing increased transcript abundance with glutamate restriction, no functional category was significantly overrepresented (Fig. 5C and Table 2). In contrast to the pattern observed in the growth phase-associated transcriptional profile, several amino acid transporter genes showed increased transcript abundance due to glutamate restriction, including gltJ and gltK, which encode glutamate transporter components. This finding suggests that the response of B. pertussis to amino acid scarcity may include enhanced scavenging of at least some amino acids even though this is not a feature of its broader stationary-phase program.
TABLE 2.
Functional categories overrepresented among genes whose transcript abundance decreased significantly in association with glutamate availabilitya
| Category | No. of array elements (% of total)
|
|
|---|---|---|
| Tohama I genome | Array elements whose transcript abundance changed in association with glutamate availability | |
| Metabolism of small molecules | 626 (16.0) | 70 (22.1) |
| Ribosome constituents | 53 (1.4) | 42 (13.2) |
| Macromolecule synthesis, modification | 206 (5.3) | 32 (10.1) |
| Virulence factors | 67 (1.7) | 15 (4.7) |
| Folding and ushering proteins | 23 (0.59) | 8 (2.5) |
P < 0.05.
Among the genes having higher transcript levels with both growth and glutamate limitation was the sigma factor gene rpoH, which is consistent with its described role in the starvation response of E. coli (15). Another shared feature of the two gene sets was an increase in transcript abundance of genes encoding iron acquisition proteins; glutamate depletion was associated with increases in transcripts for three of the alcaligin biosynthesis genes and the regulatory genes hurI and fecR. Since the glutamate-depleted medium was not iron depleted, amino acid limitation seems to serve as a signal for activation of iron-scavenging mechanisms, perhaps indicating overlapping regulation that has evolved because amino acid-restricted environments also tend to have low free iron availability.
The set of genes with lower transcript abundance in the setting of glutamate depletion exhibited similarities with the genes with decreased transcript levels during the transition from log phase to stationary phase. Significantly overrepresented in both groups were genes required for protein synthesis, with glutamate-depleted cultures having lower transcript levels for numerous ribosomal protein genes and macromolecule synthesis and modification genes, including several genes encoding RNA polymerase subunits and translation initiation and elongation factors.
Virulence factor genes showed significantly decreased transcript abundance in association with both growth and glutamate limitation. These genes included prn, tcfA, fimB/fhaD, fimD/fhaE, cyaB, vag8, and sphB1. Transcript levels for fhaB decreased 3.9-fold with glutamate restriction, which was just below the fourfold variation filter used in data analysis. The bipA gene, which exhibits peak transcript levels in the Bvg-intermediate phase but is more highly expressed in the Bvg+ phase than in the Bvg− phase (10), also exhibited lower transcript abundance in response to glutamate depletion. The virulence factor expression patterns under glutamate limitation conditions were not identical to those associated with growth phase changes. The transcript abundance for the pertussis toxin operon genes and the dermonecrotic toxin gene, dnt, for instance, decreased from log phase to stationary phase but varied little with glutamate availability.
One functional category highly represented among genes with lower transcript levels under glutamate restriction conditions but not during stationary phase was “Metabolism of small molecules,” including genes involved in carbohydrate and amino acid metabolism and cofactor and carrier biosynthesis, possibly representing a decrease in overall metabolic activity. This group included most of the members of the nuo NADH dehydrogenase I operon, perhaps indicating shifts in the abundance of respiratory electron transport chain components, a phenomenon described in E. coli and hypothesized to coordinate ATP production with changing energy requirements of log-phase cultures compared with stationary-phase cultures (40).
Role of BvgAS in the glutamate limitation response.
To assess whether the Bvg system participates in the glutamate limitation response, glutamate depletion experiments were carried out with Tohama I-Sc3. The Bvgc mutant exhibited a transcriptional profile similar to that of Tohama I, although the degree of variation for many genes, particularly those exhibiting increased transcript levels with growth, was muted (Fig. 5B). Of note was the finding that the decrease in transcript abundance of several virulence genes known to be Bvg regulated, such as prn, tcfA, sphB1, vag8, and bipA, was maintained in Tohama I-Sc3. The expression patterns for some genes appeared to be more alike under moderate and severe glutamate restriction in Tohama I-Sc3 than in Tohama I, indicating that there was induction of the glutamate limitation response in Tohama I-Sc3 at less severe levels of glutamate deprivation. In addition, Tohama I-Sc3 exhibited essentially no increase in cell density, as determined by optical density measurements, when it was grown in SSM with either 51% or 2.6% of the usual glutamate concentration (Fig. 5A). These findings indicate that BvgAS is required for optimal tolerance of glutamate depletion.
The 513 array elements displaying transcriptional variation according to glutamate availability were compared with the 352 array elements comprising the Bvg regulon, as defined by Cummings et al. (10). Only 92 array elements belonged to both groups, and exactly one-half of these elements also underwent growth phase-associated regulation in Tohama I (Fig. 4). Although BvgAS appears to play a critical role in growth phase-associated regulation and responses to glutamate restriction, its actions apparently overlap but are distinct in these two transcriptional programs and occur in cooperation with other regulatory systems.
B. pertussis adherence varies with growth phase and nutritional state.
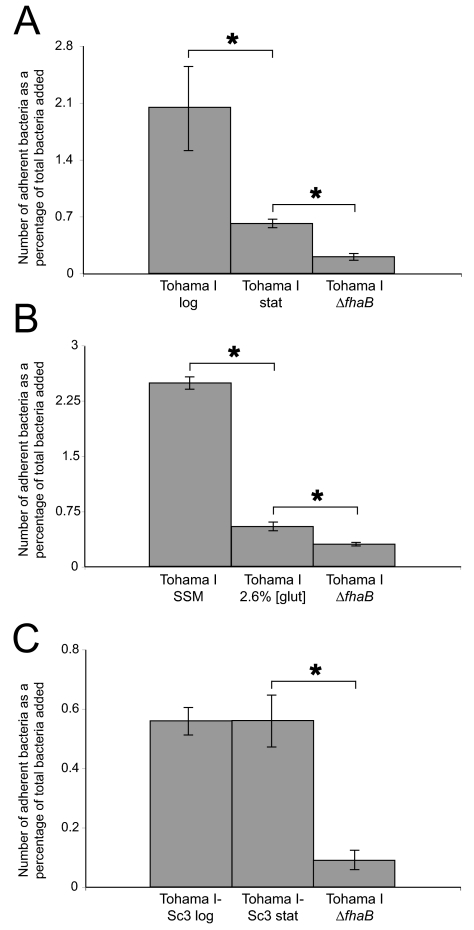
Because several of the virulence genes that exhibited decreased transcript abundance during the transition from log phase to stationary phase and under glutamate limitation conditions encode known adhesins, we investigated whether B. pertussis adherence to human cells in vitro changes with growth phase and glutamate availability. We discovered that Tohama I cells harvested in stationary phase adhere to BEAS-2B cells, a human bronchial epithelial cell line, significantly less well than cells collected in mid-log phase adhere (Fig. 6A). Likewise, Tohama I cells adhered significantly less well after growth in severely glutamate-depleted medium than after growth in glutamate-replete medium (Fig. 6B). In both sets of experiments, the adherence was still significantly greater than that exhibited by a mutant unable to produce FHA, one of the major adhesins in the bordetellae (9), suggesting that entry into stationary phase or exposure to glutamate restriction does not completely abolish expression of adhesins. Since Tohama I-Sc3 failed to exhibit variation of adhesin transcript abundance in accordance with growth phase, we performed the same adhesion assay using this strain. As predicted by its growth phase-related transcript abundance profile, the Bvgc mutant showed no difference in adherence when it was collected from stationary-phase cultures and from mid-log-phase cultures (Fig. 6C).
FIG. 6.
Variation in adherence with growth phase and glutamate availability: number of bacteria adhering to BEAS-2B cells, expressed as a percentage of the original inoculum, for Tohama I (A) and Tohama I-Sc3 (C) grown to either log or stationary (stat) phase and for Tohama I grown under glutamate-replete or glutamate-depleted conditions (B). The data in panel A are means ± standard errors of the means for nine wells (three wells in each of three independent experiments). The data in panels B and C are means ± standard errors of the means for six to eight wells (three or four wells in each of two independent experiments). An asterisk indicates that the P value is <0.05.
DISCUSSION
Detailed global transcriptional profiling of B. pertussis over the course of its growth curve revealed a striking shift in patterns of gene transcript abundance during the transition from log phase to stationary phase, including decreases in the transcript levels of most known virulence factor genes. Examination of transcriptional responses to depletion of glutamate, a key nutrient for B. pertussis, revealed general similarities but distinctive features compared to growth-related changes, suggesting that nutrient deprivation may contribute to growth phase-associated regulation but likely acts in concert with other stimuli in growing cultures. Adherence, a major virulence property, was confirmed to change with growth phase along with the transcript abundance of various adhesin genes, indicating that growth phase-associated transcriptional regulation leads to significant alterations in the virulence phenotype.
Our comparison of different growth parameters revealed the merits of using multiple approaches for measuring growth. In the analysis of growth through late stationary phase (Fig. 3), for example, optical density was useful because it provided an immediately available measure of overall cell mass while the cultures were growing. Only CFU counts, however, could show that a greater proportion of wild-type cells than cells of the Bvg+ constitutive mutant remained culturable. Measurements of DNA concentration offered the advantage of not being affected by possible cell clumping.
To our knowledge, global modulation of virulence due to nutrient limitation has not been described previously in B. pertussis, but our findings are supported by the results of prior investigations of specific individual virulence factors and the relationship of their expression levels to growth phase. One of the earliest studies, published in 1984, showed that the concentrations of protective antigens, relative to bacterial cell concentrations, in B. pertussis cultures grown for vaccine production fell rapidly at the end of logarithmic growth (2). More specifically, pertussis toxin secretion has been observed to decline after cultures enter stationary phase (4, 31). Secretion itself, rather than production or assembly of toxin subunits or the secretion apparatus, has been determined to be rate limiting, but intracellular accumulation of toxin subunit and secretion proteins also plateaus in stationary phase (31). Similarly, adenylate cyclase production has been found to peak during mid-log phase (21). A study of three B. pertussis vaccine strains by Westdijk and colleagues illustrated that FHA production also declines from mid-log to late-stationary phase (41). Our study suggests that regulation of transcript abundance is one important mechanism by which virulence factor production is coordinated with changes in growth phase.
In two studies, virulence factor production was observed to vary in different ways with respect to growth phase. In an examination of LPS production, cell-associated LPS concentrations decreased from log phase to stationary phase (13). The amount of LPS secreted per cell into culture media, however, increased during stationary phase, a phenomenon hypothesized to be due to cell death and lysis or partial cell membrane degradation (13). In addition, pertactin production was discovered to increase in three strains with growth, whereas pertussis toxin production decreased in two strains but in one strain decreased from mid-log phase to the transition from log phase to stationary phase and then increased in late stationary phase (41). These observations illustrate that posttranscriptional regulatory events also play a role in virulence factor production during the growth cycle. The demonstration by Westdijk et al. that pertussis toxin production differs according to strain suggests the possibility that there are strain-specific deviations from the general growth-associated pattern of decreased virulence factor production and the possibility that there are strain-specific differences in pathogenicity.
Studies of global gene expression patterns in other pathogenic bacteria, such as Streptococcus pneumoniae (17) and Helicobacter pylori (38), have also shown that there are major shifts in transcript profiles at the transition from log phase to stationary phase. In contrast to our findings with B. pertussis, however, in other bacterial species virulence gene transcript abundance and function generally increase from log phase to stationary phase. In H. pylori, many known virulence factor genes exhibit peak expression levels in late log or stationary phase. These genes include cagA, the cytotoxin-associated gene; napA, encoding the neutrophil-activating protein; and flaA, encoding the major flagellin subunit (38). Correspondingly, maximal levels of motility of H. pylori occur just prior to the transition from log phase to stationary phase (38). When subjected to nitrogen deprivation, Neisseria meningitidis produces increased levels of capsular polysaccharide and kills mice at lower inoculum sizes (24). In Salmonella enterica serovar Typhimurium, expression of Salmonella pathogenicity island 1 genes, which include genes for a type III secretion apparatus involved in induction of macrophage apoptosis, is highest at the transition from log phase to stationary phase, the same point in the growth cycle at which the apoptosis-inducing phenotype is maximal (22). Regarding the specific effect of nutrient limitation on this organism, another study determined that expression of hilA, which encodes a central regulator of Salmonella pathogenicity island 1 transcription, is activated by carbon source deprivation (35).
The peak in virulence factor transcript levels and phenotypes at late log or stationary phase in other organisms has been postulated to occur because bacterial pathogens sense stressful conditions, like those encountered during stationary phase, as signals of arrival in the host and use these cues to coordinate virulence functions (see, for example, the discussion in reference 35). B. pertussis may display an opposite pattern of virulence factor expression because unlike many other pathogens, it has no environmental niche outside its human host and no infection site outside the respiratory tract (25). Since B. pertussis appears to survive only in the human airway, it may have evolved to link growth phase and virulence with changing conditions in different locations of this restricted environment or during different stages of infection. Prior studies have suggested that Bvg-regulated expression of virulence factors may occur in a temporally or spatially defined sequence (10, 30). A possible model for the B. pertussis infectious cycle consists of elaboration of adhesins, toxins, and other virulence factors during early stages of infection, thus permitting colonization, evasion of host immunity, and acquisition of nutrients, followed by declining virulence factor production when bacteria reach high densities and have depleted locally available nutrients. Repression of virulence factor production during stationary phase in vivo could conserve energy in the presence of extreme nutrient scarcity, decrease antigenicity and subsequent immune responses, and decrease adherence to host cells in preparation for exit from the host.
Although microarray-based transcriptional profiling is a powerful tool for elucidating coordinated regulation of virulence and other functions relevant to pathogenicity in concordance with environmental stimuli, it does not differentiate among the mechanisms by which transcript levels are controlled. Our discovery that the Bvg system is crucial in shaping growth phase-related gene expression patterns and in helping to determine glutamate limitation responses implies that induction or repression of transcription is one important level of regulation in these response programs. Bogdan et al. demonstrated that sulfate accumulates in B. pertussis cultures grown in SSM and suggested that the organism decreases pertussis toxin production upon sensing high sulfate concentrations resulting from its metabolism of cysteine (4). This means of Bvg modulation might be an important cause of the alterations in transcript abundance that we observed during the transition from log phase to stationary phase.
Our study illustrates, however, that growth phase-associated changes in transcript abundance are not due exclusively to sulfate-mediated Bvg modulation. BvgAS does not simply alter transcription through the same pathways acted upon by sulfate or NA, for many of the genes regulated according to growth phase or glutamate availability are not part of the Bvg regulon as defined in a previous study (10) that evaluated transcriptional responses to these classical chemical modulators. At the same time, the variation in many virulence gene transcript levels according to glutamate availability in the Bvgc mutant indicates that other regulatory systems control virulence expression in cooperation with BvgAS. Although the Bvg system has been characterized extensively, our findings highlight the need for further elucidation of the signals, other than temperature, NA, or sulfate, that act upon BvgAS and of the ways in which BvgAS interacts with other regulatory pathways.
Posttranscriptional events are also likely to affect transcript abundance significantly. The stability of mRNA has been observed to vary in a gene-specific manner according to growth phase or exposure to starvation or other stressful conditions (37). During stationary phase, there is a marked decrease in Salmonella enterica serovar Dublin, for example, in the transcript decay rate for rpoS, which encodes a sigma factor that serves as a central regulator of stationary phase and osmotically controlled genes in various gram-negative bacteria (37).
It would be of interest to explore further the mechanisms by which growth phase- and glutamate restriction-related changes in transcript abundance are effected in B. pertussis. Investigation of responses to other factors that might be relevant during stationary phase in the host environment, such as restriction of iron or other nutrients, would also be informative. In E. coli, for instance, distinct regulatory and physiological responses are elicited in response to depletion of nitrogen, phosphate, or glucose or other carbon sources (12). Since B. pertussis persists in its human host for at least 4 weeks and probably longer (25), it must possess the ability to adjust to and survive under adverse conditions for prolonged periods. The stationary-phase transcriptional profile that we describe here is probably the outcome of a sophisticated and integrated response to the multitude of stressful stimuli during this phase of its life cycle.
Supplementary Material
Acknowledgments
This work was supported by NICHD grants K12-HD00850-18 (to M.M.N.), NIAID AI057188, and AI54970 (to D.A.R.).
We appreciate the technical assistance of Anthony Miller. We thank Emily Gogol for assistance with adhesion assays. Jeff F. Miller (University of California, Los Angeles) generously provided Tohama I-Sc3.
Editor: D. L. Burns
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Amstad, P., R. R. Reddel, A. Pfeifer, L. Malan-Shibley, G. E. Mark, 3rd, and C. C. Harris. 1988. Neoplastic transformation of a human bronchial epithelial cell line by a recombinant retrovirus encoding viral Harvey ras. Mol. Carcinog. 1:151-160. [DOI] [PubMed] [Google Scholar]
- 2.Bellalou, J., and E. H. Relyveld. 1984. Studies on culture conditions of Bordetella pertussis and relationship to immunogenicity of vaccines. Ann. Microbiol. (Paris) 135B:101-110. [DOI] [PubMed] [Google Scholar]
- 3.Bock, A., and R. Gross. 2001. The BvgAS two-component system of Bordetella spp.: a versatile modulator of virulence gene expression. Int. J. Med. Microbiol. 291:119-130. [DOI] [PubMed] [Google Scholar]
- 4.Bogdan, J. A., J. Nazario-Larrieu, J. Sarwar, P. Alexander, and M. S. Blake. 2001. Bordetella pertussis autoregulates pertussis toxin production through the metabolism of cysteine. Infect. Immun. 69:6823-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinig, M. M., C. A. Cummings, G. N. Sanden, P. Stefanelli, A. Lawrence, and D. A. Relman. 2006. Significant gene order and expression differences in Bordetella pertussis despite limited gene content variation. J. Bacteriol. 188:2375-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chae, C., S. Sharma, J. R. Hoskins, and S. Wickner. 2004. CbpA, a DnaJ homolog, is a DnaK co-chaperone, and its activity is modulated by CbpM. J. Biol. Chem. 279:33147-33153. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, P. A., and A. M. Jones. 2003. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol. 11:367-373. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, P. A., and J. F. Miller. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62:3381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotter, P. A., M. H. Yuk, S. Mattoo, B. J. Akerley, J. Boschwitz, D. A. Relman, and J. F. Miller. 1998. Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect. Immun. 66:5921-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummings, C. A., H. J. Bootsma, D. A. Relman, and J. F. Miller. 2006. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J. Bacteriol. 188:1775-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferenci, T. 2001. Hungry bacteria—definition and properties of a nutritional state. Environ. Microbiol. 3:605-611. [DOI] [PubMed] [Google Scholar]
- 13.Hozbor, D., M. E. Rodriguez, A. Samo, A. Lagares, and O. Yantorno. 1993. Release of lipopolysaccharide during Bordetella pertussis growth. Res. Microbiol. 144:201-209. [DOI] [PubMed] [Google Scholar]
- 14.Imaizumi, A., Y. Suzuki, S. Ono, H. Sato, and Y. Sato. 1983. Heptakis (2,6-O-dimethyl) beta-cyclodextrin: a novel growth stimulant for Bordetella pertussis phase I. J. Clin. Microbiol. 17:781-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkins, D. E., E. A. Auger, and A. Matin. 1991. Role of RpoH, a heat shock regulator protein, in Escherichia coli carbon starvation protein synthesis and survival. J. Bacteriol. 173:1992-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasuga, T., Y. Nakase, K. Ukishima, and K. Takatsu. 1954. Studies on Haemophilus pertussis. V. Relation between the phase of bacilli and the progress of the whooping-cough. Kitasato Arch. Exp. Med. 27:57-62. [PubMed] [Google Scholar]
- 17.Ko, K. S., S. Park, W. S. Oh, J. Y. Suh, T. Oh, S. Ahn, J. Chun, and J. H. Song. 2006. Comparative analysis of growth-phase-dependent gene expression in virulent and avirulent Streptococcus pneumoniae using a high-density DNA microarray. Mol. Cell 21:82-88. [PubMed] [Google Scholar]
- 18.Ko, K. S., K. R. Peck, W. S. Oh, N. Y. Lee, J. H. Lee, and J. H. Song. 2005. New species of Bordetella, Bordetella ansorpii sp. nov., isolated from the purulent exudate of an epidermal cyst. J. Clin. Microbiol. 43:2516-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolter, R., D. A. Siegele, and A. Tormo. 1993. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47:855-874. [DOI] [PubMed] [Google Scholar]
- 20.Lane, A. G. 1970. Use of glutamic acid to supplement fluid medium for cultivation of Bordetella pertussis. Appl. Microbiol. 19:512-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leusch, M. S., S. Paulaitis, and R. L. Friedman. 1990. Adenylate cyclase toxin of Bordetella pertussis: production, purification, and partial characterization. Infect. Immun. 58:3621-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundberg, U., U. Vinatzer, D. Berdnik, A. von Gabain, and M. Baccarini. 1999. Growth phase-regulated induction of Salmonella-induced macrophage apoptosis correlates with transient expression of SPI-1 genes. J. Bacteriol. 181:3433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez de Tejada, G., J. F. Miller, and P. A. Cotter. 1996. Comparative analysis of the virulence control systems of Bordetella pertussis and Bordetella bronchiseptica. Mol. Microbiol. 22:895-908. [DOI] [PubMed] [Google Scholar]
- 24.Masson, L., and B. E. Holbein. 1985. Influence of nutrient limitation and low pH on serogroup B Neisseria meningitidis capsular polysaccharide levels: correlation with virulence for mice. Infect. Immun. 47:465-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattoo, S., and J. D. Cherry. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin. Microbiol. Rev. 18:326-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattoo, S., M. H. Yuk, L. L. Huang, and J. F. Miller. 2004. Regulation of type III secretion in Bordetella. Mol. Microbiol. 52:1201-1214. [DOI] [PubMed] [Google Scholar]
- 27.McDougald, D., L. Gong, S. Srinivasan, E. Hild, L. Thompson, K. Takayama, S. A. Rice, and S. Kjelleberg. 2002. Defences against oxidative stress during starvation in bacteria. Antonie Leeuwenhoek 81:3-13. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. F., S. A. Johnson, W. J. Black, D. T. Beattie, J. J. Mekalanos, and S. Falkow. 1992. Constitutive sensory transduction mutations in the Bordetella pertussis bvgS gene. J. Bacteriol. 174:970-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 30.Prugnola, A., B. Arico, R. Manetti, R. Rappuoli, and V. Scarlato. 1995. Response of the bvg regulon of Bordetella pertussis to different temperatures and short-term temperature shifts. Microbiology 141:2529-2534. [DOI] [PubMed] [Google Scholar]
- 31.Rambow-Larsen, A. A., and A. A. Weiss. 2004. Temporal expression of pertussis toxin and Ptl secretion proteins by Bordetella pertussis. J. Bacteriol. 186:43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richard, H. T., and J. W. Foster. 2003. Acid resistance in Escherichia coli. Adv. Appl. Microbiol. 52:167-186. [DOI] [PubMed] [Google Scholar]
- 33.Serres, M. H., and M. Riley. 2000. MultiFun, a multifunctional classification scheme for Escherichia coli K-12 gene products. Microb. Comp. Genomics 5:205-222. [DOI] [PubMed] [Google Scholar]
- 34.Siegele, D. A., and R. Kolter. 1992. Life after log. J. Bacteriol. 174:345-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song, M., H. J. Kim, E. Y. Kim, M. Shin, H. C. Lee, Y. Hong, J. H. Rhee, H. Yoon, S. Ryu, S. Lim, and H. E. Choy. 2004. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J. Biol. Chem. 279:34183-34190. [DOI] [PubMed] [Google Scholar]
- 36.Speert, D. P., S. W. Farmer, M. E. Campbell, J. M. Musser, R. K. Selander, and S. Kuo. 1990. Conversion of Pseudomonas aeruginosa to the phenotype characteristic of strains from patients with cystic fibrosis. J. Clin. Microbiol. 28:188-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takayama, K., and S. Kjelleberg. 2000. The role of RNA stability during bacterial stress responses and starvation. Environ. Microbiol. 2:355-365. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, L. J., D. S. Merrell, B. A. Neilan, H. Mitchell, A. Lee, and S. Falkow. 2003. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect. Immun. 71:2643-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wackwitz, B., J. Bongaerts, S. D. Goodman, and G. Unden. 1999. Growth phase-dependent regulation of nuoA-N expression in Escherichia coli K-12 by the Fis protein: upstream binding sites and bioenergetic significance. Mol. Gen. Genet. 262:876-883. [DOI] [PubMed] [Google Scholar]
- 41.Westdijk, J., J. van den Ijssel, M. Thalen, C. Beuvery, and W. Jiskoot. 1997. Quantification of cell-associated and free antigens in Bordetella pertussis suspensions by antigen binding ELISA. J. Immunoass. 18:267-284. [DOI] [PubMed] [Google Scholar]
- 42.Zinser, E. R., and R. Kolter. 2004. Escherichia coli evolution during stationary phase. Res. Microbiol. 155:328-336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.