Abstract
Pseudomonas aeruginosa is an important opportunistic pathogen which is capable of causing both acute and chronic infections in immunocompromised patients. Successful adaptation of the bacterium to its host environment relies on the ability of the organism to tightly regulate gene expression. RsmA, a small RNA-binding protein, controls the expression of a large number of virulence-related genes in P. aeruginosa, including those encoding the type III secretion system and associated effector proteins, with important consequences for epithelial cell morphology and cytotoxicity. In order to examine the influence of RsmA-regulated functions in the pathogen on gene expression in the host, we compared global expression profiles of airway epithelial cells in response to infection with P. aeruginosa PAO1 and an rsmA mutant. The RsmA-dependent response of host cells was characterized by significant changes in the global transcriptional pattern, including the increased expression of two Kruppel-like factors, KLF2 and KLF6. This increased expression was mediated by specific type III effector proteins. ExoS was required for the enhanced expression of KLF2, whereas both ExoS and ExoY were required for the enhanced expression of KLF6. Neither ExoT nor ExoU influenced the expression of the transcription factors. Additionally, the increased gene expression of KLF2 and KLF6 was associated with ExoS-mediated cytotoxicity. Therefore, this study identifies for the first time the human transcription factors KLF2 and KLF6 as targets of the P. aeruginosa type III exoenzymes S and Y, with potential importance in host cell death.
Pseudomonas aeruginosa is an opportunistic gram-negative bacterium which can cause infections in plants, animals, and humans (33, 35). This organism causes serious respiratory, urinary tract, and skin infections in immunocompromised individuals and is the predominant pulmonary pathogen associated with morbidity and mortality of cystic fibrosis (CF) patients (12). The most common mutation associated with the cystic fibrosis transmembrane conductance regulator (CFTR) is ΔF508, and it has also been shown that CFTR can serve as a receptor for P. aeruginosa (46).
The P. aeruginosa genome consists of 6.3 million bp, 8% of which are predicted to be involved in gene regulation (56). Such a substantial proportion of regulation-associated genes is thought to have important consequences for the tightly controlled production of a diverse set of virulence factors (reviewed in reference 52). Strict control over the selected production of appropriate virulence determinants is essential to allow the organism to effectively colonize the host during different stages of infection. Regulation of gene expression in P. aeruginosa is accomplished by a number of signal transduction systems, including the two-component system GacS/GacA (16, 49) and two recently described hybrid sensor kinases, RetS and LadS (11, 60). These environmental sensors have been shown to coordinate gene expression by modulating the expression of the untranslated RNA RsmZ, which subsequently alleviates the effects of the small RNA-binding protein RsmA (2, 11, 18, 45, 60).
In P. aeruginosa, posttranscriptional regulation of a number of genes involved in virulence is mediated by RsmA. RsmA negatively affects the production of hydrogen cyanide, extracellular enzymes, and the quorum-sensing molecules C4- and 3-oxo-C12-homoserine lactone, PvdS synthesis, and antibiotic resistance (2, 18, 41, 45). However, RsmA exerts a positive regulatory effect on motility, rhamnolipid synthesis, vfr transcription, and Pseudomonas quinolone signal biosynthesis in P. aeruginosa (2, 18, 41).
Recent work in this laboratory has shown that RsmA played an important role in the interaction between P. aeruginosa and human airway epithelial cells in a cell culture model (41). An rsmA mutant failed to cause actin depolymerization and cytotoxicity in epithelial cells but displayed significantly increased invasion into epithelial cells (41). Subsequently, the rsmA mutant was demonstrated to have decreased transcription of several regulators of the type III secretion system (TTSS) and failed to secrete proteins essential for a functional TTSS (41).
The TTSS is used by a number of different animal and plant pathogens to produce and deliver bacterial effector proteins directly into the cytoplasm of eukaryotic cells (reviewed in reference 21). P. aeruginosa is known to secrete four effector proteins termed exoenzymes (Exo). The prevalence of genes encoding ExoS, ExoT, ExoU, and ExoY in isolates of P. aeruginosa has been investigated by a number of groups, and it has generally been shown that the presence of exoS and that of exoU are mutually exclusive in the same strain (5, 51). The reason for this is unclear, although P. aeruginosa PA14, which lacks a gene encoding ExoS, was recently shown to encode ExoU on a pathogenicity island (15).
ExoS and ExoT are related proteins which have N-terminal GTPase-activating protein (GAP) activity and C-terminal ADP-ribosyltransferase (ADPRT) activity (9, 10, 25, 30, 43). While the GAP activities of ExoS and ExoT act on the same set of host proteins, the ADPRT activity of ExoS inhibits intracellular signaling by acting on human Ras-related proteins (7, 17, 36, 61) and is distinct from that of ExoT, which acts on host proteins such as Crk-I and Crk-II (57). ExoU exerts phospholipase activity (53) and is a potent cytotoxic factor for a number of cell types in vitro, including epithelial cells and macrophages (6, 14). ExoY, an adenylate cyclase, elevates the intracellular concentration of cyclic AMP by interacting with an unidentified eukaryotic factor (64). Recently, ExoY was shown to cause actin depolymerization of epithelial cells and to influence invasion (3).
Because P. aeruginosa RsmA influences a number of epithelial cell phenotypes associated with the TTSS (41), we sought to investigate the role of RsmA on the global transcriptional response of human airway epithelial cells to infection with P. aeruginosa. Using such an approach, it was hypothesized that the activation/repression of host processes elicited by bacterial components under the control of RsmA would be identified. Here we characterize the genome-wide response of epithelial cells to P. aeruginosa and show that the specific induction of two members of the Kruppel-like factor (KLF) family of transcription regulators is RsmA dependent. These human transcription factors play key roles in cellular proliferation, the response to tissue injury, the regulation of immune cells, and lung development (28, 29, 31, 32, 62, 63). Induction of these genes in response to PAO1, but not the rsmA mutant, led us to identify the specific bacterial components responsible for the induction of KLF2 and KLF6. Increased expression of KLF2 was dependent on bacterial expression of ExoS, while elevated expression of KLF6 required the production of both ExoS and ExoY. These data suggest an important role for RsmA-mediated regulation of specific effector proteins of the TTSS in activating host transcription factors during the infection of airway epithelial cells with P. aeruginosa.
MATERIALS AND METHODS
Bacterial strains and human epithelial cells.
The P. aeruginosa strains used in this study are listed in Table 1. CFTE29o− cells possess a ΔF508 mutation of CFTR and were derived from human tracheobronchial epithelium (13). Cells were cultured on bovine serum albumin-collagen-fibronectin-coated plastic, using minimum essential medium supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were incubated at 37°C in a humidified 5% CO2 atmosphere. All cell culture reagents were supplied by Sigma, unless stated otherwise.
TABLE 1.
Pseudomonas aeruginosa strains used in this study
| Strain | Relevant genotype or description | Reference or source |
|---|---|---|
| PAO1 | Wild type | 20 |
| PAZH13 | rsmA deletion mutant, derivative of PAO1 | 45 |
| RP289 | PAO1 | 59 |
| RP550 | exoS deletion mutant, derivative of RP289 | 59 |
| RP555 | exoT deletion mutant, derivative of RP289 | 59 |
| RP562 | exoY deletion mutant, derivative of RP289 | 59 |
| RP580 | exoST deletion mutant, derivative of RP289 | 59 |
| RP568 | exoSY deletion mutant, derivative of RP289 | 59 |
| RP564 | exoTY deletion mutant, derivative of RP289 | 59 |
| RP576 | exoSTY deletion mutant, derivative of RP289 | 59 |
| PAK | Wild type | 11 |
| retS | retS deletion mutant, derivative of PAK | 11 |
| PA14 | Wild type | 47 |
| MH1 | Human clinical isolate | Mercy University Hospital, Cork, Ireland |
| MH2 | Human clinical isolate | Mercy University Hospital, Cork, Ireland |
Epithelial cell infection conditions.
For epithelial cell infection studies, cells were washed twice with sterile phosphate-buffered saline, detached, and seeded in antibiotic-free medium into newly coated plastic vessels 24 h prior to infection so as to achieve 80% confluence at the time of infection. Bacterial strains were cultured aerobically for 16 to 18 h in the described cell culture medium without antibiotics (infection medium) at 37°C. Bacteria were subcultured in infection medium for an additional 4 h under the growth conditions described above so that bacterial strains were in the exponential phase of growth. Following three rounds of centrifugation (4,000 × g) and phosphate-buffered saline washing to remove extracellular components, bacterial densities were adjusted so as to infect cells at a multiplicity of infection (MOI) of approximately 50:1. Serial dilutions were plated onto Luria-Bertani (LB) agar to confirm the MOI used. At the indicated times postinfection, cells which were already detached from the plastic were collected by centrifugation, and those cells remaining attached were removed with trypsin-EDTA prior to collection by centrifugation. Harvested cells were frozen immediately at −70°C for subsequent RNA isolation.
Epithelial cell infection studies using bacterially conditioned cell-free supernatant, heat-killed bacteria, or LPS.
Epithelial cells and bacterial strains were cultured as described above. For the generation of bacterially conditioned cell-free supernatant (bacterial supernatant), bacterial cells (after 4 h of subculture) were pelleted by centrifugation (4,000 × g), and the supernatant was subsequently filter sterilized (0.02 μm). To prepare heat-killed bacteria, bacterial strains (after 4 h of subculture) were pelleted by centrifugation in a plastic container which was placed in boiling water for 30 min. In each case, preparations were confirmed to be free of viable bacteria by plating aliquots of the samples on LB agar. Preparations of bacterial supernatant or heat-killed bacteria were added to the cells to reflect an MOI of 50:1. Lipopolysaccharide (LPS; diluted in infection medium) from P. aeruginosa (serotype 10) or Escherichia coli (O26:B6) (both supplied by Sigma) was added to epithelial cells at a final concentration of 10 μg/ml. Epithelial cells were harvested as described above.
RNA isolation and array sample preparation.
Total RNA was isolated from epithelial cells by using an RNeasy kit (QIAGEN) according to the manufacturer's instructions. Genomic DNA was removed by using DNA-free (Ambion), and the samples were confirmed by PCR to be free of DNA prior to cDNA synthesis. Biological samples from two independent infection experiments were used in two independent array experiments, as outlined below. Isolated RNAs were used in the preparation of fragmented cRNAs for array hybridization according to the manufacturer's protocol (Affymetrix). Briefly, 8.5 μg of RNA was converted to double-stranded cDNA by reverse transcription, using a Superscript cDNA synthesis kit (Invitrogen) and an oligo(dT) primer containing a T7 RNA polymerase site added 3′ of the poly(T) (Affymetrix). After second-strand synthesis, cDNAs were converted to biotin-labeled cRNAs by in vitro transcription (Enzo). The labeled cRNAs were purified using a Genechip clean-up module (QIAGEN). Total cRNA quality and yield were measured using a GeneQuant spectrophotometer, and 20 μg of cRNA was fragmented at 94°C for 35 min in fragmentation buffer (40 mM Tris-acetate, pH 8.1, 100 mM potassium acetate, 30 mM magnesium acetate). Full-length and fragmented cRNA sample integrity was assessed by electrophoresis on a 1% agarose-Tris-borate-EDTA gel, with visualization by staining with SYBR green. Fragmented cRNAs were sent to Geneservice (Cambridge, United Kingdom) for assessment of RNA quality, using a Bioanalyzer (Agilent), before hybridization to a human genome U133A array (Affymetrix). The arrays were washed and stained in a fluidic station and then scanned according to the manufacturer's protocols (Affymetrix). For the six array samples, the 3′/5′ ratio of the control probe sets did not reach a value of >3, indicating that sample preparation was not compromised in our study.
Microarray data analysis.
Raw image analysis was performed using Microarray Suite v5.0 software (Affymetrix). Downstream data analysis was performed using GeneSight software, version 2.7 (Biodiscovery). For all analyses, the signal intensities from two independent infection and array experiments were used. To filter the array data, analysis of variance for comparison between multiple groups of samples (uninfected cells or PAO1- or rsmA mutant-infected cells) was used to investigate significant differences (P < 0.05) in gene expression between samples. Using the filtered data, a series of Student's t tests was employed to determine significant differences (P < 0.05) in gene expression between two particular samples.
For subsequent analysis, a 1.5-fold cutoff was applied between two samples to identify different levels of gene expression.
Investigation of biological processes with significant alterations postinfection.
Expression Analysis Systematic Explorer (EASE; http://david.niaid.nih.gov/david/ease.htm) was used to facilitate the biological interpretation of lists of transcripts derived from the array analysis. The EASE score measures the statistical significance of changes in regulation of a biological process or molecular function by comparing the number of genes changed experimentally to the total number of genes spotted on the array that are annotated for that process, thus identifying categories of genes that are overrepresented under the experimental conditions. Categories with P values of <0.05 compared to uninfected control cells were considered significantly altered. Gene Ontology (http://www.geneontology.org) annotations were used to categorize the lists of biological processes.
Real-time reverse transcription-PCR analysis.
RNAs were isolated and genomic DNA was removed as described above. Five hundred nanograms of total RNA was reverse transcribed using oligo(dT) and Expand reverse transcriptase (Roche) as recommended by the manufacturer. Contaminating bacterial cDNAs were not detected using species-specific primers. cDNA standards were prepared, and real-time reverse transcription-PCR analysis was carried out using a Light Cycler apparatus (Roche) as previously described (42). MgCl2 concentrations and thermocycling conditions were optimized for each primer set (available upon request), and melting curve analysis confirmed the specificity of the reaction, as described previously (48). Normalization of gene expression between samples was carried out using hypoxanthine phosphoribosyltransferase (HPRT) as a control (4), as its expression was not significantly altered, as determined by array analysis (data not shown).
Quantification of cell cytotoxicity by measuring LDH release.
The release of lactate dehydrogenase (LDH) into culture supernatants was measured using an LDH cytotoxicity detection kit (Roche) according to the manufacturer's instructions. FBS was omitted from the infection medium, as the inclusion of FBS resulted in high background levels of LDH activity. Infection of epithelial cells was carried out as described above, using an MOI of 50:1, and allowed to proceed for the indicated times postinfection.
Microarray accession number.
The entire array data set has been deposited in the Gene Expression Omnibus (GEO) database at NCBI (http://www.ncbi.nlm.nih.gov/geo/) under GEO accession number GSE4485.
RESULTS
Comparison of significantly altered epithelial transcripts after infection with PAO1 or the rsmA mutant.
Recent work in our laboratory investigated the impact of RsmA on a number of airway epithelial cell phenotypes, including actin depolymerization, cytotoxicity, and invasion. These cellular phenotypes were influenced by the positive effect of RsmA on the expression of the TTSS in P. aeruginosa (41). To obtain a greater understanding of the role of RsmA during the interaction of P. aeruginosa with host cells, gene expression profiles were generated using U133A arrays for airway epithelial cells infected with PAO1 or the rsmA mutant and compared to those for uninfected cells. It was hypothesized that a 1.5-h infection time, coupled with an MOI of 50:1, would enable the identification of host genes which were differentially expressed during the early stages of infection, prior to a host transcriptional response associated with cell death.
The identification of transcripts with significantly altered expression levels was performed by using a combination of statistical approaches, as described in Materials and Methods. Infection with PAO1 was responsible for 241 transcripts with increased expression levels and 112 transcripts with decreased expression levels compared to those in uninfected cells at 1.5 h postinfection (see Table S1 in the supplemental material). By comparison, infection with the rsmA mutant resulted in 151 and 123 transcripts with increased and decreased transcription levels, respectively, compared to those in uninfected cells (see Table S2 in the supplemental material).
In the current study, the rsmA mutant failed to significantly alter the expression of a number of transcripts which were significantly increased following infection with PAO1. A number of these PAO1-regulated transcripts were also identified by other investigators following infection of epithelial cells with P. aeruginosa. These included PAO1-induced expression of the phosphatases DUSP1 (37) and DUSP10 (23) and the Rho protein RHOB (37). McMorran et al. (37) showed that infection with P. aeruginosa PA103 caused induction of the transcriptional regulator NR4A1, while in the current study, PAO1 infection resulted in the detection of elevated expression levels of the related nuclear receptor NR4A2 (see Table S1 in the supplemental material). Additionally, PAO1 infection increased the expression of the transcriptional regulator Jun (see Table S1 in the supplemental material), as previously observed in three separate studies (22, 24, 50). In our study, infection with PAO1 increased the expression of two related transcriptional regulators, KLF2 and KLF6 (see Table S1 in the supplemental material). In supplementary material provided by Hybiske et al. (22), increased expression of KLF6, but not KLF2, was observed following infection of a CF nasal cell line with P. aeruginosa PAK.
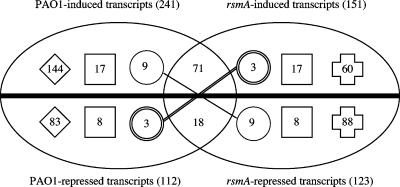
To differentiate between the responses of epithelial cells to infections with PAO1 and the rsmA mutant, significantly altered transcripts (relative to those in uninfected control cells) were grouped so as to distinguish those that were similarly (see Table S3 in the supplemental material), differentially (see Table S4 in the supplemental material), and reciprocally (see Table S5 in the supplemental material) regulated by PAO1 and the rsmA mutant (Fig. 1). Fourteen percent of the total number of significantly altered transcripts were regulated in a similar manner by PAO1 and the rsmA mutant compared to those in uninfected cells (71 and 18 transcripts with increased and decreased transcription levels, respectively) (Fig. 1; see Table S3 in the supplemental material). Those with increased transcription levels included two members of the solute carrier family (SLC13A3 and SLC26A3). Infection with PAO1 and the rsmA mutant resulted in decreased expression of the cell surface receptor CD44, which is involved in the activation of cytotoxic T cells (38).
FIG. 1.
Analysis of epithelial gene regulation after infection with PAO1 and the rsmA mutant. The Venn diagram illustrates the number of significantly altered transcripts, as identified by array analysis, which were similarly, differentially, or reciprocally regulated after infection with PAO1 or the rsmA mutant. The thick horizontal line represents those transcripts that were not significantly altered compared to those in uninfected cells. Values above and below the line represent transcripts with increased and decreased expression levels, respectively, compared to those in uninfected cells. Values in the intersecting region of the Venn diagram represent the numbers of transcripts which were similarly regulated by PAO1 and the rsmA mutant. Values in circles represent those transcripts which were reciprocally regulated; values in squares represent identical transcripts which were differentially regulated due to a significant difference (≥1.5-fold) in expression level between PAO1 and the rsmA mutant (t test; P < 0.05); and values in diamonds and crosses indicate transcripts significantly altered by PAO1 and the rsmA mutant, respectively. The total numbers of transcripts with increased and decreased expression levels in response to infection with PAO1 or the rsmA mutant are indicated in parentheses.
Those transcripts that were differentially regulated by infection were subdivided into the following two groups: (i) transcripts whose expression levels were significantly altered by either PAO1 or the rsmA mutant compared to those in uninfected cells and (ii) transcripts whose expression levels were significantly altered by both PAO1 and the rsmA mutant compared to those in uninfected cells but which showed significant differences in expression between the two infected samples.
Infection with PAO1 resulted in 144 and 83 transcripts with increased and decreased transcription levels, respectively, which were not significantly altered by infection with the rsmA mutant (Fig. 1; see Table S4 in the supplemental material). These transcripts included those for the transcription factors KLF2 and KLF6 (PAO1 induced), the immunity-related surface receptor HLA-DRB1 (PAO1 repressed), and the chemokine CCL19 (PAO1 repressed). In contrast, infection with the rsmA mutant resulted in 60 and 88 transcripts with increased and decreased transcription levels, respectively, which were not significantly altered by infection with PAO1 (Fig. 1; see Table S4 in the supplemental material). Included in this list were the genes for INFA8 (rsmA induced), which is known to be a target of the transcriptional regulator IRF5, and the cell adhesion molecules CCL11, ICAM2, ISLR, and LAMA1 (rsmA repressed).
The second group of differentially regulated transcripts, which were significantly altered by both strains but showed significant differences in the levels of expression between the two infected samples, was made up of 17 and 8 transcripts with increased and decreased transcription levels, respectively, compared to those in uninfected cells (Fig. 1; see Table S4 in the supplemental material). Included in this group were the genes for the signal transduction molecule MAP2K1, which was induced to a higher level as a result of infection with PAO1 than that in rsmA mutant-infected cells, and the cytokine interleukin-2, which showed a lower level of transcription as a result of infection with the rsmA mutant than that in PAO1-infected cells.
A small number of transcripts exhibited reciprocal regulation when cells were infected with PAO1 or the rsmA mutant (Fig. 1; see Table S5 in the supplemental material). Included in the list of nine transcripts shown to be PAO1 induced but rsmA repressed compared to those in uninfected cells were transcripts involved in the regulation of transcription (RORA, SMARCA2, and HOXA9). The three transcripts which were PAO1 repressed but rsmA induced were those for DAPK1, KCNJ5, and SAMD4, all of which are involved in ion transport.
Infection with PAO1 or the rsmA mutant alters a distinct set of biological processes.
EASE software (http://david.niaid.nih.gov/david/ease.htm) with Gene Ontology annotations (http://www.geneontology.org) was employed to investigate the biological processes (Table 2) that were significantly altered during infection of epithelial cells with PAO1 or the rsmA mutant. This statistical analysis revealed clear differences in the biological processes that were altered as a result of infections of epithelial cells with PAO1 and the rsmA mutant. PAO1, but not the rsmA mutant, caused significant alterations in biological processes such as cell growth, positive regulation of cell proliferation, and immune cell and lymphocyte activation. Furthermore, PAO1 infection resulted in significant changes in pathways related to transcription and the regulation of transcription at 1.5 h postinfection which were not changed after infection with the rsmA mutant (Table 2). Additionally, genes involved in immune and defense responses were overrepresented among the genes that were significantly altered postinfection with PAO1 but not with the rsmA mutant. In contrast, the rsmA mutant was responsible for the alteration of processes involving cell communication and the metabolism of lipids, aromatic amino acids, and amino acid derivatives (Table 2).
TABLE 2.
Biological processes significantly altered in response to infection with PAO1 and the rsmA mutant
| Gene Ontology annotation categorya | EASE scoreb
|
Targets chosen for reverse transcription-PCR | |
|---|---|---|---|
| Uninfected vs PAO1-infected cells | Uninfected vs rsmA mutant-infected cells | ||
| Development | 0.00327259 | 0.044995631 | FGF18, KLF6c |
| Cell growth | 0.003632335 | NS | |
| Regulation of transcription | 0.004455422 | NS | Jun, FGF18, KLF2, KLF6 |
| Immune cell activation | 0.004748091 | NS | KLF6 |
| Cell activation | 0.004748091 | NS | KLF6 |
| Regulation of transcription, DNA dependent | 0.005062711 | NS | Jun, FGF18, KLF2, KLF6 |
| Lymphocyte activation | 0.010608842 | NS | KLF6 |
| Transcription, DNA dependent | 0.012742898 | NS | Jun, FGF18, KLF2, KLF6 |
| Response to biotic stimulus | 0.0163397 | NS | KLF6 |
| Transcription | 0.016418748 | NS | Jun, FGF18, KLF2, KLF6 |
| Immune response | 0.017303871 | NS | KLF6 |
| Response to external stimulus | 0.018355915 | NS | KLF6 |
| Regulation of cellular process | 0.019197391 | NS | FGF18 |
| Regulation of biological process | 0.021607717 | NS | FGF18 |
| Regulation of neurotransmitter levels | 0.022346738 | 0.009804003 | |
| Organogenesis | 0.031143844 | NS | FGF18, KLF6 |
| Regulation of cell growth | 0.031228117 | NS | |
| Signal transduction | 0.043411124 | NS | FGF18 |
| Positive regulation of cell proliferation | 0.046373762 | NS | FGF18 |
| Morphogenesis | 0.047113929 | NS | FGF18, KLF6 |
| Phenol metabolism | 0.048148555 | NS | |
| Defense response | 0.049987952 | NS | KLF6 |
| Cell communication | NS | 0.004495031 | |
| Lipid metabolism | NS | 0.020087184 | |
| Cell adhesion | NS | 0.021125648 | |
| Lipid catabolism | NS | 0.029643693 | |
| Amino acid derivative metabolism | NS | 0.037618284 | |
| Aromatic amino acid family metabolism | NS | 0.043814181 | |
| Cellular process | NS | 0.044110669 | |
Gene Ontology categories for biological processes (http://www.geneontology.org) determined by EASE to be significantly overrepresented after infection with PAO1 or the rsmA mutant.
The EASE score measures the statistical significance of changes in the regulation of biological processes by comparing the number of genes that were seen to change experimentally to the total number of genes spotted on the U133A array that are annotated for that biological process. NS, not significantly altered (EASE score of >0.05 for uninfected versus infected cells).
While the biological process “development” was significantly overrepresented in response to infection with both PAO1 and the rsmA mutant, FGF18 and KLF6, which belong to this functional category, were significantly altered in response to infection with PAO1 only, not with the rsmA mutant.
Confirmation of array data using real-time reverse transcription-PCR.
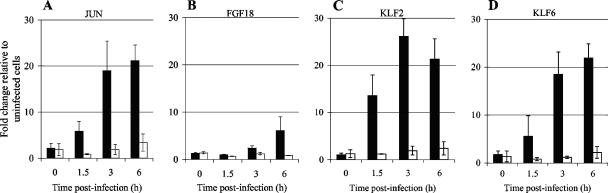
Four genes (those encoding Jun, fibroblast growth factor 18 [FGF18], KLF2, and KLF6) were chosen for verification of the array data because they are involved in cell growth/proliferation and transcriptional regulation. These genes were also chosen because they showed altered transcription in response to PAO1, but not the rsmA mutant, compared to that in uninfected cells. In addition, KLF2 and KLF6 expression has been shown to be induced by both gram-positive (40) and gram-negative (1, 19, 22, 54) bacteria. However, the mechanism(s) behind the induction of these genes has not yet been elucidated.
PAO1 caused increased expression of Jun at 1.5, 3, and 6 h postinfection, with maximal induction of approximately 20-fold (compared to the case in uninfected cells) observed at 6 h postinfection. In contrast, the rsmA mutant failed to increase the expression level of Jun more than 3.3-fold at 6 h postinfection (Fig. 2). Increased expression (sixfold) of FGF18 was only observed at 6 h postinfection with PAO1, while the rsmA mutant failed to increase the expression of FGF18 compared to that in uninfected cells (Fig. 2). Increased expression of KLF2 was detected as early as 1.5 h postinfection with PAO1. Maximal induction of KLF2, to a level approximately 25-fold higher than that in uninfected cells, was observed at 3 h postinfection (Fig. 2). KLF6 expression levels also increased in response to PAO1, with the most induction (21-fold) observed at 6 h postinfection (Fig. 2). In contrast, the rsmA mutant failed to alter the expression of either of these transcription factors more than 2.4-fold compared to that in uninfected cells for up to 6 h postinfection (Fig. 2).
FIG. 2.
Real-time reverse transcription-PCR analysis of epithelial cell gene expression after infection with PAO1 and the rsmA mutant. The expression of (A) Jun, (B) FGF18, (C) KLF2, and (D) KLF6 following infection with PAO1 (black bars) and the rsmA mutant (white bars) was investigated at 0, 1.5, 3, and 6 h postinfection (with a starting MOI of 50:1). Gene expression is presented as the x-fold change relative to expression in uninfected cells and was normalized for cDNA content by using the housekeeping gene encoding HPRT. Data (means ± standard deviations [SD]) are representative of three independent biological experiments.
Thus, for these four genes, temporal expression analysis confirmed the array data, which showed increased expression of these genes in response to PAO1 but not in response to the rsmA mutant. The real-time reverse transcription-PCR analysis indicated that early induction of Jun, KLF2, and KLF6 was observed in response to PAO1, yet it was also evident that sustained transcriptional stimulation was observed for up to 6 h postinfection for these three genes (Fig. 2).
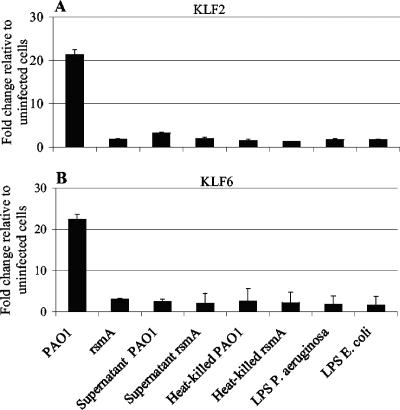
Cell-associated or secreted factors are insufficient to alter the expression of KLF2 or KLF6.
Gene expression analysis was conducted to identify if cell-associated or secreted factors were capable of increasing the expression of KLF2 or KLF6. Exposure of epithelial cells for 3 h to bacterially conditioned cell-free supernatant, heat-killed bacteria, or LPS from P. aeruginosa or E. coli was insufficient to alter the expression of either gene (Fig. 3). In the same assay, infection with PAO1, but not the rsmA mutant, resulted in increased expression of both genes compared to that in uninfected cells, as previously observed (Fig. 3). These data showed that gene expression of KLF2 and KLF6 was regulated in a more specific manner.
FIG. 3.
Real-time reverse transcription-PCR analysis of KLF2 and KLF6 expression in response to live bacteria, bacterially conditioned cell-free supernatant, heat-killed bacteria, and LPS. The expression of (A) KLF2 and (B) KLF6 following 3 h of exposure to live bacteria or selected bacterial preparations (with a starting MOI of 50:1 for live bacteria and equivalent MOIs for bacterial preparations) is shown. LPS was used at a final concentration of 10 μg/ml. Gene expression is presented as the x-fold change relative to expression in uninfected cells and was normalized for cDNA content by using the housekeeping gene encoding HPRT. Data (means ± SD) are representative of three independent biological experiments.
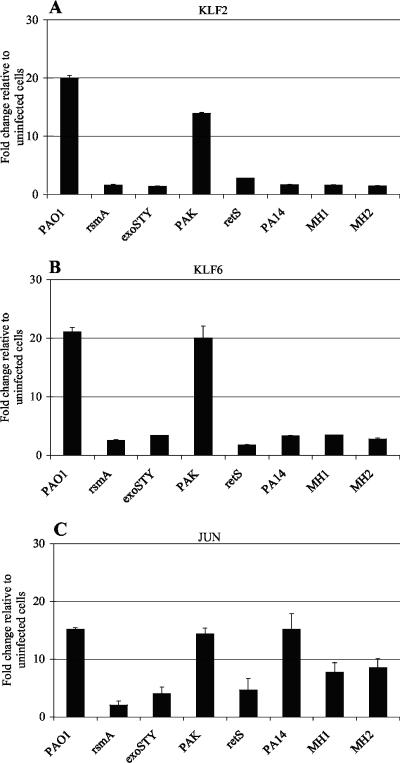
Increased expression of KLF2 and KLF6 is not observed in response to infection with P. aeruginosa strains lacking ExoS.
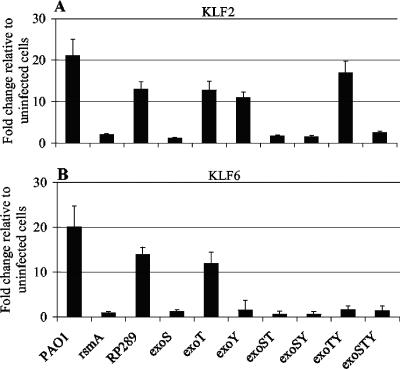
A number of well-characterized P. aeruginosa strains and mutants as well as clinical isolates were investigated for the ability to alter the gene expression of KLF2 and KLF6. Gene expression analysis indicated that PAO1 and PAK induced the expression of KLF2 and KLF6. However, elevated gene expression of KLF2 and KLF6 was not observed following infection with the rsmA mutant, an exoSTY mutant (PAO1 lacking the effector proteins ExoS, ExoT, and ExoY) (59), an retS mutant, or the clinical isolates PA14, MH1, and MH2 (Fig. 4). P. aeruginosa PA14 is known to secrete ExoT and ExoU but lacks the gene encoding ExoS and does not produce detectable levels of ExoY (39). The P. aeruginosa clinical isolates MH1 and MH2 display similar secretion profiles, in that they also secrete ExoT and ExoU but do not secrete detectable levels of ExoS and ExoY (H. Mulcahy, unpublished data). Thus, increased gene expression of KLF2 and KLF6 was only associated with strains capable of secreting ExoS (PAO1 and PAK) and was not observed following infection with strains lacking ExoS (rsmA, exoSTY, and retS mutants, PA14, MH1, and MH2). The pattern of gene regulation observed for Jun was not the same as that observed for KLF2 and KLF6, but appeared to be a more generalized response to infection. Compared to that in uninfected cells, a modest increase in expression of Jun was observed in response to the exoSTY and retS mutants (Fig. 4). Furthermore, increased expression of Jun was observed after infection with PA14, MH1, and MH2 compared to that in uninfected cells (Fig. 4).
FIG. 4.
Epithelial cell gene expression in response to a series of P. aeruginosa strains. Gene expression of (A) KLF2, (B) KLF6, and (C) Jun at 3 h postinfection (with a starting MOI of 50:1) is presented as the x-fold change relative to expression in uninfected cells and was normalized for cDNA content by using the housekeeping gene encoding HPRT. Data (means ± SD) are representative of three independent biological experiments.
ExoS-dependent induction of KLF2 and ExoS- and ExoY-dependent induction of KLF6.
In order to determine the potential impact of ExoS, ExoT, and ExoY on gene expression of KLF2 and KLF6, a series of isogenic mutants of PAO1 (59) were employed in epithelial cell infection experiments, and real-time reverse transcription-PCR was performed. At 3 h postinfection, increased gene expression of KLF2 was only observed in response to strains capable of producing the effector protein ExoS (Fig. 5). Separately, it was shown that increased gene expression of KLF6 depended on the production of both ExoS and ExoY (Fig. 5). This showed that production of ExoS alone and of ExoS and ExoY in combination was necessary for increased gene expression of KLF2 and KLF6, respectively.
FIG. 5.
Influence of ExoS, ExoT, and ExoY on gene expression of KLF2 and KLF6, using real-time reverse transcription-PCR. Gene expression of (A) KLF2 and (B) KLF6 at 3 h postinfection (with a starting MOI of 50:1) is presented as the x-fold change relative to expression in uninfected cells and was normalized for cDNA content by using the housekeeping gene encoding HPRT. Data (means ± SD) are representative of three independent biological experiments. The single, double, and triple mutants of exoS, exoT, and exoY are derivatives of a parental strain of P. aeruginosa PAO1 (RP289), as described previously (59).
Increased KLF2 and KLF6 expression is associated with ExoS-mediated cytotoxicity.
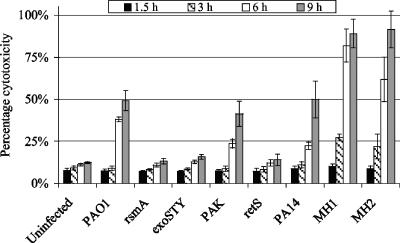
Previous work in our laboratory has shown that the rsmA mutant fails to cause cytotoxicity in epithelial cells, in a manner similar to that of an exoSTY mutant of PAO1 (41). Other investigators have shown a distinction between ExoS- and ExoU-mediated mammalian cell death (6, 8, 14, 34, 44, 58). Therefore, we sought to determine whether ExoS- or ExoU-mediated cytotoxicity was associated with the expression of KLF2 and KLF6 by exposing epithelial cells to a series of strains capable of producing these effector proteins. PAO1 and PAK secrete ExoS, ExoT, and ExoY but not ExoU, and they were shown to cause increased cell death compared to that of uninfected cells (Fig. 6). However, the rsmA, exoSTY, and retS mutants, which do not secrete any effector proteins, failed to induce cytotoxicity compared to their respective parental P. aeruginosa strains (Fig. 6). Compared to those for uninfected cells, increased levels of cell death were observed following infection with PA14, MH1, and MH2. In fact, the cytotoxicity observed in response to the clinical isolates MH1 and MH2 was more rapid and more potent than that observed with all the other strains tested (Fig. 6). These results suggested that the increased gene expression of KLF2 and KLF6 observed in response to P. aeruginosa infection was associated with ExoS- but not ExoU-mediated cell death.
FIG. 6.
Epithelial cell cytotoxicity in response to a series of P. aeruginosa strains. The release of LDH into culture supernatants was measured using an LDH cytotoxicity detection kit (Roche) at the indicated times postinfection (with a starting MOI of 50:1). Cytotoxicity is expressed as a percentage of the total amount of LDH released from cells treated with 1% Triton X-100. Data (means ± SD) are representative of two independent biological experiments.
DISCUSSION
In this study, the host transcriptional response to P. aeruginosa infection, specifically those transcripts altered as a consequence of RsmA-mediated regulation, was investigated. In the model proposed by Goodman et al. (11), environmental signals during chronic infection lead to decreased levels of free RsmA. A number of investigators have also shown that mutation of rsmA results in a number of phenotypes associated with isolates from chronically infected patients, including a lack of motility (2, 18), increased production of homoserine lactones (18), increased antibiotic resistance (41), and a lack of a functional TTSS (41). Thus, we sought to investigate the differential response of epithelial cells to PAO1 and the rsmA mutant.
Microarray analysis demonstrated the significance of RsmA in modulating the host response to infection. RsmA influenced the expression of a large number of epithelial transcripts in a distinct manner. This was evident in the number of differentially regulated transcripts uniquely regulated by PAO1 or the rsmA mutant. The rsmA mutant failed to alter transcripts involved in controlling a number of biological processes relevant to microbial infection, including immune and defense responses and immune cell and lymphocyte activation. Mining of the array data also pointed to the importance of genes involved in the regulation of transcription at 1.5 h postinfection. Temporal expression analysis confirmed the array data, which showed increased expression of Jun, FGF18, KLF2, and KLF6 in response to PAO1 but not in response to the rsmA mutant. PAO1-induced expression of KLF2 and KLF6 was also observed in a non-CF tracheal epithelial cell line (our unpublished data). However, since this was not an isogenically derived cell line, we cannot conclude definitively regarding the significance of this result.
As members of the KLF transcription factor family, KLF2 and KLF6 play important roles in cell proliferation and the host response to tissue injury. KLF2 prevents apoptosis of T lymphocytes (32) and has been shown to be important for the development of blood vessels, as KLF2 knockout mice die at embryonic day 12 due to hemorrhage (31). KLF6 binding has been shown to alter the transcription of a number of genes with roles in cell proliferation, development, and tissue repair (28, 29, 63). Increased expression of KLF2 following infection of murine macrophages with Yersinia enterocolitica has been reported (1, 19, 54). Additionally, induction of both KLF2 and KLF6 has been documented following infection of tracheal epithelial cells with Staphylococcus aureus (40). While P. aeruginosa-mediated induction of KLF6 has previously been reported as supplementary data by Hybiske et al. (22), to our knowledge this is the first report indicating increased expression of KLF2 in response to P. aeruginosa infection and the first to identify the specific bacterial component(s) responsible for the induction of KLF2 and KLF6.
KLF2 and KLF6 expression was significantly induced in response to infection with PAO1, but not the rsmA mutant, indicating a role for downstream targets of RsmA in eliciting this specific host response. Subsequent investigations indicated that live bacterial infection was required for the induction of KLF2 and KLF6, as bacterially conditioned cell-free supernatant, heat-inactivated bacteria, or LPS was insufficient for the induction of KLF2 or KLF6. These data, in conjunction with our recent data showing the absence of a functional TTSS in the rsmA mutant (41), prompted us to investigate whether the type III effectors were possible modulators of KLF2 and KLF6 expression.
Elevated transcriptional expression of KLF2 was observed only in response to infections with strains capable of secreting ExoS. Since the ADPRT, but not GAP, activities of ExoS and ExoT differ in terms of substrate specificity, this suggests that the ADPRT activity of ExoS was responsible for the observed increase in gene expression of KLF2 during infection. It has also been shown that the ADPRT activity of ExoS, not that of ExoT, is required to induce cytotoxicity (44) and apoptosis (26, 27) in cultured mammalian cells. Recently, it was shown that the ADPRT activity of ExoS is essential for virulence in a mouse model of acute pneumonia (55). To clarify the specific roles of these domains, expression analysis of KLF2 in response to ExoS with a mutated GAP and/or ADPRT domain would be necessary. Induction of KLF6 was detected only in response to strains capable of secreting ExoS in combination with ExoY. This suggested that the specific ADPRT activity of ExoS, in combination with bacterial production of ExoY, was required for the induction of KLF6, although this has not yet been investigated. Synergistic effects between the type III effector proteins in eliciting a distinct host transcriptional response to P. aeruginosa infection were shown recently (23). However, the study by Ichikawa et al. did not highlight synergy, specifically that between ExoS and ExoY, in influencing the host response to infection, as seen with KLF6 in our study. While differential expression of KLF2 or KLF6 was not detected (23), this may be attributable to the use of different cell lines, P. aeruginosa strains, and infection conditions.
Epithelial cell death due to the presence of ExoS but not ExoU was associated with elevated gene expression of KLF2 and KLF6. Three clinical isolates of P. aeruginosa, including PA14, failed to increase the expression of either KLF2 or KLF6, and this was attributed to the lack of production of both ExoS and ExoY by these strains. However, we cannot rule out the possibility that these strains did not fail in their ability to increase the expression of these genes, but rather acted to suppress induction, a condition which may ultimately favor the bacterium. Whether the induction of KLF2 and KLF6 is a mechanism of host defense or serves to benefit the bacterium is not currently known. This issue is the subject of current investigations.
In this study, we have shown that regulation of the TTSS mediated by the posttranscriptional regulator RsmA has important effects on the host transcriptional response to P. aeruginosa infection. Significant alterations in the expression of genes involved in the regulation of transcription were observed early in the infection process. ExoS and ExoY were shown to specifically increase the expression of selected human transcription factors, and this increased gene expression was associated with ExoS-mediated cytotoxicity in epithelial cells. The host transcription factors KLF2 and KLF6 have important roles in the regulation of genes associated with cell proliferation, lung development, and tissue injury. Delineation of the molecular mechanisms by which ExoS and ExoY influence the expression of these genes may help to identify the importance of exoenzyme-mediated modulation of host gene expression during infection with P. aeruginosa.
Supplementary Material
Acknowledgments
We acknowledge kind gifts of cells and strains from James Clair, Andrew L. Goodman, Dieter C. Gruenert, Russell E. Vance, Marvin Whiteley, and Paul Williams. We thank Pat Higgins and Maurice O'Donoghue for excellent technical assistance, Fiona Riordan for help with preliminary experiments, and Kathleen O'Sullivan for invaluable advice concerning statistical analysis.
This work was supported in part by grants awarded by the Higher Education Authority of Ireland (PRTLI programs to F.O.), The Science Foundation of Ireland (SFI 02/IN.1/B1261, 04/BR/B0597 to F.O.), the European Commission (QLK3-CT-2000-31759, QLTK3-CT-2001-0010, and QLK5-CT-2002-0091 to F.O.), and the Health Research Board (RP76/2001 and RP/2004/145 to F.O.).
Editor: V. J. DiRita
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Bohn, E., S. Muller, J. Lauber, R. Geffers, N. Speer, C. Spieth, J. Krejci, B. Manncke, J. Buer, A. Zell, and I. B. Autenrieth. 2004. Gene expression patterns of epithelial cells modulated by pathogenicity factors of Yersinia enterocolitica. Cell. Microbiol. 6:129-141. [DOI] [PubMed] [Google Scholar]
- 2.Burrowes, E., C. Baysse, C. Adams, and F. O'Gara. 2006. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152:405-418. [DOI] [PubMed] [Google Scholar]
- 3.Cowell, B. A., D. J. Evans, and S. M. Fleiszig. 2005. Actin cytoskeleton disruption by ExoY and its effects on Pseudomonas aeruginosa invasion. FEMS Microbiol. Lett. 250:71-76. [DOI] [PubMed] [Google Scholar]
- 4.de Kok, J. B., R. W. Roelofs, B. A. Giesendorf, J. L. Pennings, E. T. Waas, T. Feuth, D. W. Swinkels, and P. N. Span. 2005. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Investig. 85:154-159. [DOI] [PubMed] [Google Scholar]
- 5.Feltman, H., G. Schulert, S. Khan, M. Jain, L. Peterson, and A. R. Hauser. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147:2659-2669. [DOI] [PubMed] [Google Scholar]
- 6.Finck-Barbancon, V., J. Goranson, L. Zhu, T. Sawa, J. P. Wiener-Kronish, S. M. Fleiszig, C. Wu, L. Mende-Mueller, and D. W. Frank. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25:547-557. [DOI] [PubMed] [Google Scholar]
- 7.Fraylick, J. E., J. R. La Rocque, T. S. Vincent, and J. C. Olson. 2001. Independent and coordinate effects of ADP-ribosyltransferase and GTPase-activating activities of exoenzyme S on HT-29 epithelial cell function. Infect. Immun. 69:5318-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frithz-Lindsten, E., Y. Du, R. Rosqvist, and A. Forsberg. 1997. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III-dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol. Microbiol. 25:1125-1139. [DOI] [PubMed] [Google Scholar]
- 9.Garrity-Ryan, L., B. Kazmierczak, R. Kowal, J. Comolli, A. Hauser, and J. N. Engel. 2000. The arginine finger domain of ExoT contributes to actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infect. Immun. 68:7100-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goehring, U. M., G. Schmidt, K. J. Pederson, K. Aktories, and J. T. Barbieri. 1999. The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase-activating protein for Rho GTPases. J. Biol. Chem. 274:36369-36372. [DOI] [PubMed] [Google Scholar]
- 11.Goodman, A. L., B. Kulasekara, A. Rietsch, D. Boyd, R. S. Smith, and S. Lory. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745-754. [DOI] [PubMed] [Google Scholar]
- 12.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruenert, D. C., M. Willems, J. J. Cassiman, and R. A. Frizzell. 2004. Established cell lines used in cystic fibrosis research. J. Cyst. Fibros. 3(Suppl. 2):191-196. [DOI] [PubMed] [Google Scholar]
- 14.Hauser, A. R., S. Fleiszig, P. J. Kang, K. Mostov, and J. N. Engel. 1998. Defects in type III secretion correlate with internalization of Pseudomonas aeruginosa by epithelial cells. Infect. Immun. 66:1413-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He, J., R. L. Baldini, E. Deziel, M. Saucier, Q. Zhang, N. T. Liberati, D. Lee, J. Urbach, H. M. Goodman, and L. G. Rahme. 2004. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. USA 101:2530-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 14:1351-1363. [DOI] [PubMed] [Google Scholar]
- 17.Henriksson, M. L., C. Sundin, A. L. Jansson, A. Forsberg, R. H. Palmer, and B. Hallberg. 2002. Exoenzyme S shows selective ADP-ribosylation and GTPase-activating protein (GAP) activities towards small GTPases in vivo. Biochem. J. 367:617-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heurlier, K., F. Williams, S. Heeb, C. Dormond, G. Pessi, D. Singer, M. Camara, P. Williams, and D. Haas. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186:2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann, R., K. van Erp, K. Trulzsch, and J. Heesemann. 2004. Transcriptional responses of murine macrophages to infection with Yersinia enterocolitica. Cell. Microbiol. 6:377-390. [DOI] [PubMed] [Google Scholar]
- 20.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hybiske, K., J. K. Ichikawa, V. Huang, S. J. Lory, and T. E. Machen. 2004. Cystic fibrosis airway epithelial cell polarity and bacterial flagellin determine host response to Pseudomonas aeruginosa. Cell. Microbiol. 6:49-63. [DOI] [PubMed] [Google Scholar]
- 23.Ichikawa, J. K., S. B. English, M. C. Wolfgang, R. Jackson, A. J. Butte, and S. Lory. 2005. Genome-wide analysis of host responses to the Pseudomonas aeruginosa type III secretion system yields synergistic effects. Cell. Microbiol. 7:1635-1646. [DOI] [PubMed] [Google Scholar]
- 24.Ichikawa, J. K., A. Norris, M. G. Bangera, G. K. Geiss, A. B. van't Wout, R. E. Bumgarner, and S. Lory. 2000. Interaction of Pseudomonas aeruginosa with epithelial cells: identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proc. Natl. Acad. Sci. USA 97:9659-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iglewski, B. H., J. Sadoff, M. J. Bjorn, and E. S. Maxwell. 1978. Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyltransferase distinct from toxin A. Proc. Natl. Acad. Sci. USA 75:3211-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia, J., M. Alaoui-El-Azher, M. Chow, T. C. Chambers, H. Baker, and S. Jin. 2003. c-Jun NH2-terminal kinase-mediated signaling is essential for Pseudomonas aeruginosa ExoS-induced apoptosis. Infect. Immun. 71:3361-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaufman, M. R., J. Jia, L. Zeng, U. Ha, M. Chow, and S. Jin. 2000. Pseudomonas aeruginosa mediated apoptosis requires the ADP-ribosylating activity of exoS. Microbiology 146:2531-2541. [DOI] [PubMed] [Google Scholar]
- 28.Kim, Y., V. Ratziu, S. G. Choi, A. Lalazar, G. Theiss, Q. Dang, S. J. Kim, and S. L. Friedman. 1998. Transcriptional activation of transforming growth factor beta1 and its receptors by the Kruppel-like factor Zf9/core promoter-binding protein and Sp1. Potential mechanisms for autocrine fibrogenesis in response to injury. J. Biol. Chem. 273:33750-33758. [DOI] [PubMed] [Google Scholar]
- 29.Kojima, S., S. Hayashi, K. Shimokado, Y. Suzuki, J. Shimada, M. P. Crippa, and S. L. Friedman. 2000. Transcriptional activation of urokinase by the Kruppel-like factor Zf9/COPEB activates latent TGF-beta1 in vascular endothelial cells. Blood 95:1309-1316. [PubMed] [Google Scholar]
- 30.Krall, R., G. Schmidt, K. Aktories, and J. T. Barbieri. 2000. Pseudomonas aeruginosa ExoT is a Rho GTPase-activating protein. Infect. Immun. 68:6066-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo, C. T., M. L. Veselits, K. P. Barton, M. M. Lu, C. Clendenin, and J. M. Leiden. 1997. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 11:2996-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo, C. T., M. L. Veselits, and J. M. Leiden. 1997. LKLF: a transcriptional regulator of single-positive T cell quiescence and survival. Science 277:1986-1990. [DOI] [PubMed] [Google Scholar]
- 33.Lau, G. W., D. J. Hassett, and B. E. Britigan. 2005. Modulation of lung epithelial functions by Pseudomonas aeruginosa. Trends Microbiol. 13:389-397. [DOI] [PubMed] [Google Scholar]
- 34.Lee, V. T., R. S. Smith, B. Tummler, and S. Lory. 2005. Activities of Pseudomonas aeruginosa effectors secreted by the type III secretion system in vitro and during infection. Infect. Immun. 73:1695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051-1060. [DOI] [PubMed] [Google Scholar]
- 36.McGuffie, E. M., J. E. Fraylick, D. J. Hazen-Martin, T. S. Vincent, and J. C. Olson. 1999. Differential sensitivity of human epithelial cells to Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 67:3494-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMorran, B., L. Town, E. Costelloe, J. Palmer, J. Engel, D. Hume, and B. Wainwright. 2003. Effector ExoU from the type III secretion system is an important modulator of gene expression in lung epithelial cells in response to Pseudomonas aeruginosa infection. Infect. Immun. 71:6035-6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNally, J. M., D. Dempsey, R. M. Wolcott, R. Chervenak, and S. R. Jennings. 1999. Phenotypic identification of antigen-dependent and antigen-independent CD8 CTL precursors in the draining lymph node during acute cutaneous herpes simplex virus type 1 infection. J. Immunol. 163:675-681. [PubMed] [Google Scholar]
- 39.Miyata, S., M. Casey, D. W. Frank, F. M. Ausubel, and E. Drenkard. 2003. Use of the Galleria mellonella caterpillar as a model host to study the role of the type III secretion system in Pseudomonas aeruginosa pathogenesis. Infect. Immun. 71:2404-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreilhon, C., D. Gras, C. Hologne, O. Bajolet, F. Cottrez, V. Magnone, M. Merten, H. Groux, E. Puchelle, and P. Barbry. 2005. Live Staphylococcus aureus and bacterial soluble factors induce different transcriptional responses in human airway cells. Physiol. Genomics 20:244-255. [DOI] [PubMed] [Google Scholar]
- 41.Mulcahy, H., J. O'Callaghan, E. P. O'Grady, C. Adams, and F. O'Gara. 2006. The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect. Immun. 74:3012-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulcahy, H., K. P. O'Rourke, C. Adams, M. G. Molloy, and F. O'Gara. 2006. LST1 and NCR3 expression in autoimmune inflammation and in response to IFN-gamma, LPS and microbial infection. Immunogenetics 57:893-903. [DOI] [PubMed] [Google Scholar]
- 43.Olson, J. C., E. M. McGuffie, and D. W. Frank. 1997. Effects of differential expression of the 49-kilodalton exoenzyme S by Pseudomonas aeruginosa on cultured eukaryotic cells. Infect. Immun. 65:248-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pederson, K. J., A. J. Vallis, K. Aktories, D. W. Frank, and J. T. Barbieri. 1999. The amino-terminal domain of Pseudomonas aeruginosa ExoS disrupts actin filaments via small-molecular-weight GTP-binding proteins. Mol. Microbiol. 32:393-401. [DOI] [PubMed] [Google Scholar]
- 45.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. Holden, M. Camara, D. Haas, and P. Williams. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pier, G. B., M. Grout, and T. S. Zaidi. 1997. Cystic fibrosis transmembrane conductance regulator is an epithelial cell receptor for clearance of Pseudomonas aeruginosa from the lung. Proc. Natl. Acad. Sci. USA 94:12088-12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 48.Rajeevan, M. S., S. D. Vernon, N. Taysavang, and E. R. Unger. 2001. Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. J. Mol. Diagn. 3:26-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 50.Reiniger, N., J. K. Ichikawa, and G. B. Pier. 2005. Influence of cystic fibrosis transmembrane conductance regulator on gene expression in response to Pseudomonas aeruginosa infection of human bronchial epithelial cells. Infect. Immun. 73:6822-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 52.Sadikot, R. T., T. S. Blackwell, J. W. Christman, and A. S. Prince. 2005. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, M. Lei, R. M. Long, J. Wiener-Kronish, and T. Sawa. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sauvonnet, N., B. Pradet-Balade, J. A. Garcia-Sanz, and G. R. Cornelis. 2002. Regulation of mRNA expression in macrophages after Yersinia enterocolitica infection. Role of different Yop effectors. J. Biol. Chem. 277:25133-25142. [DOI] [PubMed] [Google Scholar]
- 55.Shaver, C. M., and A. R. Hauser. 2004. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect. Immun. 72:6969-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 57.Sun, J., and J. T. Barbieri. 2003. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 regulator of kinase (Crk) proteins. J. Biol. Chem. 278:32794-32800. [DOI] [PubMed] [Google Scholar]
- 58.Sundin, C., M. L. Henriksson, B. Hallberg, A. Forsberg, and E. Frithz-Lindsten. 2001. Exoenzyme T of Pseudomonas aeruginosa elicits cytotoxicity without interfering with Ras signal transduction. Cell. Microbiol. 3:237-246. [DOI] [PubMed] [Google Scholar]
- 59.Vance, R. E., A. Rietsch, and J. J. Mekalanos. 2005. Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. Infect. Immun. 73:1706-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ventre, I., A. L. Goodman, I. Vallet-Gely, P. Vasseur, C. Soscia, S. Molin, S. Bleves, A. Lazdunski, S. Lory, and A. Filloux. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. USA 103:171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vincent, T. S., J. E. Fraylick, E. M. McGuffie, and J. C. Olson. 1999. ADP-ribosylation of oncogenic Ras proteins by Pseudomonas aeruginosa exoenzyme S in vivo. Mol. Microbiol. 32:1054-1064. [DOI] [PubMed] [Google Scholar]
- 62.Wani, M. A., S. E. Wert, and J. B. Lingrel. 1999. Lung Kruppel-like factor, a zinc finger transcription factor, is essential for normal lung development. J. Biol. Chem. 274:21180-21185. [DOI] [PubMed] [Google Scholar]
- 63.Warke, V. G., M. P. Nambiar, S. Krishnan, K. Tenbrock, D. A. Geller, N. P. Koritschoner, J. L. Atkins, D. L. Farber, and G. C. Tsokos. 2003. Transcriptional activation of the human inducible nitric-oxide synthase promoter by Kruppel-like factor 6. J. Biol. Chem. 278:14812-14819. [DOI] [PubMed] [Google Scholar]
- 64.Yahr, T. L., A. J. Vallis, M. K. Hancock, J. T. Barbieri, and D. W. Frank. 1998. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 95:13899-13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.