Abstract
Vibrio vulnificus is a pathogenic bacterium that causes gastroenteritis and primary septicemia. To identify factors involved in microbial adherence to the host cells, we investigated bacterial proteins capable of binding to fibronectin, one of the main components comprised of the extracellular matrix of mammalian cells. A protein of ∼35 kDa was purified from the extracts of V. vulnificus by its property to bind to immobilized fibronectin. This protein was identified as OmpU, one of the major outer membrane proteins of V. vulnificus. In binding assays using immobilized fibronectin, the number of ompU mutant cells bound to fibronectin was only 4% of that of wild-type cells bound to fibronectin. In addition, the exogenous addition of antibodies against OmpU resulted in a decreased ability of wild-type V. vulnificus to adhere to fibronectin. The ompU mutant was also defective in its adherence to RGD tripeptide (5% of the adherence of the wild type to RGD), cytoadherence to HEp-2 cells (7% of the adherence of the wild type to HEp-2), cytotoxicity to cell cultures (39% of the cytotoxicity of the wild type), and mortality in mice (10-fold increase in the 50% lethal dose). The ompU mutant complemented with the intact ompU gene restored its abilities for adherence to fibronectin, RGD tripeptide, and HEp-2 cells; cytotoxicity to HEp-2 cells; and mouse lethality. This study indicates that OmpU is an important virulence factor involved in the adherence of V. vulnificus to the host cells.
Vibrio vulnificus is a gram-negative pathogenic bacterium that is encapsulated, motile, and invasive. This pathogen causes primary septicemia, necrotizing wound infections, and gastroenteritis, especially in humans with a heavy alcohol drinking habit or hepatic diseases (24). Several virulence factors have been proposed for V. vulnificus: lipopolysaccharide (2, 19), capsular polysaccharide (42), a cytolytic hemolysin (10), an elastase (22), iron availability (43), and phospholipase A2 (38). Genetic studies using knockout mutants of V. vulnificus vvpA (cytolysin) or vvpE (elastase) did not provide any evidence supporting that they were key virulence factors causing lethality of mice or death of HEp-2 cells (12, 35, 40). However, the cytolysin, but not the metalloprotease, was essential for causing damage in tissues of the infected mice (7). On the contrary, an important role of capsular polysaccharide was demonstrated by measuring attenuated mouse lethality of noncapsulated mutant V. vulnificus (45). Type IV pilin was also confirmed to be involved in the virulence of V. vulnificus via a genetic deletion of pilD or pilA gene (26, 27). In addition, motility was discovered to be a virulence determinant of V. vulnificus (15, 17).
Besides the structural apparatus, several transcriptional regulators were reported to be important for the pathogenesis of V. vulnificus. An alternative sigma factor, RpoS, was shown to be important for both the survival and virulence of this pathogenic bacterium (11, 29). An experiment employing in vivo-induced antigen technology identified the hlyU gene to be induced during the infection process of V. vulnificus, which encodes an activator for hemolysin production in V. cholerae (39). In that study, deletion of this gene was found to cause a dramatic decrease in bacterial cytotoxicity (16).
The first step in the microbial infection of host cells is mediated primarily by the interaction of the pathogen with connective tissues or epithelial cells (6). Fibronectin (FN) is considered the most important extracellular matrix (ECM) protein involved in cellular adherence (33), and it is one of the receptors on epithelial cells for bacterial adherence (5). Regarding the bacterial interaction with FN, gram-positive bacteria such as Staphylococcus aureus and Streptococcus pyogenes have been extensively investigated (reviewed in reference 34). In these microorganisms, FN-binding proteins (FNBPs) with conserved domains mediated bacterial adhesion to and invasion of host cells. Deletion analysis of FNBPs of S. aureus localized primary FN-binding sites, such as repeats of 35 to 40 residues in the carboxyl-terminal part of the protein (8). Assays of invasion of S. aureus cells into human corneal epithelial cells indicated that inhibitors specifically blocking actin polymerization or tyrosine kinase of host cells abolished bacterial entry into host cells (13). This result suggested a model in which the invasion of S. aureus into host cells required the activation of a signal cascade including actin polymerization and tyrosine kinase and that FNBP may serve as a trigger for the activation of this signaling pathway.
In this study, we examined whether V. vulnificus may interact with the ECM, and specifically with FN, and demonstrated the adherence of V. vulnificus to immobilized FN. We then defined the nature of a bacterial surface protein(s) interacting with FN.
MATERIALS AND METHODS
Strains, plasmids, and culture cultivation.
The strains and plasmids used in this study are listed in Table 1. Escherichia coli strains used for plasmid DNA preparation and the conjugational transfer of plasmid were grown in Luria-Bertani (LB) broth or on LB plates containing 1.5% (wt/vol) agar. V. vulnificus strains were grown in LB medium supplemented with 2% (wt/vol) NaCl (LBS), unless stated otherwise. Ampicillin was added to the medium at 100 μg ml−1 for the maintenance of plasmids in E. coli or at 300 μg ml−1 for the selection of V. vulnificus exconjugants. All medium components were purchased from Difco, and the chemicals and antibiotics were obtained from Sigma.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory collection |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Muλpir; OriT of RP4 Kmr; conjugational donor | 20 |
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) [F′ traD36 proAB′ lacIqZΔM15] | 44 |
| V. vulnificus | ||
| MO6-24/O | Clinical isolate | 41 |
| DK1 | ΔompU mutant of MO6-24/O | This study |
| Plasmids | ||
| pGEMT-Easy | Vector used for cloning of PCR product | Promega |
| pDK1 | pGEMT-Easy ompU+ | 28 |
| pTrcHisB | Expression vector for a histidine-tagged protein | Invitrogen |
| pTrcompU | pTrcHisB ompU′ | 28 |
| pBluescript SK(−) | OriR6K OriT AprrpsL | Stratagene |
| pBS-DK1 | pBluescript SK(−) ompU+ | This study |
| pBS-dU | pBS-DK1 ΔompU | This study |
| pDM4 | Suicide vector; OriR6K Cmr | 21 |
| pDM-dU | pDM4 ΔompU | This study |
| pCOS5 | OriV OriT Apr Cmrcos | 3 |
| pJH0311 | pCOS5, multicloning sites; Apr | S. H. Choi |
| pSM1 | pJH0311 ompU+ | This study |
Assay of binding to the ECM.
V. vulnificus strain MO6-24/O (41) was freshly grown up to an optical density at 600 nm (OD600) of 1.0 in LBS at 30°C, harvested by centrifugation at 7,000 × g, and resuspended in phosphate-buffered saline (PBS) (150 mM NaCl, 17 mM NaPO4, 1 mM EGTA, pH 7.0). Ninety-six-well plates were coated with one of the ECM components (FN, collagen, or laminin; Sigma) at a concentration of 100 μg ml−1 in PBS (4°C, 18 h). After blocking the plates with bovine serum albumin (BSA) (1% for 1 h) at room temperature (RT), 1 × 105 V. vulnificus cells were added to each of the coated wells. After being washed four times with PBS-1% BSA, bound bacteria were retrieved by resuspension in PBS-0.1% Triton X-100 at RT for 10 min and counted by plating onto LBS agar. The 96-well plate with BSA instead of the ECM components was used as a control.
Purification of a fibronectin-binding protein of V. vulnificus.
Three hundred milliliters of V. vulnificus MO6-24/O at an OD600 of 1.0 was resuspended in 10 ml of PBS and lysed with a sonicator. Forty-eight-well culture plates were coated by treatment with 50 μg of FN at 4°C for 18 h and washed three times with PBS. After blocking with PBS-1% BSA for 1 h, the plate was then washed twice with PBS. Two hundred microliters of bacterial lysates (an equivalent of ∼2 mg protein) was added to each well coated with FN, incubated for 3 h, and washed six times with PBS to remove unbound proteins. Bound proteins on 48 wells were released in 100 μl of PBS-1% sodium dodecyl sulfate (SDS), pooled, and concentrated fivefold with Centricon (Amicon). The eluted proteins were separated by 10% SDS-polyacrylamide gel electrophoresis (PAGE). The proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane and stained with 0.5% Ponceau (Sigma). The retrieved band was identified by amino-terminal sequencing with a protein sequencing system (Milligen 6600).
Formation of OmpU-specific antibodies.
Recombinant OmpU was overexpressed in Escherichia coli as described previously by Park et al. (28). Briefly, two primers, ompU R-F (5′-ATGGGCCCAAAGTCATCTTTGG-3′) and ompU R-R (5′-GCGGATCCAATCTATCTAGAAT-3′), were used to amplify a 1.63-kb DNA fragment containing the ompU gene and a 513-bp upstream region from the genomic DNA of V. vulnificus MO6-24/O. This DNA was then cloned into the pGEMT-Easy vector (Promega) to produce pDK1. Using two EcoRI sites of pDK1 (one at position 232 from the start codon of ompU and the other in the multicloning site), a 791-bp DNA fragment containing the 3′-terminal portion of ompU was isolated and cloned into pTrcHisB (Invitrogen), resulting in a tagging of OmpU with six histidines. After being transformed into JM109, recombinant OmpU was overexpressed and purified with a His-Bind kit (Novagen).
Six-week-old female BALB/c mice were immunized intraperitoneally with 10 μg of recombinant OmpU, which was emulsified 1:1 in Freund complete adjuvant. The animals were boosted twice at 3-week intervals with the same amount of OmpU protein emulsified in Freund incomplete adjuvant. A week after the third immunization, blood samples of mice were pooled and used for further experiments as polyclonal antibodies against recombinant V. vulnificus OmpU (anti-OmpU). Blood samples were also obtained from nonimmunized mice and then used as a control.
Assay of binding to RGD.
Each well of 96-well culture plates was coated with 20 μg of RGD tripeptide (NH2-Arg-Gly-Asp-COOH) (Takara) in PBS at 4°C for 18 h, blocked with PBS-1% BSA at RT for 1 h, and then washed three times with PBS. A total of 1 × 105 cells of freshly grown V. vulnificus (OD600 of 0.7) were added to each of the RGD-coated wells and incubated at RT for 30 min. After being washed four times with PBS-1% BSA, the bound bacteria were retrieved, serially 10-fold diluted, and then enumerated by plating onto LBS agar.
Treatment of V. vulnificus with OmpU-specific antibodies.
To confirm the role of OmpU in bacterial binding to FN or to host cells as well as in bacterial cytotoxicity, wild-type V. vulnificus cells were preincubated with anti-OmpU (20 ∼ 40 μg ml−1) before being used for binding assays. As a control, wild-type V. vulnificus cells were treated with the same concentrations of mouse preimmune serum instead of anti-OmpU.
Construction of ompU mutant and complementation strains.
Using SphI-SpeI sites present in pDK1, a 1,630-bp DNA fragment of pDK1 was subcloned into pBluescript SK(−) (Stratagene) to yield pBS-DK1. After a 204-bp PstI DNA fragment was deleted in the ompU gene, the SphI-SpeI DNA fragment of the ompU-deleted plasmid (1,426 bp) was then cloned into the suicide vector pDM4 (21). The resultant construct, pDM-dU, was used to mutate the ompU gene in wild-type V. vulnificus by allelic exchange to yield an ompU knockout V. vulnificus strain, DK1. Deletion of the PstI fragment within the ompU gene caused the loss of a portion of the protein (amino acid residues 15 to 82).
A 1,312-bp DNA was amplified from V. vulnificus genomic DNA using two primers, ompU-F (5′-CTAGGAGCTCCACAATCGAACAGTGTTCATA-3′ [underlined bases indicate the SacI site]) and ompU-R (5′-CTAGCCCGGGCATAGTGACAACCCAATCTAT-3′ [underlined bases indicate the SmaI site]), which contains an open reading frame and a 307-bp upstream region of the ompU gene. This DNA fragment was then cloned into the corresponding site of the broad-host-range vector pJH0311 to produce pSM1. This ompU-containing plasmid was mobilized in V. vulnificus strain DK1 via conjugation. Strain DK1 carrying pJH0311 was also constructed in the same manner to serve as a control.
Western blot analysis.
Lysates of various V. vulnificus cells (wild type, DK1, DK1 with pJH0311, and DK1 with pSM1) were prepared by resuspending the harvested cells in PBS and disrupting them with a sonicator. After centrifugation at 12,000 × g for 10 min, portions of the supernatants (50 μg of protein per well) were subjected to SDS-PAGE and then transferred onto a PVDF membrane. The membranes were blocked with 5% nonfat dry milk in Tris-buffered saline with 0.1% Tween 20 for 1 h at RT and then treated with OmpU antibodies at 4°C overnight. Upon incubation with horseradish peroxidase-conjugated anti-mouse immunoglobulin G, immunoreactive bands were visualized using an enhanced chemiluminescence system (Cell Signaling).
Cell line adherence assay.
Adherence assays were performed with the HEp-2 cell line derived from human laryngeal epithelial cells (Korean cell line number 10023) in 24-well culture plates. Each well of the culture plates was seeded with 105 HEp-2 cells and grown overnight at 37°C in the presence of 5% CO2. After removing the medium and washing the cells twice with PBS, 1 ml of serum-free Dulbecco's modified Eagle medium (Gibco-BRL) was added to the HEp-2 cells. Various V. vulnificus cells (wild type, DK1, DK1 carrying pJH0311, and DK1 with pSM1) were grown at 30°C overnight in LBS broth. Cell monolayers were then inoculated in triplicate with 50 μl of the diluted bacteria to give a multiplicity of infection (MOI) of 10 and incubated at 37°C in 5% CO2 for 30 min. The monolayer was then washed five times with prewarmed PBS to remove nonadherent bacteria. Following the last wash, the HEp-2 cells were lysed with 0.1% Triton X-100 treatment for 15 min. The bacteria were recovered from these cells with PBS, serially 10-fold diluted, and then plated onto LBS agar.
Cytotoxicity assays.
Cytotoxicity of V. vulnificus to HEp-2 cells was measured using the CytoTox 96 Non-Radioactive Cytotoxicity assay kit (Promega). This cytotoxicity kit measures the lactate dehydrogenase (LDH) activity released into the culture medium by lysed cells. To measure the total LDH activity of the cell lines used in the assays, Triton X-100 was added to a final concentration of 1.0% (vol/vol) to lyse the host cells. The LDH activity was then determined by colorimetric assay according to the manufacturer's instructions and is represented as a percentage of LDH activities relative to the total LDH activities of the cells treated with 1% Triton X-100.
As a second assay to measure the cytotoxicity of V. vulnificus, the viability of HEp-2 cells was also checked using propidium iodide (PI), which stains nucleic acids of dead cells with disrupted membranes. Flow cytometric analysis to estimate the percentage of cells stained with PI was performed on at least 5,000 cells from each sample by fluorescence-activated cell sorting (FACScan; BD Biosciences).
LD50 determination.
For 50% lethal dose (LD50) determinations, specific-pathogen-free, 7-week-old, female ICR mice were used without pretreatment with iron-dextran. Cultures of bacterial strains grown overnight in LB medium with 0.86% (wt/vol) NaCl were freshly cultivated in the same medium up to an OD600 of 0.7, harvested, washed once in PBS, and then resuspended in PBS-0.01% gelatin. One hundred microliters of serial dilutions of the bacterial suspension was then injected intraperitoneally into six mice per dilution group. The numbers of dead mice were determined 24 h after the injection, and the LD50 was calculated using the equation described previously by Reed and Muench (32).
Statistical analyses.
Results were expressed as the means ± standard deviations from three independent experiments. Statistical analysis was performed using Student's t test (SYSTAT program, SigmaPlot version 9; Systat Software Inc.). Differences were considered significant if P values were <0.05. Data with P values of <0.01 are indicated with two asterisks, whereas data with P values between 0.01 and 0.05 are indicated with one asterisk.
RESULTS
Binding of V. vulnificus to ECM components.
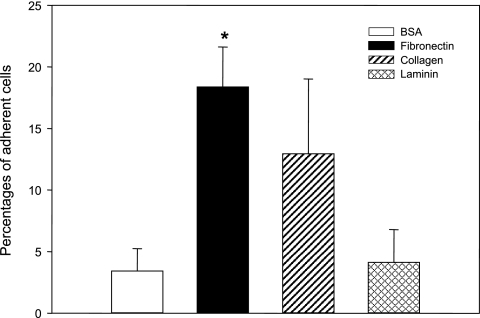
We examined the ability of V. vulnificus to adhere to three components of ECM, collagen, fibronectin, and laminin (Fig. 1). In contrast to the low number of bacterial cells bound to the coated BSA (3.4% of the added bacteria), higher numbers of bacteria were retained on coated fibronectin and collagen. However, the percentage of bacteria that bound to laminin was similar to that of bacteria that adhered to BSA (4.1% of the added V. vulnificus cells). Despite the higher bacterial affinity for collagen (13% of the added bacteria), the number of bound bacteria was not statistically different from that of bacteria bound to BSA. Numbers of bacteria that bound to FN (18% ± 3.3% of the added bacteria) were significantly higher (P = 0.002) than the numbers of bacteria that bound to BSA.
FIG. 1.
Adherence of Vibrio vulnificus to various components of the extracellular matrix. Wild-type V. vulnificus cells (1 × 105) were added to a multiwell plate coated with one of three extracellular matrix proteins at a concentration of 100 μg ml−1 and incubated for 10 min. Assay of binding of V. vulnificus to each component was performed eight times. The percentages of bacterial cells that adhered to wells coated with BSA (an open bar), fibronectin (a black bar), collagen (a hatched bar), or laminin (a crosschecked bar) were determined by plating the eluted bacteria onto LBS agar. Error bars represent the means ± standard deviations from three independent experiments. The asterisk indicates a binding level that was significantly different from that of the BSA-coated control by Student's t test.
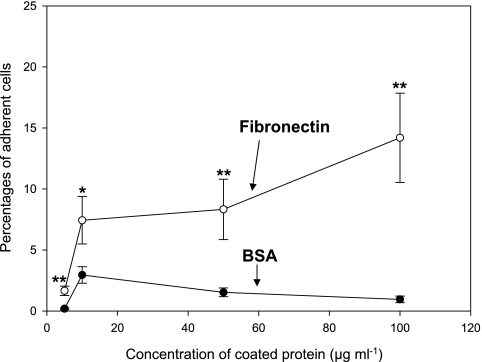
Binding of V. vulnificus to immobilized FN was further examined in subsequent experiments. The numbers of bacteria that bound to an FN-coated surface increased in a dose-dependent manner (Fig. 2). The percentage of bacteria that bound to the immobilized FN was increased up to 14% at 100 μg ml−1 of FN. On the other hand, less than 3% of added bacteria were retained on the surface coated with BSA regardless of the amount of BSA used.
FIG. 2.
Adherence of Vibrio vulnificus to fibronectin- or BSA-coated surfaces. Wild-type V. vulnificus cells were added to the multiwell plate coated with various concentrations (5, 10, 50, and 100 μg ml−1) of fibronectin or BSA and incubated for 10 min. The percentages of bacterial cells that adhered to wells coated with BSA (filled circles) or fibronectin (open circles) were determined by counting CFU on LBS agar plates. Assay of binding of V. vulnificus to fibronectin or BSA at each concentration was performed eight times. Error bars represent the means ± standard deviations from three independent experiments. Asterisks indicate binding levels that were significantly different from those of the BSA-coated control at the corresponding concentration by the Student's t test. Data with P values of <0.01 are indicated with two asterisks, whereas data with P values of between 0.01 and 0.05 are indicated with one asterisk.
Identification of the OmpU protein as a fibronectin-binding protein of Vibrio vulnificus.
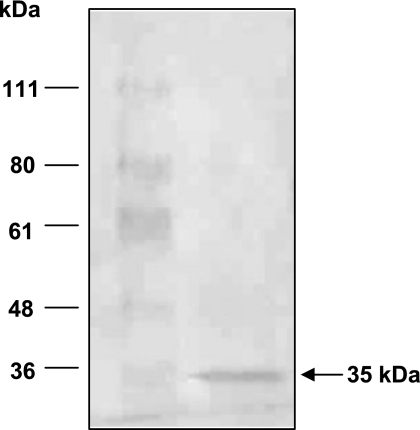
Binding of V. vulnificus to FN led us to identify a bacterial protein involved in this process. A protein of about 35 kDa was retrieved from lysates of V. vulnificus MO6-24/O by its affinity for FN-coated surfaces (Fig. 3). The sequence of the first 21 amino acids at the amino terminus of the isolated protein was AELYNQDGTSLDMGGRAEARL. A TBlast search using this partial sequence showed complete identity to the OmpU proteins of both V. vulnificus strain CMPC6 (GenBank accession number AE016802.1) and V. vulnificus strain YJ016 (accession number BA000037.2). In the analysis of the proteins isolated from FN-binding assays, we could not observe the protein bands corresponding to FN (200 ∼ 250 kDa [plasma FN] or 550 kDa [cellular FN]) on a PVDF membrane, possibly due to the low efficiency of blotting of these proteins with high molecular weights.
FIG. 3.
Identification of a fibronectin-binding protein of Vibrio vulnificus. A crude lysate of wild-type V. vulnificus was added to a multiwell plate coated with 50 μg of fibronectin and incubated for 3 h. After being washed with PBS, the proteins bound to the fibronectin-coated surface were eluted with 1% sodium dodecyl sulfate and concentrated fivefold with Centricon as described in Materials and Methods. Upon separation by 10% SDS-PAGE and transfer to a PVDF membrane, the eluted proteins were visualized by Ponceau. A protein of 35 kDa was excised and subjected to N-terminal amino acid sequencing.
Role of OmpU protein in binding of V. vulnificus to the immobilized FN and to immobilized RGD tripeptide.
The function of OmpU in the FN binding of V. vulnificus was assessed using an ompU knockout mutant. The V. vulnificus ompU mutant was made by removing a PstI DNA fragment of 204 bp, which is present within an open reading frame of the ompU gene. The mutant results in a loss of a portion of the amino acids from residues 15 to 82 of OmpU.
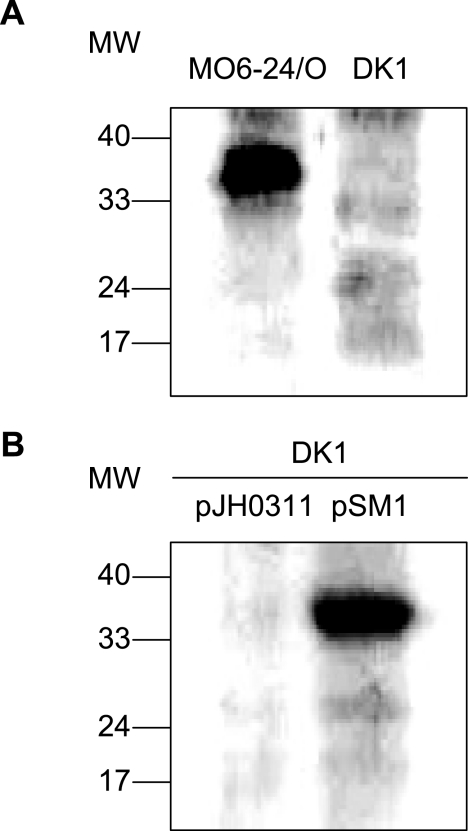
ompU mutant strain DK1 was verified by Western blot analysis using anti-OmpU (Fig. 4A). While OmpU was detected in lysates of wild-type V. vulnificus strain MO6-24/O, the immunoreactive band was not present in lysates of DK1. The intact ompU gene was added back to ompU mutant strain V. vulnificus DK1 as a pJH0311-based plasmid, pSM1. Strain DK1, containing pJH0311, was also constructed as a control strain. Expression of the OmpU protein in the complemented strain was also shown in Western blots using anti-OmpU, whereas the OmpU protein was not detected in the control strain, DK1 harboring pJH0311 (Fig. 4B).
FIG. 4.
Western blot analysis using polyclonal antibodies against the recombinant OmpU protein (anti-OmpU). (A) Confirmation of ompU mutant V. vulnificus by Western blot analysis. Lane 1, lysate of wild-type V. vulnificus MO6-24/O; lane 2, lysate of DK1, an ompU mutant of V. vulnificus. (B) Confirmation of the ompU complementation strain by Western blot analysis. Lane 1, lysate of DK1 harboring a broad-host-range vector, pJH0311; lane 2, lysate of DK1 harboring pSM1, a complementation plasmid of the wild-type ompU gene. Lysates of four different strains of V. vulnificus were prepared as described in Materials and Methods. Fifty micrograms of protein was subjected to SDS-PAGE per each sample and transferred onto a membrane. After being blocked with 5% nonfat dry milk, the membrane was incubated with mouse antibodies against the recombinant OmpU and was then incubated with horseradish peroxidase-conjugated anti-mouse immunoglobulin G. Immunoreactive proteins were visualized using an enhanced chemiluminescence system. MW, molecular weight (in thousands).
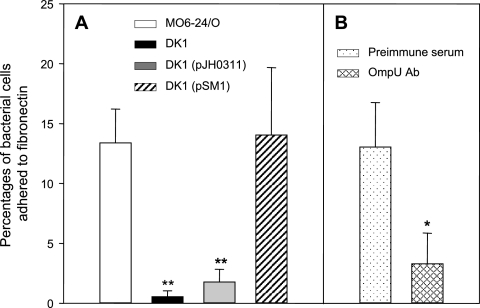
The ability of this ompU mutant to bind to FN was compared with that of the wild-type strain (Fig. 5A). While a significant portion of the added wild type was bound to FN in a dose-dependent manner (∼13%), the ability to bind to FN was significantly disrupted in the ompU mutant strain; that is, 0.55% of the added mutant bacteria were bound to the FN-coated surface.
FIG. 5.
Role of the OmpU protein in adherence of Vibrio vulnificus to immobilized fibronectin. (A) Adherence of various V. vulnificus strains to fibronectin. V. vulnificus cells (wild-type V. vulnificus MO6-24/O [open bar], ompU mutant strain DK1 [closed bar], DK1 carrying pJH0311 [gray bar], and DK1 carrying a complementation plasmid, pSM1 [hatched bar]) were added to a 96-well plate coated with fibronectin. The percentage of bacterial cells that adhered to fibronectin-coated wells was determined by plating bound bacteria onto LBS agar. Error bars represent the means ± standard deviations from three independent experiments. Assay of the binding of each V. vulnificus strain to fibronectin was performed in quadruplicate for each experiment. Asterisks indicate binding levels that were significantly different from that of wild-type V. vulnificus by the Student's t test. Data with P values of <0.01 are indicated with two asterisks, whereas data with P values of between 0.01 and 0.05 are indicated with one asterisk. (B) Inhibition of fibronectin binding of wild-type V. vulnificus by anti-OmpU. The percentage of bacterial cells that adhered to fibronectin-coated wells was determined for wild-type V. vulnificus preincubated with mouse preimmune serum (dotted bar) or with anti-OmpU (cross-hatched bar) at 20 μg ml−1. Error bars represent the means ± standard deviations from three independent experiments. Assay of binding of V. vulnificus to fibronectin was performed in quadruplicate for each experiment. The asterisk indicates a binding level that was significantly different from that of wild-type V. vulnificus pretreated with mouse preimmune serum by Student's t test (0.01 < P < 0.05). Ab, antibody.
Evidence for the involvement of OmpU of V. vulnificus in FN binding was strengthened by an additional experiment using anti-OmpU. Preincubation of wild-type V. vulnificus with anti-OmpU at concentration of 20 μg ml−1 significantly decreased the bacterial binding of 3.3% of the added bacteria to FN (Fig. 5B). On the other hand, pretreatment of wild-type V. vulnificus with the same concentration of mouse preimmune serum did not result in any alteration of the bacterial binding to FN (∼13%).
The FN-binding ability of the ompU-complemented strain, DK1(pSM1), was also examined (Fig. 5A). The ability of the bacteria to adhere to FN was fully restored when the intact ompU gene was added to the ompU mutant strain. As expected, the presence of a vector plasmid, pJH0311, did not alter the inability of the ompU knockout strain to adhere to FN.
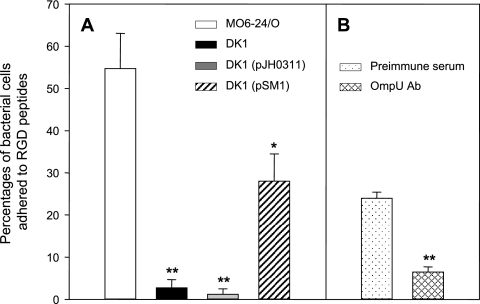
It is well known that FN-binding proteins bind to FN by specific interactions with the RGD repeat of FN (9). When we examined whether V. vulnificus could bind to immobilized RGD tripeptide, we found that a significant portion of V. vulnificus cells (55% of the added bacteria) was bound to the RGD tripeptide (Fig. 6A). On the other hand, the ability of bacteria to adhere to RGD was almost abolished in the case of ompU knockout mutant DK1 (2.7% of the added bacteria). Wild-type V. vulnificus was treated with 20 μg ml−1 of anti-OmpU or mouse preimmune serum and then tested for binding to RGD tripeptides (Fig. 6B). Twenty-four percent of V. vulnificus cells adhered to RGD when they were treated with preimmune serum. On the contrary, only 6.5% of bacteria were bound to RGD if they were incubated with anti-OmpU. In the same manner, an addition of the intact ompU gene into strain DK1 also endowed binding ability to RGD tripeptide to up to 52% of that of wild-type V. vulnificus (Fig. 6A).
FIG. 6.
Role of OmpU protein in adherence of Vibrio vulnificus to immobilized RGD tripeptide. (A) Adherence of various V. vulnificus strains to the RGD tripeptide. V. vulnificus cells (wild-type V. vulnificus MO6-24/O [open bar], ompU mutant DK1 [closed bar], DK1 carrying pJH0311 [gray bar], and DK1 carrying a complementation plasmid, pSM1 [hatched bar]) were added to the 96-well plate coated with RGD tripeptide. Each well of the culture plate was coated with 20 μg of RGD tripeptide in PBS at 4°C for 18 h. V. vulnificus cells (1 × 105) were added to each of the RGD-coated wells and incubated at RT for 30 min. The percentage of bacterial cells that adhered to RGD-coated wells was determined by counting CFU of the eluted bacteria on LBS agar plates. Error bars represent the means ± standard deviations from three independent experiments. Assay of binding of V. vulnificus to RGD tripeptide was performed in quadruplicate for each experiment. Asterisks indicate binding levels that were significantly different from that of wild-type V. vulnificus by Student's t test. Data with P values of <0.01 are indicated with two asterisks, whereas data with P values of between 0.01 and 0.05 are indicated with one asterisk. (B) Inhibition of RGD binding of wild-type V. vulnificus by anti-OmpU. The percentage of bacterial cells that adhered to RGD-coated wells was determined for wild-type V. vulnificus preincubated with mouse preimmune serum (dotted bar) or with anti-OmpU (cross-hatched bar) at 20 μg ml−1. Error bars represent the means ± standard deviations from three independent experiments. Assay of binding of V. vulnificus to RGD tripeptide was performed in quadruplicate for each experiment. Asterisks indicate binding levels that were significantly different (P < 0.01) from that of wild-type V. vulnificus pretreated with mouse preimmune serum by Student's t test. Ab, antibody.
Role of OmpU in cytoadherence of V. vulnificus.
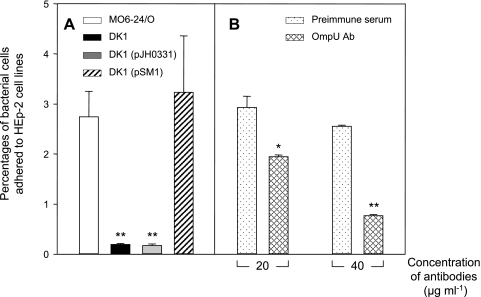
Based on the observation that OmpU may serve as a ligand for FN, we extended our experiments to reveal the function of this protein in interactions with host cells. The first assay that we performed was an examination of the role of OmpU in bacterial adherence to the HEp-2 epithelial cell line (Fig. 7A). When the ratio of bacteria to HEp-2 cells was 10:1, the percentage of adhered bacteria of the initially added bacteria was 2.7% in the case of the assay of HEp-2 cells with wild-type V. vulnificus. In the case of the assay with the ompU mutant, the portion of the bacterial cells that adhered to the HEp-2 cells was quite low, e.g., 0.18% of the added bacteria.
FIG. 7.
Role of OmpU protein in adherence of Vibrio vulnificus to HEp-2 cells. (A) Cytoadherence of various V. vulnificus strains to HEp-2 cells. V. vulnificus cells (wild-type V. vulnificus MO6-24/O [open bar], ompU mutant DK1 [closed bar], DK1 carrying pJH0311 [gray bar], and DK1 carrying a complementation plasmid, pSM1 [hatched bar]) were added to HEp-2 cells at an MOI of 10 and incubated for 30 min. Following five washes with PBS, the HEp-2 cells were treated with 0.1% Triton X-100 for 15 min. The percentage of bacterial cells that adhered to HEp-2 was determined by measuring CFU of the retrieved bacteria on LBS agar plates. Assay of adherence of various V. vulnificus cells to HEp-2 was performed in quadruplicate, and data are presented with error bars, which are standard deviations of three independent experiments. Asterisks indicate binding levels that were significantly different from that of wild-type V. vulnificus by Student's t test. Data with P values of <0.01 are indicated with two asterisks, whereas data with P values of between 0.01 and 0.05 are indicated with one asterisk. (B) Inhibition of cytoadherence of wild-type V. vulnificus cells to HEp-2 cells by anti-OmpU. Prior to the cytoadherence test, wild-type V. vulnificus cells were treated with either antibodies against recombinant OmpU (OmpU Ab) (cross-hatched bars) or mouse preimmune serum (dotted bars) for 30 min at two different concentrations (20 or 40 μg ml−1). The percentage of bacterial cells that adhered to HEp-2 cells was determined by measuring CFU of the adhered bacteria on LBS plates. Each cytoadherence assay was done in quadruplicate, and data are presented along with standard deviations of three independent experiments. Asterisks indicate binding levels that were significantly different (P < 0.01) from that of wild-type V. vulnificus pretreated with mouse preimmune serum by the Student's t test.
The role of OmpU in the cytoadherence of wild-type V. vulnificus was confirmed using anti-OmpU (Fig. 7B). Wild-type V. vulnificus cells were treated with either 20 or 40 μg ml−1 of anti-OmpU prior to the adherence tests while the ratio of bacteria to HEp-2 cells was kept at 10:1. As a control, wild-type V. vulnificus cells were also incubated with mouse preimmune serum at the same concentrations and used for the adherence tests. In the case of wild-type V. vulnificus cells preincubated with anti-OmpU, they showed decreased levels of cytoadherence in a dose-dependent manner, down to 0.77% of the anti-OmpU-treated cells. In contrast, 2.6 or 2.9% of the added V. vulnificus cells adhered to the cell lines when they were treated with 20 or 40 μg ml−1 preimmune serum, respectively.
In addition, the cytoadherence of strain DK1 to HEp-2 cells was fully recovered to wild-type levels when it carried pSM1, an ompU+ plasmid, whereas the introduction of pJH0311 to strain DK1 did not cause any change in bacterial adherence to HEp-2 cells (Fig. 7A).
Role of OmpU in cytotoxicity of V. vulnificus.
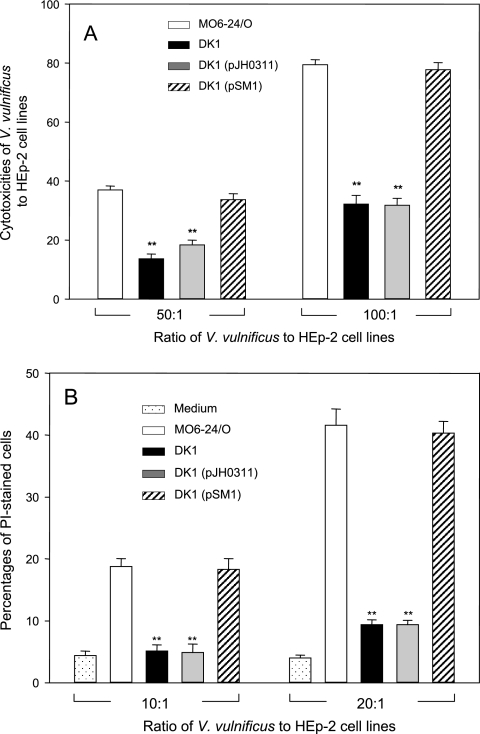
In a subsequent experiment, we asked whether the defects in FN binding as well as in cytoadherence of the ompU mutant could affect bacterial virulence toward host cells. Incubation of wild-type V. vulnificus cells at an MOI of 50 or 100 resulted in 37% or 79% cytotoxicity of host cells, respectively, when determined by LDH release assay (Fig. 8A). On the other hand, the V. vulnificus ompU mutant was significantly less cytotoxic than wild-type V. vulnificus (14% and 32% at MOIs of 50 and 100, respectively).
FIG. 8.
Role of the OmpU protein of Vibrio vulnificus in bacterial cytotoxicity to HEp-2 cells. (A) Determination of cytotoxicity of various V. vulnificus strains by estimating the activity of LDH. Using a CytoTox96 assay kit, LDH released from the HEp-2 cells was measured upon incubation with wild-type V. vulnificus (open bars), ompU mutant DK1 (black bars), DK1 carrying pJH0311 (gray bars), or DK1 carrying pSM1 (hatched bars). HEp-2 cells (1 × 105) were incubated with V. vulnificus cells for 30 min at two different MOIs, 50 and 100. Each assay was performed in quadruplicate and repeated three times. The data are shown with error bars, which are standard deviations of three independent experiments. Asterisks indicate enzyme activities that were significantly different (P < 0.01) from that of wild-type V. vulnificus by Student's t test. (B) Determination of the cytotoxicities of various V. vulnificus strains by staining with PI. The percentages of dead HEp-2 cells during incubation with wild-type V. vulnificus (open bars), ompU mutant strain DK1 (black bars), DK1 carrying pJH0311 (gray bars), or DK1 carrying pSM1 (hatched bars) were measured at two different MOIs, 10 and 20. HEp-2 cells not exposed to V. vulnificus were also stained with PI (dotted bars). Error bars represent the means ± standard deviations from three independent experiments. Asterisks indicate percentages of PI-stained cells that were significantly different (P < 0.01) from that of wild-type V. vulnificus cells by Student's t test.
Upon incubation with either wild-type or ompU V. vulnificus, the viability of HEp-2 cells was also checked by staining the dead cells with PI (Fig. 8B). Without the addition of V. vulnificus, 4.0 to 4.4% of HEp-2 cells were stained with PI, whereas the percentages of PI-stained cells exposed to wild-type V. vulnificus increased to 18 and 42% at MOIs of 10 and 20, respectively. When HEp-2 cells were incubated with the V. vulnificus ompU mutant at MOIs of 10 and 20, only 5.1 and 9.4% of HEp-2 cells, respectively, were stained with PI. These values are significantly less than those found with the wild type.
In the subsequent experiments, we examined whether attenuated phenotypes of strain DK1 in cytotoxicities to HEp-2 cell lines could be recovered to the wild-type phenotype by introducing pSM1. In the assays for measuring bacterial cytotoxicities, strain DK1 carrying pSM1 showed 91 to 98% of the cytotoxicities of wild-type V. vulnificus (Fig. 8A and B). On the other hand, the control strain (strain DK1 with a vector, pJH0311) showed cytotoxicities similar to that of strain DK1 in both LDH and PI staining assays.
Role of OmpU in lethality of V. vulnificus to mice.
We examined the role of the V. vulnificus OmpU protein in pathogenesis by using a mouse infection model. Upon intraperitoneal injection of various numbers of bacterial cells into mice, the numbers of dead mice were determined 24 h after the injection (Table 2). One of the experiments was presented as a representative experiment. Mice infected with the wild type showed an LD50 of 3.4 × 103 cells, whereas mice injected with the ompU mutant showed a 10-fold-higher LD50, i.e., 3.4 × 104 cells.
TABLE 2.
LD50s of Vibrio vulnificus
| Strain | Genotype | No. of dead mice/no. of total mice for no. of injected bacteria
|
LD50 (CFU) | |||
|---|---|---|---|---|---|---|
| 3.4 × 10 | 3.4 × 102 | 3.4 × 103 | 3.4 × 104 | |||
| MO6-24/O | ompU+ | 1/6 | 3/6 | 5/6 | 6/6 | 3.4 × 103 |
| DK1 | ΔompU | 0/6 | 0/6 | 3/6 | 6/6 | 3.4 × 104 |
| DK1(pJH0311) | ΔompU | 0/6 | 0/6 | 1/5 | 5/1 | 1.0 × 105 |
| DK1(pSM1) | ompU+ | 0/6 | 0/6 | 4/6 | 6/6 | 1.9 × 104 |
The complemented and the control strains were also tested for their LD50 values in a mouse model. A higher number of the control strain, DK1 carrying pJH0311, was required to achieve 50% killing (1.0 × 105 cells), as the ompU mutant strain did. To obtain 50% lethality of mice by the complemented strain, 1.9 × 104 cells (which is 5.5 times more wild-type cells) were need, suggesting that the introduction of the wild-type ompU gene into DK1 caused a partial complementation in mouse virulence.
DISCUSSION
The capability of bacterial adherence to the host cells is an important virulence factor for many pathogens, including V. vulnificus. The severe and rapid cytopathological characteristics of V. vulnificus infection make this organism a model system to investigate host-pathogen interactions. In adhesion assays using immobilized ECM components, V. vulnificus showed an ability to bind to FN (Fig. 1), suggesting that it may possess a surface protein(s) that interacts with this host component. In a subsequent experiment to isolate the adherence factor of V. vulnificus binding to FN, the OmpU protein, one of the outer membrane proteins, was identified (Fig. 3).
OmpU has been extensively studied in V. cholerae, a pathogen that colonizes the human intestine and produces cholera disease (25). The expression of ompU, encoding outer membrane porin in V. cholerae, is positively regulated by ToxR, the key regulator of cholera toxin production and other virulence determinants (4). Through a genetic construction of V. cholerae overexpressing OmpT, the other major outer membrane porin, the ToxR-dependent modulation of two outer membrane porins, OmpU and OmpT, was found to be important for the expression of virulence factors and intestinal colonization by V. cholerae (30). Interestingly, V. cholerae OmpU was previously found to be an FN-binding protein and was suggested to be a potential adherence factor in V. cholerae (37). In that study, OmpU protein selectively bound to FN and to an arginine-glycine-asparagine (RGD) tripeptide. Subsequent studies, however, indicated that OmpU was not involved in the adherence of V. cholerae to the brush border of the rabbit small intestine (25, 31). In contrast, OmpU was reported to be an important colonization factor in Vibrio fischeri to establish mutualistic symbiosis within a light organ of its squid host, Euprymna scolopes; however, its affinity for FN was not reported (1). It is likely that OmpU proteins in Vibrio spp. may function as a component of the adhesion apparatus via their affinity for FN. Sequence comparison of OmpU proteins indicates that this protein is conserved in various Vibrio spp. including V. cholerae, V. parahaemolyticus, V. fischeri, V. splendidus, and V. vulnificus. The identities of amino acid sequences range from 59 to 76% among OmpU-homologous proteins.
Our study using an ompU knockout V. vulnificus mutant and antibodies against OmpU clearly shows that the OmpU protein of V. vulnificus is involved in FN binding (Fig. 4). Adherence and cytotoxicity assays (Fig. 5, 6, and 7) demonstrated that V. vulnificus cells interact with the host cell epithelium via a direct interaction between bacterial OmpU and FN of the host cells at the early stage of infection. Failure of the ompU mutant of V. vulnificus to adhere to host cells apparently results in defects in bacterial cytotoxicity against host cells. In gram-positive bacteria, several FNBPs had been reported as major virulence factors: FNBPA and FNBPB for S. aureus (14, 36), Sfb1 for Streptococcus pyogenes (23), and CshA for Streptococcus gordonii (18). A current model for the function of these FNBPs suggested that the binding of FNBP with multiple FNs resulted in the localization of integrin binding sites, which induce the clustering of integrins in the membrane of host cells (34). The clustering of integrin in host cells may trigger the phosphorylation of tyrosine kinase and actin rearrangement, which are essential for the internalization of bacteria into host cells (13). In the case of Streptococcus pyogenes, two distinct cellular mechanisms were proposed for Sfb1-negative and Sfb1-positive strains with respect to the involvement of integrin in bacterial invasion (23). In contrast, nothing is known about the exact mechanism(s) by which OmpU facilitates the pathogenesis of V. vulnificus, and further investigations should be performed to evaluate the contribution of the OmpU-FN interaction to the processes causing disease in humans by V. vulnificus.
Acknowledgments
This study was supported by the 21C Frontier Microbial Genomics and Application Center Program, Ministry of Science and Technology (grant MG05-0201-5-0 to S.-J.P. and K.-H.L.), Republic of Korea.
Editor: V. J. DiRita
REFERENCES
- 1.Aeckersberg, F., C. Lupp, B. Feliciano, and E. G. Ruby. 2001. Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J. Bacteriol. 183:6590-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahrani, K., and J. D. Oliver. 1990. Studies on the lipopolysaccharide of virulent and avirulent strains of Vibrio vulnificus. Biochem. Cell Biol. 68:547-551. [DOI] [PubMed] [Google Scholar]
- 3.Connell, T. D., A. J. Martone, and R. K. Holmes. 1995. A new mobilizable cosmid vector for use in Vibrio cholerae and other gram-negative bacteria. Gene 153:85-87. [DOI] [PubMed] [Google Scholar]
- 4.Crawford, J. A., J. B. Kaper, and V. J. DiRita. 1998. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol. Microbiol. 29:235-246. [DOI] [PubMed] [Google Scholar]
- 5.Di Martino, P. 2001. Effects of antibiotics on adherence of Pseudomonas aeruginosa and Pseudomonas fluorescens to human fibronectin. Chemotherapy 47:344-349. [DOI] [PubMed] [Google Scholar]
- 6.Di Martino, P., J. Rebiere-Huet, and C. Hulen. 2000. Effects of antibiotics on adherence of Pseudomonas aeruginosa and Pseudomonas fluorescens to A549 pneumocyte cells. Chemotherapy 46:129-134. [DOI] [PubMed] [Google Scholar]
- 7.Fan, J. J., C. P. Shao, Y. C. Ho, C. K. Yu, and L. I. Hor. 2001. Isolation and characterization of a Vibrio vulnificus mutant deficient in both extracellular metalloprotease and cytolysin. Infect. Immun. 69:5943-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flock, J. I., G. Froman, K. Jonsson, B. Guss, C. Signas, B. Nilsson, G. Raucci, M. Hook, T. Wadstrom, and M. Lindberg. 1987. Cloning and expression of the gene for a fibronectin-binding protein from Staphylococcus aureus. EMBO J. 6:2351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fowler, T., E. R. Wann, D. Joh, S. Johansson, T. J. Foster, and M. Hook. 2000. Cellular invasion by Staphylococcus aureus involves a fibronectin bridge between the bacterial fibronectin-binding MSCRAMMs and host cell β1 integrins. Eur. J. Cell Biol. 79:672-679. [DOI] [PubMed] [Google Scholar]
- 10.Gray, L. D., and A. S. Kreger. 1987. Mouse skin damage caused by cytolysin from Vibrio vulnificus and by V. vulnificus infection. J. Infect. Dis. 155:236-241. [DOI] [PubMed] [Google Scholar]
- 11.Hülsmann, A., T. M. Rosche, I. S. King, H. M. Hassan, D. M. Beam, and J. D. Oliver. 2003. RpoS-dependent stress response and exoenzyme production in Vibrio vulnificus. Appl. Environ. Microbiol. 69:6114-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong, K. C., H. S. Jeong, J. H. Rhee, S. E. Lee, S. S. Chung, A. M. Starks, G. M. Escudero, P. A. Gulig, and S. H. Choi. 2000. Construction and phenotypic evaluation of a Vibrio vulnificus vvpE mutant for elastolytic protease. Infect. Immun. 68:5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jett, B. D., and M. S. Gilmore. 2002. Host-parasite interactions in Staphylococcus aureus keratitis. DNA Cell Biol. 21:397-404. [DOI] [PubMed] [Google Scholar]
- 14.Jonsson, K., C. Signas, H. P. Muller, and M. Lindberg. 1991. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur. J. Biochem. 202:1041-1048. [DOI] [PubMed] [Google Scholar]
- 15.Kim, Y. R., and R. H. Rhee. 2003. Flagellar basal body flg operon as a virulence determinant of Vibrio vulnificus. Biochem. Biophys. Res. Commun. 304:405-410. [DOI] [PubMed] [Google Scholar]
- 16.Kim, Y. R., S. E. Lee, C. M. Kim, S. Y. Kim, E. K. Shin, D. H. Shin, S. S. Chung, H. E. Choy, A. Progulske-Fox, J. D. Hillman, M. Handfield, and R. H. Rhee. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, J.-H., J. B. Rho, K.-J. Park, C. B. Kim, Y.-S. Han, S. H. Choi, K.-H. Lee, and S.-J. Park. 2004. Role of flagellum and motility in pathogenesis of Vibrio vulnificus. Infect. Immun. 72:4905-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNab, R., A. R. Holmes, J. M. Clarke, G. W. Tannock, and H. F. Jenkinson. 1996. Cell surface polypeptide CshA mediates binding of Streptococcus gordonii to other oral bacteria and to immobilized fibronectin. Infect. Immun. 64:4204-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McPherson, V. L., J. A. Watts, L. M. Simpson, and J. D. Oliver. 1991. Physiological effects of the lipopolysaccharide of Vibrio vulnificus on mice and rats. Microbios 67:141-149. [PubMed] [Google Scholar]
- 20.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicidal vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milton, D. L., R. O'Toole, O. Horstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyoshi, S., H. Nakazawa, K. Kawata, K. Tomochita, K. Tobe, and S. Shinoda. 1998. Characterization of the hemorrhagic reaction caused by Vibrio vulnificus metalloprotease, a member of the thermolysin family. Infect. Immun. 66:4851-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molinari, G., M. Rohde, C. A. Guzman, and G. S. Chhatwal. 2000. Two distinct pathways for the invasion of Streptococcus pyogenes in non-phagocytic cells. Cell. Microbiol. 2:145-154. [DOI] [PubMed] [Google Scholar]
- 24.Morris, J. G. 1988. Vibrio vulnificus—a new monster of the deep? Ann. Intern. Med. 109:261-263. [DOI] [PubMed] [Google Scholar]
- 25.Nakasone, N., and M. Iwanaga. 1998. Characterization of outer membrane protein OmpU of Vibrio cholerae O1. Infect. Immun. 66:4726-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paranjpye, R. N., J. C. Lara, J. C. Pepe, C. M. Pepe, and M. S. Strom. 1998. The type IV leader peptidase/N-methyltransferase of Vibrio vulnificus controls factors required for adherence to HEp-2 cells and virulence in iron-overloaded mice. Infect. Immun. 16:5659-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paranjpye, R. N., and M. S. Strom. 2005. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 73:1411-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, D.-K., K.-E. Lee, C.-H. Baek, I. H. Kim, J.-H. Kwon, W. K. Lee, K.-H. Lee, B.-S. Kim, S. H. Choi, and K.-S. Kim. 2006. Cyclo(Phe-Pro) modulates the expression of ompU in Vibrio spp. J. Bacteriol. 188:2214-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, K.-J., M.-J. Kang, S. H. Kim, H.-J. Lee, J.-K. Lim, S. H. Choi, S.-J. Park, and K.-H. Lee. 2004. Isolation and characterization of rpoS from a pathogenic bacterium, Vibrio vulnificus: role of σS in survival of exponential-phase cells under oxidative stress. J. Bacteriol. 186:3304-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Provenzano, D., and K. E. Klose. 2000. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. USA 97:10220-10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Provenzano, D., C. M. Lauriano, and K. E. Klose. 2001. Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in Vibrio cholerae virulence. J. Bacteriol. 183:3652-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reed, L. J., and H. Muench. 1938. A simple method of estimating the 50% endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 33.Ruoslathi, E. 1999. Fibronectin and its integrin receptors in cancer. Adv. Cancer Res. 76:1-20. [DOI] [PubMed] [Google Scholar]
- 34.Schwarz-Linek, U., M. Hook, and J. R. Potts. 2004. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 52:631-641. [DOI] [PubMed] [Google Scholar]
- 35.Shao, C. P., and L. I. Hor. 2000. Metalloprotease is not essential for Vibrio vulnificus virulence in mice. Infect. Immun. 68:3569-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Signas, C., G. Raucci, K. Jonsson, P.-E. Lindgren, G. M. Anantharamaiah, M. Hook, and M. Lindberg. 1989. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc. Natl. Acad. Sci. USA 86:699-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sperandio, V., J. A. Giron, W. D. Silveira, and J. B. Kaper. 1995. The OmpU outer membrane protein, a potential adherence factor of Vibrio cholerae. Infect. Immun. 63:4433-44438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Testa, J., L. W. Daniel, and A. S. Kreger. 1984. Extracellular phospholipase A2 and lysophospholipase produced by Vibrio vulnificus. Infect. Immun. 45:458-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams, S. G., S. R. Attridge, and P. A. Manning. 1993. The transcriptional activator HlyU of Vibrio cholerae: nucleotide sequence and role in virulence gene expression. Mol. Microbiol. 9:751-760. [DOI] [PubMed] [Google Scholar]
- 40.Wright, A. C., and J. G. Morris, Jr. 1991. The extracellular cytolysin of Vibrio vulnificus: inactivation and relationship to virulence in mice. Infect. Immun. 59:192-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright, A. C., J. G. Morris, Jr., D. R. Maneval, Jr., K. Richardson, and J. B. Baker. 1985. Cloning of the cytotoxin-hemolysin gene of Vibrio vulnificus. Infect. Immun. 50:922-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright, A. C., J. L. Powell, M. K. Tanner, L. A. Ensor, A. B. Karpas, J. G. Morris, Jr., and M. B. Sztein. 1999. Differential expression of Vibrio vulnificus capsular polysaccharide. Infect. Immun. 67:2250-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright, A. C., L. M. Simpson, and J. D. Oliver. 1981. Role of iron in the pathogenesis of Vibrio vulnificus. Infect. Immun. 34:503-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 45.Zuppardo, A. B., and R. J. Siebeling. 1998. An epimerase gene essential for capsule synthesis in Vibrio vulnificus. Infect. Immun. 66:2601-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]