Abstract
Unlike most other cells, hyphae of filamentous fungi permanently elongate and lack nonpolar growth phases. We identified AgBoi1/2p in the filamentous ascomycete Ashbya gossypii as a component required to prevent nonpolar growth at hyphal tips. Strains lacking AgBoi1/2p frequently show spherical enlargement at hyphal tips with concomitant depolarization of actin patches and loss of tip-located actin cables. These enlarged tips can repolarize and resume hyphal tip extension in the previous polarity axis. AgBoi1/2p permanently localizes to hyphal tips and transiently to sites of septation. Only the tip localization is important for sustained elongation of hyphae. In a yeast two-hybrid experiment, we identified the Rho-type GTPase AgRho3p as an interactor of AgBoi1/2p. AgRho3p is also required to prevent nonpolar growth at hyphal tips, and strains deleted for both AgBOI1/2 and AgRHO3 phenocopied the respective single-deletion strains, demonstrating that AgBoi1/2p and AgRho3p function in a common pathway. Monitoring the polarisome of growing hyphae using AgSpa2p fused to the green fluorescent protein as a marker, we found that polarisome disassembly precedes the onset of nonpolar growth in strains lacking AgBoi1/2p or AgRho3p. AgRho3p locked in its GTP-bound form interacts with the Rho-binding domain of the polarisome-associated formin AgBni1p, implying that AgRho3p has the capacity to directly activate formin-driven actin cable nucleation. We conclude that AgBoi1/2p and AgRho3p support polarisome-mediated actin cable formation at hyphal tips, thereby ensuring permanent polar tip growth.
Polarization is a fundamental requirement for diverse cellular processes such as vectorial transport in epithelial cells, directed cell movement in amoeba, or cell shape development during early embryogenesis (17, 58). A specialized form of polarization is found in filamentous fungi. There, polarization is established with the emergence of each new hypha, and then polar growth is permanently maintained at the hyphal tip, a primary requirement for fungal mycelial development (26, 36). This growth mode enables fungi to rapidly colonize surfaces in order to reach nutrients or mating partners. Filamentation is also of importance in virulence. Plant pathogens use hyphae to enter the host through stomatal pores or via lesions (18), and the human pathogens Candida albicans or Aspergillus fumigatus can initiate hyphae in order to escape from macrophages or to penetrate into host tissues (10, 32).
Ashbya gossypii is an attractive model organism to study permanent polarization. It was originally isolated as a plant pathogen and belongs to the ascomycetes (5). The genome is completely sequenced (15), and efficient molecular tools like replicative plasmids and PCR-based gene targeting have been established (53, 59, 64). Further, for more than 95% of the genes in A. gossypii a homologue is found in the yeast Saccharomyces cerevisiae (13, 15), which allows comparisons of the role of homologous genes in the respective cellular environments.
Mycelial development of A. gossypii starts with an isotropic growth phase in the middle of the needle-shaped spore. This is the only stage in which isotropic growth is observed in A. gossypii development. Next, a first germ tube emerges from the germ bubble perpendicular to the axis of the spore. This first polarization event is comparable to bud emergence during early G1 phase in S. cerevisiae. A striking difference between the yeast-like and filamentous morphogeneses is the temporal organization of polar growth. Whereas in S. cerevisiae extension occurs only during the early phase of bud growth and then switches to isotropic growth at the G2/M transition, the hyphal tip in A. gossypii remains permanently polarized. During this sustained growth of a hypha, lateral branches and septa are established, resulting in multiple polarization sites in a common cytoplasm, while in S. cerevisiae only one polarization site is maintained at a time (36, 40, 41, 63).
Several proteins have been characterized that are involved in polarized growth of A. gossypii. AgCdc42p and AgCdc24p, a Rho-type GTPase and its GTP exchange factor, are necessary for the initial polarization during germ tube emergence. In the absence of AgBem2p, the putative GTPase-activating protein of AgCdc42p, germ bubbles establish multiple polarization sites and hyphal tips can swell (60). Swelling at hyphal tips has also been observed in hyphae lacking AgRho3p, implicating a role of this GTPase in maintaining polar growth (61). The polarisome component AgSpa2p remains permanently localized to hyphal tips once it has localized at sites of germ tube emergence or lateral branch initiation (30). The Ras-like GTPase AgRsr1p guides hyphal growth, i.e., hyphae lacking this GTPase frequently deviate from the growth axis and often pause during tip extensions due to disassembly of the polarisome (7). The formin AgBni1p, an essential component of the polarisome, is responsible for the formation of actin cables required for tip-directed vesicle transport (47). Work in other fungi has revealed several polarity factors which are orthologues of the A. gossypii proteins mentioned above (12, 26, 27, 37, 48, 49, 56).
It is widely accepted that the ancestral growth form of fungi is filamentous (33). Why elongating hyphae of filamentous fungi do not switch from polar to nonpolar growth like budding yeast cells is not known. The simple explanation that genes important for such a switch are missing in A. gossypii is very likely incorrect, because the A. gossypii genome has homologues for all S. cerevisiae genes involved in bud growth (15, 28). We aimed at identifying A. gossypii genes encoding proteins important to prevent nonpolar growth at hyphal tips. Here we report the identification of the SH3/PH domain-containing protein AgBoi1/2p and present experimental evidence that AgBoi1/2p interacts with the Rho-type GTPase AgRho3p to prevent nonpolar growth at hyphal tips, most likely via a reinforcement of the polarisome-mediated actin cable nucleation.
MATERIALS AND METHODS
Strains and media.
General molecular cloning procedures were performed according to reference 46 or the manufacturer's instructions. Unless otherwise indicated, the A. gossypii Agleu2 ΔAgthr4Δ strain(4) was used for all strain constructions, and it is designated “wild type” throughout this work. Culture conditions and transformation protocols for A. gossypii and S. cerevisiae were adapted from references 59 and 2, respectively. For each newly generated strain, at least two independent transformants were included in the analysis. Strains used in this study are listed in Table 1.
TABLE 1.
Strains used in this study
| Name | Genotype | Reference or source |
|---|---|---|
| Wild type | Agleu2Δ Agthr4Δ | 4 |
| Agboi1/2Δ | Agboi1/2Δ::GEN3 Agleu2Δ Agthr4Δ or Agboi1/2Δ::NAT1 Agleu2Δ Agthr4Δ | This study |
| Agrho3Δ | Agrho3Δ::GEN3 Agleu2Δ Agthr4Δ | 61 |
| Agboi1/2Δrho3Δ | Agboi1/2::GEN3 Agrho3Δ::NAT1 Agleu2Δ Agthr4Δ or Agboi1/2::NAT1 Agrho3Δ::GEN3 Agleu2Δ Agthr4Δ | This study |
| AgBOI1/2-GFP | AgBOI1/2-GFP::GEN3 Agleu2Δ Agthr4Δ | This study |
| AgBOI1/2-GFPcyk1Δ | AgBOI1/2-GFP::GEN3 Agcyk1Δ Agleu2Δ Agthr4Δ | This study |
| AgBOI11/2ΔN-GFP | AgBOI11/2Δ(21-721)-GFP::GEN3 Agleu2Δ Agthr4Δ | This study |
| AgBOI11/2ΔC-GFP | AgBOI11/2Δ(742-964)-GFP::GEN3 Agleu2Δ Agthr4Δ | This study |
| AgSPA2-GFP | AgSPA2-GFP::GEN3 Agleu2Δ Agthr4Δ | 30 |
| AgSPA2-GFPboi1/2Δ | AgSPA2-GFP::GEN3 Agboi1/2Δ::NAT1 Agleu2Δ Agthr4Δ | This study |
| AgSPA2-GFPrho3Δ | AgSPA2-GFP::GEN3 Agrho3Δ::NAT1 Agleu2Δ Agthr4Δ | This study |
| AgBOI1/2-GFPrho3Δ | AgBOI1/2-GFP::GEN3 Agrho3Δ::NAT1 Agleu2Δ Agthr4Δ | This study |
Microscopy.
The microscope setup used in this study and subsequent image- processing settings were the same as those described previously (30). Rhodamine-phalloidin staining was done as described previously (30).
Strain and plasmid constructions.
Strains generated in this study are listed in Table 1, and oligonucleotides used are listed in Table 2. AgBOI1/2 was deleted by PCR-based gene targeting using either the oligonucleotides AgBOI1/2-S1 and AgBOI1/2-S2 on the geneticin resistance marker GEN3 (59), generating Agboi1/2Δ from Agleu2Δthr4Δ, or the oligonucleotides AgBOI1/2-N1 and AgBOI1/2-N2 on the nourseothrycin resistance marker NAT1 (24), generating Agboi1/2Δ from Agleu2Δthr4Δ, AgSPA2-GFPboi1/2Δ from AgSPA2-GFP, and Agboi1/2Δrho3Δ from Agrho3Δ (61). As verification primers served AgBOI1/2-G1, AgBOI1/2-G4, AgBOI1/2-I, and, for the NAT1 marker, V2-NAT1 and V3-NAT1. AgRHO3 was deleted using the oligonucleotides AgRHO3-N1 and AgRHO3-N2 on NAT1, generating Agboi1/2Δrho3Δ from Agboi1/2Δ, AgBOI1/2-GFPrho3Δ from AgBOI1/2-GFP, and AgSPA2-GFPrho3Δ from AgSPA2-GFP. As verification oligonucleotides served AgRHO3-G1, AgRHO3-G4, and AgRHO3-I.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence |
|---|---|
| AgBOI1/2-S1 | AGGATGAGTTTTTCGTTCAAGAAGGCGCTGAAGCTGACCGGGATGGCTAGGGATAACAGGGTAAT |
| AgBOI1/2-S2 | TCTTTCGTGTTTATAATACAGACTTTTCAATAGCACTTGAAATCAAGGCATGCAAGCTTAGATCT |
| AgBOI1/2-N1 | TCTTTCGTGTTTATAATACAGACTTTTCAATAGCACTTGAAATCAGGGGCAGGGCATGCTCATGTAG |
| AgBOI1/2-N1 | TCTTTCGTGTTTATAATACAGACTTTTCAATAGCACTTGAAATCAGGGGCAGGGCATGCTCATGTAG |
| AgBOI1/2-G1 | GAGGACTCTTGAAGGCACGG |
| AgBOI1/2-G4 | AATTGACTCCTAACGAAGCTCC |
| AgBOI1/2-I | AACCAGGCCGATTCTGCGG |
| V2-NAT1 | GTGGTGAAGGACCCATCCAG |
| V3-NAT1 | ACATGAGCATGCCCTGCCCC |
| AgRHO3-N1 | CTGGCATCAGAGGAAGCTCCCACCACCAAGCTCTACAAACACAAGATGGGTACCACTCTTGACGAC |
| AgRHO3-N2 | ATATTAGTATAGTCTAAAGTTGCAGGCAGTGGGTATTAAAGTTCAGGGGCAGGGCATGCTCATGTAG |
| AgRHO3-G1 | CACGCGCAGCACGCGAGCTG |
| AgRHO3-G4 | CGCCGTTCCGCCTCGGCCGG |
| AgRHO3-I | CTCCGAGGATGACGATCTTG |
| AgBOI1/2-GFP1 | TTATTTATTAATCTGGCAGGACTGCTCTTTCGTGTTTATAATACAAGGGACCTGGCACGGAGC |
| AgBOI1/2-GFP2 | AACGAGTACTATGGCCTAGATCCGAAATATATGGGTGAGAAGATTGGTGCAGGCGCTGGAGCTG |
| AgCYK1-N1 | GTAACGTATCGCTAGTTCAACCTACCAGCAGTGCATCGGGAGCTACTAGGATGGGTACCACTCTTGACGAC |
| AgCYK1-N2 | CGTATTAATTATTATTGTTGTCCTTATCCATCGTTACGTAAGTATAATTAGGGGCAGGGCATGCTCATGTAG |
| AgCYK-G1 | TAGAGACCACGGCATTTG |
| AgCYK-G4 | GGCTGCTTTCTCCTATTG |
| 5SmaI-AgBOI1/2 | TCCCCCGGGTATGAGTTTTTCGTTCAAGAAGG |
| 3BamHI-AgBOI1/2 | CGGGATCCTCAAATCTTCTCACCCATATATTTCG |
| 5EcoRI-AgRHO3 | GATCGAATTCATGCCTCTGTGTGGGTCGAG |
| 3BamHIAgRHO3 | CGACGGATCCTAACTGCTGCTTTCGGCCTCGG |
| AgRHO3_G24Vfor | GTGGCTTGCGGGAAGACGTCGCTG |
| AgRHO3_(G24V)rev | GTCTCCGAGGATGACGATCTTG |
| AgRHO3_T29Nfor | AACTCGCTGTTGAACGTGTTCAC |
| AgRHO3_(T29N)rev | CTTCCCGCAAGCACCGTCTCC |
| AgRHO3_Q71Lfor | CTGCACATCACGCTGAGCCTGTG |
| AgRHO3_(Q71L)rev | GTTGTCCACGAAGATGTCATGG |
| AgBOI1/2ΔCfor | GCGAATGGCAATGCTAACGAGTAC |
| AgBOI1/2ΔCrev | CTTACTCATCCAACCCGAGCACGC |
| AgBOI1/2ΔNfor | CGGAACGTTACAGTAAATGACGCTG |
| AgBOI1/2ΔNrev | ACTGTCGTCGCCCTGCTCCATCCCG |
The 2,952-bp AgBOI1/2 open reading frame (ORF), including 917-bp upstream and 279-bp downstream sequences, was identified on a plasmid from a library (kindly provided by K.-P. Stahmann, Forschungszentrum Jülich, Germany) and subcloned with PstI/BamHI into pRS415 (51), generating pAGBOI1/2. The AgBOI1/2 ORF was tagged with the green fluorescent protein (GFP) on pAGBOI1/2 at the C- terminal coding region in a PCR-based yeast cotransformation using the oligonucleotides AgBOI1/2-GFP-S1 and AgBOI1/2-GFP-S2 on pGUG (30), generating pAGBOI1/2-GFP. The EcoRI/BamHI fragment carrying the GUG with flanking AgBOI1/2 regions was subcloned into pUC19 (65). The resulting pTCBOI1/2-GFP was amplified in Escherichia coli, digested with EcoRI/BamHI, and transformed into Agleu2Δthr4Δ, generating the AgBOI1/2-GFP strain. The AgBOI1/2-GFP Agcyk1Δ strain was generated from AgBOI1/2-GFP by PCR-based gene targeting using the oligonucleotides AgCYK1-N1 and AgCYK1-N2 on NAT1 and verified using the oligonucleotides AgCYK1-G1 and AgCYK1-G4.
Truncated AgBOI1/2-GFP alleles were generated by PCR. First, the 3,562-bp SacI/NdeI(blunt) fragment from pAGBOI1/2-GFP was subcloned into the SacI/SmaI site of pUC19, generating pNCDBOI. Oligonucleotides AgBOI1/2ΔCfor and AgBOI1/2ΔCrev were then used for PCR on pNCDBOI. The product was self ligated and transformed into E. coli. The 543-bp NcoI/BglII(blunt) fragment from this plasmid was cloned into the NcoI/EcoRI(blunt) site of pTCBOI1/2-GFP. The resulting plasmid, pTCBOI1/2ΔC-GFP, was digested with BamHI/EcoRI and transformed into the Agleu3Δ Agthr4Δ strain, generating the AgBOI1/2ΔC-GFP strain. Oligonucleotides AgBOI1/2ΔNfor and AgBOI1/2ΔNrev were used for PCR on pNCDBOI. The product was self ligated and transformed into E. coli. The 2,780-bp MscI/BamHI fragment from pTCBOI1/2-GFP was cloned into the MscI/BamHI site of this plasmid, generating pTCBOI1/2ΔN-GFP, which was digested with BamHI/SpeI and transformed into the Agleu3Δ Agthr4Δ strain, generating the AgBOI1/2ΔN-GFP strain.
For the two-hybrid assay, pGADT7 bearing the activation domain and pGBKT7 bearing the binding domain (Invitrogen) served as vectors. The AgBOI1/2 ORF was amplified by PCR from pAGBOI1/2 with the oligonucleotides 5SmaI-AgBOI1/2 and 3BamHI-AgBOI1/2. The PCR product was then cloned into pUC19, generating p2HBOI1/2, and the BamHI/SmaI fragment was subcloned into pGADT7, generating pAD-AgBOI1/2. The AgRHO3 ORF was amplified with the oligonucleotides 5EcoRI-AgRHO3 and 3BamHI-AgRHO3 from pAG11320 (15). The PCR product was cloned into pUC19, generating p2HRHO3. The EcoRI/BamHI fragment was then subcloned into pGBKT7, generating pBD-AgRHO3. Point mutations for the dominant AgRHO3 alleles were generated by PCR using the oligonucleotide pairs AgRHO3_G24Vfor and AgRHO3_(G24V)rev, AgRHO3_T29Nfor and AgRHO3_(T29N)rev, and AgRHO3_Q71Lfor and AgRHO3_(Q71L)rev on p2HRHO3. The PCR products self ligated and transformed into E. coli, and the respective EcoRI/BamHI fragments were then subcloned into pGBKT7. Truncated AgBOI1/2 alleles for two-hybrid experiments were generated as the truncated AgBOI1/2-GFP alleles. p2HBOI1/2 was amplified by PCR with the oligonucleotides AgBOI1/2ΔCfor and AgBOI1/2ΔCrev, and the PCR product self ligated and transformed into E. coli. The SacII/BamHI fragment was subcloned into the SacII/BamHI site of pAD-AgBOI1/2, generating pAD-AgBOI1/2ΔC. p2HBOI1/2 was amplified by PCR with AgBOI1/2ΔCfor and AgBOI1/2ΔCrev. The PCR product self ligated and transformed into E. coli. The SmaI/BamHI fragment was cloned into pGADT7, generating pAD-AgBOI1/2ΔN. To test interactions, the two-hybrid plasmids were cotransformed into the yeast strain PJ69-4a (29) and plated on synthetic complete medium without leucine and tryptophanedropout plates. Interactions were determined qualitatively with a 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) overlay or quantitatively (see below).
Biochemical assays.
The protocol for Western blotting was adapted from previous works (31, 54). Briefly, A. gossypii was cultured in full medium at 30°C for 16 h. The mycelium was harvested by filtration and washed twice in cold phosphate-buffered saline (PBS); 0.5 ml wet volume of mycelium was then mixed with 0.5 ml NP-40 lysis buffer (50 mM Tris-Cl, pH 7.5; 150 mM NaCl; 5 mM EDTA; 1% NP-40; 2 mM NaPPi), 1/40 volume protease inhibitor (Roche), and 0.5 ml glass beads in a 2-ml screw-cap tube. The mycelium was vortexed six times for 30 s in a FastPrep (Qbiogene) and centrifuged at 14 krpm for 10 min. The protein concentration from the supernatant was determined in a Bradford assay (Bio-Rad). Equal amounts of protein were boiled in 2× sample buffer (100 mM Tris-Cl, pH 6.8; 4% sodium dodecyl sulfate [SDS]; 0.2% bromphenol blue; 20% glycerol; 100 mM dithiothreitol). The extracts were separated on an SDS-polyacrylamide gel electrophoresis gel. Proteins were immobilized to nitrocellulose in an electrotransfer chamber. The membrane was washed with PBS, blocked, incubated at 1/1,000 with the primary anti-GFP antibody (Roche catalog no. 11814460001), washed, and incubated with the secondary antibody. The secondary antibody was detected with an ECL kit (Amersham).
The beta-galactosidase activity for quantitative yeast two-hybrid experiments was determined as described previously (2).
RESULTS
AgBoi1/2p prevents nonpolar growth at hyphal tips and is required for maximal tip extension rates.
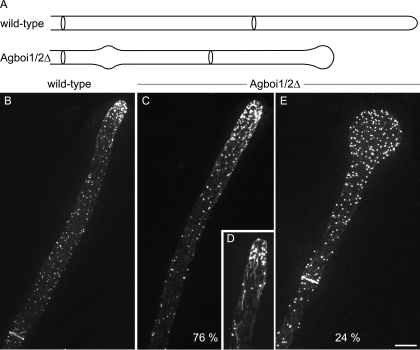
By screening A. gossypii gene knockout strains, we found that the Agboi1/2Δ mutant displayed alternating phases of hyphal tip swelling and polar growth, indicating transient defects in sustained polarization (Fig. 1A). Twenty-four hours after inoculation of mutant spores in liquid medium at 30°C, 24% ± 9% of hyphal tips were enlarged. When incubated at 20°C, 13% ± 3% hyphal tips were enlarged (standard deviations [SD]; n > 500). In wild-type hyphae, less than 1% of tips were enlarged at either of these temperatures. Septation and branching seemed not to be affected in the mutant strain.
FIG. 1.
Spherical enlargement of hyphal tips in Agboi1/2Δ. (A) Schematic representation of the tip polarization defect in Agboi1/2Δ. (B to E) Rhodamine-phalloidin stainings of wild-type and Agboi1/2Δ hyphae. Panels B, C, and E represent single hyphae from >20-h cultures, whereas panel D represents a hypha from a <16-h culture. More actin patches accumulate at hyphal tips in mature hyphae than in hyphae at younger stages. As the signal intensity of accumulated actin patches cross-fades the signal from the weaker actin cables, the actin cable network is better seen in younger hyphae. Refer also to Fig. 1A in reference 30. Bar, 10 μm.
Using rhodamine-phalloidin stainings of Agboi1/2Δ and wild-type mycelia, differences in the actin cytoskeletons were investigated. In polarized Agboi1/2Δ hyphae, the localization of actin patches and actin cables appeared like in the wild type (Fig. 1B, C, and D). In spherically enlarged Agboi1/2Δ tips, actin cables were not detected any more and actin patches were evenly distributed within the tip (Fig. 1E). As in the wild type, actin rings were established in subapical regions of Agboi1/2Δ mycelia (Fig. 1B and E).
When Agboi1/2Δ strains were grown on solid full medium at 30°C, the radial colony growth speed was 108 μm/h ± 6 μm/h compared to 196 μm/h ± 6 μm/h for the wild type (SD; n > 20). In order to determine whether this reduced radial growth of mutant colonies was solely the consequence of frequent growth arrests caused by hyphal tip enlargements or whether Agboi1/2Δ hyphae suffered in addition from reduced tip extension rates, we monitored individual hyphae by time-lapse microscopy. Further, we wanted to know how frequently spherically enlarged tips could reinitiate polar growth.
The maximal elongation speed of A. gossypii hyphae on solid full medium is reached after 48 h and stays constant until the nutrient supply decreases (30). For Agboi1/2Δ, we determined a maximal tip extension rate of 125 μm/h ± 19 μm/h compared to 240 μm/h ± 15 μm/h in the wild type at the edge of 3-day-old colonies grown at 29°C (SD; n > 30; Table 3). These numbers for mutant and wild-type hyphae are slightly higher, as determined from the radial growth speed of colonies, because individual hyphae do not grow exactly radially at the edge of the colonies.
TABLE 3.
Summary of polar growth phenotypes
| Strain | Maximum tip extension speed (μm/h) on solid complete medium at 29°C | % Tips with nonpolar growth (1-day-old mycelium; 30°C; n > 500) | Repolarization in original growth axis after nonpolar tip growth | % Lysis at 37°C as germ bubble | % Lysis at 37°C as unipolar germling |
|---|---|---|---|---|---|
| Wild type | 240 ± 14 | <1 | <1 | <1 | |
| Agboi1/2Δ | 125 ± 19 | 24 ± 9 | 15 out of 20 | 33 ± 2 | 67 ± 2 |
| Agrho3Δ | 127 ± 9 | 26 ± 8 | 16 out of 23a | 33 ± 4 | 67 ± 4 |
| Agboi1/2Δrho3Δ | 123 ± 14 | 23 ± 7 | NDb | 34 ± 1 | 66 ± 1 |
This datum was taken from reference 61.
ND, not determined.
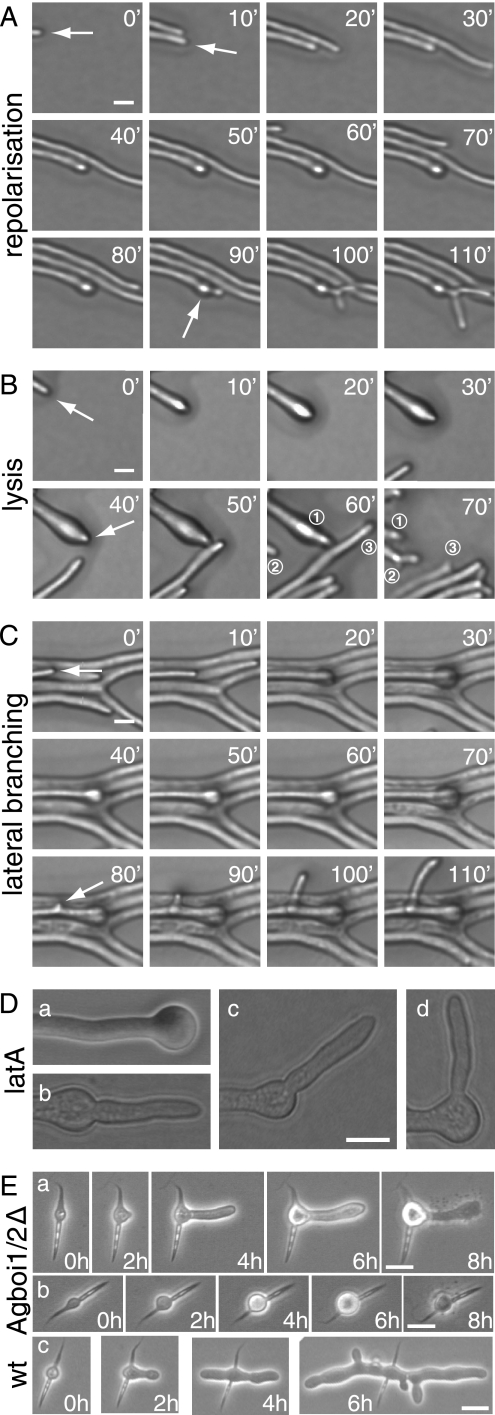
From 20 hyphae displaying spherically enlarged tips, 15 repolarized in the original growth axis within a 3-h observation time. Eleven of these repolarized hyphae generated apical branches within 10 to 40 min after repolarization, the predominant branching type in mature hyphae (Fig. 2A). Two tips continued to elongate without an apical branch, and two repolarized hyphae lysed (Fig. 2B). Growth of repolarized hyphae accelerated to the speed prior to depolarization within 20 to 30 min. Five of the 20 observed swollen tips did not repolarize within the observation time. Of these, four developed a lateral branch within 50 μm subapical of the enlarged tip (Fig. 2C), and one lysed. Treating A. gossypii mycelia for 20 min with 20 μM of the actin-depolymerizing drug latrunculin A (6) also causes spherical enlargement of hyphal tips, but repolarization upon removal of the drug occurs in different directions. Seventy percent of the depolarized hyphae repolarized at an angle of 180° to 145°; 30% did so at 145° to 90° (n > 200; Fig. 2D). Hyphal tips stopped elongating but did not start swelling when mycelia were treated with 200 μM latrunculin A.
FIG. 2.
Spherical enlargement and repolarization of Agboi1/2Δ hyphae (A to D) and polarity establishment in germinating Agboi1/2Δ spores (E). The three sequences of images in panels A, B, and C represent examples of fast-elongating hyphae at the edge of a colony grown on solid full medium for 3 days at 29°C. Images were acquired every 10 min, and the elapsed time is indicated for each frame. (A) Hypha elongating with 120 μm/h until the tip starts enlarging (second frame). After 80 min the hypha continues to grow in the original polarity axis (white arrow) followed by a tip branching within 10 to 15 min. This series of events was observed in 11 of 15 repolarizing hyphae; only the respective time intervals varied. (B) Enlargement of a hypha (second frame), a repolarization in the original polarity axis after 40 min, and tip lysis 20 to 30 min after repolarization. For better orientation, three hyphae of the frames at 60 and 70 min are numbered. Hypha “1” lysed between the two acquisitions and was pushed backwards; only the most apical part can be seen. Hypha “2” formed an apical branch, and hypha “3” was delocalized due to the abrupt lysis of hypha “1”. Two of the 15 repolarizing hyphae lysed and two continued to grow with 100 to 120 μm/h during the observation period. (C) Enlargement at the tip of a fast-growing hypha, which did not repolarize within 2 h but established a new lateral branch subapically (white arrow, frame 80 min). Four of five nonrepolarizing hyphae developed a lateral branch close to the swollen tip, and one lysed during swelling. (D) Treatment of wild-type hyphae with latrunculin A. Mycelia cultured from spores in liquid full medium for 18 h were treated with 20 μM latrunculin A for 20 min, resulting in more than 99% spherically enlarged tips (a). The drug was then washed out, and the mycelia were incubated on solid full medium. After 3 h, 77% of hyphae were repolarized. (b and c) Tips with a repolarization angle between 180° and 145°. (d) Repolarization angle between 145° and 90°. Bar, 10 μM. (E) The two sequences a and b represent examples of germinating Agboi1/2Δ spores at 37°C. (c) A wild-type spore at 37°C. Images were acquired every 2 h, and the elapsed time is indicated for each frame. Germination starts in the middle of the needle-shaped spores (time = 0 h). (a) The germ bubble establishes a polar growth site (2 h) and a germ tube, the first hypha, emerges (4 h). The germ tube enlarges at the tip (6 h) and subsequently lyses (8 h). (b) The germling does not establish a polar growth site, and lysis occurs at the stage of a germ bubble (8 h). The wild type (c) establishes two polar growth sites from the germ bubble in the first 4 h and initiates lateral branches. Bars, 10 μm. wt, wild type. latA, latrunculin A.
We further found that AgBOI1/2 is essential for the formation of mycelia at elevated temperatures. When Agboi1/2Δ spores were incubated at 37°C on solid full medium, less than 3% (n > 200) formed mycelia, compared to over 90% at 20°C. At 37°C, the germinating Agboi1/2Δ spores lysed either as unipolar germlings (67% ± 2%) or as germ bubbles (33% ± 2%) (Fig. 2E). Germination efficiency of wild-type spores was over 90% at 20°C, 30°C, and 37°C.
Taken together, these data show that AgBoi1/2p is required to prevent nonpolar growth at hyphal tips and for maximal hyphal tip extension. The majority of spherically enlarged tips repolarized after 10 to 80 min in the original axis, indicating that directional cues at hyphal tips are not lost during enlargement. Actin patches in enlarged Agboi1/2Δ tips are still present but are delocalized, and actin cables are no longer present at the tip. The maintenance of directionality observed in repolarized Agboi1/2Δ hyphae is not seen when latrunculin A-treated hyphal tips repolarize. Apparently, the destruction of all actin cytoskeletal structures in the hyphae leads also to a loss of directional cues.
AgBoi1/2p and its fungal homologues.
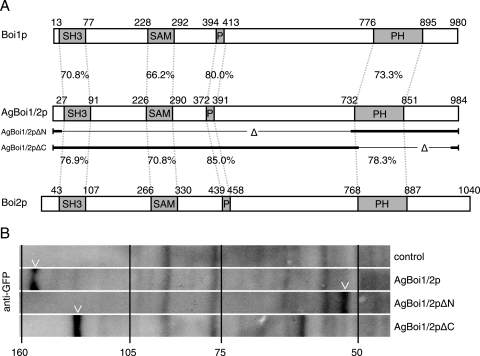
A. gossypii AgBOI1/2 encodes a protein of 984 amino acids (aa). It is the syntenic homologue of the functionally redundant S. cerevisiae genes BOI1 and BOI2, which were identified as Bem-one-interacting proteins (3, 9, 34). The overall identity between AgBoi1/2p and its S. cerevisiae homologues Boi1 and Boi2 is 36% and 38%, respectively, and between S. cerevisiae Boi1 and Boi2 is 36% (Fig. 3A). The homologue identified in Schizosaccharomyces pombe, Pob1, shares 22% identity with AgBoi1/2p (55). All four proteins carry an N-terminal SH3 domain (35), followed by a SAM domain (52), a proline-rich region (less pronounced in Pob1; 35) and a PH domain (22, 42) (Fig. 3A). A blastp search with an E value cutoff below 1e-10 against GenBank protein sequences (www.ncbi.nlm.nih.gov) revealed further hits in Kluyveromyces lactis (CAG99981), Candida albicans (XP_717921 and XP_436423), Candida glabrata (CAG58163), Magnaporthe grisea (XP_367385), Neurospora crassa (CAD21496), A. fumigatus (EAL87433), and A. nidulans (XP_662100 and XP_408633), demonstrating that the Boi proteins are evolutionarily conserved.
FIG. 3.
Alignment and expression of AgBoi1/2p. (A) Alignment of A. gossypii AgBoi1/2p with S. cerevisiae Boi1p and Boi2p. Individual domains are marked with boxes, and the identity between corresponding domains is indicated (generated with the application “needle” from “EMBOSS” [43]). The sequences of the partial proteins AgBoi1/2pΔN and AgBoi1/2pΔC are indicated below the AgBoi1/2p sequence, with the thin lines marking the respective deletions. (B) Western blot of wild-type AgBoi1/2p and truncated proteins. Proteins from 16-h cultures were extracted, separated on an SDS-polyacrylamide gel electrophoresis gel, transferred to a membrane, and hybridized with an anti-GFP antibody. Molecular masses from a standard protein marker are indicated in kilodaltons. The molecular mass of the GFP epitope tag with a linker is 28 kDa. A degradation product was detected for AgBoi1/2pΔC at ∼65 kDa.
AgBoi1/2p or truncations thereof (see below) are expressed during growth, as shown by immunoblotting with an anti-GFP antibody against a C-terminally GFP-tagged protein expressed from the endogenous promoter (Fig. 3B). The size of the wild-type fusion protein is ∼150 kDa (107 kDa for AgBoi1/2p and 28 kDa for the GFP tag).
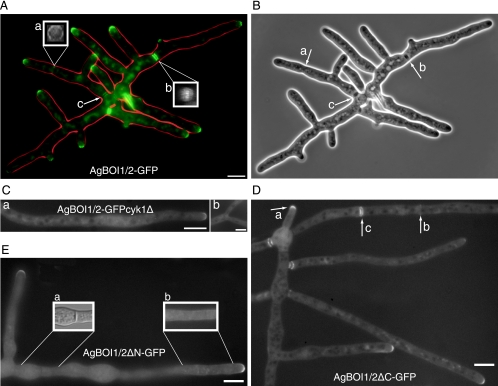
AgBoi1/2p localizes to tips and to sites of septation.
In order to localize AgBoi1/2p, the GFP was fused to the 3′ end of the AgBOI1/2 coding region. The GFP tag did not impair the function of AgBoi1/2p. Spores of AgBOI1/2-GFP were cultured in full medium, and the GFP signal was localized by fluorescence microscopy (Fig. 4A). AgBoi1/2p-GFP localized to tips as a crescent at the cortex. Time-lapse analysis showed that this localization was permanent (data not shown). AgBoi1/2p-GFP also localized to sites of septation either as a single ring in the vicinity of hyphal tips (Fig. 4A) or as double discs further distant from the tip (Fig. 4A). Single-ring localization occurred at presumptive sites of septation not yet visible in the phase-contrast image (Fig. 4B), whereas double-disc localization was clearly visible as a mature septum in the phase-contrast image (Fig. 4B), implying a transition from the single ring to the double-disc structure. We approximated 2 to 3 h for the transition from the single ring to the double-disc structure. At older septa localizing to the vicinity of the germinated spore, AgBoi1/2p-GFP was still visible as a double disc, but the signal started to fade away (Fig. 4A). Due to low GFP signal intensity in fixed AgBOI1/2-GFP strains, we were unable to visualize AgBoi1/2p-GFP together with actin. From the available data, we assume that AgBoi1/2p-GFP colocalizes with the actin ring. Upon contraction of this ring, the two discs might then be established flanking the site of contraction.
FIG. 4.
Localization of AgBoi1/2p. (A) Localization of wild-type AgBoi1/2p. Localization of the GFP signal is represented in green, and the hyphal periphery is in red. AgBoi1/2p-GFP is detected as a crescent at hyphal tips and as two different structures at sites of septation, either as a single ring at presumptive sites of septation (a) or as a double disc in mature septa (b). The two inserts (a and b) show three-dimensional reconstructed views of the indicated septal localizations. For each insert, a series of images was acquired along the z axis, and the planes were 90° rotated along the vertical axis. This gives a view along the hyphal tube onto the septum. (c) In mature septa, AgBoi1/2-GFP localization diminishes. (B) Phase-contrast image of that shown in panel A. Presumptive sites of septation identified from AgBoi1/2p-GFP ring localization are not yet visible (a), and mature septa identified from AgBoi1/2p-GFP disc localization are visible as black structures spanning the hyphae (b and c). (C) Localization of AgBoi1/2p-GFP in an Agcyk1Δ background. Septa are absent in the Agcyk1Δ strain, and AgBoi1/2p-GFP signal was observed neither in subapical regions (a) nor at sites of apical branching (b) where they are found in wild type. Bar, 10 μm. (D) Localization of AgBoi1/2pΔC. AgBoi1/2pΔC-GFP localizes at hyphal tips (a) and at septa as single-ring (b) or double-disc (c) structures as seen for the wild type. (E) Localization of AgBoi1/2pΔN. AgBoi1/2pΔN-GFP localizes only to hyphal tips and not to septa. (a) Nomarski illumination of the hyphal segment below. In the middle a septum is clearly visible but a GFP signal is lacking at the corresponding site. The GFP signal also appears to localize weakly over the entire cortex, as seen on insert b, which represents a brighter scaling of a single plane from the hyphal segment below. Bar, 10 μm.
In the Agboi1/2Δ strain, actin rings are formed and complete septa are established, indicating that AgBoi1/2p is not essential for septation. As AgBoi1/2p-GFP colocalizes with septa, we wanted to investigate whether this localization depends on septa and, if so, whether septum localization of AgBoi1/2p has an effect on polarization of hyphal tips. AgCYK1 encodes a septum-localizing protein with homology to the IQGAP family of proteins. In the absence of AgCYK1, hyphal tip extension rates are wild-type-like, but no septa are present (62). We deleted the AgCYK1 ORF in the background of AgBOI1/2-GFP. AgBoi1/2p-GFP still localized to hyphal tips but could not be detected in subapical regions where septa are expected (Fig. 4C).
Taken together, we show that AgBoi1/2p localizes to hyphal tips and to sites of septation. Septum localization of AgBoi1/2p depends on AgCyk1p, but AgCyk1p is dispensable for the permanent localization of AgBoi1/2p to hyphal tips. Moreover, the localization of AgBoi1/2p at sites of septation is not required for permanent polar growth of hyphal tips.
Functional analysis with truncated AgBOI1/2 alleles.
AgBoi1/2p localizes to tips and to sites of septation. In order to map the region in AgBoi1/2p important for localization of the protein, we expressed GFP fusions of two partial alleles of AgBoi1/2p from the endogenous promoter. In AgBOI1/2ΔC-GFP the coding region for amino acids (aa) 742 to 964 was eliminated, which includes the PH domain but leaves the 20 C-terminal amino acids. In AgBOI1/2ΔN-GFP the coding region for aa 21 to 721 was eliminated, which includes the SH3 and the SAM domains and the proline-rich region. Western blotting with an anti-GFP antibody confirmed the expression of a 59-kDa AgBoi1/2ΔN-GFP and a 111-kDa AgBoi1/2ΔC-GFP protein, respectively (including 28 kDa for the GFP tag) (Fig. 3B). The two truncated proteins, however, were not functional. Analysis of polarized tips and the radial growth speed did not reveal significant differences compared to the complete deletion strain. The AgBOI1/2ΔN strain displayed 29% ± 3% spherically enlarged tips, and AgBOI1/2ΔC displayed 23% ± 3% compared to 24% ± 9% in the complete deletion strain (SD; n > 500; see above). The radial colony growth speed at 30°C on full medium plates was 117 μm/h ± 12 μm/h for AgBOI1/2ΔN and 104 μm/h ± 17 μm/h for AgBOI1/2ΔC, versus 108 μm/h ± 28 μm/h for the complete deletion strain (SD; n > 20).
Using fluorescence microscopy, we identified the AgBoi1/2pΔC-GFP localizing as a crescent to hyphal tips, as a single ring to presumptive sites of septation, and as a double disc to mature septa (Fig. 4D). AgBoi1/2pΔN-GFP localized only to hyphal tips (Fig. 4E). At sites of septation a signal could not be observed, even though septa were still formed, as seen in Nomarski acquisitions. The absence of AgBoi1/2pΔN-GFP at septa might also explain why the corresponding band in the Western blot is less intense than the wild-type band (Fig. 3B). A weak localization was also detected along the subapical cortex.
Together, these data show that in A. gossypii both the N-terminal region containing the SH3 domain, the SAM domain, and the proline-rich region as well as the C-terminal part containing the PH domain are required for AgBoi1/2p function. Wild-type-like localization, which includes tip and septum localization, depends on the N-terminal part of the protein. Interestingly, the C-terminal part alone containing the PH domain localizes independently to tips but not to septa.
AgBoi1/2p and AgRho3p signal in the same pathway.
The deletion of AgRHO3 leads to spherically enlarged tips with delocalized actin, temperature-sensitive germ tube emergence due to early lysis of germlings, and reduced colony growth speed (61), which resembles the Agboi1/2Δ phenotype.
We therefore wanted to investigate whether the phenotype of a strain deleted for both AgBOI1/2 and AgRHO3 is additive, suggesting independent functions of AgBoi1/2p and AgRho3p, or whether a double deletion shows the same phenotype as the single deletions, suggesting a common function with the two proteins involved. For that reason, AgRHO3 was deleted in an Agboi1/2Δ strain and vice versa, and the resulting Agboi1/2Δrho3Δ strains were compared to the respective single-deletion strains. The phenotype was quantified for the rate of spherically enlarged tips, the distribution of arrested germlings at elevated temperature, and the hyphal tip extension rate (Table 3).
Tip polarization was determined after 24 h at 30°C from spores inoculated in liquid full medium. For the Agboi1/2Δ strain, we found 24% ± 9% enlarged tips; for the Agrho3Δ strain, we found 26% ± 8%; and for the double-deletion strain Agboi1/2Δrho3Δ, we found 23% ± 7% (SD; n > 500). The ratio between germ bubbles and unipolar germlings was quantified from spores streaked onto an agar layer incubated at 37°C (see above). For Agboi1/2Δ we detected 67% ± 2% unipolar germlings, for Agrho3Δ we detected 67% ± 4% unipolar germlings, and for the double-deletion strain Agboi1/2Δrho3Δ we detected 66% ± 1% unipolar germlings (SD; n > 100). The tip extension rate determined for the Agboi1/2Δ strain was 125 μm/h ± 19 μm/h (see above), the tip extension rate determined for Agrho3Δ was 127 μm/h ± 9 μm/h, and the tip extension rate determined for the double-deletion strain Agboi1/2Δrho3Δ was 123 μm/h ± 14 μm/h.
The three analyses performed did not reveal significant differences between the double-deletion strain and either of the single-deletion mutants. This suggests that AgBOI1/2 and AgRHO3 signal together in one and the same pathway to prevent depolarization of hyphal tips.
Loss of the polarisome component AgSpa2p precedes spherical enlargement in Agboi1/2Δ and in Agrho3Δ.
Spherically enlarged tips in Agboi1/2Δ strains lack tip-emanating actin cables, and actin patches localize evenly distributed in the enlarged tip. We wanted to investigate the behavior of the polarisome before and during spherical enlargement. In the wild type, AgSpa2p localizes permanently to sites of growth independently of actin (30, 47).
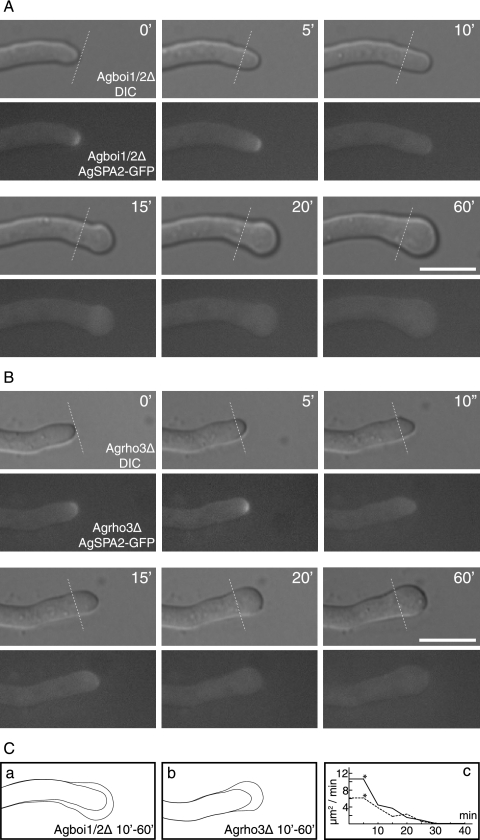
The AgBOI1/2 ORF was deleted in the AgSPA2-GFP background. AgSpa2p-GFP was followed by fluorescence time-lapse microscopy for 60 min, before and during the spherical enlargement of a hyphal tip. Images were taken every 5 min. During the polarized growth phase in Agboi1/2Δ, AgSpa2p-GFP localized to the hyphal tip, as observed in the wild type. When loss from the tip was observed, the tip stopped elongating and started to enlarge for 20 min (Fig. 5A and C). This demonstrates that AgBoi1/2p is required to keep AgSpa2p permanently polarized at hyphal tips.
FIG. 5.
AgSpa2p-GFP localization during spherical enlargement in Agboi1/2Δ and Agrho3Δ. (A and B) The respective top images represents the Nomarski acquisitions, the bottom images the fluorescence acquisitions; the time is indicated in minutes. AgSpa2p-GFP localizes to hyphal tips during polarized growth phases, as indicated in the two first acquisitions (t = 0; t = 5). Lines in the respective frame indicate the tip extension during the first 5 min. AgSpa2p-GFP is lost while the tip is not yet enlarged (t = 10). Enlargement starts in the following frame (t = 15) and lasts for 45 min (t = 20 to 60). Due to different growth media, temperatures, and developmental stages, the extension rate is decreased compared to mature hyphae of the wild type on full medium at 30°C. The entire observation lasted for more than 120 min. Bar, 10 μm. (C) The hyphal cortex prior to enlargement at 10 min and fully enlarged at 60 min for Agboi1/2Δ (a) and Agrho3Δ (b), respectively. (c) The time-dependent decrease of surface expansion inAgboi1/2Δ and Agrho3Δ is indicated. The time corresponds to that for panels A and B; an asterisk indicates the last frame with polarized AgSpa2p.
We also analyzed the dynamics of AgSpa2p-GFP in the background of an Agrho3Δ strain. Similarly, AgSpa2p-GFP was apically localized during polarized growth phases, and loss of AgSpa2p-GFP preceded spherical enlargement (Fig. 5B and C).
Figure 5A and B show representative examples chosen from three independent acquisitions each.
AgBoi1/2p-GFP localization in Agrho3Δ strains.
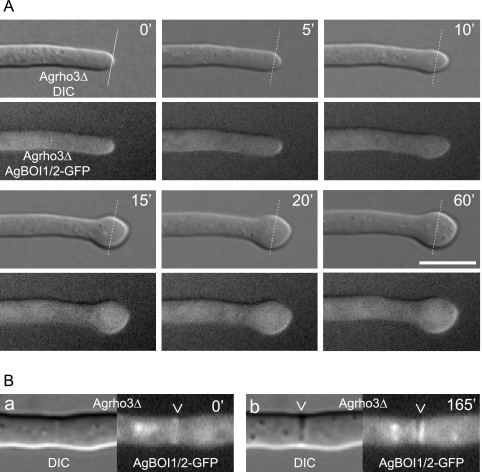
We have further analyzed the localization of AgBoi1/2p-GFP in the Agrho3Δ strain. During polarized growth phases, AgBoi1/2p-GFP localized as a crescent to hyphal tips (Fig. 6A) and in subapical regions as a single ring to presumptive sites of septation, which developed to a double disc within 2 to 3 h (Fig. 6B). This time frame is comparable to the dynamics at septa in wild-type hyphae (see above). When hyphal tips enlarged,AgBoi1/2p-GFP was still present at the hyphal tip but distributed at the enlarging cortex (Fig. 6A). Despite several different approaches, we did not obtain a functional fusion of AgRho3p to the GFP.
FIG. 6.
AgBoi1/2p-GFP localization in Agrho3Δ. (A) The respective top images represent the Nomarski acquisitions, the bottom images the fluorescence acquisitions; the time is indicated in minutes. AgBoi1/2p-GFP localizes to hyphal tips during polarized growth phases as indicated in the two first acquisitions (t = 0; t = 5). When the hyphal tip starts to enlarge, AgBoi1/2p-GFP is distributed over the enlarging cortex at the hyphal tip. (B) Localization of AgBoi1/2p-GFP in Agrho3Δ at septal sites. AgBoi1/2p-GFP is established as a ring (a) and develops to a double-disc structure (b). The presumptive site of septation identified from AgBoi1/2p-GFP ring localization is not yet visible in Nomarski (a), whereas mature septa identified from AgBoi1/2p-GFP disc localization become visible as black structures spanning the hyphae (b).
AgRho3p interacts in two-hybrid experiments with AgBoi1/2p and with AgBni1p.
AgBoi1/2p and AgRho3p act in a common pathway. We wanted to investigate whether AgBoi1/2p and AgRho3p also interact physically. Therefore, the two proteins were tested in a yeast two-hybrid experiment.
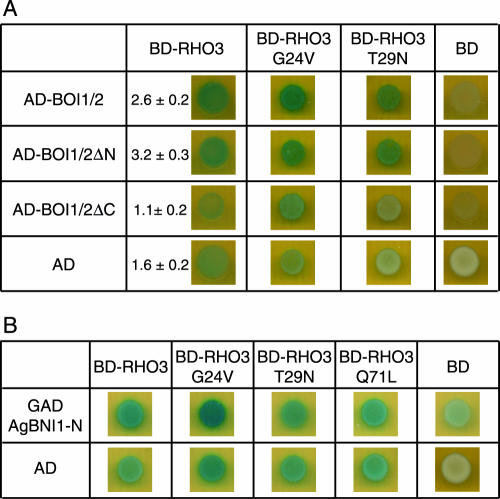
AgBoi1/2p with an N-terminal fusion to the Gal4 activation domain on pAD-AgBOI1/2 and AgRho3p with an N-terminal fusion to the Gal4 binding domain on pBD-AgRHO3 were coexpressed in the yeast two-hybrid strain PJ69-4a (29). Interaction was determined qualitatively based on blue coloration of colonies overlaid with X-Gal. This demonstrated an interaction between AgBoi1/2p and AgRho3p (Fig. 7A). A slight self activation of pBD-AgRHO3 in combination with the vector pGADT7 alone was also detected. A quantitative determination of the β-galactosidase activity revealed a significant difference between pAD-AgBOI1/2 and pBD-AgRHO3 compared to the vector controls (P < 0.002; Fig. 7A).
FIG. 7.
AgBoi1/2p-AgRho3p-AgBni1p two-hybrid interactions. (A) AgBoi1/2p-AgRho3p interaction. In the left column, the (truncated) AgBoi1/2p fused to the activation domain and the activation domain alone are listed, and in the top row the AgRho3p fused to the binding domain and the binding domain alone are listed. (B) AgRho3p-AgBni1p interaction. In the left column the AgBni1 protein fused to the activation domain (47) and the activation domain alone are listed. In the top row, the AgRho3p fused to the binding domain and the binding domain alone are listed. In each image 5 μl of a yeast culture of strain PJ69-4a with an optical density at 600 nm of 0.1 and transformed with the corresponding plasmids encoding the fusions proteins was spotted, incubated overnight, overlaid with X-Gal, and incubated for 16 to 24 h. Blue color indicates interaction. The quantitative determination of the β-galactosidase activity is given in arbitrary units. Error is the standard error of the mean. AD, activation domain; BD, binding domain.
In order to map the AgRho3p interaction site on AgBoi1/2p, we also tested the two truncated alleles on pAD-AgBOI1/2ΔC and pAD-AgBOI1/2ΔN (see above and Fig. 3A). We found a significant interaction between AgBoi1/2ΔNp and AgRho3p but not between AgBoi1/2ΔCp and AgRho3p (P < 0.0004; Fig. 7A).
We wanted to know whether the AgBoi1/2p-AgRho3p interaction was dependent on AgRho3p specifically bound to GDP or GTP. Therefore, we introduced the constitutively active AgRho3pG24V and the constitutively negative AgRho3pT29N mutations into pBD-AgRHO3 by analogy to a ras oncogene (11). We found that both the constitutively active AgRho3pG24V and the constitutively negative AgRho3pT29N proteins interact with the full-length AgBoi1/2p and the N-terminally truncated AgBoi1/2pΔN (Fig. 7A).
In spherically enlarged Agboi1/2Δ tips, actin cables were not detected (see above), which could originate from a deactivation of the formin AgBni1p. It has previously been suggested that the Rho-type GTPase AgCdc42p in its constitutive active form interacts and thereby activates AgBni1p to promote actin cables from sites of hyphal tip growth (47). We thus tested an interaction between AgRho3p and the Rho binding domain (RBD) of AgBni1p and found that constitutive active AgRho3pG24V but not constitutive negative AgRho3pT29N interacted with the RBD of AgBni1p. Neither the constitutive active mutations AgRho3pQ71L and AgRho3pQ71H as reported by Nakano et al. (38) and Schmitz et al. (47), respectively, nor the constitutive negative AgRho3pT29N interacted with the RBD of AgBni1p. An interaction between the wild- type AgRho3p allele and the RBD of AgBni1p was weakly visible.
These data show that AgBoi1/2p and AgRho3p interact in two-hybrid experiments. The C-terminal region of AgBoi1/2p is important for this interaction, and AgRho3p binds either in its GTP- or GDP-bound form. Further, AgRho3p interacts in its GTP-bound form with the RBD of the formin AgBni1p.
DISCUSSION
With AgBoi1/2p we identified a new protein in A. gossypii that acts together with the Rho-type GTPase AgRho3p to prevent nonpolar growth at hyphal tips. Both proteins have orthologues in other filamentous fungi, which suggests that the underlying mechanism of permanent polarized growth is conserved.
In strains deleted for AgBOI1/2 or AgRHO3, AgSpa2p-GFP is lost prior to spherical enlargement. Loss of AgSpa2p alone should not lead to a depolarization of the tip, since AgSpa2p is not necessary for polarizing the hyphal tip, as demonstrated in an Agspa2Δ strain (30). This implies that AgSpa2p is lost in a functional complex required to promote polar growth, and AgBoi1/2p and AgRho3p have a stabilizing function on this complex. By analogy to S. cerevisiae, this could be the polarisome, a complex including Bud6, Pea2, and Bni1 (50), which directs polarized cell growth via nucleation of actin cables at growing bud tips (19, 20, 45). For all of these components a homologue has been identified in A. gossypii (28). The formin AgBni1p in A. gossypii is required to promote actin cable formation at hyphal tips, and it is likely activated by AgCdc42p (47). Since tip-localizing actin cables are lost in enlarged tips of Agboi1/2Δ and Agrho3Δ strains, we assume that AgBoi1/2 and AgRho3p can also activate AgBni1p to prevent nonpolar growth. This is supported with the finding that constitutively active AgRho3pG24V interacts with the RBD of AgBni1p.
In addition to frequent depolarizations of hyphal tips,Agboi1/2Δ and Agrho3Δ strains also display a reduced hyphal tip extension rate while the actin cytoskeleton remains polarized. These two defects can be completely independent of each other, implying that AgBoi1/2p and AgRho3p have two (or more) separable functions, or the two defects may depend on each other. The first hypothesis would be in agreement with a model in S. cerevisiae, where Rho3 has a role in exocytosis that is distinct from its role in actin polarity (1, 44). One could argue that the reduced tip extension rate in Agboi1/2Δ and Agrho3Δ results from a secretion defect, as for the rho3-V51 mutation in S. cerevisiae, which is separable from the actin polarization defect observed only in the complete RHO3 deletion. In the second hypothesis, we propose that the AgBoi1/2p-AgRho3p complex permanently reinforces actin cable nucleation via AgRho3p-dependent activation of AgBni1p during hyphal extension. Lack of AgBoi1/2p or AgRho3p eliminates this activation, resulting in permanently reduced hyphal tip growth. AgBni1 is then probably activated by AgCdc42p alone and possibly by AgRho4p. However, the deletion of AgRHO4 does not show a growth defect (61). The frequent loss of the polarisome observed in Agboi1/2Δ or Agrho3Δ strains would suggest that the stable localization of the polarisome at the hyphal tip requires AgBni1p activation via AgRho3p and AgBoi1/2p. In this model, a stable polarisome depends on the function of the polarisome itself and suggests that reinforcement of the polarisome is essential for its stable localization in a positive feedback. AgRho3p then primarily activates AgBni1p during hyphal extension, whereas AgCdc42p is more important during germination and branching. In agreement with this hypothesis would be the distinct participation of Rho-type GTPases in polarized actin cable formation as reported for S. cerevisiae. There, Rho3p and Rho4p are essential for direct Bni1p and Bnr1p activation during bud growth, whereas Cdc42p functions in cable assembly at the initiation of bud growth (16). We favor the second hypothesis and propose that AgBoi1/2p and AgRho3p promote and strengthen AgBni1p-driven actin cable nucleation and thereby prevent nonpolar growth at hyphal tips.
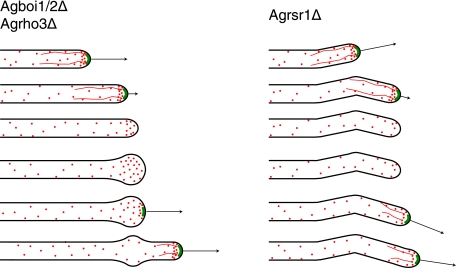
Similar to what is seen for the Agboi1/2Δ and Agrho3Δ strains, the deletion of AgRSR1 also causes frequent loss of AgSpa2p from hyphal tips (7). However, in Agrsr1Δ strains this is not associated with a spherical enlargement. Upon loss of AgSpa2p in Agrsr1Δ strains, tip extension is stopped and the tips do not enlarge. Subsequent repolarization occurs in a frequent uncoordinated manner, which is in contrast to repolarization in Agboi1/2Δ or Agrho3Δ, where the axis of growth is maintained when spherically enlarged tips are repolarized (Fig. 8).
FIG. 8.
Schematic comparison of polarisome loss in Agrsr1Δ, Agboi1/2Δ, and Agrho3Δ strains. The contours of hyphae are depicted in black, the polarisome is depicted in green, actin cables are depicted as red lines, and actin patches are depicted as red dots. Arrows indicate the growth speed of hyphae. The developmental pattern is given in descending order. The initial pattern with the first three hyphae is comparable between Agrsr1Δ and Agboi1/2Δ/Agrho3Δ. In Agboi1/2Δ/Agrho3Δ the hyphal tip enlarges, which is not observed for Agrsr1Δ. Repolarization in Agboi1/2Δ and Agrho3Δ maintains the axis of growth, whereas in Agrsr1Δ resumption of growth occurred in a frequently uncoordinated manner to the previous axis of polarity.
Deletion of the WASP-like protein AgWAL1 in A. gossypii causes a severe tip extension defect and specific clustering of cortical actin patches in subapical regions of hyphal tips instead of at the hyphal apex (57). Interestingly, in Agrsr1Δ strains 34% of hyphae do not have polarized actin (7). In the Agboi1/2Δ strain, 24% of hyphal tips are spherically enlarged, but they still carry actin patches. Thus, the absence of a spherical enlargement in Agrsr1Δ tips that have lost AgSpa2p might be due to the absence of actin patches. We therefore hypothesize that AgRsr1p mediates tip-based actin cable nucleation via AgBoi1/2p and AgRho3p, polarization of actin patches via AgWal1p, and growth guidance in an unknown way.
Deletion of both BOI1 and BOI2 in S. cerevisiae, but not single deletions alone, leads to impaired morphogenesis and poor viability; cells become round and large or lyse with buds displaying defects in bud formation and in the maintenance of cell polarity (9, 34). Pob1 in S. pombe is essential for cell growth, which is in contrast to AgBOI1/2 in A. gossypii. A temperature-sensitive Pob1 allele expressed in S. pombe leads to quick cessation of cellular elongation when shifted to the restrictive temperature. Cells start swelling in the middle and revealed a cell separation defect (55).
Boi1 and Boi2 in S. cerevisiae are localized to the periphery of buds during much of the budding cycle and to necks late in the cell cycle. Mutations in the PH domain affect localization to the bud, whereas mutations in the SH3 domains prevent a localization to the neck (25). We found that the N-terminal domain of A. gossypii AgBoi1/2p, which includes the SH3 domain, the SAM domain, and the proline-rich region, localizes efficiently to hyphal tips and to sites of septation, whereas the PH domain localizes solely to hyphal tips and is absent at septa, suggesting that localization of AgBoi1/2p is ensured by the N-terminal region and not by the PH domain. S. pombe Pob1 localizes to cell tips during interphase and translocates near the division plane at cytokinesis, where it can be seen either as a ring or as a split disc similar to AgBoi1/2p in A. gossypii (55).
In S. cerevisiae Boi1, the PH domain with its C-terminal downstream region (deleted for aa 5 to 733, which includes the SH3 domain, the SAM domain, and the proline-rich region) confers good growth in a boi1Δ boi2Δ strain (9), and boi1Δ boi2Δ cells carrying BOI2ΔN (deleted for aa 1 to 465, which includes the SH3 domain, the SAM domain, and the proline-rich region) grow as well as the wild type (34). A C-terminal region of Pob1 in S. pombe carrying the PH domain rescued pob1Δ cells, while the N-terminal region bearing the SH3 and the SAM domains did not. However, the rescued cells were not normalized in morphology (55). This is a clear-cut difference compared to A. gossypii, where the C-terminal region alone is unable to restore even partial function.
The proline-rich regions in S. cerevisiae Boi1p and Boi2p interact directly with the second SH3 domain in Bem1p (9, 34). AgBoi1/2pΔC also displays a two-hybrid interaction with AgBem1p, which is an essential protein in A. gossypii (our unpublished results), unlike Bem1 in S. cerevisiae and Scd2/Ral3 in S. pombe, which are not (8, 14, 21).
Our results together with the data from S. cerevisiae and S. pombe suggest a role for the Boi proteins in the organization of the actin cytoskeleton during cellular elongation and septation. A completely different role for S. cerevisiae Boi1 and Boi2 has been demonstrated in the NoCut pathway during cytokinesis. There, Boi1 and Boi2 regulate Ipl1-dependent inhibition of abscission via shuttling between the nucleus and the bud neck in order to prevent cytokinesis, while the division plane is not yet cleared from chromatin (39). Cytokinesis and mitosis are not linked in A. gossypii (23). We could not observe AgBoi1/2p-GFP localizing to nuclei, and the absence of AgBoi1/2p from septal sites in Agcyk1Δ strains does not affect growth. Therefore, AgBoi1/2p in A. gossypii is most likely not involved in the process of abscission during vegetative growth and therefore illustrates an example where the cellular role of homologous genes has dramatically changed. However, we cannot exclude that AgBoi1/2p may play a role in septation during sporulation. Septa are essential for formation of the ascus, and AgBoi1/2p might prevent abscission while mitosis is still incomplete.
Acknowledgments
We are grateful to Daniele Cavicchioli for sharing data on the AgBoi1/2p AgBem1p interaction, to K.-P. Stahmann for providing an A. gossypii plasmid library, to J. H. McCusker for plasmid pAG25, and to H.-P. Schmitz for plasmid pGADAgBNI1-N. We thank Marie-Pierre Gulli, Yves Barral, and Matthias Peter for helpful discussions.
This work was supported by the Swiss National Science Foundation Grant 31-55941.98 to P.P. and J.W.
Footnotes
Published ahead of print on 1 September 2006.
REFERENCES
- 1.Adamo, J. E., G. Rossi, and P. Brennwald. 1999. The Rho GTPase Rho3 has a direct role in exocytosis that is distinct from its role in actin polarity. Mol. Biol. Cell 10:4121-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, A. 1998. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Akada, R., J. Yamamoto, and I. Yamashita. 1997. Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol. Gen. Genet. 254:267-274. [DOI] [PubMed] [Google Scholar]
- 4.Altmann-Johl, R., and P. Philippsen. 1996. AgTHR4, a new selection marker for transformation of the filamentous fungus Ashbya gossypii, maps in a four-gene cluster that is conserved between A. gossypii and Saccharomyces cerevisiae. Mol. Gen. Genet. 250:69-80. [DOI] [PubMed] [Google Scholar]
- 5.Ashby, S. F., and W. Nowell. 1926. The fungi of stigmatomycosis. Ann. Botany 40:69-84. [Google Scholar]
- 6.Ayscough, K. R., J. Stryker, N. Pokala, M. Sanders, P. Crews, and D. G. Drubin. 1997. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137:399-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer, Y., P. Knechtle, J. Wendland, H. Helfer, and P. Philippsen. 2004. A Ras-like GTPase is involved in hyphal growth guidance in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 15:4622-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bender, A., and J. R. Pringle. 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bender, L., H. S. Lo, H. Lee, V. Kokojan, V. Peterson, and A. Bender. 1996. Associations among PH and SH3 domain-containing proteins and Rho-type GTPases in Yeast. J. Cell Biol. 133:879-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman, J., and P. E. Sudbery. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918-930. [DOI] [PubMed] [Google Scholar]
- 11.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117-127. [DOI] [PubMed] [Google Scholar]
- 12.Boyce, K. J., M. J. Hynes, and A. Andrianopoulos. 2005. The Ras and Rho GTPases genetically interact to coordinately regulate cell polarity during development in Penicillium marneffei. Mol. Microbiol. 55:1487-1501. [DOI] [PubMed] [Google Scholar]
- 13.Brachat, S., F. S. Dietrich, S. Voegeli, Z. Zhang, L. Stuart, A. Lerch, K. Gates, T. Gaffney, and P. Philippsen. 2003. Reinvestigation of the Saccharomyces cerevisiae genome annotation by comparison to the genome of a related fungus: Ashbya gossypii. Genome Biol. 4:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, E. C., M. Barr, Y. Wang, V. Jung, H. P. Xu, and M. H. Wigler. 1994. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell 79:131-141. [DOI] [PubMed] [Google Scholar]
- 15.Dietrich, F. S., S. Voegeli, S. Brachat, A. Lerch, K. Gates, S. Steiner, C. Mohr, R. Pohlmann, P. Luedi, S. Choi, R. A. Wing, A. Flavier, T. D. Gaffney, and P. Philippsen. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304-307. [DOI] [PubMed] [Google Scholar]
- 16.Dong, Y., D. Pruyne, and A. Bretscher. 2003. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J. Cell Biol. 161:1081-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drubin, D. G., and W. J. Nelson. 1996. Origins of cell polarity. Cell 84:335-344. [DOI] [PubMed] [Google Scholar]
- 18.Esser, K. 1994. The Mycota, a comprehensive treatise on fungi as experimental systems for basic and applied research. Springer-Verlag, Berlin, Germany.
- 19.Evangelista, M., D. Pruyne, D. C. Amberg, C. Boone, and A. Bretscher. 2002. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4:260-269. [DOI] [PubMed] [Google Scholar]
- 20.Fujiwara, T., K. Tanaka, A. Mino, M. Kikyo, K. Takahashi, K. Shimizu, and Y. Takai. 1998. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell 9:1221-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukui, Y., and M. Yamamoto. 1988. Isolation and characterization of Schizosaccharomyces pombe mutants phenotypically similar to ras1. Mol. Gen. Genet. 215:26-31. [DOI] [PubMed] [Google Scholar]
- 22.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gladfelter, A. S., A. K. Hungerbuehler, and P. Philippsen. 2006. Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 172:347-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 25.Hallett, M. A., H. S. Lo, and A. Bender. 2002. Probing the importance and potential roles of the binding of the PH-domain protein Boi1 to acidic phospholipids. BMC Cell Biol. 3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris, S. D., and M. Momany. 2004. Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 41:391-400. [DOI] [PubMed] [Google Scholar]
- 27.Harris, S. D., N. D. Read, R. W. Roberson, B. Shaw, S. Seiler, M. Plamann, and M. Momany. 2005. Polarisome meets spitzenkorper: microscopy, genetics, and genomics converge. Eukaryot. Cell 4:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermida, L., S. Brachat, S. Voegeli, P. Philippsen, and M. Primig. 2005. The Ashbya Genome Database (AGD)-a tool for the yeast community and genome biologists. Nucleic Acids Res. 33:D348-D352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knechtle, P., F. Dietrich, and P. Philippsen. 2003. Maximal polar growth potential depends on the polarisome component AgSpa2 in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 14:4140-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, Y. J., and B. D. Hall. 2004. Body plan evolution of ascomycetes, as inferred from an RNA polymerase II phylogeny. Proc. Natl. Acad. Sci. USA 101:4507-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsui, Y., R. Matsui, R. Akada, and A. Toh-e. 1996. Yeast src homology region 3 domain-binding proteins involved in bud formation. J. Cell Biol. 133:865-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayer, B. J. 2001. SH3 domains: complexity in moderation. J. Cell Sci. 114:1253-1263. [DOI] [PubMed] [Google Scholar]
- 36.Momany, M. 2002. Polarity in filamentous fungi: establishment, maintenance and new axes. Curr. Opin. Microbiol. 5:580-585. [DOI] [PubMed] [Google Scholar]
- 37.Momany, M., P. J. Westfall, and G. Abramowsky. 1999. Aspergillus nidulans swo mutants show defects in polarity establishment, polarity maintenance and hyphal morphogenesis. Genetics 151:557-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakano, K., J. Imai, R. Arai, E. A. Toh, Y. Matsui, and I. Mabuchi. 2002. The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J. Cell Sci. 115:4629-4639. [DOI] [PubMed] [Google Scholar]
- 39.Norden, C., M. Mendoza, J. Dobbelaere, C. V. Kotwaliwale, S. Biggins, and Y. Barral. 2006. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell 125:85-98. [DOI] [PubMed] [Google Scholar]
- 40.Philippsen, P., A. Kaufmann, and H. P. Schmitz. 2005. Homologues of yeast polarity genes control the development of multinucleated hyphae in Ashbya gossypii. Curr. Opin. Microbiol. 8:370-377. [DOI] [PubMed] [Google Scholar]
- 41.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. J. Cell Sci. 113:571-585. [DOI] [PubMed] [Google Scholar]
- 42.Rebecchi, M. J., and S. Scarlata. 1998. Pleckstrin homology domains: a common fold with diverse functions. Annu. Rev. Biophys. Biomol. Struct. 27:503-528. [DOI] [PubMed] [Google Scholar]
- 43.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 44.Roumanie, O., H. Wu, J. N. Molk, G. Rossi, K. Bloom, and P. Brennwald. 2005. Rho GTPase regulation of exocytosis in yeast is independent of GTP hydrolysis and polarization of the exocyst complex. J. Cell Biol. 170:583-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sagot, I., A. A. Rodal, J. Moseley, B. L. Goode, and D. Pellman. 2002. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4:626-631. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Schmitz, H.-P., A. Kaufmann, M. Köhli, P. P. Laissue, and P. Philippsen. 2006. From function to shape: a novel role of a formin in morphogenesis of the fungus Ashbya gossypii. Mol. Biol. Cell 17:130-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seiler, S., and M. Plamann. 2003. The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell 14:4352-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaw, B. D., and M. Momany. 2002. Aspergillus nidulans polarity mutant swoA is complemented by protein O-mannosyltransferase pmtA. Fungal Genet. Biol. 37:263-270. [DOI] [PubMed] [Google Scholar]
- 50.Sheu, Y. J., B. Santos, N. Fortin, C. Costigan, and M. Snyder. 1998. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18:4053-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stapleton, D., I. Balan, T. Pawson, and F. Sicheri. 1999. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat. Struct. Biol. 6:44-49. [DOI] [PubMed] [Google Scholar]
- 53.Steiner, S., J. Wendland, M. C. Wright, and P. Philippsen. 1995. Homologous recombination as the main mechanism for DNA integration and cause of rearrangements in the filamentous ascomycete Ashbya gossypii. Genetics 140:973-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toya, M., Y. Iino, and M. Yamamoto. 1999. Fission yeast Pob1p, which is homologous to budding yeast Boi proteins and exhibits subcellular localization close to actin patches, is essential for cell elongation and separation. Mol. Biol. Cell 10:2745-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Virag, A., and S. D. Harris. 2006. Functional characterization of Aspergillus nidulans homologues of Saccharomyces cerevisiae Spa2 and Bud6. Eukaryot. Cell 5:881-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walther, A., and J. Wendland. 2004. Apical localization of actin patches and vacuolar dynamics in Ashbya gossypii depend on the WASP homolog Wal1p. J. Cell Sci. 117:4947-4958. [DOI] [PubMed] [Google Scholar]
- 58.Weiner, O. D. 2002. Regulation of cell polarity during eukaryotic chemotaxis: the chemotactic compass. Curr. Opin. Cell Biol. 14:196-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wendland, J., Y. Ayad-Durieux, P. Knechtle, C. Rebischung, and P. Philippsen. 2000. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene 242:381-391. [DOI] [PubMed] [Google Scholar]
- 60.Wendland, J., and P. Philippsen. 2000. Determination of cell polarity in germinated spores and hyphal tips of the filamentous ascomycete Ashbya gossypii requires a rhoGAP homolog. J. Cell Sci. 113:1611-1621. [DOI] [PubMed] [Google Scholar]
- 61.Wendland, J., and P. Philippsen. 2001. Cell polarity and hyphal morphogenesis are controlled by multiple rho-protein modules in the filamentous ascomycete Ashbya gossypii. Genetics 157:601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wendland, J., and P. Philippsen. 2002. An IQGAP-related protein, encoded by AgCYK1, is required for septation in the filamentous fungus Ashbya gossypii. Fungal Genet. Biol. 37:81-88. [DOI] [PubMed] [Google Scholar]
- 63.Wendland, J., and A. Walther. 2005. Ashbya gossypii: a model for fungal developmental biology. Nat. Rev. Microbiol. 3:421-429. [DOI] [PubMed] [Google Scholar]
- 64.Wright, M. C., and P. Philippsen. 1991. Replicative transformation of the filamentous fungus Ashbya gossypii with plasmids containing Saccharomyces cerevisiae ARS elements. Gene 109:99-105. [DOI] [PubMed] [Google Scholar]
- 65.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]