Abstract
Staphylococcus simulans secretes lysostaphin, a bacteriolytic enzyme that specifically binds to the cell wall envelope of Staphylococcus aureus and cleaves the pentaglycine cross bridges of peptidoglycan, thereby killing staphylococci. The study of S. aureus mutants with resistance to lysostaphin-mediated killing has revealed biosynthetic pathways for cell wall assembly. To identify additional genes involved in cell wall envelope biosynthesis, we have screened a collection of S. aureus strain Newman transposon mutants for lysostaphin resistance. Bursa aurealis insertion in SAV2335, encoding a polytopic membrane protein with predicted protease domain, caused a high degree of lysostaphin resistance, similar to the case for a previously described femAB promoter mutant. In contrast to the case for this femAB mutant, transposon insertion in SAV2335, herein named lyrA (lysostaphin resistance A), did not cause gross alterations of cell wall cross bridges such as truncations of pentaglycine to tri- or monoglycine. Also, inactivation of LyrA in a methicillin-resistant S. aureus strain did not precipitate a decrease in β-lactam resistance as observed for fem (factor essential for methicillin resistance) mutants. Lysostaphin bound to the cell wall envelopes of lyrA mutants in a manner similar to that for wild-type staphylococci. Lysostaphin resistance of lyrA mutants is attributable to altered cell wall envelope properties and may in part be due to increased abundance of altered cross bridges. Other lyr mutants with intermediate lysostaphin resistance carried bursa aurealis insertions in genes specifying GTP pyrophosphokinase or enzymes of the purine biosynthetic pathway.
The gram-positive microbe Staphylococcus aureus is a commensal of the human skin and nares (33). Breaches in dermal or mucosal epithelia as well as defects in host immunity precipitate invasive S. aureus infections with a wide spectrum of human disease, ranging from localized or systemic abscesses, septicemia, and endocarditis to septic emboli (1, 33). The cell wall envelope of S. aureus functions as a protective bacterial surface organelle and is comprised of peptidoglycan (murein) with attached polysaccharides (capsular polysaccharide as well as poly-N-glucosamine expolysaccharide), teichoic acids, and proteins that are covalently or noncovalently immobilized within the murein sacculus (39). Many antistaphylococcal therapies are based on molecules that inhibit cell wall biosynthesis or that destroy the physiological functions of the envelope in maintaining bacterial integrity and mediating evasion from host immune responses (6). These therapies include small molecules such as β-lactams and glycopeptides (inhibitors of peptidoglycan synthesis) as well as lysostaphin, a bacteriocin that degrades the bacterial cell wall envelope (6, 8, 28, 43).
Staphylococcal peptidoglycan consists of linear glycan strands with repeating disaccharide units, N-acetylmuramic acid (β1-4)-N-acetylglucosamine (MurNAc-GlcNAc), of variable length (17, 18). Wall peptides, l-Ala-d-iGln-(NH2-Gly5)-l-Lys-d-Ala-d-Ala, are linked via an amide bond between the d-lactyl moiety of MurNAc and the amino group of l-Ala at position one. Staphylococcal wall peptides harbor pentaglycines (NH2-Gly5) at the ɛ-amino group of lysine (l-Lys) at position three (19, 38, 58). Within fully assembled staphylococcal peptidoglycan, about 95% of wall peptides are cross-linked via an amide bond between the carboxyl group of d-Ala at position four and the amino group of a pentaglycine cross bridge attached to a neighboring wall peptide (16, 52, 55). This cross-linking organizes linear peptidoglycan strands into a single, large macromolecule (murein sacculus) with a diameter of 50 to 100 nm, thereby protecting staphylococci from osmotic lysis and providing a scaffold for cell wall envelope assembly (20, 31).
The gram-positive bacterium Staphylococcus simulans secretes lysostaphin, a glycyl-glycine endopeptidase with bacteriolytic activity against staphylococci harboring pentaglycine cross bridges (49). The envelope of S. simulans contains serine/glycine cross bridges that provide immunity to this bacteriocin (10, 47). Lysostaphin is secreted as a proenzyme. Proteolytic cleavage of 13 N-terminal tandem repeats generates mature lysostaphin with two functionally separable domains (25, 46). The N-terminal domain with glycyl-glycine endopeptidase activity cleaves pentaglycine cross bridges (49), whereas the C-terminal cell wall-targeting domain promotes bacteriocin binding to staphylococcal peptidoglycan (2, 22).
Mutations in any one of three fem genes (factors essential for methicillin resistance), fmhB (femX), femA, and femB, confer high-level lysostaphin resistance on mutant staphylococci but decreased resistance to β-lactam compounds such as methicillin and oxacillin (48, 54). Fem factors catalyze nonribosomal synthesis of pentaglycine cross bridges (15, 34, 48, 51). FmhB utilizes charged glycyl-tRNA and lipid II [C55-PP-MurNAc-(l-Ala-d-iGln-l-Lys-d-Ala-d-Ala)-(β1-4)-GlcNAc] as substrates and adds the first glycine to the ɛ-amino of l-Lys (48, 51). In contrast to the case for fmhB, an essential gene, null mutations in femA and femB do not abolish staphylococcal growth (48). Using the product of the FmhB-catalyzed reaction [C55-PP-MurNAc-(l-Ala-d-iGln-(NH2-Gly1)-l-Lys-d-Ala-d-Ala)-(β1-4)-GlcNAc], FemA and FemB also utilize charged glycyl-tRNAs and promote addition of Gly2/Gly3 (FemA) or Gly4/Gly5 (FemB) (15, 26, 34). Mutations in fem genes cause truncations (Gly1 or Gly3) in staphylococcal cross bridges, which precipitates the observed increase in lysostaphin resistance of staphylococci (15, 54).
In this study, we screened S. aureus transposon mutants with defined insertion sites in approximately two-thirds of all S. aureus genes for increased lysostaphin resistance. One of these mutants displayed dramatically increased resistance to lysostaphin without a concomitant decrease in β-lactam resistance. This mutant contained a transposon insertion in the hitherto-uncharacterized gene SAV2335. Several other mutations caused an intermediate lysostaphin resistance phenotype.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All bacterial strains used in this study are listed in Table 1. Escherichia coli strains were grown with aeration at 37°C in Luria-Bertani broth (LB). S. aureus strains Newman (14) and RN4220 (29), methicillin resistant S. aureus strain BB270 (4), and S. simulans strain TNK1 (2) were propagated in tryptic soy broth (TSB) at 37°C with aeration. The transposon mutant collection, Phoenix (ΦΗΞ) library, used in this study was described by Bae et al. (3) and was grown in TSB medium containing 10 μg/ml erythromycin. Other antibiotics were used at the following concentrations: 10 μg/ml chloramphenicol for selection of pHTT4 (59) and 5 or 7.5 μg/ml chloramphenicol for selection of pCL55 (32) and derivatives thereof in S. aureus; 100 μg/ml ampicillin for selection of plasmids pCL55, pGEX2TK (Amersham Biosciences, Uppsala, Sweden), pGST-K-CWT (2), and pMCSG7 (53) in E. coli; and 30 μg/ml kanamycin for pHTT2 (59) selection in E. coli. E. coli strain CA8000 (24) was used for glutathione S-transferase (GST) expression and purification (pGEX2TK). All other proteins were expressed in E. coli strain BL21(DE3) (56). E. coli strains XL1-Blue (Stratagene, La Jolla, CA) and DH5α (23) were used for cloning purposes.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant features | Reference or source |
|---|---|---|
| Staphylococcus aureus strains | ||
| BB270 | Methicillin-resistant S. aureus strain | 4 |
| BB308 | BB270 with transposon insertion in femAB promoter region | 5 |
| ANG365 | BB270 with same transposon insertion as in strain BB308, but transposon was transduced via strain ANG133 | This study |
| ANG366 | BB270 with transposon insertion in SAV2335 (lyrA); transposon derived from Phoenix library strain ΦΗΞ12843 | This study |
| RN4220 | Transformable laboratory strain | 29 |
| ANG244 | RN4220 with transposon insertion in intergenic region; transposon insertion derived from phoenix library strain ΦΗΞ17 | This study |
| ANG245 | RN4220 with transposon insertion in SAV2335 (lyrA); N-terminal transposon derived from Phoenix library strain ΦΗΞ12843 | This study |
| ANG246 | RN4220 with transposon insertion in SAV2335 (lyrA); C-terminal transposon derived from Phoenix library strain ΦΗΞ09166 | This study |
| ANG266 | RN4220 with integrated plasmid pCL55 | This study |
| ANG267 | ANG245 with integrated plasmid pCL55 | This study |
| ANG268 | ANG245 with integrated complementation plasmid pCL-lyrA | This study |
| ANG273 | ANG245 with integrated complementation plasmid pCL-lyrA-E135A | This study |
| ANG274 | ANG245 with integrated complementation plasmid pCL-lyrA-R139A | This study |
| ANG275 | ANG245 with integrated complementation plasmid pCL-lyrA-H210A | This study |
| ANG276 | ANG245 with integrated complementation plasmid pCL-lyrA-R196A | This study |
| OS2(pHTT4) | OS2 with pHTT4 for expression of cell wall anchored seb reporter construct | 59 |
| Newman | Human clinical isolate | 14 |
| ANG142 | Newman with transposon insertion in intergenic region; transposon derived from Phoenix library strain ΦΗΞ17 | 3 |
| ANG133 | Newman with transposon insertion in femAB promoter region; transposon derived from strain BB308 | This study |
| ANG144 | Newman with transposon insertion in SAV2335 (lyrA); N-terminal transposon derived from Phoenix library strain ΦΗΞ12843 | This study |
| ANG249 | Newman with transposon insertion in SAV2335 (lyrA); C-terminal transposon derived from Phoenix library strain ΦΗΞ09166 | This study |
| ΦΗΞ02817 | Newman strain with transposon insertion in SAV2332 | 3 |
| ΦΗΞ11235 | Newman strain with transposon insertion in SAV2332 | 3 |
| ΦΗΞ11894 | Newman strain with transposon insertion in SAV2332 | 3 |
| ΦΗΞ12577 | Newman strain with transposon insertion in SAV2332 | 3 |
| ΦΗΞ06569 | Newman strain with transposon insertion in SAV2333 | 3 |
| ΦΗΞ09451 | Newman strain with transposon insertion in SAV2333 | 3 |
| ΦΗΞ03723 | Newman strain with transposon insertion in SAV2334 | 3 |
| ΦΗΞ09166 | Newman strain with transposon insertion in SAV2335 (C terminal) | 3 |
| ΦΗΞ12843 | Newman strain with transposon insertion in SAV2335 (N terminal) | 3 |
| ΦΗΞ02438 | Newman strain with transposon insertion in SAV2336 | 3 |
| ΦΗΞ12541 | Newman strain with transposon insertion in SAV2236 | 3 |
| ΦΗΞ01722 | Newman strain with transposon insertion in SAV2337 | 3 |
| ΦΗΞ04698 | Newman strain with transposon insertion in SAV2337 | 3 |
| ΦΗΞ05245 | Newman strain with transposon insertion in SAV2337 | 3 |
| ΦΗΞ08112 | Newman strain with transposon insertion in SAV2337 | 3 |
| ΦΗΞ08290 | Newman strain with transposon insertion in SAV2337 | 3 |
| ΦΗΞ00669 | Newman strain with transposon insertion in SAV2338 | 3 |
| ΦΗΞ07753 | Newman strain with transposon insertion in SAV2338 | 3 |
| ΦΗΞ08527 | Newman strain with transposon insertion in SAV2338 | 3 |
| ΦΗΞ08910 | Newman strain with transposon insertion in SAV2338 | 3 |
| ΦΗΞ10459 | Newman strain with transposon insertion in SAV2338 | 3 |
| Phoenix library mutants with intermediate lysostaphin resistance | ||
| ΦΗΞ05368 | Newman strain with transposon insertion in SAV1001 | 3 |
| ΦΗΞ06682 | Newman strain with transposon insertion in SAV1002 | 3 |
| ΦΗΞ02380 | Newman strain with transposon insertion in SAV1064 (purE) | 3 |
| ΦΗΞ05091 | Newman strain with transposon insertion in SAV1065 (purK) | 3 |
| ΦΗΞ02924 | Newman strain with transposon insertion in SAV1066 (purC) | 3 |
| ΦΗΞ12894 | Newman strain with transposon insertion in SAV1067 (purS) | 3 |
| ΦΗΞ05399 | Newman strain with transposon insertion in SAV1068 (purQ) | 3 |
| ΦΗΞ03038 | Newman strain with transposon insertion in SAV1070 (purF) | 3 |
| ΦΗΞ03863 | Newman strain with transposon insertion in SAV1071 (purM) | 3 |
| ΦΗΞ03896 | Newman strain with transposon insertion in SAV1074 (purD) | 3 |
| Escherichia coli strains | ||
| XL1-Blue | Stratagene | |
| DH5α | 23 | |
| BL21(DE3) | 56 | |
| CA8000 | 24 | |
| CA8000(pGEX2TK) | GST expression strain | Amersham Biosciences |
| BL21(DE3)(pGEX-K-CWT) | GST-CWT expression strain | 2 |
| XL1-Blue(pCL55) | S. aureus single-site integration vector | 32 |
| ANG265 | XL1-Blue with pCL-lyrA; lyrA (SAV2335) complementation construct | This study |
| ANG269 | XL1-Blue with pCL-lyrA-E135A; lyrA (SAV2335) complementation construct with E135A substitution | This study |
| ANG270 | XL1-Blue with pCL-lyrA-R139A; lyrA (SAV2335) complementation construct with R139A substitution | This study |
| ANG271 | XL1-Blue with pCL-lyrA-H210A; lyrA (SAV2335) complementation construct with H210A substitution | This study |
| ANG272 | XL1-Blue with pCL-lyrA-R196A; lyrA (SAV2335) complementation construct with R196A substitution | This study |
| XL1-Blue(pMCSG7) | Ligation-independent cloning vector for expression of His-tagged proteins | 53 |
| ANG237 | DH5α with pMCSG7-LSTΔCWT for expression of lysostaphin glycyl-glycine endopeptidase lacking the cell wall-targeting domain | This study |
| ANG238 | BL21(DE3) with pMCSG7-LSTΔCWT for expression of lysostaphin glycyl-glycine endopeptidase lacking the cell wall-targeting domain | This study |
| BL21(DE3)(pHTT2) | Φ11 hydrolase expression stain | 59 |
| Staphylococcus simulans TNK1 | 2 |
Phage transduction and determination of transposon insertion sites.
Phage 85 was used to move transposons from original transposon mutant strains into Newman, RN4220, or BB270 strain backgrounds. Transductants were streaked twice for single colonies on tryptic soy agar (TSA)-10 μg/ml erythromycin or TSA-10 μg/ml erythromycin-40 mM sodium citrate plates. Transposon insertion sites of ΦΗΞ library transductants were confirmed by inverse PCR, which was performed as described by Bae et al. (3). Strain ANG133 (lysostaphin-resistant control strain) was generated by transducing the transposon of strain BB308 (5) located within the femAB promoter region into strain Newman. The transposon insertion site in ANG133 was confirmed by arbitrarily primed PCR as described by O'Toole et al. (42). Primer set ARB1 (GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT) and Tn-Transpos-far (GATCCCGAAGTAACTAAGTATATG) was used for the first round of PCR, while primer set ARB2 (GGCCACGCGTCGACTAGTAC) and Tn-Transpos (TCTATTCCTAAACACTTAAGA) was used for the second round of PCR and primer Tn-Transpos was used for subsequent sequence determination of the transposon-chromosome junction site. Strains ANG366 (lyrA) and ANG365 (femAB; phenotypically identical to BB308) were constructed by moving transposons from strains ANG144 and ANG133 into the methicillin-resistant S. aureus strain BB270 (4). If not otherwise stated, all lyrA (SAV2335) mutant strains were derived from Phoenix library mutant ΦΗΞ12843.
Lysostaphin lysis assay.
Lysostaphin was purchased from AMBI Products LLC (Lawrence, NY) and stored frozen as a 2-mg/ml stock solution in 0.02 M sodium acetate buffer, pH 4.5. S. aureus strains were grown overnight in TSB at 37°C with aeration. The following day, 0.4-ml culture aliquots were diluted with 0.6 ml fresh TSB and cell densities determined (typically the optical density at 600 nm [OD600] was between 1.5 and 2.1). This value was set as 100% and 0 min. Lysostaphin (final concentration, 50 μg/ml lysostaphin in 20 mM Tris-HCl, pH 7.5) was added, OD600 values read at timed intervals, and data plotted as percent OD600 of the initial reading. For quantitative assessments of lysostaphin resistance, overnight cultures of staphylococci were washed twice with 50 mM Tris-HCl, pH 8.0, and suspended at an OD600 of ∼1.6. Following addition of 0.1 ml of a 100-, 50-, 25-, 12.5-, or 6.25-μg/ml lysostaphin stock solution in 50 mM Tris-HCl, pH 8.0 (final concentrations of ∼10 μg/ml to 0.625 μg/ml) to 1 ml of culture, the decline in optical density was recorded. For determination of lysostaphin sensitivity of purified cell wall and peptidoglycan, samples were suspended in 1 ml 50 mM Tris-HCl, pH 8.0, to an OD600 of ∼1.8. Lysis was monitored following the addition of 0.1 ml of 12.5-μg/ml or 50-μg/ml lysostaphin in 50 mM Tris-HCl, pH 8.0 (final concentration of ∼1.25 μg/ml or 5 μg/ml). Isolated cell wall samples of wild-type, femAB mutant, and lyrA mutant Newman strains were prepared and analyzed in triplicate. The slope in the linear area of each lysis curve was determined, the average and standard deviation for triplicate samples were calculated as positive values, and statistical significance was determined using the t test. For lysis assays with purified lysostaphin lacking the cell wall-targeting domain (LSTΔCWT), S. aureus overnight cultures were washed once with 50 mM Tris-HCl, pH 8.0, and suspended at an OD600 of ∼1.9. The decline in OD600 was followed after the addition of 0.1 ml of lysostaphin lacking its cell wall-targeting domain at a concentration of 8 mg/ml (∼800-μg/ml final concentration) to 1 ml of washed S. aureus cells.
Complementation and alanine substitution mutagenesis of lyrA.
For the complementation of bursa aurealis insertion in SAV2335 (lyrA), S. aureus lyrA was cloned with its native promoter into the E. coli/S. aureus single-site integration vector pCL55 (32). The lyrA open reading frame and 300 bp of upstream nucleotide sequence were PCR amplified with primer pair 5-BamHI-319bp-SAV2335 (CGGGATCCATAAATTTGGTTCACCTTGCTTGTACAC) and 3-KpnI-SAV2355 (GGGGTACCGAGGTACTAGCAAGCGCTTTGTTATTA), using strain Newman chromosomal DNA as the template. The resulting PCR product was cut with the restriction enzymes BamHI and KpnI and ligated with plasmid pCL55 that had been cut with the same enzymes, generating pCL-lyrA. The plasmid was electroporated into S. aureus strain ANG245 (RN4220 with transposon insertion in SAV2335 transduced from strain ΦΗΞ12843) and selected on TSA plates containing 5 or 7.5 μg/ml chloramphenicol. As controls, pCL55 without insert was integrated into the chromosomes of RN4220 and ANG245. SAV2335 (lyrA) complementation was measured with lysostaphin lysis assays. The QuikChange method (Stratagene, La Jolla, CA) was used to change amino acids Glu135, Arg139, His210, and Arg196 to alanines, using plasmid pCL-lyrA as the template and primer pairs 5-SAV2335-E135A (CTGATGGCGTTCGTAGTAGCATTCGGATTCCGTTCATAC) and 3-SAV2335-E135A (GTATGAACGGAATCCGAATGCTACTACGAACGCCATCAG) for E135A substitution, 5-SAV2335-R139A (GTAGTAGAATTCGGATTCGCATCATACTTACAAAATATTG) and 3-SAV2335-R139A (CAATATTTTGTAAGTATGATGCGAATCCGAATTCTACTAC) for R139A substitution, 5-SAV2335-H210A (TATATTGCAACGACATTCGCAGCTTCAATGACATTCGGA) and 3-SAV2335-H210A (TCCGAATGTCATTGAAGCTGCGAATGTCGTTGCAATATA) for H210A substitution, and 5-SAV2335-R196A (GATTCTTGGTGAATTAATTGCAGCGACTAAAGGACGTACAA) and 3-SAV2335-R196A (TTGTACGTCCTTTAGTCGCTGCAATTAATTCACCAAGAATC) for R196A substitution. The resulting plasmids pCL-lyrA-E135A, pCL-lyrA-R139A, pCL-lyrA-H210A, and pCL-lyrA-R196A were introduced into strain ANG245. All plasmids were initially cloned in E. coli strain XL1-Blue and DNA sequences of inserts verified by fluorescent automated sequencing.
Determination of MICs.
MICs were determined by the broth microdilution method according to document M7-A7 from CLSI (9). MICs were determined for lysostaphin, vancomycin, and nisin in Mueller-Hinton broth (BD Diagnostic Systems), pH 7.2, containing 25 mg/liter Ca2+ and 10 mg/liter Mg2+; for daptomycin in medium containing 50 mg/liter Ca2+; and for oxacillin in medium supplemented with 2% NaCl. One hundred microliters of medium containing twofold dilutions of different antibiotics was inoculated with ∼5 × 104 bacteria, and OD600 values were determined after 24 h of growth at 37°C by using a 96-well plate reader (Tecan, Austria). MICs were determined in triplicate for strains Newman, ANG133 (Newman femAB), ANG144 (Newman lyrA), BB270 (methicillin-resistant strain), ANG365 (BB270 femAB; BB308) and ANG366 (BB270 lyrA).
Purification of cell wall envelopes and peptidoglycan.
Cell walls and peptidoglycan were purified from strains Newman, ANG133, and ANG144 essentially as described by de Jonge et al. (11). Briefly, S. aureus overnight cultures were diluted 1:200 and grown in 1 liter TSB for 3 h at 37°C. Bacteria were sedimented by centrifugation, extracted by boiling in 4% sodium dodecyl sulfate (SDS), and extensively washed with H2O to remove all SDS. Bacterial suspensions were mixed 1:1 with 0.1-mm glass beads (Biospec Products, Inc., Bartesville, OK) and lysed either using a Bead Beater (for reverse-phase high-pressure liquid chromatography [rpHPLC] analysis of muropeptides) or by vigorous vortexing twice for 15 min. Lysed cells were treated with α-amylase, DNase, RNase, and finally trypsin to digest carbohydrates, nucleic acids, and proteins (11). This material was termed the purified cell wall fraction and was analyzed in lysostaphin lysis assays. The purified cell wall material was treated with 48% hydrofluoric acid and finally with alkaline phosphatase (11), thereby generating the purified peptidoglycan fraction. Purified peptidoglycan was used for lysostaphin lysis assays and, following mutanolysin digestion and reduction, analyzed by rpHPLC (11, 21).
Analysis of muropeptides.
Five milligrams of purified peptidoglycan was digested in a final volume of 1.24 ml in 12.5 mM phosphate buffer, pH 5.9, with 50 μg mutanolysin from Streptomyces globisporus (Sigma, St. Louis, MO) at 37°C for 17 h. Digested peptidoglycan was reduced, and 1 to 3 mg peptidoglycan was separated by rpHPLC (11, 21). A 3-μm-particle-size octyldecyl silane Hypersil 250 × 4.6 C18 column equipped with a 10- by 4-mm guard column made of the same material (Thermos Electron Corporation, Bellefonte, PA) were used for this experiment. Twenty-microliter aliquots of HPLC fractions were subjected to ion spray mass spectrometry at the Proteomics Core Laboratory at the University of Chicago. An LC/MSD Trap XCT (Agilent Technologies, Palo Alto, CA) was used in the nanospray mode to determine the mass-to-charge (m/z) ratios of muropeptides. For separation, buffer A (0.1% formic acid in deionized water) and buffer B (0.1% formic acid in acetonitrile) were used with the following gradient: 8 min of 3% B, within-32-min linear increase from 8% to 60% B, and within-10-min linear increase from 60% B to 90% B. Muropeptides eluted between 20 and 25 min, and the LC/MSD Trap software 3.2 was used to analyze data.
Purification and structure determination of Φ11 hydrolase-released surface protein anchor peptides.
Plasmid pHTT4 (59) expressing Seb-MH6-CWS was electroporated into strain RN4220 or ANG245, and fresh transformants were used for the experiment. E. coli strain BL2(DE3)(pHTT2) (59) expressing histidine-tagged Φ11 hydrolase was used for purification of the recombinant enzyme. Φ11 hydrolase purified from a 2-liter culture was used to digest the cell wall of each strain analyzed. Enzyme purification was performed by Ni-nitrilotriacetic acid (Ni-NTA) chromatography under denaturing conditions as previously described with some modifications (40). Proteins were purified over a 2-ml bed volume column of nickel nitrilotriacetic acid resin preequilibrated with 30 ml denaturing buffer A (6 M guanidine hydrochloride, 0.1 M Na2PO4, 0.01 M Tris-HCl, pH 8.0). Lysates from Φ11 hydrolase-expressing E. coli strains were applied, and columns were washed with 30 ml equilibration buffer, 30 ml buffer Q1 (pH 8.0) (8 M urea, 0.1 M Na2PO4, 0.01 M Tris-HCl), and 30 ml buffer Q1 (pH 6.5) and eluted with 9 ml buffer Q1 (pH 4.5). Eluted proteins were dialyzed against 2 liters of 1 M urea, 150 mM NaCl, and 50 mM Tris-HCl (pH 7.5) at 4°C for 4 h and again overnight against 2 liters of fresh buffer lacking urea. Two-liter S. aureus cultures were used to purify anchor peptides as described previously (59). rpHPLC and matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometric analysis were performed as previously described (36).
Cell wall targeting assays.
GST-CWT, harboring a fusion of the lysostaphin cell wall-targeting domain (CWT) to the C terminus of GST (2), and a GST control were purified by affinity chromatography using glutathione-Sepharose 4B resin (Bioworld, Dublin, OH). Strains BL21(DE3)(pGST-K-CWT) and CA8000(pGEX2TK) were used for GST-CWT and GST overexpression, respectively, and purification was performed as previously described (7, 22). Two-milliliter fractions containing the purified proteins were dialyzed twice against 1 liter of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 30% glycerol buffer, and proteins were subsequently stored at −20°C. Protein concentrations were determined using the bicinchoninic acid protein assay reagent kit from Pierce (Rockford, IL). For binding assays, S. aureus strains Newman and ANG144 (lyrA) were grown overnight in TSB medium at 37°C with aeration. The following day, cultures were diluted 1:100 and grown in 20 to 30 ml fresh TSB for 3 h at 37°C. Mid-log-phase cultures were washed twice with 50 mM Tris-HCl (pH 8.0) and suspended at OD600s of 10 and twofold dilutions thereof. Fifty microliters of bacterial suspensions were mixed with 11.6 μg GST-CWT or GST in a final reaction volume of 100 μl and incubated for 10 min at room temperature. This resulted in a final OD600 of 5 for the most concentrated bacterial suspensions. GST-CWT (or GST) bound to staphylococci was collected by centrifugation for 5 min at 16,000 × g, and supernatant with unbound protein was removed. Bacterial sediments were suspended in 20 μl sample buffer, and 10 μl of boiled sample was analyzed by 10% SDS-polyacrylamide gel electrophoresis and Coomassie brilliant blue staining. The abundances of GST-CWT and GST were quantified using the spot demo function of the FluorChem software (Alpha Innotech Cooperation, San Leandro, CA). To determine background GST-CWT binding, 11.6 μg GST-CWT protein was incubated and centrifuged in a 100-μl reaction volume without staphylococci.
Purification of lysostaphin without cell wall-targeting domain.
His-tagged lysostaphin (residues 1 to 154, encompassing the glycyl-glycine endopeptidase but lacking the CWT) was generated and purified with the ligation-independent expression vector pMCSG7. Chromosomal DNA of S. simulans strain TNK1 and primer pair 5-Lyso-LIC (TACTTCCAATCCAATGCTGCTGCAACACATGAACATTCAG) and 3-LysoDelta CWT-LIC (TTATCCACTTCCAATGTCAACCTGTATTCGGCGTTGGAGT) were used to amplify the lysostaphin endopeptidase domain. The resulting PCR product was treated with T4 polymerase in the presence of dCTP, while the SspI-cut and gel-purified vector pMCSG7 was treated with T4 polymerase in the presence of dGTP for 15 min at 12°C. The plasmid vector and PCR product were gel purified, mixed, and incubated for 5 min at room temperature. Following the addition of EDTA (pH 8.0) to a final concentration of 6.25 mM and 5 min of incubation at room temperature, reaction products were transformed into E. coli strain DH5α. Isolated pMCSG7-LSTΔCWT was transformed into E. coli BL21(DE3). Protein expression and purification via Ni-NTA chromatography were performed as previously described (22). Protein concentrations were determined with the bicinchoninic acid reaction kit from Pierce (Rockford, IL).
RESULTS
Screening for lysostaphin-resistant mutants.
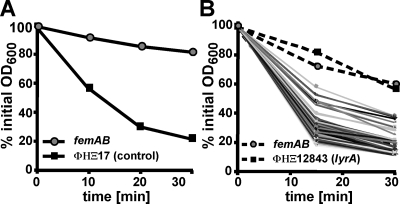
Previous work constructed the Phoenix library (ΦΗΞ), an ordered, nonredundant collection of bursa aurealis transposon insertions in 1,812 open reading frames of S. aureus strain Newman, a human clinical isolate (3). The Phoenix library covers approximately two-thirds of all staphylococcal genes. Here we assayed these mutants for lysostaphin resistance, with the goal of discovering novel factors that contribute to cell wall biosynthesis. Our assay system was based on the decline in optical density of S. aureus cultures upon addition of lysostaphin (30). To calibrate our assay, we used the lysostaphin-sensitive Newman strain and a lysostaphin-resistant femAB mutant Newman strain containing a transposon insertion in the femAB promoter region (5). As control for a lysostaphin-sensitive bursa aurealis variant, we also used ΦΗΞ17, harboring a transposon insertion in an intergenic region that does not affect lysostaphin resistance (Fig. 1A). Typically, batches of 100 ΦΗΞ mutants and control strains were assayed in parallel; a representative graph of this analysis is shown in Fig. 1B. As expected, the vast majority of mutants remained sensitive to lysostaphin, as their optical densities declined in a fashion similar to that of the ΦΗΞ17 control strain. Some ΦΗΞ mutants displayed a slight increase in resistance, whereas very few mutants (ΦΗΞ12843) showed a dramatic increase in lysostaphin resistance similar to that observed for the femAB mutant control strain (Fig. 1B).
FIG. 1.
Screening for S. aureus Newman bursa aurealis insertion mutants with increased resistance to lysostaphin. (A) Calibration of lysostaphin lysis assay with strains that are resistant (ANG133 [femAB]) or sensitive (ΦΗΞ17 [bursa aurealis insertion in an irrelevant intergenic region]) to this bacteriocin. Overnight cultures of S. aureus ANG133 and ΦΗΞ17 were treated with lysostaphin, and lysis was measured as a decline in OD600 over time. (B) Mutants of the Phoenix library (1,812 strains with nonredundant bursa aurealis insertions in open reading frames) were screened with this assay. The graph displays the lysostaphin lysis profiles of 100 ΦΗΞ mutants. ANG133 (femAB) and ΦΗΞ12843 displayed a lysostaphin-resistant phenotype.
Classes of mutants with increased lysostaphin resistance.
Bursa aurealis insertions in genes of the purine biosynthesis cluster (SAV1064, SAV1065, SAV1066, SAV1067, SAV1068, SAV1070, SAV1071, and SAV1074 [Table 1]) generated mutants with intermediate levels of lysostaphin resistance. Further, bursa aurealis insertions in SAV1001 and SAV1002 (Table 1) also resulted in slowed lysostaphin-mediated lysis, similar to that observed for purine biosynthesis mutants. In the S. aureus MU50 genome sequence, SAV1001 and SAV1002 were annotated as two separate genes due to a sequencing error. Recently, this chromosomal region has been resequenced, and only one gene, annotated as encoding GTP pyrophosphokinase, was found to be carried in this region (60). A BLAST search with SA0860, the S. aureus strain N315 homolog of SAV1001/1002, revealed that proteins with significant homology can be found in other gram-positive bacteria, including Staphylococcus epidermidis, several Bacillus spp., and Listeria spp. A COG2761 domain was found in SA0860, which is also found in FrnE, a predicted dithiol-disulfide isomerase involved in polyketide biosynthesis. Finally, ΦΗΞ12843, a mutant carrying a single bursa aurealis insertion in SAV2335, displayed a strong lysostaphin-resistant phenotype and was analyzed further in this study. The affected gene was named lyrA for lysostaphin resistance A.
lyrA.
SAV2335 (lyrA) encodes a 419-amino-acid polypeptide with unknown function. The protein is predicted to assemble as a polytopic membrane protein with eight transmembrane segments within its N-terminal part (TMHMM server v. 2.0) (Fig. 2A). BLAST analysis revealed that proteins with significant homology across the whole length of the protein are present only in other S. aureus strains and in all sequenced staphylococci, i.e., Staphylococcus haemolyticus, Staphylococcus saprophyticus, and Staphylococcus epidermidis. An Abi (abortive infection) domain or COG1226 domain was found within the C-terminal portion of the transmembrane domain (Fig. 2B). This Abi domain is also found in type II CAAX prenyl endopeptidases, which are eukaryotic membrane metalloproteases that truncate the C-terminal ends of farnesylated or geranyl-geranylated proteins (44). Known bacterial proteins containing an Abi domain are encoded by Lactococcus lactis plasmid pCI750 and are located upstream of abiG, a gene involved in abortive bacteriophage infection (41, 57). Lactobacillus plantarum carries a chromosomal gene specifying a polytopic membrane protein involved in bacteriocin production (12). Nevertheless, the biochemical function of bacterial Abi domain-containing proteins is not yet known.
FIG. 2.
Predicted membrane topology and domain structure of LyrA. (A) The membrane topology of LyrA (SAV2335) was predicted with the TMHMM v 2.0 program (http://www.cbs.dtu.dk/services/TMHMM/). The 419-amino-acid protein encompasses eight transmembrane domains; amino acid positions of membrane-spanning regions are indicated at the tops and bottoms of boxes. An Abi domain (shaded) is positioned between residues 121 and 221 (Conserved Domain Database and Search Service, v2.02) (35). Positions of alanine substitutions analyzed for their contribution to lysostaphin resistance (see text) are indicated. (B) Sequence alignment of Abi domain consensus sequence (top sequence, amino acids 1 to 94) and LyrA (bottom sequence, amino acids 121 to 221). Three conserved motifs as identified by Pei and Grishin (44) are overlined. Five semi-invariant residues within these motifs are indicated by black symbols, where the square indicates the predicted catalytic residue. Three of five semi-invariant residues are found in SAV2335 and were replaced with alanines (see text).
Complementation analysis of lyrA.
Although the lyrA gene does not appear to be part of an operon, we interrogated bursa aurealis insertions in flanking nucleic acid sequences for lysostaphin-resistant phenotypes by using two or more independent mutations, with the exception of SAV2334, for which only one mutant was available (Table 1). Mutations in SAV2332, SAV2333, SAV2334, SAV2336, SAV2337, and SAV2338 did not alter lysostaphin resistance of staphylococci. Two independent transposon insertions within SAV2335 (lyrA), ΦΗΞ12843 (mentioned above) containing a transposon insertion 124 nucleotides after the start codon and ΦΗΞ09166 with a transposon insertion 906 nucleotides after the start codon, both displayed a lysostaphin-resistant phenotype (data not shown). We transduced both bursa aurealis insertions in lyrA into the parent Newman strain or RN4220, a laboratory strain widely used for genetic manipulations. Lysostaphin resistance phenotypes were observed for all transductants carrying bursa aurealis insertions in lyrA (data not shown).
Full-length lyrA, including 300 bp of upstream nucleotide sequences with a putative promoter, was cloned into the S. aureus single-site integration vector pCL55 (32). The product, plasmid pCL-lyrA, was electroporated into ANG245 (lyrA), resulting in strain ANG268. This complementation strain was examined together with control strains for lysostaphin resistance. Figure 3 shows that strain ANG267, an lyrA mutant harboring an empty vector insertion, is resistant to lysostaphin, whereas complementation strain ANG268, carrying a wild-type copy of lyrA, is sensitive. Thus, transposon insertions in lyrA confer lysostaphin resistance on staphylococcal variants.
FIG. 3.
S. aureus lyrA mutants are resistant to lysostaphin-mediated killing. Overnight cultures of S. aureus strain ANG266 (wild-type [wt]parent with empty vector pCL55), ANG267 (lyrA mutant strain with empty vector pCL55), complementation strain ANG268 (lyrA mutant strain with a wild-type copy of the lyrA gene inserted into the chromosome), or ANG273 (lyrA mutant strain with lyrA gene carrying the amino acid substitution E135A) were treated with lysostaphin. Lysostaphin-mediated lysis of staphylococci was measured as a decline in OD600 over time.
Alanine substitutions in the ABI domain of lyrA do not affect lysostaphin resistance.
The ABI domains encompasses three motifs with five semiconserved residues, three of which are found in the ABI domain of LyrA, including Glu135 (the proposed active-site residue of CAAX proteases) (44), Arg139, and His210 (Fig. 2B). We changed Glu135, Arg139, and His210 as well as a nonconserved residue outside of these motifs (Arg196) to alanines and assayed LyrA function after chromosomal integration of lyrA variants by using the single-site integration vector pCL55. LyrA variants harboring single alanine substitutions restored the lysostaphin-sensitive phenotype, as shown for strain ANG273, expressing lyrA E135A (Fig. 3 and data not shown). This result indicates that individual conserved residues of the ABI domain of LyrA are not required for lysostaphin resistance.
Lysostaphin resistance of lyrA mutants is a property of the cell wall envelope and distinct from that of fem mutants.
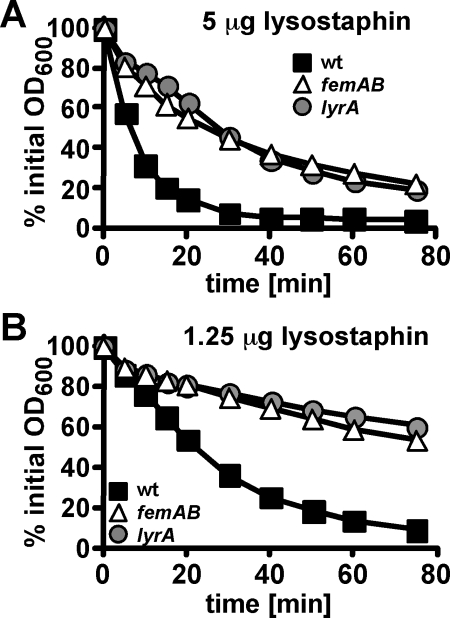
As a quantitative assessment of lysostaphin resistance, we performed bacteriocin sensitivity assays with washed overnight cultures and decreasing amounts of lysostaphin. S. aureus ANG144 (lyrA) and ANG133 (femAB) displayed similar levels of resistance: fourfold higher lysostaphin concentrations (5 μg/ml for mutants compared to 1.25 μg/ml for wild-type Newman) were required to obtain 50% lysis within 20 min (Fig. 4A and B). Consistent with this observation, a fourfold increase in lysostaphin MIC in broth culture was observed for strains ANG144 (lyrA) and ANG133 (femAB) compared to wild-type strain Newman (Table 2).
FIG. 4.
lyrA mutants display lysostaphin resistance similar to that of a femAB mutant control strain. S. aureus strain Newman (wild type [wt]) and transposon mutants ANG133 (femAB) and ANG144 (lyrA) were grown overnight at 37°C in TSB medium. Bacteria were harvested by centrifugation, washed twice with Tris buffer, and suspended to an initial OD600 of ∼1.6. Lysostaphin lysis assays were performed with 5 μg/ml (A) or 12.5 μg/ml (B) bacteriocin.
TABLE 2.
MICs for wild-type, femAB mutant, and lyrA mutant strains in Newman and methicillin-resistant BB270 strain backgrounds
| Strain | Genotype | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|---|
| Lysostaphin | Oxacillin | Vancomycin | Nisin | Daptomycin | ||
| Newman | 0.5 | 0.0625 | 1 | 32 | 0.5 | |
| ANG133 | femAB | 2 | 0.0625 | 1 | 32 | 1 |
| ANG144 | lyrA | 2 | 0.0625 | 2 | 32 | 0.5 |
| BB270 | mec | 0.5 | 128 | 0.5 | 16 | 0.5 |
| ANG365 | mec femAB | 2 | 0.0625 | 0.125 | 4 | 0.125 |
| ANG366 | mec lyrA | 1 | 64 | 1 | 16 | 0.25 |
The lysostaphin resistance of known fem mutants is caused by structural alterations in pentaglycine cross bridges, the substrates for lysostaphin (54). These changes lead to a concomitant reduction in β-lactam resistance in methicillin-resistant S. aureus strains such as BB270 (4, 54). To test whether inactivation of lyrA leads to decreased β-lactam resistance (similar to the case for fem mutants), we constructed strain ANG366 (lyrA mutation in the methicillin-resistant strain BB270). In contrast to the case for the femAB mutant, inactivation of lyrA does not significantly affect β-lactam resistance. This is reflected by oxacillin MICs of 128 and 64 μg/ml for wild-type BB270 and lyrA mutant strains, compared to an oxacillin MIC of 0.0625 μg/ml for strain ANG365 (BB270 femAB) (Table 2). Additional MIC determinations revealed that the lyrA mutants in Newman and BB270 backgrounds are slightly (twofold) more resistant towards the cell wall-active antibiotic vancomycin. Differences in MICs for the membrane-acting antibiotics nisin and dactomycin were minimal. A twofold decrease in daptomycin resistance was observed only in the methicillin-resistant BB270 background for the lyrA mutant (Table 2).
To obtain greater insight into the molecular defect of lyrA mutants, we purified peptidoglycan and assayed isolated material for lysostaphin sensitivity at different steps during the purification process. Purified cell wall, consisting mainly of peptidoglycan and wall teichoic acid (see Materials and Methods), of strain ANG144 (lyrA) and control strains was incubated with 1.25 μg/ml lysostaphin and any decline in optical density analyzed (Fig. 5A). Lysis assays were performed in triplicate and the slope of each lysis curve calculated. The lysis slope for cell walls of wild-type strain Newman was 1.15 ± 0.12, whereas the lysis slopes for both mutants were significantly shallower (0.38 ± 0.09 for femAB [P < 0.01 compared to wild type] and 0.39 ± 0.02 for lyrA [P < 0.01 compared to wild type]). Cell wall material was further treated with hydrofluoric acid and alkaline phosphatase, a protocol that removes cell wall constituents with phosphodiester linkages, for example, teichoic acids. Isolated peptidoglycan was incubated with 5 μg/ml lysostaphin and analyzed as described above. The purified peptidoglycan of the lyrA variant was only slightly more lysostaphin resistant than that of the wild-type parent (Fig. 5B). The calculated lysis slopes for the wild type and the lyrA mutant had similar values of 0.52 ± 0.09 and 0.41 ± 0.08, respectively, while the lysostaphin-resistant femAB mutant gave a significantly smaller value of 0.09 ± 0.04 (P < 0.01 compared to wild type and lyrA mutant). Finally, purified peptidoglycan was digested with mutanolysin, a muramidase that cuts the repeating disaccharide MurNAc-GlcNAc, and the resulting muropeptides were separated by rpHPLC. Mutanolysin-digested peptidoglycan from a wild-type strain generated the expected muropeptide profile (Fig. 6A). For example, ion spray mass spectrometry confirmed m/z 1254 (M + H) for peak 5 (observed in the wild-type and the lyrA and femAB mutants), representing disaccharide-pentapeptide-pentaglycine muropeptide [MurNAc-(l-Ala-d-iGln-(NH2-Gly5)-l-Lys-d-Ala-d-Ala)-(β1-4)-GlcNAc] (peaks are numbered as reported by de Jonge et al. [11]). In agreement with the observations documented in Fig. 5B, only minor differences in the muropeptide profile were obtained with peptidoglycan isolated from lyrA mutant staphylococci (Fig. 6B). Among these differences in composition is the early-eluting additional peak X (observed only in lyrA mutant peptidoglycan) and an overall decrease in peptidoglycan cross-linking (judged by the decrease in UV-absorbent material during later portions of the chromatogram [fraction 110 and later]). In contrast, clear differences were observed in the muropeptide profile of the femAB mutant strain. For instance, a dramatic increase in the abundance of peak 4 in comparison to peak 5 was seen (Fig. 6C). We measured m/z 1026 (M + H) for peak 4, consistent with the mass of disaccharide-pentapeptide harboring a single glycine cross bridge [MurNAc-(l-Ala-d-iGln-(NH2-Gly1)-l-Lys-d-Ala-d-Ala)-(β1-4)-GlcNAc] (11).
FIG. 5.
Lysostaphin resistance of lyrA mutants is a cell wall envelope property. (A) Equal amounts of purified cell wall envelope of S. aureus Newman (wild type [wt]) and femAB or lyrA mutant strains were incubated with 1.25 μg/ml lysostaphin, and the decline in OD600 was measured over time. Positive values for lysis slopes in the linear range are 1.15 ± 0.12 for the wild type, 0.38 ± 0.09 for the femAB mutant (P < 0.01 compared to wild type), and 0.39 ± 0.02 for the lyrA mutant (P < 0.01 compared to wild type). (B) A similar experimental approach as in panel A, using purified peptidoglycan (without polysaccharide, protein, and teichoic acids) and 5 μg/ml lysostaphin. Lysis slopes are 0.52 ± 0.09 for the wild type, 0.41 ± 0.08 for the lyrA mutant, and 0.09 ± 0.04 for the femAB mutant (P < 0.01 compared to wild type and lyrA mutant). Experiments were performed in triplicate.
FIG. 6.
Peptidoglycan analysis of lyrA mutant staphylococci. Purified peptidoglycan from S. aureus strain Newman (wild type [wt]) (A) or isogenic lyrA (B) and femAB mutant (C) staphylococci was digested with mutanolysin, and muropeptides were separated by rpHPLC. The peaks in chromatogram in panel A are labeled as reported by de Jonge et al. (11). Ion signals of m/z 1254 for compounds in peak number 5 (A to C) and m/z 1026 for peak number 4 (C, femAB mutant) were measured by ion spray mass spectrometry. These measurements are consistent with compounds of the structure MurNAc-(l-Ala-d-iGln-(NH2-Gly5)-l-Lys-d-Ala-d-Ala)-(β1-4)-GlcNAc (m/z 1254) and MurNAc-(l-Ala-d-iGln-(NH2-Gly1)-l-Lys-d-Ala-d-Ala)-(β1-4)-GlcNAc (m/z 1026).
Cell wall cross bridges of lyrA mutant peptidoglycan.
To generate a high-resolution analysis of peptidoglycan cross bridges, we exploited an experimental scheme used previously by this laboratory to determine the cell wall anchor structures of surface proteins (59). Sortase A cleaves the cell wall sorting signals of surface proteins between the threonine and glycine residues of the LPXTG motif (50). The C-terminal carboxyl of threonine is subsequently amide linked to the pentaglycine cross bridges of lipid II molecules (45) and incorporated into the cell wall. SEB-MH6-CWS, an engineered surface protein substrate, is linked to staphylococcal cell wall cross bridges by sortase A (59) (Fig. 7A). Following surface protein solubilization (Φ11 hydrolase cleavage of N-acetylmuramoyl-l-Ala and d-Ala-Gly bonds within the murein sacculus [Fig. 7B]), surface proteins can be purified by affinity chromatography on Ni-NTA. CNBr cleaves at methionyl residues and cuts the C-terminal peptide of SEB-MH6-CWT to generate H6-AQALPET peptides with linked cross bridges. These compounds are purified by an additional affinity chromatography step on Ni-NTA and eluted compounds analyzed by mass spectrometry.
FIG. 7.
Cell wall cross bridges of lyrA mutant peptidoglycan. (A) Schematic representation of the engineered sortase A substrate Seb-MH6-CWS. Enterotoxin B (seb) is fused to the protein A cell wall-anchoring signal (sorting signal) with a methionyl-six-histidyl tag inserted in the fusion site. N-terminal signal sequence, cyanogen bromide (CNBr), and sortase A cleavage sites are indicated. (B) Illustration of staphylococcal peptidoglycan structure with sortase-anchored Seb-MH6-CWS and cleavage sites for Φ11 hydrolase. GN and MN, N-acetylglucosamine and N-acetylmuramic acid, respectively. (C and D) MALDI-MS spectra of rpHPLC-purified anchor peptides from wild-type (wt) (C) and lyrA mutant (D) staphylococci. Surface protein anchor peptides with m/z 2336 are consistent with a peptide of the structure NH2-Ala-γ-Gln-Lys (NH2-H6AQALPET-Gly5)-Ala-COOH. See Table 3 for a complete listing of observed and calculated masses of anchor peptides in wild-type and lyrA mutant strains.
To analyze peptidoglycan cross bridges, plasmid pHTT4, encoding Seb-MH6-CWS, was electroporated into S. aureus RN4220 and ANG245 (lyrA). Cell walls of these S. aureus strains were solubilized with Φ11 hydrolase, Seb-MH6-CWS was purified and cleaved with CNBr, and C-terminal anchor peptides were purified by another affinity chromatography step and then separated by rpHPLC (59). rpHPLC-purified anchor peptides were subjected to MALDI-TOF mass spectrometric analysis. The predominant anchor peptide of surface protein in the parent strain RN4220 generated an m/z 2236.13 compound. This compound has been previously reported for Φ11 hydrolase-released anchor peptides in S. aureus strain OS2 (59), and its mass is in agreement with the predicted mass of 2236.39 for the tetrapeptide-anchor peptide structure NH2-Ala-γ-Gln-Lys-(NH2-H6AQALPET-Gly5)-Ala-COOH (Fig. 7C and Table 3). In addition to m/z 2236.13, a weaker signal, m/z 2307.16, was also detected. This compound represents pentapeptide tethered to surface protein [NH2-Ala-γ-Gln-Lys-(NH2-H6AQALPET-Gly5)-Ala-Ala-COOH] (calculated mass of 2307.47). Additional signals at m/z 2264.21, 2279.1, 2235.17, and 2350.17 represent formylated and carbamylated tetra- and pentapeptide anchor structures, respectively (see Table 3 for a listing of observed and calculated masses). The same cell wall compounds were identified when we analyzed surface protein anchor structures of the lyrA mutant strain (Fig. 7D). However, in addition to major anchor peptides, several smaller compounds with aberrant structure were detected. Some of these compounds display m/z values consistent with only mono- and diglycyl cross bridges (Table 3). Thus, the vast majority of cell wall cross bridges in the parent strain RN4220 represent pentaglycine compounds (we could not detect aberrant cross bridges), whereas the peptidoglycan of lyrA mutants harbors small amounts of truncated cross bridges or cross bridges with aberrant structure.
TABLE 3.
Surface protein anchor peptides and their structures in S. aureus RN4220 and its isogenic lyrA mutant
| S. aureus strain | Anchor peptide properties
|
||
|---|---|---|---|
|
m/z
|
Predicted structure | ||
| Observed | Calculated | ||
| RN4220 | 2236.13 | 2236.39 | NH2-Ala-Gln-Lys-(NH2-H6AQALPET-Gly5)-Ala-COOH |
| 2264.12 | 2264.40 | 2236.39 + formyl | |
| 2279.14 | 2279.42 | 2236.39 + carbamyl | |
| 2307.16 | 2307.47 | NH2-Ala-Gln-Lys-(NH2-H6AQALPET-Gly5)-Ala-Ala-COOH | |
| 2335.17 | 2335.48 | 2307.47 + formyl | |
| 2350.18 | 2350.50 | 2307.47 + carbamyl | |
| ANG245 (lyrA) | 2007.00 | 2008.18 | NH2-Ala-Gln-Lys-(NH2-H6AQALPET-Gly1)-Ala-COOH |
| 2035.98 | 2036.19 | 2008.18 + formyl | |
| 2064.00 | 2065.24 | NH2-Ala-Gln-Lys-(NH2-H6AQALPET-Gly2)-Ala-COOH | |
| 2093.00 | 2093.25 | 2065.24 + formyl | |
| 2078.98 | 2079.26 | NH2-Ala-Gln-Lys-(NH2-H6AQALPET-Gly1)-Ala-Ala-COOH | |
| 2107.00 | 2107.27 | 2079.26 + formyl | |
| 2122.03 | 2122.30 | NH2-Ala-Gln-Lys-(NH2-H6AQALPET-Gly3)-Ala-COOH | |
| 2151.05 | 2150.31 | 2122.30 + formyl | |
| 2136.04 | 2136.32 | NH2-Ala-Gln-Lys-(NH2-H6AQALPET-Gly2)-Ala-Ala-COOH | |
| 2163.03 | 2164.33 | 2136.33 + formyl | |
| 2178.04 | 2179.36 | NH2-Ala-Gln-Lys-(NH2-H6AQALPET-Gly4)-Ala-COOH | |
| 2206.05 | 2207.37 | 2179.36 + formyl | |
| 2236.08 | 2236.39 | NH2-Ala-Gln-Lys-(NH2-H6AQALPET-Gly5)-Ala-COOH | |
| 2264.07 | 2264.40 | 2236.39 + formyl | |
| 2279.09 | 2279.42 | 2236.39 + carbamyl | |
| 2307.11 | 2307.47 | NH2-Ala-Gln-Lys-(NH2-H6AQALPET-Gly5)-Ala-Ala-COOH | |
| 2335.12 | 2335.48 | 2307.47 + formyl | |
| 2350.12 | 2350.50 | 2307.47 + carbamyl | |
lyrA mutations do not affect lysostaphin binding to the cell wall envelope.
Removal of the cell wall-targeting domain of lysostaphin greatly diminishes the ability of the mutant bacteriocin to kill staphylococci without interfering with its glycyl-glycine endopeptidase activity (2). To test whether lyrA mutations affect lysostaphin binding and thereby confer bacteriocin resistance, we used GST-CWT (a fusion between GST and the lysostaphin cell wall-targeting domain) or GST alone (2). Wild-type and lyrA mutant staphylococci were adjusted to similar optical densities, and serial dilutions of staphylococci were mixed with reporter protein. Following a brief incubation step, staphylococci and bound protein were sedimented by centrifugation and unbound protein removed with the supernatant. Bound protein was solubilized by boiling in SDS and visualized on Coomassie stained SDS-polyacrylamide gels. No obvious difference in binding of GST-CWT between wild-type and lyrA mutant bacteria was observed (Fig. 8A and B). As a control, no significant binding of GST alone to staphylococcal cells was observed (data not shown).
FIG. 8.
Bacteriocin binding to the cell wall envelopes of wild-type and lyrA mutant staphylococci. (A) Staphylococci from S. aureus strains Newman (wild type [wt]) and its isogenic lyrA mutant were suspended at an OD600 of 10. Twofold bacterial dilutions were mixed with 11.6 μg GST-CWT, resulting in final OD600 readings of between 5 and 0.31. Reaction mixtures were incubated for 10 min at room temperature and subsequently centrifuged. GST-CWT bound to staphylococci solubilized by boiling in sample buffer and were analyzed on 10% Coomassie blue-stained SDS-polyacrylamide gels. Sizes of protein standards (lane M) are indicated in kilodaltons. (B) Graphic representation of GST-CWT binding to staphylococci. The abundance of GST-CWT bound to the cell wall of staphylococci (A) was quantified and averaged, and standard deviations for 11 independent experiments were recorded. Readings on the y axis represent arbitrary units. GST-CWT sedimentation in samples without staphylococci was used to determine nonspecific binding and x-axis values. (C) Staphylococci from S. aureus strains Newman (wild type) and isogenic femAB and lyrA mutants were incubated with LSTΔCWT, an engineered lysostaphin enzyme with glycyl-glycine endopeptidase activity but lacking the C-terminal cell wall-targeting domain. Staphylococcal lysis was measured as a decline in OD600 over time.
To specifically address the requirement of the cell wall-targeting domain for the observed resistance phenotype, we cloned and purified lysostaphin containing only the glycyl-glycine endopeptidase domain but lacking the cell wall-targeting domain (LSTΔCWT). Wild-type S. aureus Newman and the lyrA femAB mutants were incubated with LSTΔCWT. Eight independently grown cultures of each strain were analyzed, and representative lysis curves are shown in Fig. 8C. The lyrA mutant strain retained its resistance properties when incubated with LSTΔCWT, indicating that the resistance phenotype is independent of the cell wall binding domain.
DISCUSSION
In this study, a search for S. aureus mutants with increased lysostaphin resistance identified genes of the purine biosynthesis pathway as being required for high-level sensitivity to this bacteriocin. We have not yet studied the involvement of purine biosynthesis genes in detail, as our initial goal was to focus on genes that promote high-level resistance. It seems noteworthy, however, that comparative transcriptional analysis of vancomycin-resistant and -sensitive S. aureus strains showed that the majority of purine biosynthesis genes are upregulated in the resistant strain (37). Bursa aurealis insertion within SAV1001/SAV1002, encoding a hitherto-unknown GTP-pyrophosphokinase (distinct from that encoded by relA), also caused increased resistance. We recently observed that the display of sortase-anchored surface proteins is drastically reduced in this mutant (A. C. DeDent, and O. Schneewind, unpublished results). Finally, bursa aurealis insertion in the previously uncharacterized gene lyrA (SAV2335) caused high-level lysostaphin resistance, similar to transposon insertion in the femAB promoter region. In contrast to mutations in femAB, which cause gross alterations of pentaglycine cross bridges, lyrA mutations caused seemingly minor defects in peptidoglycan and pentaglycine cross bridge structure (Fig. 6 and 7). Among these changes is a slightly altered muropeptide profile detected for mutanolysin-digested peptidoglycan, with the appearance of an additional peak X and slightly decreased peptidoglycan cross-linking (Fig. 7). Structural analysis of surface protein anchor peptides identified peptidoglycan cross bridges with a reduced number of glycine residues. Although these changes are certainly consistent with lysostaphin resistance, it is at this time not possible to attribute the bacteriocin phenotype solely to these structural changes as can be accomplished with control data collected for the femAB mutant strain. It is certainly conceivable that lyrA mutations trigger additional changes in the staphylococcal cell wall envelope that simply escaped our phenotypic analysis.
Addition of bacteriocin to purified peptidoglycan (stripped of its carbohydrate, teichoic acid, and protein constituents) revealed that femAB, but not lyrA mutant, peptidoglycan retained the lysostaphin resistance properties measured for intact staphylococci. The simplest explanation for these data is that only some, but certainly not all, of the lysostaphin resistance properties can be attributed to structural changes in peptidoglycan. What are those presumed alterations in cell wall envelope properties (nonpeptidoglycan structural changes) that can contribute to lysostaphin resistance? Secondary wall polymers or modifications thereof come to mind. Previous work failed to identify a correlation between lysostaphin sensitivity and the abundance of capsular material produced by staphylococci (22, 27). Our preliminary work suggests that teichoic acids are produced by lyrA mutants at physiological levels. Inactivation of tagO, specifying a factor essential for wall teichoic biosynthesis, did not abrogate the lysostaphin resistance phenotype of lyrA mutants (unpublished results).
Bioinformatic analysis revealed that LyrA contains eight hydrophobic domains within its N-terminal part, strongly suggesting that this protein is a multispanning membrane protein. Proteins with high homology to LyrA are found only in other staphylococcal species. An Abi domain was found within the hydrophobic part of this protein. It has been suggested that Abi-containing proteins are metallo-dependent membrane proteases required for protein processing. This domain is found in the eukaryotic type II CaaX proteases, and residues important for proteolytic activity have been identified in a mutagenesis study of the yeast CaaX protease Rce1 (13). Even though proteins which show a high degree of homology to LyrA are found only in staphylococcal strains, proteins annotated to contain Abi domains are found in many bacterial genomes. Indeed, LyrA and four additional proteins containing an Abi domain are encoded within the Mu50 genome (SAV1030, SAV1780, SAV2031, and SAV2313). No function has been assigned to any of these proteins or, by extension, to any bacterial protein annotated to harbor an Abi domain. Interestingly, several Abi domain-encoding genes are found within a chromosomal locus responsible for bacteriocin production in Lactobacillus plantarum C11 (12), and the S. aureus gene encoding the Abi domain-containing protein SAV1030 is in close proximity to two genes which show homology to the bacteriocin lactococcin 972 and its immunity factor. A previous bioinformatic analysis of Abi-containing proteins identified thee conserved motifs with five semi-invariant residues (Fig. 2B) (44). Three of these residues are also present in LyrA. However, when we changed these residues to alanines, the protein was still functional as judged by complementation of the lysostaphin resistance phenotype (Fig. 3). Assuming that the previous bioinformatic analysis correctly predicted the active-site glutamic acid residue, protease activity in LyrA does not seem to be required for lysostaphin resistance.
The mature form of lysostaphin encompasses two domains, the glycyl-glycine endopeptidase domain and the C-terminal cell wall-targeting domain that is required for the binding of the molecule to the envelope of S. aureus (2). GST-CWT reporter protein was used to compare binding affinities of the cell wall-targeting domain to the envelopes of wild-type and lyrA mutant bacteria. Using this assay, we were not able to detect a statistically significant difference in binding (Fig. 8A and B). Furthermore, deletion of the cell wall-targeting domain from recombinant lysostaphin enzymes did not affect the resistance phenotype of lyrA mutants (Fig. 8C). Thus, unlike the case for a mutant with complete deletion of femAB, which displays both CWT binding and substrate property defects for lysostaphin (22), lyrA-mediated resistance points again to nonpeptidoglycan attributes of mutant staphylococci. Future work on the envelope structure of staphylococci is needed to explore the biosynthetic pathways of peptidoglycan and its secondary polymers and with it explain the phenotypes of lyr mutants reported here.
Acknowledgments
We thank members of our laboratory for helpful discussions.
This work was supported in part by a grant from Biosynexus Inc. The study of lysostaphin provides important insight into assembly of surface proteins into the staphylococcal cell wall, work which is supported by U.S. Public Health Service grants AI38897 and AI52474 from the National Institute of Allergy and Infectious Diseases, Division of Microbiology and Infectious Diseases, to O.S.
REFERENCES
- 1.Archer, G. L., and M. W. Climo. 2001. Staphylococcus aureus bacteremia—consider the source. N. Engl. J. Med. 344:55-56. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., and O. Schneewind. 1996. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 15:4789-4797. [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, T., A. K. Banger, A. Wallace, E. M. Glass, F. Aslund, O. Schneewind, and D. M. Missiakas. 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. USA 101:12312-12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger-Bächi, B. 1983. Increase in transduction efficiency of Tn551 mediated by the methicillin resistance marker. J. Bacteriol. 154:533-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger-Bächi, B., L. Barberis-Maino, A. Strassle, and F. H. Kayser. 1989. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol. Gen. Genet. 219:263-269. [DOI] [PubMed] [Google Scholar]
- 6.Bradley, J. S. 2005. Newer antistaphylococcal agents. Curr. Opin. Pediatr. 17:71-77. [DOI] [PubMed] [Google Scholar]
- 7.Cambronne, E. D., J. A. Sorg, and O. Schneewind. 2004. Binding of SycH chaperone to YscM1 and YscM2 activates effector yop expression in Yersinia enterocolitica. J. Bacteriol. 186:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Climo, M. W., R. L. Patron, B. P. Goldstein, and G. L. Archer. 1998. Lysostaphin treatment of experimental methicillin-resistant Staphylococcus aureus aortic valve endocarditis. Antimicrob. Agents Chemother. 42:1355-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CLSI. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 7th ed. M7-A7. CLSI, Wayne, Pa.
- 10.DeHart, H. P., H. E. Heath, L. S. Heath, P. A. LeBlanc, and G. L. Sloan. 1995. The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl. Environ. Microbiol. 61:1475-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Jonge, B. L., Y. S. Chang, D. Gage, and A. Tomasz. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 267:11248-11254. [PubMed] [Google Scholar]
- 12.Diep, D. B., L. S. Havarstein, and I. F. Nes. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178:4472-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolence, J. M., L. E. Steward, E. K. Dolence, D. H. Wong, and C. D. Poulter. 2000. Studies with recombinant Saccharomyces cerevisiae CaaX prenyl protease Rce1p. Biochemistry 39:4096-4104. [DOI] [PubMed] [Google Scholar]
- 14.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 15.Ehlert, K., W. Schroder, and H. Labischinski. 1997. Specificities of FemA and FemB for different glycine residues: FemB cannot substitute for FemA in staphylococcal peptidoglycan pentaglycine side chain formation. J. Bacteriol. 179:7573-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gally, D., and A. R. Archibald. 1993. Cell wall assembly in Staphylococcus aureus: proposed absence of secondary crosslinking reactions. J. Gen. Microbiol. 139:1907-1913. [DOI] [PubMed] [Google Scholar]
- 17.Ghuysen, J. M., and J. L. Strominger. 1963. Structure of the cell wall of Staphylococcus aureus, strain Copenhagen. I. Preparation of fragments by enzymatic hydrolysis. Biochemistry 338:1110-1119. [DOI] [PubMed] [Google Scholar]
- 18.Ghuysen, J. M., and J. L. Strominger. 1963. Structure of the cell wall of Staphylococcus aureus, strain Copenhagen. II. Separation and structure of disaccharides. Biochemistry 338:1119-1125. [DOI] [PubMed] [Google Scholar]
- 19.Ghuysen, J. M., D. J. Tipper, and J. L. Strominger. 1965. Structure of the cell wall of Staphylococcus aureus, strain Copenhagen. IV. The teichoic acid-glycopeptide complex. Biochemistry 10:474-485. [DOI] [PubMed] [Google Scholar]
- 20.Giesbrecht, P., T. Kersten, H. Maidhof, and J. Wecke. 1998. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 62:1371-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glauner, B., J.-V. Höltje, and U. Schwarz. 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 263:10088-10095. [PubMed] [Google Scholar]
- 22.Gründling, A., and O. Schneewind. 2006. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J. Bacteriol. 188:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 24.Hayes, W. 1953. The mechanism of genetic recombination in Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 18:75-93. [DOI] [PubMed] [Google Scholar]
- 25.Heinrich, P., R. Rosenstein, M. Bohmer, P. Sonner, and F. Götz. 1987. The molecular organization of the lysostaphin gene and its sequences repeated in tandem. Mol. Gen. Genet. 209:563-569. [DOI] [PubMed] [Google Scholar]
- 26.Henze, U., T. Sidow, J. Wecke, H. Labischinski, and B. Berger-Bächi. 1993. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J. Bacteriol. 175:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King, B. F., M. L. Biel, and B. J. Wilkinson. 1980. Facile penetration of the Staphylococcus aureus capsule by lysostaphin. Infect. Immun. 29:892-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kokai-Kun, J. F., S. M. Walsh, T. Chanturiya, and J. J. Mond. 2003. Lysostaphin cream eradicates Staphylococcus aureus nasal colonization in a cotton rat model. Antimicrob. Agents Chemother. 47:1589-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 30.Kusuma, C. M., and J. F. Kokai-Kun. 2005. Comparison of four methods for determining lysostaphin susceptibility of various strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3256-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labischinski, H., and H. Maidhof. 1994. Bacterial peptidoglycan: an overview and evolving concepts, p. 23-38. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall, vol. 27. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 32.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101-105. [DOI] [PubMed] [Google Scholar]
- 33.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 34.Maidhof, H., B. Reinicke, P. Blumel, B. Berger-Bächi, and H. Labischinski. 1991. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Bacteriol. 173:3507-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marraffini, L. A., and O. Schneewind. 2005. Anchor structure of staphylococcal surface proteins. V. Anchor structure of the sortase B substrate IsdC. J. Biol. Chem. 280:16263-16271. [DOI] [PubMed] [Google Scholar]
- 37.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz, E., J. M. Ghuysen, and H. Heymann. 1967. Cell walls of Streptococcus pyogenes, type 14. C polysaccharide-peptidoglycan and G polysaccharide-peptidoglycan complexes. Biochemistry 6:3659-3670. [DOI] [PubMed] [Google Scholar]
- 39.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarre, W. W., H. Ton-That, K. F. Faull, and O. Schneewind. 1999. Multiple enzymatic activities of the murein hydrolase from staphylococcal phage phi11. Identification of a d-alanyl-glycine endopeptidase activity. J. Biol. Chem. 274:15847-15856. [DOI] [PubMed] [Google Scholar]
- 41.O'Connor, L., A. Coffey, C. Daly, and G. F. Fitzgerald. 1996. AbiG, a genotypically novel abortive infection mechanism encoded by plasmid pCI750 of Lactococcus lactis subsp. cremoris UC653. Appl. Environ. Microbiol. 62:3075-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 43.Patron, R. L., M. W. Climo, B. P. Goldstein, and G. L. Archer. 1999. Lysostaphin treatment of experimental aortic valve endocarditis caused by a Staphylococcus aureus isolate with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 43:1754-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pei, J., and N. V. Grishin. 2001. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem. Sci. 26:275-277. [DOI] [PubMed] [Google Scholar]
- 45.Perry, A. M., H. Ton-That, S. K. Mazmanian, and O. Schneewind. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J. Biol. Chem. 277:16241-16248. [DOI] [PubMed] [Google Scholar]
- 46.Recsei, P. A., A. D. Gruss, and R. P. Novick. 1987. Cloning, sequence, and expression of the lysostaphin gene from Staphylococcus simulans. Proc. Natl. Acad. Sci. USA 84:1127-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson, J. M., J. K. Hardman, and G. L. Sloan. 1979. Relationship between lysostaphin endopeptidase production and cell wall composition in Staphylococcus staphylolyticus. J. Bacteriol. 137:1158-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohrer, S., K. Ehlert, M. Tschierske, H. Labischinski, and B. Berger-Bächi. 1999. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc. Natl. Acad. Sci. USA 96:9351-9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schindler, C. A., and V. T. Schuhardt. 1964. Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc. Natl. Acad. Sci. USA 51:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneewind, O., A. Fowler, and K. F. Faull. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103-106. [DOI] [PubMed] [Google Scholar]
- 51.Schneider, T., M. M. Senn, B. Berger-Bächi, A. Tossi, H. G. Sahl, and I. Wiedemann. 2004. In vitro assembly of a complete, pentaglycine interpeptide bridge containing cell wall precursor (lipid II-Gly5) of Staphylococcus aureus. Mol. Microbiol. 53:675-685. [DOI] [PubMed] [Google Scholar]
- 52.Snowden, M. A., H. R. Perkins, A. W. Wyke, M. V. Hayes, and J. B. Ward. 1989. Cross-linking and O-acetylation of newly synthesized peptidoglycan in Staphylococcus aureus H. J. Gen. Microbiol. 135:3015-3022. [DOI] [PubMed] [Google Scholar]
- 53.Stols, L., M. Gu, L. Dieckman, R. Raffen, F. R. Collart, and M. I. Donnelly. 2002. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25:8-15. [DOI] [PubMed] [Google Scholar]
- 54.Stranden, A. M., K. Ehlert, H. Labischinski, and B. Berger-Bächi. 1997. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strominger, J. L., K. Izaki, M. Matsuhashi, and D. J. Tipper. 1967. Peptidoglycan transpeptidase and d-alanine carboxypeptidase: penicillin-sensitive enzymatic reactions. Fed. Proc. 26:9-22. [PubMed] [Google Scholar]
- 56.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 57.Tangney, M., and G. F. Fitzgerald. 2002. Effectiveness of the lactococcal abortive infection systems AbiA, AbiE, AbiF and AbiG against P335 type phages. FEMS Microbiol. Lett. 210:67-72. [DOI] [PubMed] [Google Scholar]
- 58.Tipper, D. J., J. L. Strominger, and J. C. Ensign. 1967. Structure of the cell wall of Staphylococcus aureus, strain Copenhagen. VII. Mode of action of the bacteriolytic peptidase from Myxobacter and the isolation of intact cell wall polysaccharides. Biochemistry 6:906-920. [DOI] [PubMed] [Google Scholar]
- 59.Ton-That, H., K. F. Faull, and O. Schneewind. 1997. Anchor structure of staphylococcal surface proteins. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J. Biol. Chem. 272:22285-22292. [DOI] [PubMed] [Google Scholar]
- 60.Wootton, M., M. B. Avison, P. M. Bennett, R. A. Howe, A. P. MacGowan, and T. R. Walsh. 2004. Genetic analysis of 17 genes in Staphylococcus aureus with reduced susceptibility to vancomycin (VISA) and heteroVISA. J. Antimicrob. Chemother. 53:406-407. [DOI] [PubMed] [Google Scholar]