Abstract
Helicobacter pylori has a highly variable genome with ongoing diversification via inter- and intragenomic recombination and spontaneous mutation. DNA repair genes modulating mutation and recombination rates that influence diversification have not been well characterized for H. pylori. To examine the role of putative base excision repair ung and mutY glycosylase and xthA apurinic/apyrimidinic endonuclease genes in H. pylori, mutants of each were constructed in strain JP26 by allelic exchange. Spontaneous mutation frequencies of JP26 mutY mutants, assessed by rifampin resistance, were consistently higher (26-fold) than that of the wild type, whereas the ung and xthA mutants showed smaller increases. In trans complementation of the JP26 mutY mutant restored spontaneous mutation frequencies to wild-type levels. In cross-species studies, H. pylori mutY complemented an Escherichia coli mutY mutant and vice versa. In contrast, the ung and mutY mutants did not show higher frequencies of intergenomic recombination or greater sensitivity to UV-induced DNA damage than the wild type. The H. pylori mutY open reading frame contains an eight-adenine homonucleotide tract; we provide evidence that this is subject to slipped-strand mispairing, leading to frameshifts that eliminate gene function. Our findings indicate that H. pylori possesses phase-variable base excision repair, consistent with a tension between repair and mutation.
DNA bases may be damaged either spontaneously or by bioactive molecules, such as reactive oxygen species (3, 36). In Escherichia coli, base excision repair (BER) plays a major role in removing mutation-inducing DNA lesions produced by base modifications (9). In BER, a damage-specific DNA glycosylase first recognizes and excises the damaged bases, detaching them from the DNA backbone, leaving apurinic/apyrimidinic (AP) sites. Then, an AP endonuclease cuts the DNA strand 5′ of the AP site and removes the sugar, and a repair patch of nucleotides is catalyzed by general-purpose DNA polymerases and ligases (50). Some DNA glycosylases that recognize oxidative damage also have AP endonuclease activity (63).
E. coli possesses several DNA glycosylases, including Ung, MutY, Fpg, and Nei, that each recognize a specific type of damaged DNA base (12). The Ung glycosylase removes uracil that is accidentally incorporated into DNA when cytosine becomes deaminated (40). MutY is responsible for excising adenine in A · G and A · C mispairs and opposite 8-oxoguanine, one of the most stable products of oxidative DNA damage (2, 16, 24, 35). Although many prokaryotic species possess putative glycosylase and AP endonuclease homologs, BER pathways are not fully conserved across all species, and the specific abnormal bases that the systems repair differ greatly (12).
Helicobacter pylori is a species of gram-negative, microaerophilic bacteria that persistently colonize the human stomach (56); the inflammation induced predisposes to peptic ulceration and gastric adenocarcinomas (4, 34, 37). Compared to many other organisms, H. pylori has a high level of genetic diversity (23, 46, 53) and has high-frequency spontaneous mutation and intergenomic recombination (25, 57). The relative lack of DNA repair homologs in H. pylori (60) is consistent with an intrinsic genomic plasticity that may be adaptive in relatively inhospitable human gastric niches (5, 30, 60). For example, the methyl-directed mismatch repair pathway is not present (60), and H. pylori MutS2 does not exhibit MutS1 functions (21).
High mutation rates may be advantageous to organisms under stressful conditions but can otherwise be costly (41, 54). Because of this tension, we studied the roles that DNA repair and, more specifically, base excision repair play in H. pylori survival. From phylogenomic analysis, putative ung and mutY glycosylase genes and an xthA AP endonuclease gene have been annotated in the H. pylori genome. In this report, we provide evidence of the primary importance of mutY in H. pylori DNA repair of spontaneous mutations, while ung and xthA play smaller roles. The mutY open reading frame (ORF) has a homonucleotide tract of eight adenines; we provide evidence that implies that frameshifts in this tract create subpopulations of cells that differ in mutation rates.
MATERIALS AND METHODS
Amino acid alignment and phylogenetic analyses of MutY homologs.
Amino acid sequences of MutY homologs were retrieved from the Comprehensive Microbial Resource from the TIGR website (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi). Conserved domains were identified with Pfam (14) and the Simple Modular Architectural Research Tool (SMART) program (http://smart.embl-heidelberg.de/) (28), which encompasses several protein domain databases. Amino acid sequences were aligned using CLUSTAL_X (58) and visualized with Genedoc (www.psc.edu/biomed/genedoc) in conservation shading mode. Phylogenetic trees were constructed using MEGA 2.1 (26) according to the neighbor-joining method (45), with 1,000 bootstrap replicates.
Bacterial strains and plasmids.
The E. coli and H. pylori strains and plasmids used in this study are listed in Table 1. E. coli CC104 wild-type and CC104 mutM mutY mutant strains were routinely grown on LB agar plates at 37°C. H. pylori strains were grown at 37°C in 5% CO2 on Trypticase soy agar (TSA), with antibiotics added as described below for individual experiments.
TABLE 1.
Plasmids and bacterial strains used in this study
| Plasmid or strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Plasmids | ||
| pMutYKm | mutY::aphA in pGEMT-Easy | This work |
| pUngCat | ung::cat in pGEMT-Easy | This work |
| pXthACat | xthA::cat in pGEMT-Easy | This work |
| pADC | ureAB fragment in pUC18 with cat cassette | 21 |
| pADC-HpMutY | ureAB fragment in pUC18 with H. pylori mutY and cat cassette | This work |
| pADC-EcMutY | ureAB fragment in pUC18 with E. coli mutY and cat cassette | This work |
| Strains | ||
| H. pylori | ||
| JP26 | Wild-type strain | 22 |
| JP26 mutY::aphA | ΔmutY with aphA insertion | This work |
| JP26 mutYHpcomp | mutY::aphA complemented with H. pylori mutY in ureAB locus | This work |
| JP26 mutYEccomp | mutY::aphA complemented with E. coli mutY in ureAB locus | This work |
| JP26 ung::cat | Δung with cat insertion | This work |
| JP26 xthA::cat | ΔxthA with cat insertion | This work |
| JP26 mutY::aphA ung::cat | ΔmutY with aphA insertion, Δung with cat insertion | This work |
| JP26 mutY::aphA xthA::cat | ΔmutY with aphA insertion, ΔxthA with cat insertion | This work |
| E. coli | ||
| CC104 | Wild-type strain | 31 |
| CC104 mutM mutY | ΔmutM with mini-Kan insertion, ΔmutY with mini-Tn10 insertion | 31 |
| CC104 mutM mutY + pADC | ΔmutM ΔmutY transformed with control plasmid pADC | This work |
| CC104 mutM mutY + pADC-EcMutY | ΔmutM ΔmutY transformed with pADC-EcMutY | This work |
| CC104 mutM mutY + pADC-HpMutY | ΔmutM ΔmutY transformed with pADC-HpMutY | This work |
Construction of H. pylori mutants.
Fragments of the HP0142 (mutY homolog), HP1347 (ung homolog), and HP1526 (xthA homolog) open reading frames from strain 26695 were amplified by PCR using primers based on sequenced H. pylori strain 26695 (Table 2). Products were cloned into pGEMT-Easy (Promega, Madison, WI) to create pUng, pMutY, and pXthA, respectively. Next, pMutY, with an internal BamHI site, was digested with BamHI and ligated to an aphA cassette conferring kanamycin resistance to create pMutYKm. Inverse PCR was performed on pUng (using primers unginvBamHI-F and unginvBamHI-R) and pXthA (with primers xthAinvBamHI-F and xthAinvBamHI-R) to introduce BamHI sites. The PCR products were digested and each subsequently ligated with a cat cassette conferring chloramphenicol resistance to create pUngCat and pXthACat. H. pylori strain JP26 was then transformed with these plasmids to achieve kanamycin resistance, chloramphenicol resistance, or both, and transformants were selected on appropriate media to obtain the JP26 mutY::aphA, JP26 ung::cat, and JP26 xthA::cat mutants and JP26 mutY::aphA ung::cat and JP26 mutY::aphA xthA::cat double mutants (Table 1). Chromosomal DNA was isolated from the transformants, and the correct insertion of the aphA and/or cat cassette was confirmed by specific PCR in each case.
TABLE 2.
Oligonucleotide primers used in this study
| Organism and primer designation | Primer genomic location (nt)a | Primer sequence (5′→3′)b | PCR product size (bp)c |
|---|---|---|---|
| H. pylori | |||
| mutY-F | 154662-154683 | TGGTATGAAGAATTTGGGCGCA | 911 |
| mutY-R | 153773-153794 | AGCGTCATAGAGCTTATGGGTA | |
| mutYXbaI-F | 154687-154712 | TCTAGATGGAAACTTTACACAACGCCCTTTTA | 1,067 |
| mutYSmaI-R | 153646-153662 | CCCGGGAGTTGTGATCAGCGCGA | |
| mutYhomoA-F | 154470-154489 | AGTCCGAATTCTTTATTGCTCTGGCGAGGGC | 112 |
| mutYhomoA-R | 154388-154410 | CGAGCTTCTAGATCATTGGGTAGTTGTGAGTGGTG | |
| mutYnoshift-F | 154349-154369 | AGTCCGAATTCACTCCCAGGGATTGGCGCATA | 112 |
| mutYnoshift-R | 154268-154291 | CGAGCTTCTAGACTTAAAAGCACGCGCTTGATATTA | |
| mutYnoshift-F+1 | 154349-154370 | AGTCCGAATTCAACTCCCAGGGATTGGCGCATA | 113 |
| mutYnoshift-R | 154268-154291 | CGAGCTTCTAGACTTAAAAGCACGCGCTTGATATTA | |
| ung-F | 1408140-1408161 | AAGGCACTAGAGGCAAAGGAGG | 1,269 |
| ung-R | 1406893-1406915 | CGATATTTAGGGTTCTATCAGCG | |
| unginvBamHI-F | 1407297-1407316 | AGCTGGATCCCCAAAAACAAACACATCATC | 4,017 |
| unginvBamHI-R | 1407604-1407621 | AGCTGGATCCACGCTAAAGCTCAACCCC | |
| xthA-F | 1605981-1606003 | CGTGTGGTATGTGTCAAAAAGGG | 1,230 |
| xthA-R | 1604774-1604793 | TTGCTGAAAACACATGGACG | |
| xthAinvBamHI-F | 1605263-1605284 | AGCTGGATCCGGCTTTAGCGATGAAGAGAGAG | 4,018 |
| xthAinvBamHI-R | 1605532-1605553 | AGCTGGATCCATATTAATACCATAGCTCACGC | |
| E. coli | |||
| Ec-mutYXbaI-F | 3101002-3101020 | TCTAGATGCCCCCAACAACAGTGAA | 1,157 |
| Ec-mutYSmaI-R | 3102139-3102158 | CCCGGGACCTTCTGCTTCACGTTGCA |
Locations for H. pylori primers are based on the sequenced H. pylori strain 26695 (60), and locations for E. coli primers are based on the sequenced E. coli strain K-12 MG1655 (6). nt, nucleotides.
The following restriction sites are underlined: BamHI (GGATCC), XbaI (TCTAGA), SmaI (CCCGGG), and EcoRI (GAATTC).
PCR product sizes apply to primer pairs.
Assay to examine recovery from UV-induced DNA damage.
H. pylori cells to be tested were grown on TSA plates for 48 h and suspended in phosphate-buffered saline (PBS), and 100 to 500 CFU was inoculated to TSA plates. Cells were exposed to UV radiation (0 to 1,380 kJ/m2) at a wavelength of 312 nm (Stratagene transilluminator) and incubated at 37°C in 5% CO2 for 96 h. Colonies were counted and survival fractions calculated.
Assay to examine intergenomic recombination.
H. pylori strains grown on TSA plates for 48 h were harvested in 1 ml of PBS, and 30 μl of the suspension was combined with 60 ng of donor DNA by spotting onto a TSA plate. The TSA plate was incubated for 18 h at 37°C in 5% CO2. Donor DNA was an 800-bp PCR product of H. pylori rpsL from streptomycin-resistant strain JP26 with an A128G mutation (19). The transformation mixture was then harvested in 1 ml of PBS, and 100-μl portions of the appropriate serial dilutions were plated onto either TSA or brucella agar (BA) plates containing 10% newborn calf serum and 20 μg/ml streptomycin. The plates were incubated for 5 days at 37°C in 5% CO2, and the total recombination frequency was determined by dividing the number of streptomycin-resistant colonies by the total CFU.
Assay of spontaneous mutation frequencies.
Since single point mutations in rpoB confer H. pylori resistance to rifampin (18), rifampin resistance was used to assess spontaneous mutation frequencies. Rifampin-sensitive H. pylori strains were diluted onto TSA plates and grown for 5 days at 37°C in 5% CO2. Colonies were picked and expanded onto nonselective TSA plates for 48 h of growth and then harvested in PBS and serially diluted onto TSA plates or BA plates containing 10% newborn calf serum and rifampin (10 μg/ml). Plates were incubated at 37°C in 5% CO2 for 96 h, colonies counted, and spontaneous mutation frequencies calculated.
Complementation of the JP26 mutY::aphA mutant.
Primers mutYXbaI-F and mutYSmaI-R (Table 2) were used to amplify ORF HP0142 (mutY) with JP26 genomic DNA as the template, and the products were ligated into pGEMT-Easy to create pMutYComp. Next, pMutYComp was digested with XbaI and SmaI to obtain the HP0142 fragment, which was ligated with XbaI- and SmaI-digested pADC containing cat in the H. pylori ureAB locus downstream of a ureA promoter (21). ORF HP0142 was thus inserted downstream of the ureA promoter, creating pADC-HpMutY. The HP0142 ORF was introduced in trans into the ureAB locus of JP26 mutY::aphA cells via natural transformation with pADC-HpMutY, creating JP26 mutYHpcomp. Transformants were selected for chloramphenicol resistance, and the correct insertion of HP0142 into ureA downstream of the ureA promoter was confirmed by PCR of the chromosomal DNA. The JP26 mutY::aphA strain complemented with E. coli mutY (JP26 mutYEccomp) was created using parallel methods. Primers Ec-mutYXbaI-F and Ec-mutYSmaI-R were used to amplify mutY from E. coli strain CC104, and then all other steps were completed as described above.
Cross-species complementation of MutY.
To determine whether H. pylori mutY (HP0142) can functionally complement in E. coli, an E. coli mutY mutM mutant (31) was studied. H. pylori MutY from shuttle plasmid pADC-HpMutY was expressed using the ureA promoter. To assess spontaneous mutation frequencies, cells of rifampin-sensitive E. coli strains were diluted onto TSA plates and grown for 1 day at 37°C, and colonies were expanded onto TSA, allowing mutations to occur. After an additional 24 h of growth, cells were harvested into saline and serially diluted onto LB plates with and without rifampin (50 μg/ml). Plates were incubated at 37°C for 24 h, colonies counted, and spontaneous mutation frequencies calculated.
Shifts in H. pylori mutY homopolymeric tract.
The 987-bp H. pylori 26695 and J99 mutY ORFs each include an eight-adenine homonucleotide tract beginning at position 273 (1, 60). Since partial mutY sequences of 413 H. pylori strains were available on http://pubmlst.org/helicobacter/, the homonucleotide region was examined for polymorphisms (database accessed March 2006). To determine whether frameshifts occur in the mutY homonucleotide tract in a population of cells growing on plate culture, a blue-white screen was developed. An ∼100-bp fragment containing the H. pylori mutY homonucleotide tract was PCR amplified from JP26 genomic DNA by use of primers mutYhomoA-F and mutYhomoA-R with 5′ EcoRI and XbaI sites, respectively (Table 2). The products were ligated into pUC18 to create pMutYhomotract, which was then transformed into DH5α cells, inoculated to carbenicillin, IPTG (isopropyl-β-d-thiogalactopyranoside), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates, and incubated for 37°C overnight. After color development at 4°C, white and blue colonies, representative of out-of-frame and in-frame products, respectively, were counted. Representative white colonies were expanded, DNA prepared, and the pMutYhomotract inserts sequenced. A similarly sized fragment from mutY, but from a region without homopolymeric tracts, was amplified as a control. Primers mutYnoshift-F and mutYnoshift-R were used to generate the positive-control in-frame product, and primers mutYnoshift-F+1 and mutYnoshift-R were used to generate the negative-control out-of-frame product. The products were ligated into pUC18 to create pMutYnoshift and pMutYnoshift+1, and the plasmids were screened as described above.
Statistical analyses.
Student's t test, unpaired with equal variance, was used to determine statistical significance in all cases. A P value of <0.05 was defined as statistically significant.
RESULTS
Analyses of MutY family homologs.
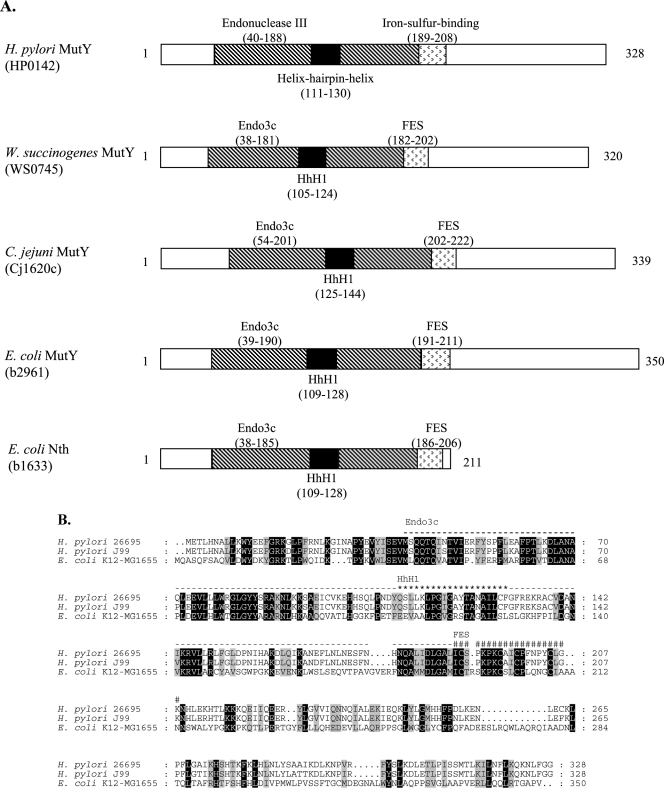
The conserved domains in MutY, based on Pfam (14) and the SMART database (http://smart.embl-heidelberg.de) (28), are shown in schematic form for representative species in Fig. 1A. Protein structure is highly conserved. The 20-amino-acid helix-hairpin-helix is the canonical DNA binding domain shared by all BER glycosylases (10). Studies of E. coli MutY implicate the iron-sulfur-binding domain in substrate recognition and protein stability (31, 44). MutY orthologs possess an endonuclease III domain with homology to E. coli Nth, a glycosylase that has AP endonuclease activity, consistent with the reported MutY AP endonuclease activity (32, 33). On the amino acid level, the aligned H. pylori 26695 and J99 MutY deduced products show 94.8% identity and 97.3% similarity, and the H. pylori 26695 and E. coli K-12 MG1655 MutY deduced products share 36.7% identity and 59.1% similarity (Fig. 1B). The phylogenetic position of MutY from H. pylori is closest to that of other Campylobacterales and is also related to that of Thermus thermophilus, an extremophile (Fig. 1C).
FIG.1.
Genetic analyses of MutY homologs. (A) Schematic of MutY homologs and E. coli endonuclease III (Nth). Conserved protein domains were identified using Pfam (14) and the SMART program (http://smart.embl-heidelberg.de) (28). (B) MutY amino acid alignment. H. pylori 26695 MutY and J99 MutY show 94.8% identity and 97.3% similarity, and H. pylori 26695 MutY and E. coli K-12 MG1655 MutY show 36.7% identity and 59.1% similarity. The helix-hairpin-helix (HhH1; ★), endonuclease (Endo3c; -), and iron-sulfur-binding (FES; #) domains of H. pylori MutY are labeled above the sequences. Residues are shaded as follows: black, 100% conservation; gray, similarity groups. (C) Phylogenetic analysis of MutY homologs. A phylogenetic tree was constructed with MutY amino acid sequences obtained from the Comprehensive Microbial Resource (http://cmr.tigr.org). The program MEGA 2.1 (26) was used to construct the phylogeny via the neighbor-joining method (45), with 1,000 bootstrap replicates. S. enterica, Salmonella enterica; Y. pestis, Yersinia pestis; H. influenzae, Haemophilus influenzae; V. cholerae, Vibrio cholerae; V. vulnificus, Vibrio vulnificus; N. meningitidis, Neisseria meningitidis; A. tumefaciens, Agrobacterium tumefaciens; S. aureus, Staphylococcus aureus; B. subtilis, Bacillus subtilis; B. anthracis, Bacillus anthracis; C. glutamicum, Corynebacterium glutamicum; A. aeolicus, Aquifex aeolicus.
H. pylori mutY shifts in homonucleotide tract.
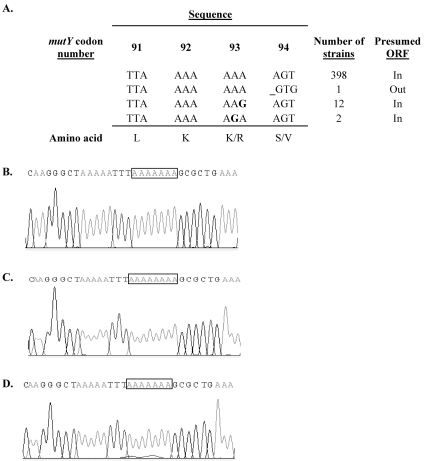
Of the 413 mutY sequences available in the H. pylori multilocus sequence typing database, polymorphisms involving the adenine homonucleotide tract were observed to occur in 15 strains (Fig. 2A). One (ALA15) of the 413 strains (2.4 × 10−3) showed a frameshift with a 1-bp deletion in the eight-adenine mutY homonucleotide tract (Fig. 2B). There also were 14 (3.4 × 10−2) substitution mutations at codon 93 (AAA) encoding lysine: 12 with synonymous (G) substitutions in the third position and 2 with nonsynonymous substitutions (G) in the second position, encoding arginine. In the blue-white screen for shifts in the H. pylori mutY homonucleotide tract, the positive control, with an in-frame nonhomonucleotide mutY fragment, gave 97% blue colonies and the negative control, with an out-of-frame nonhomonucleotide mutY fragment, gave 96.6% white colonies (data not shown). From the screen with the mutY homonucleotide fragment, two white colonies showed a shift from eight adenines (Fig. 2C) to seven (Fig. 2D), providing further evidence that natural phase variation occurs in mutY. Since these studies show that H. pylori subpopulations differing in expression of the product encoded by mutY exist, we sought to compare phenotypes of the on and off versions by creating defined mutants.
FIG. 2.
Polymorphisms in mutY. (A) Genotypes of 413 mutY homonucleotide tracts from the H. pylori multilocus sequence typing database (http://pubmlst.org/helicobacter/). Substitutions are shown in bold, and the frameshift is depicted by _. (B) Chromatogram of mutY homonucleotide tract from strain ALA15. (C) Chromatogram of blue colony from mutY blue-white screen (eight adenines). (D) Chromatogram of white colony from mutY blue-white screen (seven adenines).
H. pylori glycosylase and AP endonuclease mutants show increased frequencies of spontaneous mutation.
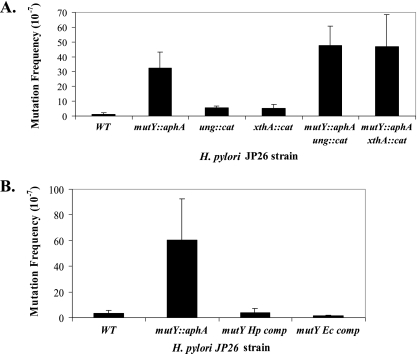
Based on the homologies and phylogenies (Fig. 1 and data not shown), we hypothesized that the ung and mutY homologs in H. pylori encode DNA glycosylases, that xthA encodes an AP endonuclease, and that these genes play a role in H. pylori BER. To test these hypotheses, H. pylori mutants were created to examine frequencies of spontaneous rifampin mutations due to BER impairment, which permits retention of errors throughout the genome. Errors occurring in rpoB may lead to alterations in the rifampin binding site of RNA polymerase, rendering the cells resistant to rifampin (18). The H. pylori ung and mutY glycosylase mutants and the xthA AP endonuclease mutant each showed increased spontaneous mutation frequencies (Fig. 3A). Compared to the wild type, the mutY mutant showed an ∼26-fold increase, but for the ung and xthA mutants, the differences were fourfold. The mutY ung and mutY xthA double mutants had nearly identical increases in spontaneous mutations (37-fold) compared to that of wild-type cells. The results further indicate that MutY activity in H. pylori is not limited by lack of Ung or XthA base excision repair functions.
FIG. 3.
Spontaneous mutation frequencies of H. pylori cells. (A) Wild-type (WT) and mutant strains. H. pylori mutY mutants showed a 26-fold increase in spontaneous mutation frequency (P < 0.05), and the ung and xthA mutants showed fourfold increases, as detected by resistance to rifampin. mutY ung and mutY xthA double mutants had 37-fold increases in the frequency of spontaneous mutation. Bars represent means ± standard deviations for ≥6 replicate experiments. (B) Complementation of H. pylori mutY mutant phenotype. H. pylori mutY mutants were complemented by either H. pylori or E. coli mutY. Spontaneous mutation frequencies returned to wild-type levels. Bars represent means ± standard deviations for ≥4 replicate experiments.
Complementation of the H. pylori mutY mutant with mutY in trans.
The mutY homolog HP0142 is present in a putative dicistronic operon in strains 26695 and J99 with no downstream genes (60). To determine whether the spontaneous mutation phenotype was specific to the mutY mutation and not due to polar effects or artifact, the H. pylori mutY::aphA mutant was complemented in trans by expressing mutY downstream of a strong ureA promoter in the distant ureAB chromosomal locus. Complementation restored the spontaneous mutation frequency to wild-type levels (Fig. 3B), confirming that the observed phenotype is specific to the mutY mutation and not due to polar effects. The H. pylori mutY::aphA mutant was also complemented in trans by E. coli mutY in the identical locus to a comparable degree.
H. pylori MutY complements E. coli MutY mutant.
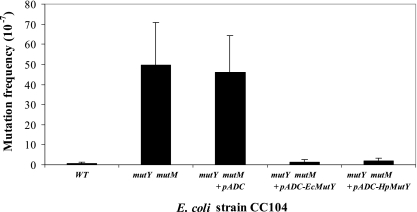
To determine whether the H. pylori MutY protein could complement a MutY defect in E. coli, the E. coli CC104 mutM mutY mutant strain (31) was transformed with H. pylori mutY in a shuttle plasmid (pADC-HpMutY), with the vector alone (pADC), or with the E. coli mutY (pADC-EcMutY) as a positive control. Compared to the wild type, the mutM mutY mutant showed a 73-fold increase in the frequency of spontaneous mutation, which was not complemented by the vector alone. However, both E. coli and H. pylori mutY nearly completely complemented the defect (Fig. 4). In total, the cross-species complementation studies provide further evidence for MutY function encoded by HP0142.
FIG. 4.
Cross-species complementation of E. coli mutY mutant. The E. coli mutY mutant was complemented by H. pylori mutY expressed from plasmid pADC-HpMutY. Transformation with the control plasmid pADC had no effect, and the positive control (pADC-EcMutY) showed full complementation. Bars represent means ± standard deviations for ≥11 replicate experiments. WT, wild type.
H. pylori ung and mutY glycosylase and xthA AP endonuclease mutants are no more sensitive to UV-induced DNA damage than the wild type.
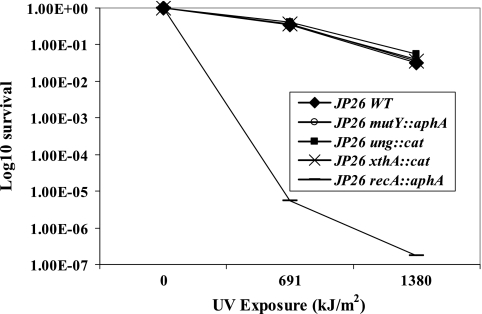
Exposure to UV generates primarily cyclobutane pyrimidine dimers and 6-4 pyrimidine-pyrimidone photoproducts (7), substrates that are not recognized by E. coli Ung and MutY. As such, we hypothesized that the H. pylori glycosylase mutants are no more sensitive to UV-induced DNA damage than wild-type cells. Furthermore, UV radiation creates interstrand cross-links in DNA as well as strand breaks that can halt replication fork progression; the DNA template can be restored by recombination repair recognizing and replacing damaged DNA. As expected, H. pylori recA mutants were more susceptible to UV exposure than wild-type cells (Fig. 5) (59). However, the ung, mutY, and xthA mutants did not show increased susceptibility to UV, providing evidence that these constituents of BER are not involved in recombination repair of damaged DNA in H. pylori.
FIG. 5.
Survival of H. pylori wild-type (WT) and mutant strains after UV exposure. H. pylori wild-type and mutant strains were exposed to 312-nm UV (0 to 1,380 kJ/m2), and survival fractions were determined. As expected, the positive control, the JP26 recA mutant, showed >4-log10 sensitivity. However, there were no significant differences between the wild type and the mutY, ung, and xthA mutants.
H. pylori glycosylase mutants do not show elevated frequencies of recombination.
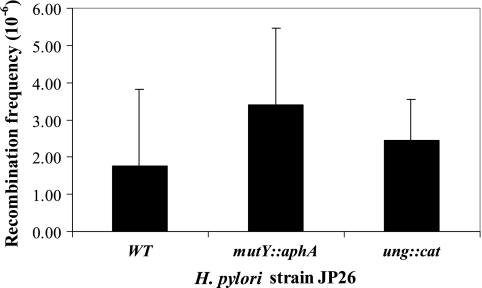
Glycosylases function not only in BER but also in homologous recombination. Nitric oxide is a potent DNA damaging agent, and NO−-induced homologous recombination in E. coli is promoted by DNA glycosylases (55). Processing of DNA base damage by glycosylases leads to the creation of AP sites and single-strand breaks, which in turn may be converted into double-strand breaks, providing substrate for recombination. However, the H. pylori ung and mutY mutants did not show elevated frequencies of intergenomic recombination (Fig. 6).
FIG. 6.
Intergenomic recombination frequencies of H. pylori wild-type (WT) and mutant strains. Ability to be transformed by an 800-bp PCR product that confers streptomycin resistance was used to determine recombination frequency. H. pylori JP26 mutY and ung mutants did not show elevated frequencies of intergenomic recombination compared to the wild-type JP26 strain. Bars represent means ± standard deviations for ≥4 replicate experiments.
Homonucleotide tracts in other bacterial glycosylase genes.
To determine whether the presence of homonucleotide tracts in the H. pylori glycosylase genes may be more prevalent, using the whole-organism genomic sequences, we conducted informatic analysis of other members of Campylobacterales and other eubacteria (Table 3). Among the other Campylobacterales, homonucleotide tracts of ≥7 nucleotides were common in ORFs annotated as mutY, ung, or nth in Campylobacter jejuni and Helicobacter hepaticus but not Wolinella succinogenes. Similarly, although homonucleotide tracts were common in other phyla, they were not universally present, and in certain organisms, including E. coli, Mycobacterium tuberculosis, and Streptococcus pyogenes, none were observed.
TABLE 3.
Homonucleotide tracts in glycosylase genes of different organisms
| Phylum | Class | Order | Organism and strain | ORF designation/tract characteristic(s)a for glycosylase gene
|
||
|---|---|---|---|---|---|---|
| mutY | ung | nth | ||||
| Proteobacteria | Epsilonproteobacteria | Campylobacterales | Helicobacter pylori 26695 | HP0142/A8; A7 | HP1347/A7 × 3 | HP0585/— |
| Helicobacter pylori 26695 | HP0602/A8; A7; T7 × 2 | |||||
| Helicobacter pylori J99 | jhp0130/A8; A7 | jhp1266/A7 × 3 | jhp0532/— | |||
| Helicobacter pylori J99 | jhp0549/A7 × 2; T7 × 2 | |||||
| Helicobacter hepaticus ATCC 51449 | HH1242/A8; A7 | HH1249/— | HH1575/A8; A7 | |||
| Campylobacter jejuni NCTC 11168 | Cj1620c/A8; A7 × 3; T7 × 2 | Cj0086c/A8; A7 | Cj0595c/T7 | |||
| Campylobacter jejuni RM1221 | CJE1792/A8; A7 × 3; T7 × 2 | CJE0081/A8; A7 | CJE0698/T7 | |||
| Wolinella succinogenes DSMZ 1740 | WS0745/— | WS0557/— | WS1060/A7 | |||
| Gammaproteobacteria | Enterobacteriales | Escherichia coli K-12 MG1655 | b2961/— | b2580/— | b1633/— | |
| Salmonella enterica serovar Paratyphi ATCC 9150 | SPA2973/A7 | SPA0271/— | SPA1400/— | |||
| Yersinia pestis KIM | y3339/A7 | y1281/C7 | y2080/— | |||
| Pasteurellales | Haemophilus influenzae KW20 Rd | HI0759/— | HI0018/A8 | HI1689/— | ||
| Vibrionales | Vibrio cholerae El Tor N16961 | VC0452/— | VC2359/— | VC1101/— | ||
| Legionellales | Legionella pneumophila Lens | lpl0930/A9; A8; T8; T7 | * | lpl2793/A7 | ||
| Betaproteobacteria | Neisseriales | Neisseria meningitidis MC58 | NMB1396/A7 | NMB1222/— | NMB0533/— | |
| Chlamydiae | Chlamydiae | Chlamydiales | Chlamydia abortus S26 3 | CAB380/A7 | CAB954/— | CAB898/A9; T7 |
| Deinococcus-Thermus | Deinococci | Thermales | Thermus thermophilus HB8 | TTHA1898/— | * | TTHA0112/— |
| Actinobacteria | Actinobacteria | Actinomycetales | Streptomyces avermitilis MA-4680 | SAV4707/— | SAV1527/— | SAV4593/— |
| Mycobacterium tuberculosis H37Rv | Rv3589/— | Rv2976c/— | Rv3674c/— | |||
| Firmicutes | Bacilli | Bacillales | Listeria innocua CLIP 11262 | lin1797/A7 | lin0408/— | lin2008/A7; T7 |
| Bacillus subtilis 168 | * | Bsu3793/— | Bsu2233/— | |||
| Bacillus anthracis Ames | BA0522/— | BA5648/T7 | BA1570/— | |||
| Lactobacillales | Streptococcus pneumoniae R6 | spr1108/A7 | spr1055/— | spr1157/A7 | ||
| Streptococcus pyogenes MGAS315 | SpyM3_1584/— | SpyM3_0621/— | SpyM3_0642/— | |||
Tract composition, size, and occurrence (e.g., × 3 denotes three occurrences) are shown. —, no tract with ≥7 of the same nucleotide; *, no annotated ORF.
DISCUSSION
HP0142 was annotated as mutY based on the strong homology of its deduced product with known MutY proteins. We show that the protein sequence possesses conserved domains and amino acid properties characteristic of MutY and that the cloned ORF complements the mutY deficiency in an E. coli mutant. Interrupting mutY in H. pylori resulted in elevated frequencies of spontaneous mutation, consistent with a role in BER, and in trans complementation with either the H. pylori or the E. coli mutY confirmed that this phenotype was not due to a polar or extraneous event. Moreover, the mutant's lack of an effect on UV repair, as expected, indicates the specificity of its role. Thus, HP0142 is an authentic mutY, but although H. pylori mutY was able to complement the E. coli mutY mutant, the two proteins may not have all of the same functions. The C-terminal domain of E. coli MutY is involved in both DNA binding and glycosylase activities (29), and future experiments should define the properties of the nonconserved C-terminal domain of H. pylori MutY. The close proximity of the phylogenetic position of H. pylori MutY with that of the extremophile T. thermophilus suggests convergent evolution due to parallel environmental constraints.
Exposure to UVB (wavelength range, 280 to 320 nm) is known to generate singlet oxygen, superoxide ions, and other free radicals, which subsequently increase 8-oxoguanine levels in cells (17, 42, 64). In mice lacking Ogg1, a mammalian glycosylase responsible for removing 8-oxoguanine from DNA, UVB exposure increases susceptibility to skin carcinogenesis (27). The E. coli xthA mutant is sensitive to inactivation by broad-spectrum near-UV (300- to 400-nm) radiation (49, 51). Therefore, the H. pylori BER mutants might be similarly sensitive to UV, as glycosylases and AP endonucleases repair the DNA damaged by oxidative by-products. Under our experimental conditions, in which 312-nm UV light was used, neither the mutY nor the xthA H. pylori mutant differed from wild-type cells in phenotype. It is possible that the experimental conditions did not generate sufficient 8-oxoguanine in the cells for a phenotype to be observed, that H. pylori has pathways to catabolize 8-oxoguanine, or that 8-oxoguanine is not a major substrate for mutY and xthA in H. pylori.
The elevated spontaneous mutation frequencies of H. pylori strains in which the genes encoding the glycosylases Ung and MutY and AP endonuclease XthA have been interrupted can be explained in several ways. First, loss of glycosylase and AP endonuclease function impairs the BER pathway, leading to increased spontaneous mutation frequency as errors are retained throughout the genome. However, the fact that the mutY strain has a greater effect (26-fold) than the ung or xthA strains (fourfold) implies that MutY has greater in vivo glycosylase activity than Ung in H. pylori and/or that the MutY substrate is generated more frequently than is the Ung substrate and that the BER products generated by H. pylori MutY activity may be substrates for AP endonucleases other than XthA. If the products generated by H. pylori MutY were substrates for XthA only, then mutations in the downstream gene in the pathway (xthA) also should yield a similar (26-fold) increase in spontaneous mutation frequency. That the xthA mutation resulted in a smaller (fourfold) difference in mutation frequency suggests the presence of other AP endonucleases in H. pylori. Alternatively, it is possible that H. pylori MutY has its own AP endonuclease activity, as has been shown for the E. coli MutY (32, 33), which has structural similarity to H. pylori MutY (Fig. 1B). Furthermore, that MutY may function independently of Ung and XthA is supported by the enhanced mutation frequency in the mutY ung and mutY xthA double mutants.
Based on phylogenomic studies, H. pylori has had loss of DNA repair pathway components, including the methyl-directed mismatch repair system found in E. coli (12). Nevertheless, it is possible that H. pylori has an unidentified mismatch repair system (8). E. coli MutY is responsible not only for repair of oxidative DNA damage but for mismatch repair as well. MutY processes at least some A · G mispairs in E. coli (2), and the lack of MutY activity generates G · C → T · A transversions, resulting in rpoB mutations that confer rifampin resistance (24). That H. pylori MutY was capable of complementing the E. coli mutY mutant suggests that it can play a similar role in mismatch repair preventing G · C → T · A transversion mutations, if such a system does exist in H. pylori.
Phase variation, a common mechanism used by gram-negative bacteria to generate intrastrain diversity, permits phenotypic variation important to adaptation to niches or to changing environmental conditions (48, 62). Translational phase variation reversibly switches gene expression via insertion or deletion of DNA repeat sequences within the coding region, altering the reading frame, which ultimately leads to a premature stop codon (8, 62). H. pylori strains vary both in the presence of repeats in particular genes and in length (48). That homopolymeric tracts are present in both mutY and ung (Table 3) suggests the possibility of phase variation, and the existence of a wild-type H. pylori strain (ALA15) with a seven-adenine tract in mutY, along with experimental evidence of frameshift, indicates that this is an actual mechanism for H. pylori to control DNA repair rates. Variation in repeat length may be generated by polymerase slippage during PCR and/or sequencing reactions, and PCR has the greatest potential to artifactually generate such variation (20). However, studies of length variation in the homonucleotide tract of H. pylori ORF HP0619 in strain JP96-9, possessing nine cytosines, provide evidence that for repeats of <11 nucleotides the observed polymorphisms are attributable to repeats of different lengths in the template and not to a PCR artifact (48). The phenomenon of phase variation in H. pylori has been explored previously for genes pldA, babB, and rfaJ (47). In recent work using enzyme-linked immunosorbent assay techniques in addition to the previously described E. coli reporter assay, we have examined the phenotypes associated with phase variation involved with a homopolymeric tract in futC, which controls Lewisy expression in H. pylori. This provides a measurable phenotype and illustrates the effect of the phase variation in an H. pylori background, as well as the efficacy of the E. coli assay (E. L. Sanabria-Valentín et al., Abstr. 106th Gen. Meet. Am. Soc. Microbiol., abstract D-001, 2006).
Phase variation of mutY offers a model for H. pylori adaptation. Although mutations are generally deleterious, spontaneous mutY frameshifts create subpopulations of hypermutators that can generate beneficial mutations under appropriate selection conditions. As hypermutators acquire the mutations needed to adapt to changing environments, the increasing number of deleterious mutations makes hypermutability selectively disadvantageous (11, 13, 15, 61). Therefore, persistent H. pylori colonization can be viewed as comprising two populations, mutator and nonmutator, living in an equilibrium determined by the fitness cost and benefit of particular mutations. Since phase variation may occur independently for mutY and ung, there may be at least four different populations (and possibly more in relation to other genes) in terms of “mutator” status. This is a more complex model than that appreciated for E. coli (38, 39, 52), but it might reflect the differing selective pressures on H. pylori.
Slipped-strand mispairing in homonucleotide tracts of DNA repair genes (e.g., mutY) also can allow for out-of-frame, hypermutator populations that have already acquired the beneficial mutations to regenerate nonhypermutator populations. Since the advantage of mutators is conditional, because mutator clones easily accumulate deleterious mutations after passage through bottlenecks (13), the reversion of an out-of-frame mutY to in frame after a beneficial mutation is acquired might be advantageous. Ultimately, the existence of homonucleotide tracts in mutY (and other BER genes) reflects second-order selection. More than just selection for better adaptation to a specific environment, second-order selection acts on the regulation of the processes of genetic adaptation to new environments (43). DNA repair genes with homonucleotide tracts reflect second-order selection because they give rise to hypermutator alleles that themselves are the substrates for selection. That H. pylori mutY possesses a phase-variable homonucleotide tract indicates that expression of the products of DNA repair genes is under the influence of replication genes, themselves subject to mutation. This nonlinear system for DNA repair, exemplified in H. pylori, could be a model for certain organisms but not others (Table 3).
Acknowledgments
We thank Felix Peng for technical assistance and Edgardo L. Sanabria-Valentín and Irina Derkatch for guidance in genetic screening. We thank A-Lien Lu (University of Maryland) for the E. coli mutants and Mark Achtman (Max-Planck-Institut für Infektionsbiologie) for the ALA15 chromatogram.
This study was supported by an Undergraduate Research Fellowship from the American Society for Microbiology, by the Diane Belfer Program in Human Microbial Ecology, and by R01 GM63270 and the Medical Scientist Training Program of the National Institutes of Health.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Au, K. G., S. Clark, J. H. Miller, and P. Modrich. 1989. Escherichia coli mutY gene encodes an adenine glycosylase active on G-A mispairs. Proc. Natl. Acad. Sci. USA 86:8877-8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckman, K. B., and B. N. Ames. 1997. Oxidative decay of DNA. J. Biol. Chem. 272:19633-19636. [DOI] [PubMed] [Google Scholar]
- 4.Blaser, M. J. 1998. Helicobacter pylori and gastric diseases. BMJ 316:1507-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser, M. J., and J. C. Atherton. 2004. Helicobacter pylori persistence: biology and disease. J. Clin. Investig. 113:321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Chandrasekhar, D., and B. Van Houten. 2000. In vivo formation and repair of cyclobutane pyrimidine dimers and 6-4 photoproducts measured at the gene and nucleotide level in Escherichia coli. Mutat. Res. 450:19-40. [DOI] [PubMed] [Google Scholar]
- 8.Cooke, C. L., J. L. Huff, and J. V. Solnick. 2005. The role of genome diversity and immune evasion in persistent infection with Helicobacter pylori. FEMS Immunol. Med. Microbiol. 45:11-23. [DOI] [PubMed] [Google Scholar]
- 9.David, S. S., and S. D. Williams. 1998. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem. Rev. 98:1221-1262. [DOI] [PubMed] [Google Scholar]
- 10.Denver, D. R., S. L. Swenson, and M. Lynch. 2003. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol. Biol. Evol. 20:1603-1611. [DOI] [PubMed] [Google Scholar]
- 11.de Visser, J. A. 2002. The fate of microbial mutators. Microbiology 148:1247-1252. [DOI] [PubMed] [Google Scholar]
- 12.Eisen, J. A., and P. C. Hanawalt. 1999. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 435:171-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funchain, P., A. Yeung, J. L. Stewart, R. Lin, M. M. Slupska, and J. H. Miller. 2000. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics 154:959-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasteiger, E., E. Jung, and A. Bairoch. 2001. SWISS-PROT: connecting biomolecular knowledge via a protein database. Curr. Issues Mol. Biol. 3:47-55. [PubMed] [Google Scholar]
- 15.Giraud, A., I. Matic, O. Tenaillon, A. Clara, M. Radman, M. Fons, and F. Taddei. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606-2608. [DOI] [PubMed] [Google Scholar]
- 16.Gu, Y., A. Parker, T. M. Wilson, H. Bai, D. Y. Chang, and A. L. Lu. 2002. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J. Biol. Chem. 277:11135-11142. [DOI] [PubMed] [Google Scholar]
- 17.Hattori, Y., C. Nishigori, T. Tanaka, K. Uchida, O. Nikaido, T. Osawa, H. Hiai, S. Imamura, and S. Toyokuni. 1996. 8-Hydroxy-2′-deoxyguanosine is increased in epidermal cells of hairless mice after chronic ultraviolet B exposure. J. Investig. Dermatol. 107:733-737. [DOI] [PubMed] [Google Scholar]
- 18.Heep, M., S. Odenbreit, D. Beck, J. Decker, E. Prohaska, U. Rieger, and N. Lehn. 2000. Mutations at four distinct regions of the rpoB gene can reduce the susceptibility of Helicobacter pylori to rifamycins. Antimicrob. Agents Chemother. 44:1713-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Israel, D. A., A. S. Lou, and M. J. Blaser. 2000. Characteristics of Helicobacter pylori natural transformation. FEMS Microbiol. Lett. 186:275-280. [DOI] [PubMed] [Google Scholar]
- 20.Jennings, M. P., D. W. Hood, I. R. Peak, M. Virji, and E. R. Moxon. 1995. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol. Microbiol. 18:729-740. [DOI] [PubMed] [Google Scholar]
- 21.Kang, J., S. Huang, and M. J. Blaser. 2005. Structural and functional divergence of MutS2 from bacterial MutS1 and eukaryotic MSH4-MSH5 homologs. J. Bacteriol. 187:3528-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang, J., D. Tavakoli, A. Tschumi, R. A. Aras, and M. J. Blaser. 2004. Effect of host species on recG phenotypes in Helicobacter pylori and Escherichia coli. J. Bacteriol. 186:7704-7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kersulyte, D., H. Chalkauskas, and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 31:31-43. [DOI] [PubMed] [Google Scholar]
- 24.Kim, M., T. Huang, and J. H. Miller. 2003. Competition between MutY and mismatch repair at A · C mispairs in vivo. J. Bacteriol. 185:4626-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraft, C., A. Stack, C. Josenhans, E. Niehus, G. Dietrich, P. Correa, J. G. Fox, D. Falush, and S. Suerbaum. 2006. Genomic changes during chronic Helicobacter pylori infection. J. Bacteriol. 188:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 27.Kunisada, M., K. Sakumi, Y. Tominaga, A. Budiyanto, M. Ueda, M. Ichihashi, Y. Nakabeppu, and C. Nishigori. 2005. 8-Oxoguanine formation induced by chronic UVB exposure makes Ogg1 knockout mice susceptible to skin carcinogenesis. Cancer Res. 65:6006-6010. [DOI] [PubMed] [Google Scholar]
- 28.Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks, J. Schultz, C. P. Ponting, and P. Bork. 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32:D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, L., and A. L. Lu. 2003. The C-terminal domain of Escherichia coli MutY is involved in DNA binding and glycosylase activities. Nucleic Acids Res. 31:3038-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Logan, R. P., and D. E. Berg. 1996. Genetic diversity of Helicobacter pylori. Lancet 348:1462-1463. [DOI] [PubMed] [Google Scholar]
- 31.Lu, A. L., and P. M. Wright. 2003. Characterization of an Escherichia coli mutant MutY with a cysteine to alanine mutation at the iron-sulfur cluster domain. Biochemistry 42:3742-3750. [DOI] [PubMed] [Google Scholar]
- 32.Lu, A. L., D. S. Yuen, and J. Cillo. 1996. Catalytic mechanism and DNA substrate recognition of Escherichia coli MutY protein. J. Biol. Chem. 271:24138-24143. [DOI] [PubMed] [Google Scholar]
- 33.Manuel, R. C., and R. S. Lloyd. 1997. Cloning, overexpression, and biochemical characterization of the catalytic domain of MutY. Biochemistry 36:11140-11152. [DOI] [PubMed] [Google Scholar]
- 34.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311-1315. [DOI] [PubMed] [Google Scholar]
- 35.Michaels, M. L., J. Tchou, A. P. Grollman, and J. H. Miller. 1992. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry 31:10964-10968. [DOI] [PubMed] [Google Scholar]
- 36.Nohmi, T., S. R. Kim, and M. Yamada. 2005. Modulation of oxidative mutagenesis and carcinogenesis by polymorphic forms of human DNA repair enzymes. Mutat. Res. 591:60-73. [DOI] [PubMed] [Google Scholar]
- 37.Nomura, A., G. N. Stemmermann, P. H. Chyou, G. I. Perez-Perez, and M. J. Blaser. 1994. Helicobacter pylori infection and the risk for duodenal and gastric ulceration. Ann. Intern. Med. 120:977-981. [DOI] [PubMed] [Google Scholar]
- 38.Notley-McRobb, L., R. Pinto, S. Seeto, and T. Ferenci. 2002. Regulation of mutY and nature of mutator mutations in Escherichia coli populations under nutrient limitation. J. Bacteriol. 184:739-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Notley-McRobb, L., S. Seeto, and T. Ferenci. 2002. Enrichment and elimination of mutY mutators in Escherichia coli populations. Genetics 162:1055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olsen, L. C., R. Aasland, H. E. Krokan, and D. E. Helland. 1991. Human uracil-DNA glycosylase complements E. coli ung mutants. Nucleic Acids Res. 19:4473-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostrowski, E. A., D. E. Rozen, and R. E. Lenski. 2005. Pleiotropic effects of beneficial mutations in Escherichia coli. Evol. Int. J. Org. Evol. 59:2343-2352. [PubMed] [Google Scholar]
- 42.Peak, M. J., and J. G. Peak. 1980. Protection by glycerol against the biological actions of near-ultraviolet light. Radiat. Res. 83:553-558. [PubMed] [Google Scholar]
- 43.Pennisi, E. 1998. How the genome readies itself for evolution. Science 281:1131, 1133-1134. [DOI] [PubMed] [Google Scholar]
- 44.Porello, S. L., M. J. Cannon, and S. S. David. 1998. A substrate recognition role for the [4Fe-4S]2+ cluster of the DNA repair glycosylase MutY. Biochemistry 37:6465-6475. [DOI] [PubMed] [Google Scholar]
- 45.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 46.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salaun, L., S. Ayraud, and N. J. Saunders. 2005. Phase variation mediated niche adaptation during prolonged experimental murine infection with Helicobacter pylori. Microbiology 151:917-923. [DOI] [PubMed] [Google Scholar]
- 48.Salaun, L., B. Linz, S. Suerbaum, and N. J. Saunders. 2004. The diversity within an expanded and redefined repertoire of phase-variable genes in Helicobacter pylori. Microbiology 150:817-830. [DOI] [PubMed] [Google Scholar]
- 49.Sammartano, L. J., and R. W. Tuveson. 1983. Escherichia coli xthA mutants are sensitive to inactivation by broad-spectrum near-UV (300- to 400-nm) radiation. J. Bacteriol. 156:904-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scharer, O. D., and J. Jiricny. 2001. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays 23:270-281. [DOI] [PubMed] [Google Scholar]
- 51.Serafini, D. M., and H. E. Schellhorn. 1999. Endonuclease III and endonuclease IV protect Escherichia coli from the lethal and mutagenic effects of near-UV irradiation. Can. J. Microbiol. 45:632-637. [PubMed] [Google Scholar]
- 52.Shaver, A. C., P. G. Dombrowski, J. Y. Sweeney, T. Treis, R. M. Zappala, and P. D. Sniegowski. 2002. Fitness evolution and the rise of mutator alleles in experimental Escherichia coli populations. Genetics 162:557-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smeets, L. C., N. L. Arents, A. A. van Zwet, C. M. Vandenbroucke-Grauls, T. Verboom, W. Bitter, and J. G. Kusters. 2003. Molecular patchwork: chromosomal recombination between two Helicobacter pylori strains during natural colonization. Infect. Immun. 71:2907-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sniegowski, P. D., P. J. Gerrish, T. Johnson, and A. Shaver. 2000. The evolution of mutation rates: separating causes from consequences. Bioessays 22:1057-1066. [DOI] [PubMed] [Google Scholar]
- 55.Spek, E. J., L. N. Vuong, T. Matsuguchi, M. G. Marinus, and B. P. Engelward. 2002. Nitric oxide-induced homologous recombination in Escherichia coli is promoted by DNA glycosylases. J. Bacteriol. 184:3501-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suerbaum, S., and P. Michetti. 2002. Helicobacter pylori infection. N. Engl. J. Med. 347:1175-1186. [DOI] [PubMed] [Google Scholar]
- 57.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson, S. A., and M. J. Blaser. 1995. Isolation of the Helicobacter pylori recA gene and involvement of the recA region in resistance to low pH. Infect. Immun. 63:2185-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 61.Trobner, W., and R. Piechocki. 1984. Selection against hypermutability in Escherichia coli during long term evolution. Mol. Gen. Genet. 198:177-178. [DOI] [PubMed] [Google Scholar]
- 62.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watanabe, T., J. O. Blaisdell, S. S. Wallace, and J. P. Bond. 2005. Engineering functional changes in Escherichia coli endonuclease III based on phylogenetic and structural analyses. J. Biol. Chem. 280:34378-34384. [DOI] [PubMed] [Google Scholar]
- 64.Yamamoto, F., S. Nishimura, and H. Kasai. 1992. Photosensitized formation of 8-hydroxydeoxyguanosine in cellular DNA by riboflavin. Biochem. Biophys. Res. Commun. 187:809-813. [DOI] [PubMed] [Google Scholar]