Abstract
Rhodopseudomonas palustris is a purple, facultatively phototrophic bacterium that uses hydrogen gas as an electron donor for carbon dioxide fixation during photoautotrophic growth or for ammonia synthesis during nitrogen fixation. It also uses hydrogen as an electron supplement to enable the complete assimilation of oxidized carbon compounds, such as malate, into cell material during photoheterotrophic growth. The R. palustris genome predicts a membrane-bound nickel-iron uptake hydrogenase and several regulatory proteins to control hydrogenase synthesis. There is also a novel sensor kinase gene (RPA0981) directly adjacent to the hydrogenase gene cluster. Here we show that the R. palustris regulatory sensor hydrogenase HupUV acts in conjunction with the sensor kinase-response regulator protein pair HoxJ-HoxA to activate hydrogenase expression in response to hydrogen gas. Transcriptome analysis indicated that the HupUV-HoxJA regulatory system also controls the expression of genes encoding a predicted dicarboxylic acid transport system, a putative formate transporter, and a glutamine synthetase. RPA0981 had a small effect in repressing hydrogenase synthesis. We also determined that the two-component system RegS-RegR repressed expression of the uptake hydrogenase, probably in response to changes in intracellular redox status. Transcriptome analysis indicated that about 30 genes were differentially expressed in R. palustris cells that utilized hydrogen when growing photoheterotrophically on malate under nitrogen-fixing conditions compared to a mutant strain that lacked uptake hydrogenase. From this it appears that the recycling of reductant in the form of hydrogen does not have extensive nonspecific effects on gene expression in R. palustris.
Nitrogenases convert nitrogen gas to ammonia, with the concomitant obligate production of hydrogen, a biofuel (16, 38). The purple, facultatively photosynthetic bacterium Rhodopseudomonas palustris has promise for eventual use in a biological process for hydrogen production because it has three nitrogenase isozymes, can generate the ATP needed for hydrogen production by photophosphorylation, and can access acetate and lignin monomers as sources of electrons for hydrogen synthesis (25, 29). A potential complication of using R. palustris for hydrogen production is that it has a propensity to take up and utilize any available external hydrogen for a variety of different metabolic functions. It takes up hydrogen to supply reductant needed for ammonia production by the process of nitrogen fixation. It uses hydrogen gas as an electron donor for photoautotrophic growth with sodium bicarbonate. In addition, by analogy with work with other purple nonsulfur phototrophs, we would expect R. palustris to use hydrogen gas as an electron supplement to allow the reduction of oxidized organic compounds during photoheterotrophic growth (21). In this way, cells can completely assimilate into biomass compounds such as malate that are relatively more oxidized than cell material.
Hydrogen utilization is typically mediated by uptake hydrogenases that catalyze the oxidation of hydrogen. Electrons derived from this process are then transferred to ferredoxins and cytochromes (43). R. palustris encodes a membrane-bound nickel-iron uptake hydrogenase and many accessory proteins for the synthesis and assembly of this enzyme (25). Manipulating R. palustris to produce hydrogen efficiently will require us to understand how it regulates its uptake hydrogenase. It will also be important to know whether mutations that block hydrogen uptake also cause changes in gene expression related to the inability to recycle reducing power. Finally, we were interested to know whether the regulatory system that directly controls uptake hydrogenase gene expression may also control the expression of other R. palustris genes outside the hydrogenase gene cluster.
We discovered early in our studies that the sequenced strain of R. palustris (strain CGA009) behaved as if it were a hydrogen uptake mutant. Careful inspection of its genome sequence subsequently revealed that strain CGA009 has a frameshift mutation in its hupV gene, which encodes one of the protein components of a predicted HupUV hydrogen sensor protein. To characterize uptake hydrogenase expression in more detail in R. palustris, we repaired the hupV frameshift mutation and generated a strain that was wild type with respect to hydrogenase synthesis. We then used a promoter-lacZ transcriptional fusion to examine hydrogenase gene expression in R. palustris cells growing in several different metabolic modes, and we investigated the effects of several different regulatory mutations on uptake hydrogenase expression. Microarray experiments comparing a hupS structural gene mutant with wild-type cells showed that hydrogen uptake and utilization by cells growing under nitrogen-fixing conditions on malate has a significant effect on the expression of about 30 genes. Other microarray experiments with the hupV mutant suggest that the HupUV hydrogen sensor protein regulates the expression of several sets of genes in addition to hydrogenase uptake genes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. During the course of mutant constructions, R. palustris strains were grown and manipulated aerobically in defined mineral medium (PM) (23) containing 10 mM succinate as a carbon and energy source, with other additions as indicated. R. palustris was grown anaerobically under light at 30°C in PM supplemented with 1 μM NiCl2. For nitrogen-fixing conditions, (NH4)2SO4 was omitted from the PM and cells were grown in sealed tubes with a nitrogen gas headspace. Hydrogen gas was added to the headspace to a concentration of 4 mM (10% hydrogen) or 40 mM (100% hydrogen) as indicated below. For photoheterotrophic growth, sodium malate was added to PM at a final concentration of 10 mM. For photoautotrophic growth, cultures were supplied with 20 mM sodium bicarbonate as the source of inorganic carbon and hydrogen gas (40 mM), or sodium thiosulfate (50 mM final) was supplied as the source of electrons. Escherichia coli strains DH5α and S17-1 were grown at 37°C in Luria-Bertani medium. Where indicated, R. palustris was grown with 100 μg per ml gentamicin (Gm), 100 μg per ml kanamycin (Km), and 10% sucrose. E. coli was grown with 100 μg per ml ampicillin (Ap), 80 μg per ml spectinomycin (Sp), or 20 μg per ml Gm.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Genotype, phenotype, or sequence of primera (5′ to 3′) | Reference, origin, or description |
|---|---|---|
| R. palustris strains | ||
| CGA009 | hupV mutant; spontaneous frameshift (4-bp deletion) | 25 |
| CGA010 | hupV repaired derivative of CGA009; 4 bp inserted in hupV | This study |
| CGA553 | ΔhoxJ mutant of CGA010; 1,095 bp deleted | This study |
| CGA554 | ΔRPA0981 mutant of CGA010; 2,217 bp deleted | This study |
| CGA550 | hupS mutant of CGA010; lacZ-Km cassette inserted in hupS | This study |
| CGA555 | rpoN mutant of CGA010; rpoN::Tn5 | 17 |
| CGA2023 | ΔregSR mutant of CGA010 | F. R. Tabita |
| RCH100 | Environmental isolate of R. palustris | Iowa City, IA |
| RCH200 | Environmental isolate of R. palustris | Iowa City, IA |
| RCH350 | Environmental isolate of R. palustris | Iowa City, IA |
| RCH500 | Environmental isolate of R. palustris | Woods Hole, MA |
| RCH550 | Environmental isolate of R. palustris | Woods Hole, MA |
| E. coli strains | ||
| DH5α | F− λ−recA1 Δ(lacZYA-argF)U169 hsdR17 thi-1 gyrA96 supE44 endA1 relA1 φ80lacZΔM15 | GIBCO-BRL |
| S17-1 | thi pro hdsR hdsM+recA; chromosomal insertion of RP4-2 (Tc::Mu Km::Tn7) | 37 |
| Plasmids | ||
| pJQ200KS | Gmr, sacB; mobilizable suicide vector | 33 |
| pUC19 | Apr; high-copy-number cloning vector | 44 |
| pUTmini-Tn5lacZ1 | Apr, Kmr; delivery plasmid for mini-Tn5 lacZ1 | 7 |
| pHRP309 | Gmr, IncQ; lacZ transcriptional fusion vector | 31 |
| pHRP314 | Apr, Smr, Spr; cohort cloning vector containing a 5.3-kb lacZ-Km cassette | 31 |
| pHRP316 | Apr, Smr, Spr; cohort cloning vector for use with pHRP309 | 31 |
| pHRP316-PhupS | Apr, Smr, Spr; hupS promoter region cloned into BamHI and SmaI sites of pHRP316 | This study |
| pHRP309-PhupS | Gmr, Smr, Spr; hupS-lacZ transcriptional fusion cloned into SmaI and XbaI sites of pHRP309 | This study |
| pUC-ΔhoxJ | Apr; 2.2-kb fragment containing a 1,095 bp in-frame deletion of hoxJ constructed by PCR and cloned into XbaI sites of pUCI9 | This study |
| pJQ-ΔhoxJ | Gmr; 2.2-kb fragment containing a 1,095 bp in-frame deletion of hoxJ constructed by PCR and cloned into XbaI sites of pJQ200KS | This study |
| pUC-hupV | Apr; 1.4-kb fragment containing repaired hupV gene cloned into BamHI sites of pUC19 | This study |
| pJQ-hupV | Gmr; 1.4-kb fragment containing repaired hupV gene cloned into BamHI sites of pJQ200KS | This study |
| pUC-ΔRPA0981 | Apr; 2.1-kb fragment containing a 2,217 bp in-frame deletion of RPA0981 constructed by PCR and cloned into XbaI site of pUC19 | This study |
| pJQ-ΔRPA981 | Gmr; 2.1-kb fragment containing a 1,095 bp in-frame deletion of hoxJ constructed by PCR and cloned into the XbaI site of pJQ200KS | This study |
| pUC-hupS | Apr; 1.3-kb fragment containing hupS and contiguous DNA cloned into the XbaI site of pUC19 | This study |
| pUC-hupS::lacZKm | Apr, Kmr; pUC-hupS containing lacZ-Km cassette from pHRP314 into the BsmI site of hupS | This study |
| pJQ-hupS::lacZKm | Gmr, Kmr; hupS::lacZ-Km fragment cloned into the XbaI site of pJQ200KS | This study |
| Primers | ||
| hupVS1 | AGGATGCACCGCTGTTTGCGG | Sequencing hupV forward primer |
| hupVS2 | ACAATCGCGGATCAGCGGATC | Sequencing hupV reverse primer |
| hupVP1-BamHI | CGGGATCCCGATGACGCGACGGATCACGGTCGG | hupV upstream primer |
| hupVP2-BamHI | CGGGATCCCGTTAGTGCGCGGTGCACACCATGCA | hupV downstream primer |
| hupV-I1 | GCCAGTGCGGCGCGGACGTCGTTCGATCCTGCCGCGATCACCGAGGATGTGG | hupV 4-bp insertion forward primer |
| hupV-I2 | CCACATCCTCGGTGATCGCGGCAGGATCGAACGACGTCCGCGCCGCACTGGC | hupV 4-bp insertion reverse primer |
| hupS-XbaI-1 | GCTCTAGACAGCGCCGCAACCGGCTTATCA | hupS upstream primer |
| hupS-XbaI-2 | GCTCTAGATTAAGCCGACTTGCCGTTGGAG | hupS downstream primer |
| hupS1-BamHI | CGGGATCCCGCTCGAACAAGCGCTGGTTGGCACC | hupS promoter forward primer |
| hupS2-SmaI | TCCCCCGGGGGAGGTGCATTCCAGCCCGTGCATCCA | hupS promoter reverse primer |
| hoxJP1-XbaI | GCTCTAGAGCAAGCGTGCTGCCGAATT | hoxJ upstream primer |
| hoxJP2-XbaI | GCTCTAGAGCCAGGATCGCGCGGAGCT | hoxJ downstream primer |
| hoxJD1 | GTTTACGCCGACCTGCTGCGCACCGGGCTCGGCTTGTGGATC | hoxJ in-frame deletion forward primer |
| hoxJD2 | GATCCACAAGCCGAGCCCGGTGCGCAGCAGGTCGGCGTAAAC | hoxJ in-frame deletion reverse primer |
| RPA0981P1-XbaI | GCTCTAGAGCTGGCGCCACCCGCTGCGA | RPA0981 upstream primer |
| RPA0981P2-XbaI | GCTCTAGAGCAACTGCCTCTAAAGAGCA | RPA0981 downstream primer |
| RPA0981D1 | GAAGTTCAGCGCATCGCCAAATACAGCAGCGTGCTCGCCGAG | RPA0981 in-frame deletion forward primer |
| RPA0981D2 | CTCGGCGAGCACGCTGCTGTATTTGGCGATGCGCTGAACTTC | RPA0981 in-frame deletion reverse primer |
Restriction sites are underlined; the four extra bases that were added to correct the frameshift in CGA009 hupV are in bold.
DNA manipulations.
Standard protocols were used for cloning and transformations. All restriction endonucleases and DNA modification enzymes were purchased from New England Biolabs (Beverly, MA). Shrimp alkaline phosphatase was purchased from Roche Diagnostics Corp. (Indianapolis, IN). PCRs were performed with Herculase DNA polymerase (Stratagene, La Jolla, CA). Chromosomal DNA was purified with the DNeasy tissue kit (QIAGEN Inc., Chatsworth, CA). DNA fragments were excised and purified from agarose gels with the QIAquick gel extraction kit (QIAGEN Inc.). DNA was sequenced at the University of Iowa DNA core facility with standard automated sequencing technology.
Construction of R. palustris mutant strains and repair of the hupV mutation to generate CGA010.
To determine the type of mutation present in the CGA009 hupV gene, we amplified and sequenced a 250-bp fragment of the hupV gene from five different strains of R. palustris by using primers hupVS1 and hupVS2 (Table 1) and compared the sequences to the CGA009 hupV sequence. The frameshift mutation present in CGA009 hupV was repaired by two-step overlap extension PCR (19) with the following modifications. In the first step, primers hupVP1-BamHI and hupV-I2 were used to PCR amplify the region upstream from the frameshift (∼0.8 kb), and primers hupVP2-BamHI and hupV-I1 were used to amplify the region downstream from the frameshift (∼0.6 kb). Primers hupV-I1 and hupV-I2 are complementary to each other and bind to hupV at both flanking ends of the frameshift. They contain four extra bases that were added to correct the frameshift (the four inserted bases are in bold type in Table 1). In a second step, a mixture of the two PCR products (100 ng each) was used as the template in a third PCR amplification using the primers hupVP1-BamHI and hupVP2-BamHI. The product of the third amplification contained the 4-bp insertion in the hupV gene that is necessary to restore the correct reading frame. This PCR product was approximately 1.4 kb and contained engineered BamHI sites at its 5′ and 3′ ends. It was digested with BamHI and ligated into BamHI-digested pUC19, yielding the plasmid pUC-hupV. The 1.4-kb BamHI fragment was then subcloned into BamHI-digested pJQ200KS, giving pJQ-hupV. This plasmid was mobilized from E. coli S17-1 into R. palustris CGA009 by conjugation. Colonies that contained plasmids that had undergone a single recombination to become inserted into the chromosome were identified by growth on PM plus Gm. These colonies were streaked onto PM plates supplemented with 10% sucrose to identify strains that had undergone a double recombination to lose the sacB-containing vector. Colonies that contained the repaired hupV gene were confirmed by PCR and sequencing. A strategy similar to that used to repair hupV was used to generate in-frame hoxJ and RPA0981 deletion mutants. PCR primers and recombinant plasmids are described in Table 1.
To generate a hupS mutant, a 1.3-kb DNA fragment containing the hupS gene plus approximately 0.5 kb of upstream and downstream flanking DNA was generated by PCR. The amplification product contained engineered XbaI cloning sites at both ends. This product was then digested with XbaI and cloned into XbaI-digested pUC19 to generate pUC-hupS. A 5.3-kb fragment containing a lacZ-Km cassette was excised from EcoRI-digested pHRP314, treated with Klenow fragment to create blunt ends, and ligated into BsmI-digested and alkaline phosphatase-treated pUC-hupS to generate pUC-hupS::lacZ-Km. The correct orientation of the cassette within the hupS gene was verified by PCR. A 6.6-kb fragment containing hupS::lacZ-Km was excised from pUC-hupS by digestion with XbaI and cloned into XbaI-digested pJQ200KS to generate pJQ-hupS::lacZ-Km. This construct was mobilized from E. coli S17-1 into R. palustris by conjugation. Sucrose-resistant, Km-resistant colonies were selected and screened by PCR for the loss of the hupS wild-type gene and presence of hupS::lacZ-Km. An rpoN::Tn5 mutant was identified from an R. palustris mini-Tn5-lacZ1 mutant library (17).
Construction of a hupS promoter-lacZ transcriptional fusion plasmid.
A reporter plasmid containing a promoterless lacZ gene fused to the hupS promoter region was constructed by a two-step cloning procedure, as described previously (32). Briefly, a 0.6-kb DNA fragment containing the promoter region of hupS was amplified by PCR with the hupS1 and hupS2 primers. The PCR product contained engineered BamHI and SmaI sites on its 5′ and 3′ ends, respectively. This product was digested with BamHI and SmaI and directionally ligated into BamHI-SmaI-digested pHRP316, adjacent to a Ω(Smr/Spr) cassette. This resulted in pHR316-PhupS. Fragments containing the Ω(Smr/Spr) cassette and the promoter region were then cut out and inserted upstream of a promoterless lacZ gene in pHRP309 to create pHRP309-PhupS. This was confirmed by restriction analysis and sequencing. pHRP309-PhupS was first moved into E. coli S17-1 by transformation and then mobilized into R. palustris by conjugation. Transconjugants were selected by growth on PM plus Gm and confirmed by colony PCR. β-Galactosidase activity was measured as previously described (9).
Hydrogen measurements.
Hydrogen was measured with a Hewlett Packard 5890 series II gas chromatograph equipped with a thermal conductivity detector and a Molecular Sieve 13X column (80/100 mesh; inner diameter, 0.25 in. by 8 ft). The temperatures of the oven, injector, and detector were 50°C, 100°C, and 100°C, respectively. Hamilton (Reno, NV) sample lock syringes were used to inject gas samples into the gas chromatograph. Protein concentrations were determined with the Bio-Rad (Richmond, CA) protein assay kit.
Transcriptome analyses.
Transcriptome analyses were carried out with glass slide microarrays prepared as described previously (29). R. palustris strains were subcultured at least twice after initial inoculation from a plate. Cells were grown to an optical density at 660 nm of 0.25 to 0.35 (mid-logarithmic phase), chilled in an ice-water bath, harvested by centrifugation, and frozen at −80°C for RNA isolation at a later time. Thawed cells were disrupted, and RNA was isolated and its quality checked as described previously (29). Labeled cDNA was prepared by direct incorporation of either the Cy3-dCTP or Cy5-dCTP fluorophore (Amersham Biosciences) during a first-strand reverse transcription reaction. Each 45-μl reaction mixture contained 12 μg of total RNA; 13.5 μg of random primers (Invitrogen); 9 μl of 5× SuperScript II RT reaction buffer (Invitrogen); 10 mM dithiothreitol; 0.5 mM (each) dATP, dGTP, and dTTP; 0.2 mM dCTP; 40 U of RNasin (Promega); 3 μl of 1 mM Cy3- or 1 mM Cy5-dCTP; and 600 U of SuperScript II reverse transcriptase (Invitrogen). After a 2-h incubation at 42°C, 15 μl of 0.5 M EDTA (pH 8.0) and 15 μl of 1 M NaOH were added to the sample, and incubation was continued at 65°C for 30 min. The sample was then neutralized by the addition of 30 μl of 3 M sodium acetate (pH 5.2) and 45 μl of H2O to bring the volume to 150 μl. The labeled cDNA was purified with the QIAquick PCR purification kit (QIAGEN). The labeling efficiency was calculated by measuring the A260 and either the A550 for Cy3 incorporation or the A650 for Cy5 incorporation. Prior to the hybridizations, the array slides were incubated in prehybridization buffer as described previously (29). Hybridizations with fluorescently labeled cDNA were performed, slides were scanned, and data analysis was carried out as described previously (29). Genes whose ratios were greater than or equal to 2 and whose scores were less than 0.025 were considered to be expressed at higher levels (30). Genes whose ratios were less than 0.5 and whose scores were greater than 0.975 were considered to be expressed at lower levels (30).
Microarray data accession number.
The microarray data have been deposited at http://www.ncbi.nlm.nih.gov/geo under accession number GSE4320.
RESULTS
Uptake hydrogenase gene cluster in R. palustris.
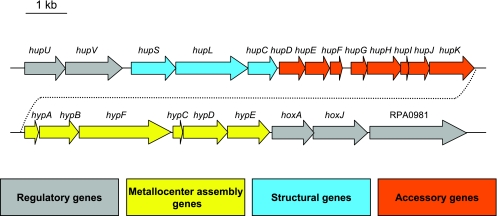
The R. palustris strain CGA009 genome has a cluster of 22 genes encoding proteins for the regulation, synthesis, and assembly of a nickel-iron membrane-bound uptake hydrogenase (Fig. 1). Three of these genes, hupS, hupL, and hupC, are predicted to encode the structural subunits of the uptake hydrogenase. Genes for accessory proteins needed for hydrogenase assembly (hup and hyp genes) are located downstream from the structural genes. A similar arrangement of genes is seen in Bradyrhizobium japonicum (4, 41). R. palustris has homologues of a set of uptake hydrogenase regulatory genes that has been characterized in Rhodobacter capsulatus (10, 11, 13), Ralstonia eutropha (3, 24, 26, 27), and B. japonicum (1, 40). It has hupU (RPA0959) and hupV (RPA0960) genes predicted to encode a hydrogen sensor protein. hoxJ (RPA0980) encodes a histidine kinase predicted to interact with HupUV, and hoxA (RPA0979) is predicted to encode a response regulator-transcription factor that is cognate to HoxJ (Fig. 1). R. palustris has an additional gene (RPA0981) adjacent to hoxJ that is a predicted sensor histidine kinase with three PAS domains. This gene is not found to be associated with hydrogenase genes in any other organism sequenced to date, although B. japonicum does have an orthologue of RPA0981.
FIG. 1.
Organization of the uptake hydrogenase gene cluster in R. palustris (RPA0959 to RPA0981). Arrows indicate the direction of transcription. Functions were assigned based on deduced similarities to known proteins (41, 43).
The sequenced R. palustris strain, CGA009, is defective in hydrogen utilization due to a hupV frameshift mutation.
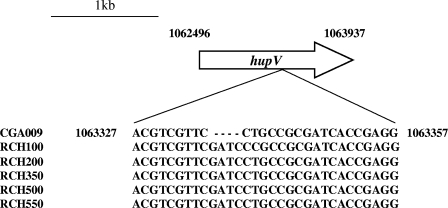
Although it has uptake hydrogenase genes, strain CGA009 was not able to grow photoautotrophically using hydrogen as an electron donor (Fig. 2A). Since CGA009 was able to grow photoautotrophically using sodium thiosulfate as an electron donor (Fig. 2B), we concluded that this strain must be defective in uptake hydrogenase activity. Careful inspection of the CGA009 genome sequence subsequently revealed a frameshift mutation in the hupV gene. Such a mutation would explain the hydrogen utilization phenotype if one assumes that the predicted R. palustris HupUV sensor hydrogenase is required to activate hydrogenase gene expression. To test this further and to test the effects of various growth conditions and other regulatory mutations on hydrogenase expression, we repaired the hupV mutation to generate an R. palustris strain that had wild-type hydrogenase activity. To determine the exact nature and extent of the mutation in CGA009, we amplified and sequenced a 250-bp fragment encompassing the region of the hupV frameshift from five different R. palustris strains that had been isolated from various soil and water samples. Alignment of these sequences indicated that the hupV gene from strain CGA009 had a 4-bp deletion (Fig. 3). Using this information, we repaired hupV in strain CGA009 to generate strain CGA010. R. palustris strain CGA010 grew photoautotrophically with hydrogen as an electron donor (Fig. 2A). It also generated less hydrogen when grown under nitrogen-fixing conditions than the hupV frameshifted strain (Table 2). Since one of the functions of uptake hydrogenases in bacteria is to recapture hydrogen produced during nitrogen fixation for use as an electron donor to the nitrogenase, the net amount of hydrogen produced by cells grown under nitrogen-fixing conditions serves as an indirect measure of uptake hydrogenase activity. The amount of hydrogen generated by the hupV mutant was similar to that of a hydrogenase structural gene mutant, CGA550 (hupS), that we constructed (Table 2). We refer to CGA010 as the wild type for the remainder of this paper.
FIG. 2.
Photoautotrophic growth of R. palustris strains CGA010 and CGA009 in the presence of hydrogen (A) or sodium thiosulfate (B). Data are representative of three different experiments.
FIG. 3.
Alignment of the R. palustris strain CGA009 hupV gene frameshifted region with the corresponding hupV gene regions from five other strains of R. palustris. Numbers indicate the location in the strain CGA009 genome sequence. The hupV frameshift was probably acquired during laboratory cultivation.
TABLE 2.
Hydrogen production, doubling times, and protein yields of R. palustris wild type and hydrogen uptake mutants grown under nitrogen-fixing conditionsa
| Carbon source utilized (concn) | O/H ratiob | H2 produced (μmol/mg protein)
|
Doubling time (h)
|
Protein yield (μg/10-ml culture)
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CGA009 (hupV) | CGA010 (wild type) | CGA550 (hupS) | CGA009 (hupV) | CGA010 (wild type) | CGA550 (hupS) | CGA009 (hupV) | CGA010 (wild type) | CGA550 (hupS) | ||
| Malate (10 mM) | 1.25 | 120 ± 20 | <1 | 110 ± 40 | 13.5 ± 1.3 | 11.5 ± 0.7 | 15.0 ± 2.3 | 500 ± 40 | 740 ± 20 | 600 ± 100 |
| Succinate (10 mM) | 1.0 | 140 ± 45 | 25 ± 35 | 125 ± 2 | 11.5 ± 0.2 | 9.5 ± 0.5 | 11.0 ± 0.7 | 740 ± 170 | 1150 ± 80 | 820 ± 200 |
| Acetate (20 mM) | 0.67 | 200 ± 45 | 110 ± 60 | 210 ± 25 | 7.5 ± 0.4 | 6.5 ± 0.8 | 8.0 ± 0.3 | 800 ± 120 | 970 ± 80 | 780 ± 250 |
Data were acquired during stationary phase after all available carbon had been utilized. Data are averages from three different experiments, plus or minus standard deviations.
The O/H ratio is the ratio of the number of oxygen atoms to the number of H atoms in the carbon source being tested.
Effects of metabolic context on hydrogenase gene expression.
We assessed the effects of various growth conditions and mutations on hydrogenase expression by measuring β-galactosidase levels in cells carrying a PhupS-lacZ reporter plasmid. The levels of PhupS-lacZ expression in wild-type cells increased as the amount of hydrogen in the headspace of cultures grown photoheterotrophically with malate was increased. As shown in Table 3, 10% and 100% hydrogen in the headspace resulted in 4-fold and 10-fold increases, respectively, in β-galactosidase activity compared to cells grown photoheterotrophically without hydrogen present. Hydrogen is not the only inducing signal, however, because PhupS-lacZ expression was activated to fourfold-higher levels in cells grown photoautotrophically with 100% hydrogen than in cells grown photoheterotrophically with 100% hydrogen. In addition, the expression levels of PhupS-lacZ were about the same in cells grown photoheterotrophically with malate under non-nitrogen-fixing conditions with 100% hydrogen present as in cells grown photoheterotrophically with malate under nitrogen-fixing conditions, a condition under which there is no net accumulation of hydrogen in the headspace of culture tubes (Table 2).
TABLE 3.
Effects of growth conditions on hupS promoter-lacZ expressiona
| Growth conditionb | β-Galactosidase activity (nmol/min/mg of protein)
|
||||
|---|---|---|---|---|---|
| CGA010 (wild type) | CGA009 (hupV) | CGA553 (hoxJ) | CGA554 (RPA0981) | CGA2023 (regSR) | |
| Photoheterotrophic | 250 ± 60 | 290 ± 120 | 6,200 ± 700 | 410 ± 100 | 1,700 ± 400 |
| Photoheterotrophic, 10% H2 | 1,100 ± 300 | 250 ± 130 | 6,500 ± 1,000 | 2,000 ± 900 | 5,800 ± 1,500 |
| Photoheterotrophic, 100% H2 | 2,800 ± 400 | 330 ± 230 | 6,500 ± 700 | 4,500 ± 100 | 9,200 ± 1,900 |
| Photoheterotrophic, N2 fixing | 2,600 ± 900 | 350 ± 110 | 6,800 ± 600 | 4,300 ± 500 | 7,200 ± 1,100 |
| Photoautotrophic, 100% H2 | 11,200 ± 1,300 | ND | 8,900 ± 200 | 12,100 ± 3,700 | 16,100 ± 600 |
β-Galactosidase activities expressed by cultures of CGA010 (wild type), CGA009 (hupV), CGA553 (hoxJ), CGA554 (ΔRPA0981), and CGA2023 (regSR) harboring pHRP309-PhupS. Data are averages of three experiments, each carried out in duplicate, plus or minus standard deviations. ND, not determined.
Photoheterotrophic cultures were grown in PM containing 10 mM sodium malate as the carbon source, without hydrogen, or with 10% or 100% hydrogen in the headspace of culture tubes. Nitrogen-fixing cultures were grown in nitrogen-fixing medium. Photoautotrophic cultures were grown in PM containing 20 mM NaHCO3 as the carbon source and 100% hydrogen as the electron donor.
The HupUV-HoxJA system responds to hydrogen to activate hydrogenase synthesis.
Whereas PhupS-lacZ expression levels were induced about 10-fold when 100% hydrogen was present in cultures of wild-type cells growing photoheterotrophically with malate as a carbon source, uninduced levels of PhupS-lacZ expression were observed in the hupV mutant strain CGA009 grown under the same conditions (Table 3). This indicates that the HupUV sensor hydrogenase is required to activate hydrogenase gene expression. Deletion of the hoxJ sensor kinase gene resulted in constitutively high levels of β-galactosidase expression in all growth modes tested (Table 3). This suggests that, in the absence of hydrogen, HoxJ plays a role in inhibiting transcription of hup genes, probably by interacting with its cognate response regulator protein, HoxA.
RpoN is required for the expression of the R. palustris uptake hydrogenase.
The R. palustris HoxA protein is a predicted RpoN-dependent transcriptional activator with a central AAA+ domain and an N-terminal helix-turn-helix DNA binding motif. We observed basal levels of PhupS-lacZ expression in rpoN mutant (strain CGA555) cells grown photoheterotrophically on malate with 10% hydrogen (data not shown). This indicates that RpoN is required for the expression of the uptake hydrogenase in R. palustris.
A novel sensor kinase protein has a small effect in repressing hydrogenase expression.
A mutant disrupted in the PAS domain-containing sensor kinase RPA0981 exhibited a small elevation of PhupS-lacZ gene expression compared to wild-type cells in some metabolic contexts. The most pronounced effect of the mutation was on hydrogenase expression in cells grown photoheterotrophically with 100% hydrogen (Table 3).
The RegS-RegR two-component regulatory system represses hydrogenase expression.
The observation that the PhupS-lacZ fusion was expressed to high levels independently of hydrogen concentration during nitrogen fixation and carbon dioxide fixation suggested that another regulatory system, in addition to the HupUV-HoxJA system, must control hydrogenase synthesis in R. palustris. Since these processes each require large amounts of reducing equivalents, we investigated the possible involvement of RegSR, an R. palustris two-component system that is homologous to the RegB-RegA and PrrB-PrrA two-component global regulatory systems that are thought to respond to intracellular redox status to regulate gene expression in the purple nonsulfur bacteria R. capsulatus and R. sphaeroides (8, 39). RegBA and PrrBA mutants have pleiotropic growth defects (15, 28). These systems have been shown to activate expression of photosynthesis, carbon fixation, and nitrogen fixation genes and to repress hupS expression (8, 14, 20). RegS is a predicted membrane-bound sensor histidine kinase, and RegR is its cognate response regulator-transcription factor. An R. palustris regSR deletion mutant grows as the wild type under photoheterotrophic, photoautotrophic, and nitrogen-fixing conditions (S. Romagnoli and F. R. Tabita, personal communication; F. E. Rey, S. K. Samanta, and C. S. Harwood, unpublished results). This indicates that, in contrast to Rhodobacter, RegSR does not play a pivotal role in global gene regulation in R. palustris. Because the regSR mutant did not have any obvious growth defects, we were able to test the effect of the RegSR regulatory system on hydrogenase gene expression in cells grown in different metabolic modes. regSR mutant cells harboring PhupS-lacZ showed higher β-galactosidase activities than did wild-type cells under all conditions tested, indicating that, as in R. capsulatus, RegSR represses hydrogenase synthesis. There was an inverse correlation between the degree of repression by RegSR and the amount of reductant used by cells in a particular growth mode (Table 3). RegSR had the smallest repressive effect on hupS expression in cells grown under nitrogen-fixing or photoautotrophic conditions.
Microarray data indicate that the HupUV-HoxJA system activates expression of a dicarboxylic acid transporter, a formate transporter, and a glutamine synthetase in addition to hydrogenase genes.
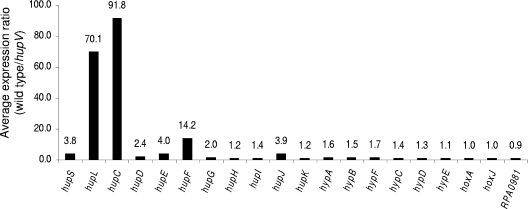
We used transcriptome analysis to identify genes that were differentially expressed between wild-type (CGA010) and hupV mutant (CGA009) cells grown photoheterotrophically with malate under nitrogen-fixing conditions. We also compared the transcriptome of a hupS mutant with that of the wild type. As expected, hupV was necessary for high levels of transcription of the hupSLC structural genes. The auxiliary genes hupDEFGHIJK are also positively regulated by this system but to a lesser degree. Furthermore, the expression of genes encoding the auxiliary Hyp proteins showed a very small dependence or no dependence at all on hupV (Fig. 4). Genes located outside the hydrogenase gene cluster that showed higher levels of expression in wild-type cells than in the hupV mutant were additional candidates that might be directly controlled by the HupUV-HoxJA regulatory system. In order to exclude genes that showed increased expression in wild-type cells due to indirect effects of hydrogen uptake and recycling, we included only genes that were not expressed at higher levels in wild-type cells than in a hupS mutant (nine candidate genes) and whose expression was dependent on RpoN (seven candidate genes) (Y. Oda and C. S. Harwood, unpublished results). Finally, we retained only those genes whose expression patterns were validated in independent experiments using an Affymetrix microarray platform that compared the wild type and the hupV mutant. Genes that met these criteria are listed in Table 4. These include genes for a putative dicarboxylic acid uptake system, a formate transporter, and a glutamine synthetase. In addition, we found one gene (RPA2297) that showed decreased expression levels in the wild type in a hupV-, rpoN-dependent fashion. RPA2297 encodes a conserved, unknown protein. Its expression level was 47-fold lower in wild-type cells than in the hupV mutant.
FIG. 4.
Average expression ratio (wild type/hupV) of genes in the uptake hydrogenase gene cluster. Cells were grown under nitrogen-fixing conditions in the presence of 10 mM malate. Data are averages of duplicates from three different experiments.
TABLE 4.
Genes unlinked to the hydrogenase gene cluster that are regulated by hupV
| RPA no. | Gene name | Avg expression ratio (wild type/hupV mutant)a | Annotation |
|---|---|---|---|
| 1976 | 5.9 (6.4) | Putative dicarboxylic acid transporter subunit | |
| 1977 | 8.5 (5.3) | Putative dicarboxylic acid transporter subunit | |
| 3201 | 2.3 (2.0) | Formate transporter | |
| 4209 | glnAII | 3.5 (2.0) | Glutamine synthetase |
Cells were grown under nitrogen-fixing conditions in the presence of 10 mM malate. Data are averages of duplicates from three different experiments. Expression changes determined in an independent Affymetrix microarray analysis are shown in parentheses.
Hydrogen uptake and recycling affects the expression of a small number of R. palustris genes.
Hydrogen recycling enables R. palustris to grow slightly faster and to reach a higher cell yield under nitrogen-fixing conditions (Table 2). However, a relatively small number of genes were differentially expressed between the wild type and the hupS mutant grown photoheterotrophically with malate under nitrogen-fixing conditions (Table 5). Similar changes in the expression of these genes were also detected in the microarray comparison of wild-type and hupV mutant strains.
TABLE 5.
Effects of hydrogen uptake on gene expression
| RPA or operon no.a | Gene name | Avg expression ratio (wild type/hupS)b | Annotation |
|---|---|---|---|
| 0274-0275 | glnK2-amtB2 | 2.3 | Nitrogen PII regulatory protein-ammonia transporter |
| 0429 | katG | 0.4 | Catalase/peroxidase |
| 0713-0718 | cobUWNO | 2.5 | Cobalamin biosynthesis |
| 1063 | 5.2 | Conserved hypothetical protein | |
| 1886 | 0.4 | Hypothetical protein | |
| 2061 | nosZ | 2.5 | Nitrous oxide reductase precursor |
| 2083-2086 | cobBM cbiG cobL | 2.1 | Cobalamin biosynthesis |
| 2094-2097 | cobTQF | 9.2 | Cobalamin biosynthesis |
| 2116 | 0.4 | Hypothetical protein | |
| 2117 | 0.4 | Putative flavodoxin | |
| 2121 | 0.5 | Conserved hypothetical | |
| 2977 | nrd | 2.3 | Ribonucleotide reductase |
| 3329 | 0.4 | Conserved hypothetical | |
| 3665-3669 | 2.5 | Urea transport | |
| 4222 | 0.5 | Hypothetical protein | |
| 4803 | 3.1 | Outer membrane siderophore receptor |
RPAs forming putative operons are grouped, and the values shown represent the gene with the highest average expression ratio.
Cells were grown under nitrogen-fixing conditions in the presence of 10 mM malate. Data are averages of duplicates from three different experiments.
DISCUSSION
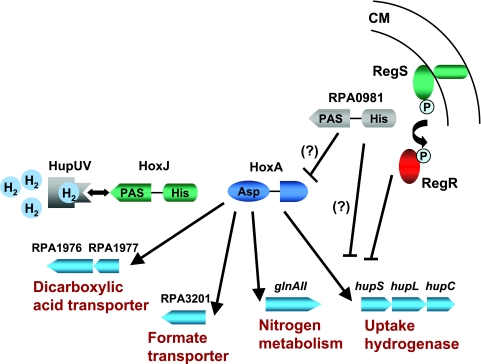
Our results suggest that the signal transduction system that regulates the expression of the uptake hydrogenase in R. palustris in response to hydrogen resembles that described for R. eutropha, an obligate aerobe (26, 27). We predict that when it binds hydrogen, the R. palustris HupUV protein transmits a signal to the histidine kinase, HoxJ, to prevent its autophosphorylation. This results in the generation of an unphosphorylated cognate response regulator (HoxA) that is proficient at activating transcription of hydrogenase genes (Fig. 5). By contrast, the HupUV sensor hydrogenase from the more closely related purple nonsulfur bacterium R. capsulatus represses expression of the uptake hydrogenase (10, 42). In the absence of hydrogen, HupUV from R. capsulatus interacts with HupT (HoxJ homologue) to increase its kinase activity. This results in phosphorylated inactive HupR (HoxA homologue) (42). This repression is relieved by the presence of hydrogen (42). In R. palustris and R. eutropha, the alternative RNA polymerase sigma factor RpoN is required to activate hydrogenase expression (36) (Oda and Harwood, unpublished data), whereas in R. capsulatus, hydrogenase expression depends on the housekeeping sigma factor sigma 70 (6).
FIG. 5.
Regulation of the uptake hydrogenase in R. palustris. The HupUV-HoxJA system activates expression of the uptake hydrogenase and other genes indicated in response to hydrogen. RegSR and, to a lesser extent, RPA0981 negatively modulate expression of the uptake hydrogenase, possibly in response to reducing conditions. The hydrogen-dependent regulatory system is similar to that of R. eutropha (27). CM, cytoplasmic membrane.
In Rhodobacter species, a redox-responsive two-component system (RegB-RegA/PrrB-PrrA) regulates expression of processes that modify the cellular redox status. The RegBA/PrrBA systems activate expression of genes for carbon dioxide fixation and nitrogen fixation, processes that utilize reducing equivalents, and the RegBA system has been shown to repress expression of genes for hydrogen oxidation, a process that generates reducing equivalents (8, 12, 14, 20). The homologous RegS-RegR system in R. palustris differs in that it does not appear to play a role in activating carbon dioxide fixation or nitrogen fixation. Our results do indicate, however, that R. palustris RegSR represses uptake hydrogenase expression in a manner that is inversely correlated with the cellular demand for reducing equivalents (Table 3). RegSR-mediated repression of uptake hydrogenase may prevent cells from becoming over-reduced.
The phenotype of an RPA0981 mutant resembles that of the regSR mutant but is less pronounced. That is, this putative sensor kinase appears to repress hydrogenase expression. The presence of PAS domains in RPA0981 suggests that it may function to sense some aspect of redox. Because of the close physical proximity of its gene on the chromosome, HoxA is a likely candidate to be the transcriptional activator cognate to RPA0981. If so, RPA0981 would be predicted to negatively modulate expression of the uptake hydrogenase by phosphorylating HoxA.
Our microarray data indicate, as expected, that the HupUV-HoxJA system regulates expression of the hydrogenase structural genes hupSLC. We were surprised to see that the average level of induction of hupS was smaller than that of hupL and hupC, because these three genes are predicted to be in an operon and the small and large subunits of the uptake hydrogenase form an heterodimer (41). One possibility is that segmental mRNA degradation is taking place (34). Expression of other genes in the hydrogenase cluster did not heavily depend on hupV (Fig. 4). The proteins encoded by these genes are likely necessary for the assembly of the metallocenters of both the sensor hydrogenase and the uptake hydrogenase (2, 5). Thus, they may need to be present at all times in order for a fully functional sensor hydrogenase to be made and ready to activate gene expression in response to hydrogen. We also found, unexpectedly, that HupUV-HoxJA regulates expression of genes for a predicted TRAP transporter for dicarboxylic acids (RPA1976-RPA1977), a formate transporter (RPA3201), and a glutamine synthetase (glnAII) (Table 4). TRAP transporters are tripartite proteins that typically include two integral membrane proteins and a periplasmic binding protein (22). A periplasmic binding protein gene is divergently transcribed from RPA1976-RPA1977 and does not appear to be controlled by HupUV-HoxJA. Formate and dicarboxylic acids, such as malate, are relatively oxidized compounds that can be completely incorporated into cell material under phototrophic growth conditions only if an exogenous reductant, such as hydrogen, is present as a cosubstrate. The induction of dicarboxylic acid and formate transport by hydrogen would be predicted to allow cells to attain higher growth yields on these compounds. In fact, we did observe that wild-type cells grew to higher cell densities on malate than did a hupV mutant when hydrogen was present (Table 2). Likewise, more reductant available from hydrogen probably results in more nitrogen converted into ammonia, which is incorporated into cell material by means of glutamine synthetases. R. palustris encodes four different glutamine synthetases (25), and GlnAII is the prevalent isozyme expressed by wild-type cells under nitrogen-fixing conditions (29). Although glnAII is mainly controlled by nitrogen starvation in an NtrB-NtrC-RpoN-dependent fashion (Oda and Harwood, unpublished results), our results suggest that the HupUV-HoxJA system also regulates its expression (Table 4).
Since hydrogen is produced along with ammonia as a product of nitrogen fixation, an uptake hydrogenase mutation results in a dramatic increase in the net amount of hydrogen that cultures accumulate under nitrogen-fixing conditions (Table 2). We reasoned that a transcriptome analysis of a hupS mutant would provide useful information about metabolic constraints that might be imposed on engineered, hydrogen-producing cells that are unable to take up and recycle hydrogen. About 30 genes were differentially expressed between wild-type and hupS mutant cells grown on malate under nitrogen-fixing conditions (Table 5). In most cases, the differences in expression levels were small. Many of the genes that were expressed at lower levels in wild-type cells than in hydrogen uptake mutant cells belonged to the hypothetical or conserved hypothetical category. Wild-type cells expressed higher levels of a high-affinity ammonium transporter (amtB), a nitrogen regulatory PII protein (glnK2), and a urea transporter (RPA3665-RPA3669) than did hupS mutant cells. This may reflect that cells sense a need to acquire more fixed nitrogen, since the ability to utilize hydrogen allows them to grow to higher cell yields with malate. Genes involved in cobalamin (vitamin B12) biosynthesis were also expressed at higher levels in the wild type, as was a gene encoding a predicted vitamin B12-dependent ribonucleotide reductase (nrd) (Table 5). Nrd converts ribonucleotide diphosphates into 2′ deoxyribonucleotides for DNA synthesis and repair (35). Since the reaction catalyzed by Nrd requires reductant (18), it seems possible that the expression of this gene is activated in response to the availability of reductant. Alternatively, the slightly faster doubling time exhibited by wild-type cells under nitrogen-fixing conditions with malate (Table 2) may result in an increased demand for deoxyribonucleotides for DNA synthesis. Finally, increased expression of the outer membrane siderophore receptor RPA4803 indicates an increased requirement for iron, possibly for uptake hydrogenase synthesis. Taken together, our results indicate that the small changes in gene expression that were seen in wild-type cells relative to the uptake hydrogenase mutant are related to the requirements for making a functional hydrogenase, or they provide a minor increase in the ability to access reducing equivalents to allow better growth on the relatively oxidized carbon source, malate. Although hydrogen recycling does not appear to have a major global effect on gene expression, the presence of hydrogen induces expression of the hydrogenase genes and several other genes that enable R. palustris to grow slightly faster and to higher yields under nitrogen-fixing conditions, especially on relatively oxidized carbon sources such as malate.
Acknowledgments
This research was supported by the Office of Science (BER), U.S. Department of Energy, grants DE-FG02-01ER63241 and DE-FG02-05ER64063, and by a predoctoral fellowship (to F.E.R.) through the Iowa Center for Biocatalysis and Bioprocessing (University of Iowa).
We thank F. Robert Tabita for sharing strain CGA2023 and Jizhong Zhou for preparing the R. palustris microarray slides.
REFERENCES
- 1.Black, L. K., C. Fu, and R. J. Maier. 1994. Sequences and characterization of hupU and hupV genes of Bradyrhizobium japonicum encoding a possible nickel-sensing complex involved in hydrogenase expression. J. Bacteriol. 176:7102-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buhrke, T., B. Bleijlevens, S. P. Albracht, and B. Friedrich. 2001. Involvement of hyp gene products in maturation of the H2-sensing [NiFe] hydrogenase of Ralstonia eutropha. J. Bacteriol. 183:7087-7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buhrke, T., O. Lenz, A. Porthun, and B. Friedrich. 2004. The H2-sensing complex of Ralstonia eutropha: interaction between a regulatory [NiFe] hydrogenase and a histidine protein kinase. Mol. Microbiol. 51:1677-1689. [DOI] [PubMed] [Google Scholar]
- 4.Casalot, L., and M. Rousset. 2001. Maturation of the [NiFe] hydrogenases. Trends Microbiol. 9:228-237. [DOI] [PubMed] [Google Scholar]
- 5.Colbeau, A., S. Elsen, M. Tomiyama, N. A. Zorin, B. Dimon, and P. M. Vignais. 1998. Rhodobacter capsulatus HypF is involved in regulation of hydrogenase synthesis through the HupUV proteins. Eur. J. Biochem. 251:65-71. [DOI] [PubMed] [Google Scholar]
- 6.Colbeau, A., and P. M. Vignais. 1992. Use of hupS::lacZ gene fusion to study regulation of hydrogenase expression in Rhodobacter capsulatus: stimulation by H2. J. Bacteriol. 174:4258-4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubbs, J. M., and F. R. Tabita. 2004. Regulators of nonsulfur purple phototrophic bacteria and the interactive control of CO2 assimilation, nitrogen fixation, hydrogen metabolism and energy generation. FEMS Microbiol. Rev. 28:353-376. [DOI] [PubMed] [Google Scholar]
- 9.Egland, P. G., and C. S. Harwood. 1999. BadR, a new MarR family member, regulates anaerobic benzoate degradation by Rhodopseudomonas palustris in concert with AadR, an Fnr family member. J. Bacteriol. 181:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsen, S., A. Colbeau, J. Chabert, and P. M. Vignais. 1996. The hupTUV operon is involved in negative control of hydrogenase synthesis in Rhodobacter capsulatus. J. Bacteriol. 178:5174-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elsen, S., A. Colbeau, and P. M. Vignais. 1997. Purification and in vitro phosphorylation of HupT, a regulatory protein controlling hydrogenase gene expression in Rhodobacter capsulatus. J. Bacteriol. 179:968-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elsen, S., W. Dischert, A. Colbeau, and C. E. Bauer. 2000. Expression of uptake hydrogenase and molybdenum nitrogenase in Rhodobacter capsulatus is coregulated by the RegB-RegA two-component regulatory system. J. Bacteriol. 182:2831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsen, S., O. Duche, and A. Colbeau. 2003. Interaction between the H2 sensor HupUV and the histidine kinase HupT controls HupSL hydrogenase synthesis in Rhodobacter capsulatus. J. Bacteriol. 185:7111-7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elsen, S., L. R. Swem, D. L. Swem, and C. E. Bauer. 2004. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol. Mol. Biol. Rev. 68:263-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eraso, J. M., and S. Kaplan. 1994. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J. Bacteriol. 176:32-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher, K., and W. E. Newton. 2002. Nitrogen fixation—a general overview, p. 1-34. In G. J. Leigh (ed.), Nitrogen fixation at the millennium. Elsevier, Amsterdam, The Netherlands.
- 17.Harrison, F. H. 2005. Peripheral pathways of anaerobic benzoate degradation in Rhodopseudomonas palustris. Ph.D. thesis. University of Iowa, Iowa City.
- 18.Holmgren, A. 1989. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 264:13963-13966. [PubMed] [Google Scholar]
- 19.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 20.Joshi, H. M., and F. R. Tabita. 1996. A global two component signal transduction system that integrates the control of photosynthesis, carbon dioxide assimilation, and nitrogen fixation. Proc. Natl. Acad. Sci. USA 93:14515-14520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelley, B. C., C. M. Meyer, C. Gandy, and P. M. Vignais. 1977. Hydrogen recycling by Rhodopseudomonas capsulata. FEBS Lett. 81:281-285. [DOI] [PubMed] [Google Scholar]
- 22.Kelly, D. J., and G. H. Thomas. 2001. The tripartite ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol. Rev. 25:405-424. [DOI] [PubMed] [Google Scholar]
- 23.Kim, M.-K., and C. S. Harwood. 1991. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol. Lett. 83:199-204. [Google Scholar]
- 24.Kleihues, L., O. Lenz, M. Bernhard, T. Buhrke, and B. Friedrich. 2000. The H2 sensor of Ralstonia eutropha is a member of the subclass of regulatory [NiFe] hydrogenases. J. Bacteriol. 182:2716-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larimer, F. W., P. Chain, L. Hauser, J. Lamerdin, S. Malfatti, L. Do, M. L. Land, D. A. Pelletier, J. T. Beatty, A. S. Lang, F. R. Tabita, J. L. Gibson, T. E. Hanson, C. Bobst, J. L. Torres, C. Peres, F. H. Harrison, J. Gibson, and C. S. Harwood. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 22:55-61. [DOI] [PubMed] [Google Scholar]
- 26.Lenz, O., M. Bernhard, T. Buhrke, E. Schwartz, and B. Friedrich. 2002. The hydrogen-sensing apparatus in Ralstonia eutropha. J. Mol. Microbiol. Biotechnol. 4:255-262. [PubMed] [Google Scholar]
- 27.Lenz, O., and B. Friedrich. 1998. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc. Natl. Acad. Sci. USA 95:12474-12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosley, C. S., J. Y. Suzuki, and C. E. Bauer. 1994. Identification and molecular genetic characterization of a sensor kinase responsible for coordinately regulating light harvesting and reaction center gene expression in response to anaerobiosis. J. Bacteriol. 176:7566-7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oda, Y., S. K. Samanta, F. E. Rey, L. Wu, X. Liu, T. Yan, J. Zhou, and C. S. Harwood. 2005. Functional genomic analysis of three nitrogenase isozymes in the photosynthetic bacterium Rhodopseudomonas palustris. J. Bacteriol. 187:7784-7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh, M. K., L. Rohlin, K. C. Kao, and J. C. Liao. 2002. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277:13175-13183. [DOI] [PubMed] [Google Scholar]
- 31.Parales, R. E., and C. S. Harwood. 1993. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for gram-negative bacteria. Gene 133:23-30. [DOI] [PubMed] [Google Scholar]
- 32.Parales, R. E., and C. S. Harwood. 1993. Regulation of the pcaIJ genes for aromatic acid degradation in Pseudomonas putida. J. Bacteriol. 175:5829-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 34.Rauhut, R., and G. Klug. 1999. mRNA degradation in bacteria. FEMS Microbiol. Rev. 23:353-370. [DOI] [PubMed] [Google Scholar]
- 35.Reichard, P. 1988. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 57:349-374. [DOI] [PubMed] [Google Scholar]
- 36.Schwartz, E., U. Gerischer, and B. Friedrich. 1998. Transcriptional regulation of Alcaligenes eutrophus hydrogenase genes. J. Bacteriol. 180:3197-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 38.Simpson, F. B., and R. H. Burris. 1984. A nitrogen pressure of 50 atmospheres does not prevent evolution of hydrogen by nitrogenase. Science 224:1095-1097. [DOI] [PubMed] [Google Scholar]
- 39.Swem, L. R., B. J. Kraft, D. L. Swem, A. T. Setterdahl, S. Masuda, D. B. Knaff, J. M. Zaleski, and C. E. Bauer. 2003. Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J. 22:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Soom, C., P. de Wilde, and J. Vanderleyden. 1997. HoxA is a transcriptional regulator for expression of the hup structural genes in free-living Bradyrhizobium japonicum. Mol. Microbiol. 23:967-977. [DOI] [PubMed] [Google Scholar]
- 41.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 42.Vignais, P. M., S. Elsen, and A. Colbeau. 2005. Transcriptional regulation of the uptake [NiFe] hydrogenase genes in Rhodobacter capsulatus. Biochem. Soc. Trans. 33:28-32. [DOI] [PubMed] [Google Scholar]
- 43.Vignais, P. M., and B. Toussaint. 1994. Molecular biology of membrane-bound H2 uptake hydrogenases. Arch. Microbiol. 161:1-10. [DOI] [PubMed] [Google Scholar]
- 44.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]