Abstract
Expansins are small extracellular proteins that promote turgor-driven extension of plant cell walls. EXPB1 (also called Zea m 1) is a member of the β-expansin subfamily known in the allergen literature as group-1 grass pollen allergens. EXPB1 induces extension and stress relaxation of grass cell walls. To help elucidate expansin's mechanism of wall loosening, we determined the structure of EXPB1 by x-ray crystallography to 2.75-Å resolution. EXPB1 consists of two domains closely packed and aligned so as to form a long, shallow groove with potential to bind a glycan backbone of ≈10 sugar residues. The structure of EXPB1 domain 1 resembles that of family-45 glycoside hydrolase (GH45), with conservation of most of the residues in the catalytic site. However, EXPB1 lacks a second aspartate that serves as the catalytic base required for hydrolytic activity in GH45 enzymes. Domain 2 of EXPB1 is an Ig-like β-sandwich, with aromatic and polar residues that form a potential surface for polysaccharide binding in line with the glycan binding cleft of domain 1. EXPB1 binds to maize cell walls, most strongly to xylans, causing swelling of the cell wall. Tests for hydrolytic activity by EXPB1 with various wall polysaccharides proved negative. Moreover, GH45 enzymes and a GH45-related protein called “swollenin” lacked wall extension activity comparable to that of expansins. We propose a model of expansin action in which EXPB1 facilitates the local movement and stress relaxation of arabinoxylan–cellulose networks within the wall by noncovalent rearrangement of its target.
Keywords: plant cell wall
Before maturation plant cells typically experience a period of prolonged cell enlargement, often resulting in a >103-fold increase in volume. The impressive height of trees, some exceeding 100 m, depends on such enlargement, which entails massive vacuolar expansion and irreversible yielding of the cellulosic cell wall. In physical terms, the rate-limiting process for cell enlargement resides within the cell wall, which must be loosened so as to allow wall stress relaxation and consequent water uptake for vacuole enlargement and stretching of the wall (1, 2). Currently, the only plant proteins shown to cause cell wall relaxation are expansins (3, 4), although xyloglucan endotransglucosylase, pectate lyase, cellulase, and other enzymes participate in cell wall restructuring during cell growth (5–8).
Expansins were originally discovered in a “fishing expedition” for catalysts of cell wall extension (9, 10). When walls are clamped in tension and incubated in acidic buffer, these proteins rapidly induce wall extension and enhance wall stress relaxation. Their biological role in promoting cell enlargement is amply supported by in vitro and in vivo experiments, as well as by studies of gene expression, gene silencing, and ectopic expression (3, 11–13). In addition to cell enlargement, expansins are also implicated in other developmental processes where wall loosening occurs, such as in fruit softening, organ abscission, seed germination, and pollen tube invasion of the grass stigma (14–17).
Two expansin families with wall-loosening activity have been identified, named α-expansins (EXPA) and β-expansins (EXPB); both are found in all groups of land plants, from mosses to flowering plants (3, 18). Although they have only ≈20% amino acid identity, EXPA and EXPB proteins are of similar size (≈27 kDa), their sequences align well with one another, and they contain a number of conserved residues and characteristic motifs distributed throughout the length of the protein. EXPA and EXPB appear to act on different cell wall components, but their native targets have not yet been well defined.
A subset of EXPBs is known in the immunological literature as group-1 grass pollen allergens (19–21). These EXPBs are abundantly and specifically expressed in grass pollen, causing hay fever and seasonal asthma in an estimated 200–400 million humans (22, 23). The extraordinary abundance of group-1 allergens [comprising up to 4% of the protein extracted from grass pollen (24)] is unique (as far as we know) in the world of expansins, which are typically found in very low abundance and tightly bound to the cell wall. The abundance of group-1 allergens in grass pollen bespeaks a unique biological role, namely to loosen the cell walls of the grass stigma and style, thereby aiding pollen tube penetration and assisting delivery of its two sperm cells to the ovule, where a double fertilization occurs, forming the diploid zygote and the triploid endosperm. Seed development follows, and, because cereal grasses provide the largest food source for humanity (e.g., rice, maize, wheat, and barley, to name but a few), the importance of these events for human welfare is hard to overestimate.
Other genes in the EXPB family are expressed in a variety of other tissues in the plant body and in general lack the specific allergenic epitopes characteristic of group-1 allergens (24, 25). These so-called “vegetative EXPBs” are thought to have cell wall loosening activity and substrate specificity similar to the group-1 allergens, but these inferences have yet to be demonstrated experimentally.
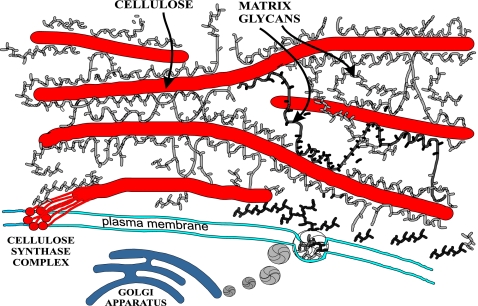
The mechanism by which expansins loosen cell walls has not yet been worked out in molecular detail. Plant cell walls consist of a scaffold of long cellulose microfibrils ≈4 nm in diameter embedded in a matrix of cellulose-binding glycans, such as xyloglucan and arabinoxylan, and gel-forming pectic polysaccharides (Fig. 1). The cellulose-binding glycans form a stable network with the cellulose microfibrils by binding to their surface via hydrogen bonds between hydroxyl groups and via van der Waals forces between the sugar rings; the network is further stabilized by calcium ions and borate diesters that link pectic polysaccharides together. Cell walls also contain small amounts of structural proteins with a reinforcing role (26, 27). Wall expansion entails rearrangement or modification of the matrix to allow turgor-driven movement or slippage of cellulose microfibrils within the matrix (1).
Fig. 1.
Schematic diagram of the plant cell wall. Cellulose microfibrils are synthesized by large complexes in the plasma membrane and are glued together by branched matrix polysaccharides synthesized in the Golgi and deposited by vesicles along the inner surface of the cell wall. The ≈4-nm-wide cellulose microfibril in cross-section consists of ≈36 β-(1→4)-d-glucans organized into a crystalline array. Polysaccharides such as arabinoxylan and xyloglucan spontaneously bind to the surface of cellulose and may also be entrapped during coalescence of the β-(1→4)-d-glucans to form the microfibril. Hydrophilic pectins and structural proteins (data not shown) also make up the matrix between cellulose microfibrils and influence the wall's physical properties (7).
Most of the biochemical work on expansins to date has focused on EXPAs, which do not hydrolyze the major structural polysaccharides of the wall and indeed are devoid of every enzyme activity assayed to date (28). Our current model proposes that EXPAs disrupt the polysaccharide complexes that link cellulose microfibrils together. The pollen EXPBs (group-1 allergens) have a marked loosening action on cell walls from grasses, but not from dicots, whereas the reverse is true for EXPAs; therefore, it seems that the two forms of expansin target different components of the cell wall (21, 24). Grass cell walls are notable for containing relatively small amounts of xyloglucan and pectin, which are replaced with β-(1→3),(1→4)-d-glucan and glucuronoarabinoxylan (29), two potential targets of EXPBs in their wall-loosening activity.
Sequence analysis suggests that expansins consist of two domains (2, 3). The putative N-terminal domain [domain 1 of EXPB1 (D1)] has distant sequence similarity (≈20% identity) to the catalytic domain of family-45 glycoside hydrolases (GH45) (http://afmb.cnrs-mrs.fr/CAZY). Despite this resemblance, EXPAs do not hydrolyze wall polysaccharides, and so the sequence similarity is enigmatic. The C-terminal domain [domain 2 of EXPB1 (D2)] has sequence similarity (from 35% to <10% identity) to another class of allergens, the group-2/3 grass pollen allergens, whose biological function is unknown (30).
In this study we present the crystal structure of a native EXPB purified from maize pollen. In the allergen field it is designated Zea m 1 isoform d, whereas by expansin nomenclature it is called EXPB1 (GenBank accession no. AAO45608). The allergen name “Zea m 1” encompasses a group of at least four pollen proteins (EXPB1, EXPB9, EXPB10, and EXPB11) in two rather divergent sequence classes (24). EXPB1 is the most abundant of the maize group-1 allergens. We also test EXPB1 for binding and activity on cell walls. At the end we discuss a molecular model of expansin action that is consistent with its structure and known biophysical and biochemical activities.
Results
EXPB1 Has Two Closely Packed Domains.
Native EXPB1 was purified from maize pollen and crystallized in 15% (wt/vol) polyethylene glycol 4000 with 0.1 or 0.2 M ammonium sulfate. Two crystals were analyzed, yielding x-ray diffraction patterns consistent with the monoclinic C2 space group. EXPB1 structure was solved and refined to 2.75-Å resolution (see Materials and Methods) with a crystallographic R-factor of 0.233 and an Rfree of 0.291 (Table 1, which is published as supporting information on the PNAS web site).
EXPB1 contains two domains [residues 19–140 (D1) and 147–245 (D2)] connected by a short linker (residues 141–146) and aligned end to end so as to make a closely packed irregular cylinder ≈66 Å long and 26 Å in diameter (Fig. 2A). At its N terminus EXPB1 has a flexible sequence (residues 1–18) containing hydroxyproline (O9) and a glycan attached to N10, part of the glycosylation consensus sequence NXT. The end of the glycan comes close to the polysaccharide-binding groove (see D1 and D2 Form a Long Polysaccharide-Binding Site) of the symmetry-related protein in the crystalline lattice, with one of the mannose residues stacking against the planar surface formed by residues Gly-39 and Gly-40 and stabilized further by two hydrogen bonds with the side chain of D37. These interactions with the symmetry-related protein account in part for the unusual ordering of the glycan, as well as the ability to crystallize the glycosylated protein.
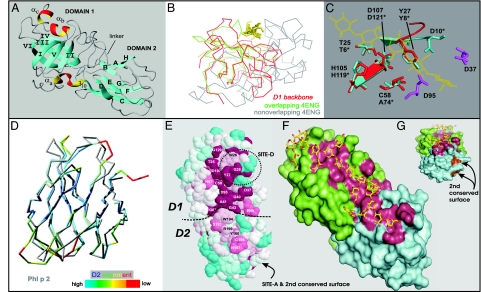
Fig. 2.
Structure of EXPB1 (PDB ID code 2HCZ). (A) Ribbon model of EXPB1 showing the overall configuration of the two domains. (B) Superposition of the peptide backbone of EXPB1 D1 (shown entirely in red) with the peptide backbone of Humicola Cel45 (PDB ID code 4ENG), colored green for regions of good alignment with EXPB1 and gray otherwise. The yellow residues indicate cellohexaose from the 4ENG model. (C) Superposition of residues making up the catalytic site of Humicola Cel45 (blue) and corresponding residues of EXPB1 (red). Other conserved acidic residues in this region of EXPB1 are shown in purple. (D) Superposition of EXPB1 D2 (colored) and Phl p 2 (gray), a group-2/3 grass pollen allergen (PDB ID code 1WHO). Coloring scale from best to poorest alignment of peptide backbones is shown at the bottom. (E) Top view of the conserved surface of EXPB1, color-coded to indicate conservation (red, most conserved; blue, least conserved; white, intermediate). Conserved residues are labeled, and the locations of two antigenic epitopes are indicated (SITE-D and SITE-A). (F) A model of glucurono-arabinoxylan (yellow and red) was manually fitted to the long open groove of EXPB1 by using the program O (66) and subsequently energy-minimized by using the program CNS (67). Green residues are from D1, cyan residues are from D2, and red residues are the conserved residues identified in E. (G) End view of same model as in F. The image in E was generated from the program CONSURF (68) by using the alignment of 80 EXPB proteins in the GenBank database and 2HCZ after removal of the N-terminal extension. The images in G and F were generated with PYMOL (DeLano Scientific) after removal of the N-terminal extension.
Based on its electron density, our model of this N-linked glycan consists of a β-(1→4)-linked backbone of GlcNac1GlcNac2Man3 with two Man residues and a Xyl residue attached to Man3 and a Fuc residue linked to GlcNac1 (Fig. 5, which is published as supporting information on the PNAS web site). Such so-called paucimannosidic-type N-linked glycans are characteristically processed in the Golgi and in post-Golgi steps (31).
Residues 1–3 were not modeled because of insufficient electron density, but N-terminal sequencing and mass spectrometry indicate their presence (24). The 24-aa signal peptide at the N terminus, predicted from the EXPB1 cDNA, was absent and was presumably excised during processing in the endoplasmic reticulum before secretion. No other posttranslational modifications, bound metals, or ligands were evident from the crystal structure.
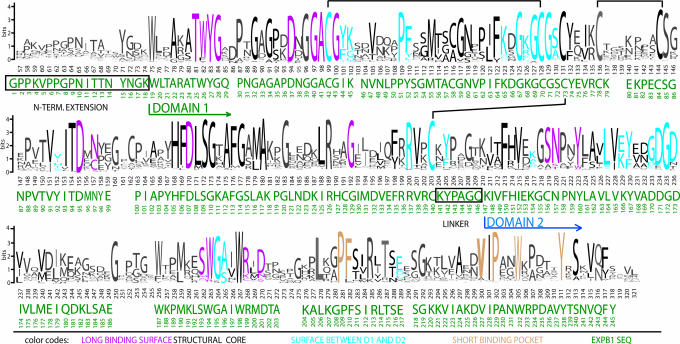
The two EXPB1 domains pack close to one another, making contact via H-bonds and salt bridges between basic residues (K65 and R137) in D1 and acidic residues (E217 and D171) in D2. These residues are highly conserved in the EXPB family (see annotated sequence logo in Fig. 3). Additional hydrogen bonding is found between S72 and D173, as well as between the peptide backbone for C42 and A196. The two domains also make contact via a hydrophobic patch consisting of I44, P51, Y52, and Y92 in D1 and L164, Y167, and the hydrocarbon chain of K166 in D2, residues that are mostly well conserved or have conservative substitutions in the EXPB family. Moreover, six highly conserved glycine residues (G43, G67, G69, G71, G172, and G195) are found at the surfaces where the two domains make contact. The lack of side chains in the glycine residues permits close packing of the two domains.
Fig. 3.
EXPB sequence logo based on 80 EXPB proteins from the GenBank database, aligned with the sequence of maize EXPB1 (green) and color-coded to indicate the structural role of the conserved residues. Residues with unspecified roles are shown in gray. The size of the one-letter amino acid code in the sequence logo indicates the degree of conservation on a logarithmic scale. The logo was generated with WebLogo (http://weblogo.berkeley.edu). Black lines between Cys residues indicate disulfide bonds.
Structure of D1.
Residues 19–140 form an irregular ovoid with rough dimensions of 35 × 30 × 24 Å. The protein fold is dominated by a six-stranded β-barrel flanked by short loops and α-helices (Fig. 2A and Fig. 6, which is published as supporting information on the PNAS web site). D1 has three disulfide bonds (Fig. 3), and the six participating cysteines are highly conserved in both EXPA and EXPB families.
Previous analysis (2, 3) indicated that D1 has distant sequence similarity to GH45, whose members have been characterized as inverting endo-β-(1→4)-d-glucanases (2, 3, 32, 33). Superposition of D1 with a GH45 enzyme [Protein Data Bank (PDB) ID code 4ENG] using the secondary structure matching algorithm in CCP4 (34) gives good overlap of the two structures for 84 residues (60%) of the peptide backbone of D1 (Fig. 2B), with an rmsd of 2.5 Å. Two of the three disulfide bonds in D1 superimpose exactly with 4ENG disulfides (the exception being C78–C84). Likewise, all of the β-strands in D1 superimpose on β-strands of 4ENG, although the β-strands in EXPB1 are generally shorter (Fig. 7, which is published as supporting information on the PNAS web site). Both structures have short α-helices, but these do not overlap in the two structures.
The GH45 enzyme is substantially larger than D1 (210 residues versus 121), and the “extra” structure in the GH45 enzyme is composed largely of loop regions and α-helices forming a large ridge and subtending structure lacking in D1 (Fig. 2B). In 4ENG this ridge makes a steep border on one side of the deep glucan-binding cleft. Because this ridge is missing in D1, the corresponding surface is more like an open groove than a deep cleft, with space to bind a large, branched polysaccharide (Fig. 2 F and G).
In addition to partial conservation of the protein fold, D1 has noteworthy, but incomplete, conservation of the catalytic site identified in GH45 enzymes (Fig. 2C). In 4ENG (residues designated with asterisks) the catalytic site is centered on aromatic residue Y8* which binds a glucose residue and is flanked by two acidic residues, D10* and D121*, serving as catalytic base and proton donor, respectively, for hydrolysis of the glycosidic bond (33, 35). D121* is flanked on one side by the hydrophobic side chains of A74* and Y8* and on the other side is part of a hydrogen-bonded network with T6*, which in turn is hydrogen-bonded to H119*. In D1 a nearly identical structure is found (Fig. 2C), where D107 corresponds to the proton donor D121*, with C58 and Y27 forming the hydrophobic pocket, while T25 and H105 overlap the corresponding residues in 4ENG. Thus, D1 possesses much of the conserved catalytic machinery for glycan hydrolysis.
What is missing in EXPB1 is a residue corresponding to D10*, the catalytic base required for glucan hydrolysis by GH45 enzymes (35). As indicated in Figs. 2C and 7, D10* is located on a loop that is not aligned with any part of EXPB1. EXPB proteins do have a conserved acidic residue, D37, which is located in a loop (residues 29–38) in the general vicinity corresponding to D10* in 4ENG. This loop is well resolved in D1. However, D37 is located too far from D107 and Y27 to function as the required base. In 4ENG, the catalytic carboxylate groups are located 8.5 Å apart, which is sufficient distance to accommodate a water molecule needed for hydrolysis (35). In D1, the carboxylates for D107 and D37 are 15 Å apart, too distant for this catalytic mechanism. Moreover, simple lateral movement of the loop to bring D37 into a correct position seems unlikely because the loop residues following D37 are rigidly held in place by several stabilizing interactions. Thus, a key part of the catalytic machinery required for hydrolytic activity of GH45 enzymes is lacking in EXPB1.
Inspection of the EXPB1 structure revealed another acidic residue, D95, which is close to D107 (the carboxylate groups are 8.5 Å away). D95 is highly conserved in group-1 allergens, as well as in EXPBs in general (Fig. 3), but not in EXPA. However, D95 is not correctly positioned relative to the D107/Y27 site and the presumed position of the glycan backbone to serve as the catalytic base for hydrolysis. D95 and D37 have an appropriate distance from each other to potentially serve in hydrolysis of a sugar residue, which might be bound to the planar hydrophobic surface made up of G39, G40, and A41 backbone atoms, but none of these residues are part of the site that is conserved with GH45 enzymes.
Enzymatic Activity.
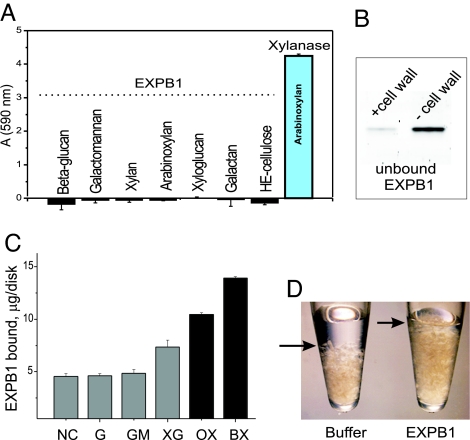
Because of the structural similarity between D1 and GH45 and the configuration of D95/D37, we tested the ability of EXPB1 to hydrolyze the major polysaccharides of the cell wall. Even with 48-h incubations, we did not detect hydrolytic activity by EXPB1 (Fig. 4A).
Fig. 4.
EXPB1 activities. (A) Hydrolytic activity of EXPB1 against various wall polysaccharides. Data are means ± SEM (n = 3). The positive control with arabinoxylan is a crude extract of maize pollen containing endoxylanase activity (69). (B) Maize cell walls bind EXPB1. After incubation of EXPB1 with or without cell wall, protein remaining in the supernatant was analyzed by SDS/PAGE and stained with SYPRO Ruby. (C) EXPB1 binding to isolated polysaccharides immobilized onto nitrocellulose membrane. NC, nitrocellulose membrane alone; G, β-(1→3),(1→4)-d-glucan; GM, glucomannan; XG, xyloglucan; OX, oat xylan; BX, birch xylan. Data are means ± SEM (n = 3). (D) Swelling of maize cell walls after 48 h of incubation with or without EXPB1. Methods were as described for the binding studies.
Taking another tack, we tested two GH45 enzymes (32, 36) and a nonenzymatic GH45-related protein named “swollenin” (37) for their abilities to catalyze cell wall extension. For these experiments, heat-inactivated walls from cucumber hypocotyls and wheat coleoptiles were clamped in tension in an extensometer, and changes in length were monitored upon addition of protein. We observed only small traces of wall extension activity for the GH45 enzymes and for swollenin (Fig. 8, which is published as supporting information on the PNAS web site). Thus, these related proteins lack significant expansin-type activity, at least with the cell walls tested here.
We conclude that, despite the structural similarity of D1 to GH45, EXPB1 does not induce wall extension via wall polysaccharide hydrolysis.
Structure of D2.
Residues 147–245 of EXPB1 make up a second domain (D2) composed of eight β-strands assembled into two antiparallel β-sheets (Fig. 2A and Fig. 9, which is published as supporting information on the PNAS web site). The two β-sheets are at slight angles to each other and form a β-sandwich similar to the Ig fold. D2 has 36% sequence identity with Phl p 2, a group-2/3 grass pollen allergen (PDB ID code 1WHO), and superposition of the two structures shows them to have identical folds (rmsd of 1.3 Å) (Fig. 2D). In comparing the two structures, we find that D2 tends to have shorter β-strands compared with Phl p 2, and the two proteins deviate slightly in the loop regions connecting the β-strands.
D1 and D2 Form a Long Potential Polysaccharide-Binding Site.
The two EXPB1 domains align so as to form a long, shallow groove with highly conserved polar and aromatic residues suitably positioned to bind a twisted polysaccharide chain of 10 xylose residues (Fig. 2 E–G). The groove extends from the conserved G129 at one end of D1, spans across a stretch of conserved residues in D1 and D2 (see numbered residues in Fig. 2E as well as annotated sequence logo in Fig. 3), and ends at N157, a distance of some 47 Å. Many of the conserved residues common to EXPA and EXPB make up this potential binding surface, including residues in the classic expansin motifs TWYG, GGACG, and HFD (see Fig. 3).
Residues that could bind a polysaccharide by van der Waals interactions with the sugar rings include W26, Y27, G40, and G44 from D1 as well as Y160 and W194 from D2. Conserved residues that might stabilize polysaccharide binding by H-bonding include T25, D37, D95, and D107 in D1 and N157, S193, and R199 in D2.
The openness of the long groove may enable EXPB1 to bind polysaccharides that are part of a bulky cell wall complex, such as on the surface of cellulose; that openness may also be important for binding branched glycans such as arabinoxylan, which itself binds to the surface of cellulose microfibrils. Because EXPB1 binds preferentially to xylans (see Binding), we have modeled an arabinoxylan, characteristic of grass cell walls, bound to the long groove of EXPB1 (Fig. 2G). From this model it is clear that the open groove of EXPB1 can accommodate the side chains found on such polysaccharides.
A second conserved surface in D2 is far removed from D1 (arrows in Fig. 2G). There is a shallow cup formed by the conserved W232 and F210. Adjacent to this pocket is a hydrophobic surface patch formed by the conserved residues P209, P229, V227, and Y238. The pocket and adjacent region could provide a second glucan binding surface for approximately three residues.
Binding.
EXPB1 bound to isolated maize cell wall (Fig. 4B). We observed that cell walls incubated with EXPB1 swelled significantly when compared with control cell walls (Fig. 4D). When purified polysaccharide fractions were immobilized onto nitrocellulose membranes, EXPB1 bound preferentially to xylans, with negligible binding to β-(1→3),(1→4)-d-glucan and glucomannan (Fig. 4C). Intermediate binding to xyloglucan was observed. Specific binding to cellulose and to nitrocellulose was also seen, although with less avidity than to xylan (A.T. and D.J.C., unpublished observations).
Discussion
With the molecular structure of EXPB1 in hand, we can examine previous inferences about expansin structure and its mechanism of cell wall loosening, but first the use of the group-1 pollen allergen for this study merits comment. Unlike other forms of expansin, which are found in very low abundance and have low solubility, the group-1 allergens are produced in copious amounts by grass pollen, from which they are readily extracted, purified, and concentrated to high levels without precipitation. Moreover, grasses produce abundant pollen, with maize being an especially liberal donor. In contrast to recombinant forms, use of the native protein ensures correct processing and posttranslational modifications. We note that expression of active expansins in various recombinant systems has proved problematic, because of improper folding, aggregation, and hyperglycosylation (M. Shieh and D.J.C., unpublished observations). Other forms of EXPB (e.g., the vegetative homologs) require harsh conditions to extract them from plant tissues (38), resulting in denatured protein; in soybean cultures an EXPB accumulates in the medium, but in a degraded and inactive form (39). EXPA proteins have been purified from various plant tissues, but in our experience they are difficult to concentrate to levels suitable for crystallization.
The high solubility and abundance of the group-1 allergens thus commends them for crystallization studies, but it should be noted that some of their biochemical properties may be specialized for their unique biological role in grass pollination. A case in point is their atypical pH dependence [maximum activity at pH 5.5 (24)], which is shifted to less acidic values than that found for other expansins. Likewise, their high solubility seems to be exceptional. Nevertheless, the general features of EXPB1 structure should prove to be common to the whole expansin family.
EXPB1 is composed of two domains. Although D1 structurally resembles GH45 and indeed has conserved much of the GH45 catalytic site, it lacks the second Asp residue (the catalytic base) required for hydrolytic activity in GH45 enzymes (33, 35). Thus, expansin's lack of wall polysaccharide hydrolytic activity, documented here for EXPB1 and in previous work for EXPA (28, 40), can be understood in structural terms as due to the lack of the required catalytic base. Furthermore, our finding that bona fide GH45 enzymes lack expansin's wall extension activity (Fig. 8) lends additional support to the conclusion that expansin does not loosen the cell wall by polysaccharide hydrolysis.
D2 as Binding Module?
We previously speculated that D2 may be a carbohydrate-binding module (CBM) (2, 4). This notion gains indirect support from the structure of D2, in which two surface aromatic residues (W194 and Y160) are in line with two aromatic residues (W26 and Y27) in D1, forming part of an extended, open, and highly conserved surface in EXPB1. D2 has an Ig-like fold. Proteins with this fold form a large superfamily of β-sandwich proteins implicated in binding interactions but lacking in enzymatic activity (41). At least 16 of the currently recognized CBM families in the Carbohydrate-Active Enzymes (CAZY) database (http://afmb.cnrs-mrs.fr/CAZY) have a β-sandwich fold. However, the specific fold topology of D2 does not match any of these CBM folds, and D2 lacks a bound metal atom, found in nearly all of the β-sandwich CBMs (42). We caution that polysaccharide binding by D2 alone has not been demonstrated experimentally, so the inference that it is a CBM should be considered speculation.
Nevertheless, from the structure of EXPB1 we expect that D2 aids glycan binding, particularly via the two surface aromatic residues W194 and Y160, aided by polar residues S193, R199, C156, and N157. These potential sugar-binding residues do not correspond to those inferred from a homology model of Lol p 1, a group-1 allergen from rye grass (43). In this model, which was based on the structure of Phl p 2, a group-2 allergen (30, 44), the authors identified two potential polysaccharide binding surfaces, one of which corresponds to the buried D2 face contacting D1.
It is notable that endoglucanases are most often found in nature as modular enzymes, coupled to a CBM via a long, highly glycosylated linker. Crystallization of intact GH45 enzymes with their CBMs has not yet been achieved, probably because the two domains do not maintain a fixed spatial relationship to each other. This difficulty of crystallization is a common experience with many CBM-coupled enzymes, and so successful crystallization of the two-domain EXPB1 is notable in this regard. In EXPB1 the linker is very short, and the multiple contacts between D1 and D2 enable close coupling of the two domains, which may function as a single unit in binding the cell wall.
Expansins as Cysteine Proteases?
A controversial hypothesis has been proposed that group-1 allergens are papain-related cysteine proteinases, with conservation of papain's active site residues C25, H159, and N175 (the “catalytic triad”) (45, 46). According to this hypothesis, C73 in EXPB1 should correspond to papain's C25. However, from the structure of EXPB1 we see that C73 participates in a disulfide bond conserved with GH45 enzymes, is relatively inaccessible, and is nowhere near the conserved surface. Moreover, the residues claimed to correspond to papain's H159 and N175 are dispersed in D2, are remote from C73, and are not conserved in expansins. We conclude that the resemblance to papain suggested by Grobe and colleagues (45, 46) is not supported by our crystallographic model of EXPB1.
The conserved surface of EXPB1 does contain two Cys residues (C58 and C156), but their environment does not resemble that of papain's active site. C58, which is conserved in about half of the EXPB family, is relatively inaccessible, being mostly buried underneath Y27 at the bottom of the extended groove. C156 is not conserved in the EXPB family but is usually replaced by serine. Experimental assays failed to detect proteinase activity in native EXPB1 (47). Moreover, the group-1 allergens are noted for their remarkable stability, which is also the case for EXPB1. We deem it likely that recombinant expression of EXPB in Pichia induced a host protease that accounted for the protein instability observed by Grobe and colleagues (45, 46). In fact, such host proteinase induction has been reported upon recombinant expression of a group-1 allergen (48).
Comparison with Vegetative EXPBs and with EXPAs.
EXPB1 is a member of the group-1 grass pollen allergens, which comprise a subset of the larger EXPB family. The EXPB family is notably larger in grasses than in other groups of land plants, and part of this expansion involved the unique evolution and radiation of the pollen allergen class of EXPBs, which are encoded by multiple genes (49). For instance, we classified 5 of the 19 EXPB genes in the rice genome as group-1 allergens (49). Multiple EXPB genes of the pollen allergen class may account in part for the numerous group-1 “isoallergens” found in grass pollen (19, 20, 50, 51).
There are minor conserved differences between the allergen class and the remaining vegetative EXPBs. These are so slight that we expect that the structural features of EXPB1 are characteristic of the vegetative EXPBs, with one exception: the N-terminal extension in EXPB1 contains a motif (VPPGPNITT) that is consistently found, with only minor variation, in group-1 grass pollen allergens but not in other EXPBs. This motif contains one or more hydroxyprolines and a glycosylated asparagine, features common to the pollen allergen class of EXPB (52). The function of this N-terminal extension is unknown, but it may play a role in protein recognition, transport, packaging, and processing by the pollen secretory apparatus. Additionally, the glycosylated extension may contribute to the exceptional solubility of the group-1 allergens (other expansins characterized to date have very low solubility) or may interact with other components of the cell wall. Although this motif is a unique hallmark of the group-1 allergens, many EXPB proteins lack an N-terminal extension altogether, and so it is not an essential part of expansin function. However, an N-terminal extension with similar posttranslational modifications was found as part of an EXPB expressed in soybean cell cultures (39). Further work will be needed to identify the function of this piece of the protein.
The good sequence alignment and conservation of motifs between the EXPB and EXPA families make it likely that EXPA proteins will have the same three-dimensional structure as reported here for EXPB1. There are two notable regions where EXPA and EXPB differ. EXPA has an additional stretch of ≈12 aa in the region corresponding to E99/P100 in EXPB1. E99 and P100 are part of a loop between β-strands IV and V in D1; these residues form part of the upraised flank to the left of the long groove identified in Fig. 2. The additional residues in EXPA may form a larger shoulder flanking this groove, stabilized by a disulfide bond between a pair of cysteines in this loop that are conserved in the EXPA family but are lacking in many EXPBs, mostly notably absent in the pollen allergens. This idea gains support from the structure of another GH45 enzyme (PDB ID code 1WC2) (53), which contains just such a loop (residues 102–114) stabilized by a disulfide bond. The loop creates a shoulder abutting the catalytic cleft. EXPAs therefore may have a steeper binding cleft than does EXPB1.
A second difference is that EXPAs lack a segment corresponding to G120–H127 in EXPB1. This segment, which contains few conserved residues, forms α-helix c and constitutes part of the surface of the pointed end of D1. This surface is remote from the conserved regions we have identified and so is unlikely to affect activity.
Allergenic Epitopes.
Allergies to grass pollen are widespread, afflicting an estimated 200–400 million people, and numerous studies have concluded that the group-1 allergens are the most important allergenic components of grass pollen (23, 23, 54, 55). Maize EXPB1 and its orthologs in turf grasses share common epitopes, as judged by antibody cross-reactivity, with the predominant epitopes found in the protein portion of the molecule and the glycosyl residues being of secondary antigenic significance (52, 56, 57). The dominant group-1 allergenic epitopes, which have been identified by epitope mapping studies, can be readily located on the surface of EXPB1. For instance, the 15-residue c98 epitope identified by Ball et al. (58) includes D107 in the conserved catalytic site of EXPB1, but it also includes residues that are exposed on the opposite side of the protein. “Site D” identified by Hiller et al. (59) overlaps part of the extended conserved groove of D1 containing the motif TWYG28 (Fig. 2E), whereas “site A” identified by Esch and Klapper (60) includes the small conserved pocket containing W232 and Y238, found on the far side of D2, as indicated in Fig. 2 E and G. This pocket is also part of “peptide 5” (22), a synthetic peptide derived from B cell epitopes of Phl p 1, the group-1 allergen of timothy grass pollen. Antibodies against peptide 5 showed great potency in reducing binding of IgEs from patients with strong grass pollen allergens, and so this peptide was considered a potentially useful component of an epitope-based vaccine for treating patients with severe allergies to grass pollen (22). With the structure of EXPB1 in hand, one may consider designing synthetic peptides that more closely resemble the natural epitopes occurring on the conserved surface of group-1 allergens. These may be of use for immunotherapy as well as mechanistic studies concerning the molecular and cellular bases for the potency of these proteins as allergens.
In view of the sequence conservation within the EXPB family, as well as within the entire expansin superfamily, it is surprising that the dominant antigenic epitopes of the group-1 allergens are not shared by vegetative EXPBs or by EXPA members. Nevertheless, this seems to be the case because antibodies raised against the group-1 allergens do not recognize other forms of expansin. This is indeed fortunate, because otherwise persons with strong allergies to grass pollen would also be allergic to fresh fruits, vegetables, grains, and other plant tissues that express members of this large gene family that is ubiquitous in plants.
A Molecular Model of Wall Loosening by Expansins.
Expansin action may be summarized as follows: the protein binds one or more wall polysaccharides and within seconds induces wall stress relaxation followed by wall extension, without hydrolysis of the wall polymers. There is no requirement for ATP or another source of chemical energy, and the wall continues to extend so long as the wall bears sufficient tension and expansin is present (that is, expansin acts catalytically, not stoichiometrically).
In the case of EXPB1, we imagine that stress relaxation begins when it binds a taut arabinoxylan tethered to a cellulose microfibril, causing local release of the arabinoxylan from the cellulose surface. Movement of the EXPB along the arabinoxylan–cellulose junction would enable it to unzip the hydrogen bonds between the polysaccharides, relaxing the taut tether and allowing turgor-driven displacement of cellulose and arabinoxylan, which may then reassociate in a relaxed state to restore wall strength. During this movement, the two expansin domains might shift in a hinge-like manner, binding and letting go of the arabinoxylan independent of each other, leading to an inchworm-like movement along the polysaccharide. We estimate that as little as a 10° shift in angle between domains could cause a one-residue dislocation of the polysaccharide along the binding surface.
To assess the feasibility of such interdomain movement, we estimated the buried surface area between the two domains using CCP4 (34). The value is 589 Å2, which is indicative of a weak interdomain interaction (61), consistent with domain movements as imagined above. A potential source of energy for these movements is the mechanical strain energy stored by the taut polysaccharide in a turgor-stretched cell wall. In this model, expansin acts as molecular device that uses the strain energy stored in a taut cellulose-binding glycan to help dissociate the glycan from the surface of cellulose.
Materials and Methods
Protein Purification, Crystallization, and Data Collection.
Native Zea m 1 was extracted from pollen of field-grown maize plants at 4°C in 0.125 M sodium carbonate and then purified to electrophoretic homogeneity in the presence of 5 mM DTT by using two chromatographic steps as described (24). With this method, four Zea m 1 isoforms were readily distinguished, and we used the most abundant isoform, Zea m 1d (EXPB1), for crystallization and activity assays. For the binding experiments, EXPB1 was further purified by HPLC on a reverse-phase column (Discovery C8, 15 cm × 4.6 mm i.d., 5 μm; Supelco, Bellefonte, PA) preequilibrated with 10% acetonitrile containing 0.1% trifluoroacetic acid. Bound protein was eluted at 1 ml·min−1 with a linear gradient of 22–90% acetonitrile in the same solution for 20 min at a flow rate of 1 ml·min−1 at 25°C. We confirmed wall extension activity of EXPB1 purified in this way.
Crystals were grown at 21°C for 9 days by using EXPB1 at 10.5 mg/ml in 100 mM Na acetate (pH 4.6) in 5-μl hanging drops, with addition of 5 μl of precipitant [15% (wt/vol) polyethylene glycol 4000 with 0.1 or 0.2 M ammonium sulfate] and with a 1-ml reservoir volume. Two crystals were analyzed, yielding diffraction patterns consistent with the monoclinic C2 space group. Crystal 1 had unit cell dimensions of a = 113.7 Å, b = 45.2 Å, and c = 70.3 Å with angles α = 90.0°, β = 124.6°, and γ = 90.0°; crystal 2 had unit cell dimensions of a = 112.6 Å, b = 44.4 Å, and c = 69.6 Å with angles α = 90.0°, β = 124.4°, and γ = 90.0°.
Data were collected by using a RU200 rotating anode x-ray generator (Rigaku) with CuKα radiation, operating at 5 kW of power (50 kV, 100 mA) (Molecular Structure, The Woodlands, TX). Three-degree oscillation frames, each exposed for 120 min, were collected on an R-AXIS IV detector. The two crystals were used to get a 93% complete data set. DENZO and SCALEPACK software suites (62) were used for data processing.
Structure Solution and Refinement.
Our final model of EXPB1 structure was based on the native crystal data set and was solved by molecular replacement calculations by using the program AMoRe (63) with the structure of Phl p 1 (PDB ID code 1N10), which has 58% amino acid identity with EXPB1 over 240 residues. EXPB1 has four more residues at its C terminus. The best molecular replacement solution in AMoRE was obtained by deleting the first 13 residues of the N terminus (attempts that included this stretch did not yield a solution) and by including all of the side chains for the rest of the protein (attempts with just the backbone atoms did not yield a good solution as well) and including all of the available data to 2.75 Å. The correlation coefficient and the R-factor for the best solution were 55.1% and 51.0%, respectively. The next best solution had an inferior correlation coefficient and R-factor of 49.3% and 53.9%, enabling us to proceed with further refinement and model building with confidence. For further refinement details and comparison with the 1N10 structure, see Supporting Text, which is published as supporting information on the PNAS web site. Coordinates and structure factors of the structure have been deposited in the Protein Data Bank [PDB ID code 2HCZ (64)]. A summary of the refinement results is given in Table 1.
Polysaccharide Hydrolysis.
Two milligrams of dye-coupled insoluble polysaccharides (AZCL polysaccharides; Megazyme, Wicklow, Ireland) were suspended in 100 μl of buffer [50 mM sodium acetate (pH 4.5) with 1 mM NaN3 and 10 mM DTT] and incubated with shaking at 30°C for 48 h with or without 30 μg of EXPB1. At the end of the incubation, 300 μl of 2.5% Trizma base was added to each tube to stop reaction, the suspension was centrifuged, and the absorbance (590 nm) of the supernatant was measured.
Binding.
Cell walls were collected from maize silks, cleaned by phenol/acetic acid/water washes (65), and lyophilized. EXPB1 was purified on a CM-Sepharose Fast Flow (Amersham Biosciences) column in a LP system (Bio-Rad) (24). EXPB1 (10 μg) was incubated with 1 mg of cell wall in 400 μl of 50 mM sodium acetate (pH 5.5) for 1 h at 25°C with agitation. After incubation, protein remaining in the supernatant was analyzed by SDS/PAGE (12% polyacrylamide) and stained with SYPRO Ruby protein gel stain (Bio-Rad).
Commercial polysaccharides [200 μg each, consisting of oat spelts xylan (Sigma), birch wood xylan (Fluka), barley β-glucan (catalog no. G-6513; Sigma), konjac glucomanna (Megazyme), or tamarind xyloglucan (Megazyme)] were dissolved in 10 μl of 20 mM sodium acetate (pH 4.5) and applied to Protran BA83 nitrocellulose membrane disks (diameter, ≈7 mm; pore size, 0.2 μm; Whatman). The disks were dried at 80°C overnight. The coated disks were incubated with blocking reagent (Roche) dissolved in 0.1 M maleic acid buffer for 1 h at room temperature to reduce nonspecific binding of EXPB1. After the blocking, the disks were washed with 20 mM Na acetate five times for 3 min each and then incubated with EXPB1 (20 μg per tube; purified by reverse-phase chromatography; see Protein Purification, Crystallization, and Data Collection) in 400 μl of 20 mM sodium acetate (pH 5.5) at 25°C for 1 h. After the incubation, the supernatant (unbound protein) was analyzed by reverse-phase chromatography (see Protein Purification, Crystallization, and Data Collection). The amount of EXPB1 bound to the coated nitrocellulose membrane disks was calculated from the reduction in the amount of unbound protein assessed by reverse-phase HPLC of the supernatant.
Supplementary Material
Acknowledgments
In memory of Paul B. Green (1921–1998). We thank Dr. Greg Farber for inestimable advice and assistance with growing the EXPB1 crystals; Dr. Javier Sampedro for useful discussions; Daniel M. Durachko, Edward Wagner, and Dr. Hemant Yennawar for expert technical assistance; Dr. Colin Mitchison (Genencor International, Palo Alto, CA) for the gift of the swollenin sample; Dr. Inés Muñoz (Uppsala University, Uppsala, Sweden) for the gift of the TrCel45 sample; and Dr. Jan-Christer Janson (Uppsala University) for the gift of the MeCel45 sample. This work was supported by Department of Energy Grant FG02-84ER13179 and National Institutes of Health Grant 5R01GM60397 (to D.J.C.).
Abbreviations
- EXPA
α-expansin
- EXPB
β-expansin
- GH45
family-45 glycoside hydrolase
- D1
domain 1 of EXPB1
- D2
domain 2 of EXPB1
- CBM
carbohydrate-binding module
- PDB
Protein Data Bank
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2HCZ).
See accompanying Profile on page 14661.
References
- 1.Cosgrove DJ. Nat Rev Mol Cell Biol. 2005;6:850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- 2.Cosgrove DJ. Plant Cell. 1997;9:1031–1041. doi: 10.1105/tpc.9.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sampedro J, Cosgrove DJ. Genome Biol. 2005;6:242.1–242.11. doi: 10.1186/gb-2005-6-12-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cosgrove DJ. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- 5.Rose JK, Braam J, Fry SC, Nishitani K. Plant Cell Physiol. 2002;43:1421–1435. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- 6.Domingo C, Roberts K, Stacey NJ, Connerton I, Ruiz-Teran F, McCann MC. Plant J. 1998;13:17–28. doi: 10.1046/j.1365-313x.1998.00002.x. [DOI] [PubMed] [Google Scholar]
- 7.Carpita NC, Gibeaut DM. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 8.Park YW, Tominaga R, Sugiyama J, Furuta Y, Tanimoto E, Samejima M, Sakai F, Hayashi T. Plant J. 2003;33:1099–1106. doi: 10.1046/j.1365-313x.2003.01696.x. [DOI] [PubMed] [Google Scholar]
- 9.McQueen-Mason S, Durachko DM, Cosgrove DJ. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z-C, Durachko DM, Cosgrove DJ. Planta. 1993;191:349–356. [Google Scholar]
- 11.Darley CP, Forrester AM, McQueen-Mason SJ. Plant Mol Biol. 2001;47:179–195. [PubMed] [Google Scholar]
- 12.Choi DS, Lee Y, Cho HT, Kende H. Plant Cell. 2003;15:1386–1398. doi: 10.1105/tpc.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pien S, Wyrzykowska J, McQueen-Mason S, Smart C, Fleming A. Proc Natl Acad Sci USA. 2001;98:11812–11817. doi: 10.1073/pnas.191380498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belfield EJ, Ruperti B, Roberts JA, McQueen-Mason SJ. J Exp Bot. 2005;56:817–823. doi: 10.1093/jxb/eri076. [DOI] [PubMed] [Google Scholar]
- 15.Cho HT, Cosgrove DJ. Proc Natl Acad Sci USA. 2000;97:9783–9788. doi: 10.1073/pnas.160276997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rose JK, Lee HH, Bennett AB. Proc Natl Acad Sci USA. 1997;94:5955–5960. doi: 10.1073/pnas.94.11.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen F, Dahal P, Bradford KJ. Plant Physiol. 2001;127:928–936. [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ. Plant Physiol. 2002;128:854–864. doi: 10.1104/pp.010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson K, Lidholm J. Int Arch Allergy Immunol. 2003;130:87–107. doi: 10.1159/000069013. [DOI] [PubMed] [Google Scholar]
- 20.Ansari AA, Kihara TK, Marsh DG. J Immunol. 1987;12:4034–4041. [PubMed] [Google Scholar]
- 21.Cosgrove DJ, Bedinger P, Durachko DM. Proc Natl Acad Sci USA. 1997;94:6559–6564. doi: 10.1073/pnas.94.12.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Focke M, Mahler V, Ball T, Sperr WR, Majlesi Y, Valent P, Kraft D, Valenta R. FASEB J. 2001;15:2042–2044. doi: 10.1096/fj.01-0016fje. [DOI] [PubMed] [Google Scholar]
- 23.Ball T, Edstrom W, Mauch L, Schmitt J, Leistler B, Fiebig H, Sperr WR, Hauswirth AW, Valent P, Kraft D, et al. FEBS J. 2005;272:217–227. doi: 10.1111/j.1432-1033.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- 24.Li LC, Bedinger PA, Volk C, Jones AD, Cosgrove DJ. Plant Physiol. 2003;132:2073–2085. doi: 10.1104/pp.103.020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Meeley RB, Cosgrove DJ. Plant Physiol. 2001;126:222–232. doi: 10.1104/pp.126.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall Q, Cannon MC. Plant Cell. 2002;14:1161–1172. doi: 10.1105/tpc.010477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribeiro JM, Pereira CS, Soares NC, Vieira AM, Feijo JA, Jackson PA. J Exp Bot. 2006;57:2025–2035. doi: 10.1093/jxb/erj153. [DOI] [PubMed] [Google Scholar]
- 28.McQueen-Mason SJ, Cosgrove DJ. Plant Physiol. 1995;107:87–100. doi: 10.1104/pp.107.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carpita NC. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:445–476. doi: 10.1146/annurev.arplant.47.1.445. [DOI] [PubMed] [Google Scholar]
- 30.De Marino S, Morelli MA, Fraternali F, Tamborini E, Musco G, Vrtala S, Dolecek C, Arosio P, Valenta R, Pastore A. Struct Folding Des. 1999;7:943–952. doi: 10.1016/s0969-2126(99)80121-x. [DOI] [PubMed] [Google Scholar]
- 31.Lerouge P, Cabanes-Macheteau M, Rayon C, Fischette-Laine AC, Gomord V, Faye L. Plant Mol Biol. 1998;38:31–48. [PubMed] [Google Scholar]
- 32.Xu B, Hellman U, Ersson B, Janson JC. Eur J Biochem. 2000;267:4970–4977. doi: 10.1046/j.1432-1327.2000.01533.x. [DOI] [PubMed] [Google Scholar]
- 33.Davies GJ, Tolley SP, Henrissat B, Hjort C, Schulein M. Biochemistry. 1995;34:16210–16220. doi: 10.1021/bi00049a037. [DOI] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project, Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 35.Davies GJ, Dodson GG, Hubbard RE, Tolley SP, Dauter Z, Wilson KS, Hjort C, Mikkelsen JM, Rasmussen G, Schulein M. Nature. 1993;365:362–364. doi: 10.1038/365362a0. [DOI] [PubMed] [Google Scholar]
- 36.Saloheimo A, Henrissat B, Hoffren AM, Teleman O, Penttila M. Mol Microbiol. 1994;13:219–228. doi: 10.1111/j.1365-2958.1994.tb00417.x. [DOI] [PubMed] [Google Scholar]
- 37.Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssonen E, Bhatia A, Ward M, Penttila M. Eur J Biochem. 2002;269:4202–4211. doi: 10.1046/j.1432-1033.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- 38.Lee Y, Choi D. Mol Cells. 2005;20:119–126. [PubMed] [Google Scholar]
- 39.Downes BP, Steinbaker CR, Crowell DN. Plant Physiol. 2001;126:244–252. doi: 10.1104/pp.126.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McQueen-Mason S, Cosgrove DJ. Proc Natl Acad Sci USA. 1994;91:6574–6578. doi: 10.1073/pnas.91.14.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bork P, Holm L, Sander C. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 42.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barre A, Rouge P. Biochem Biophys Res Commun. 2002;296:1346–1351. doi: 10.1016/s0006-291x(02)02091-0. [DOI] [PubMed] [Google Scholar]
- 44.Fedorov AA, Ball T, Valenta R, Almo SC. Int Arch Allergy Immunol. 1997;113:109–113. doi: 10.1159/000237520. [DOI] [PubMed] [Google Scholar]
- 45.Grobe K, Becker WM, Petersen A. Eur J Biochem. 1999;263:33–40. doi: 10.1046/j.1432-1327.1999.00462.x. [DOI] [PubMed] [Google Scholar]
- 46.Grobe K, Poppelmann M, Becker WM, Petersen A. Eur J Biochem. 2002;269:2083–2092. doi: 10.1046/j.1432-1033.2002.02856.x. [DOI] [PubMed] [Google Scholar]
- 47.Li LC, Cosgrove DJ. Eur J Biochem. 2001;268:4217–4226. doi: 10.1046/j.1432-1327.2001.02336.x. [DOI] [PubMed] [Google Scholar]
- 48.Poppelmann M, Becker WM, Petersen A. Electrophoresis. 2002;23:993–997. doi: 10.1002/1522-2683(200204)23:7/8<993::AID-ELPS993>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 49.Sampedro J, Lee Y, Carey RE, dePamphilis C, Cosgrove DJ. Plant J. 2005;44:409–419. doi: 10.1111/j.1365-313X.2005.02540.x. [DOI] [PubMed] [Google Scholar]
- 50.Petersen A, Becker WM, Schlaak M. J Allergy Clin Immunol. 1993;92:789–796. doi: 10.1016/0091-6749(93)90055-k. [DOI] [PubMed] [Google Scholar]
- 51.Suphioglu C, Singh MB, Knox RB. Int Arch Allergy Appl Immunol. 1993;102:144–151. doi: 10.1159/000236565. [DOI] [PubMed] [Google Scholar]
- 52.Petersen A, Becker WM, Moll H, Blumke M, Schlaak M. Electrophoresis. 1995;16:869–875. doi: 10.1002/elps.11501601144. [DOI] [PubMed] [Google Scholar]
- 53.Xu B, Janson JC, Sellos D. Eur J Biochem. 2001;268:3718–3727. doi: 10.1046/j.1432-1327.2001.02280.x. [DOI] [PubMed] [Google Scholar]
- 54.Marsh DG, Milner FH, Johnson P. Int Arch Allergy Appl Immunol. 1966;29:521–535. doi: 10.1159/000229739. [DOI] [PubMed] [Google Scholar]
- 55.Ball T, Fuchs T, Sperr WR, Valent P, Vangelista L, Kraft D, Valenta R. FASEB J. 1999;13:1277–1290. doi: 10.1096/fasebj.13.11.1277. [DOI] [PubMed] [Google Scholar]
- 56.Howlett BJ, Clarke AE. Biochem J. 1981;197:707–714. doi: 10.1042/bj1970707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petersen A, Schramm G, Schlaak M, Becker WM. Clin Exp Allergy. 1998;28:315–321. doi: 10.1046/j.1365-2222.1998.00221.x. [DOI] [PubMed] [Google Scholar]
- 58.Ball T, Vrtala S, Sperr WR, Valent P, Susani M, Kraft D, Valenta R. J Biol Chem. 1994;269:28323–28328. [PubMed] [Google Scholar]
- 59.Hiller KM, Esch RE, Klapper DG. J Allergy Clin Immunol. 1997;100:335–340. doi: 10.1016/s0091-6749(97)70246-x. [DOI] [PubMed] [Google Scholar]
- 60.Esch RE, Klapper DG. Mol Immunol. 1989;26:557–561. doi: 10.1016/0161-5890(89)90007-2. [DOI] [PubMed] [Google Scholar]
- 61.Nooren IM, Thornton JM. J Mol Biol. 2003;325:991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 62.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 63.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 64.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fry SC. The Growing Plant Cell Wall: Chemical and Metabolic Analysis. London: Longman Scientific and Technical; 1988. [Google Scholar]
- 66.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 67.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 68.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 69.Wu SS, Suen DF, Chang HC, Huang AH. J Biol Chem. 2002;277:49055–49064. doi: 10.1074/jbc.M208804200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.